In several human cancer cell lines, HSP90 inhibitors destabilize macrophage inhibitory factor protein; systemic treatment with an HSP90 inhibitor slows tumor growth and extends overall survival in a mouse model of HER2-positive human breast cancer.
Abstract
Intracellular macrophage migration inhibitory factor (MIF) often becomes stabilized in human cancer cells. MIF can promote tumor cell survival, and elevated MIF protein correlates with tumor aggressiveness and poor prognosis. However, the molecular mechanism facilitating MIF stabilization in tumors is not understood. We show that the tumor-activated HSP90 chaperone complex protects MIF from degradation. Pharmacological inhibition of HSP90 activity, or siRNA-mediated knockdown of HSP90 or HDAC6, destabilizes MIF in a variety of human cancer cells. The HSP90-associated E3 ubiquitin ligase CHIP mediates the ensuing proteasome-dependent MIF degradation. Cancer cells contain constitutive endogenous MIF–HSP90 complexes. siRNA-mediated MIF knockdown inhibits proliferation and triggers apoptosis of cultured human cancer cells, whereas HSP90 inhibitor-induced apoptosis is overridden by ectopic MIF expression. In the ErbB2 transgenic model of human HER2-positive breast cancer, genetic ablation of MIF delays tumor progression and prolongs overall survival of mice. Systemic treatment with the HSP90 inhibitor 17AAG reduces MIF expression and blocks growth of MIF-expressing, but not MIF-deficient, tumors. Together, these findings identify MIF as a novel HSP90 client and suggest that HSP90 inhibitors inhibit ErbB2-driven breast tumor growth at least in part by destabilizing MIF.
In normal cells, heat shock chaperones guide proper folding of nascent polypeptide clients into mature proteins, assist in multimeric complex assembly, and regulate cellular levels of clients by promoting their degradation. Importantly, during oncogenesis the normal chaperone function becomes subverted to allow malignant transformation and enable cancer cell survival. Cancer cells are in a constant state of proteotoxic stress, both from an adverse microenvironment (hypoxia and acidosis) and from within (conformationally aberrant oncoproteins, high levels of ROS, high levels of DNA damage, and genomic instability). Thus, their proteins, and in particular their oncoproteins, require constant massive chaperone support to prevent protein aggregation and promote tumor cell survival (Whitesell and Lindquist, 2005; Taipale et al., 2010; Trepel et al., 2010). Hence, in addition to their oncogene addiction, cancer cells also require activated heat shock proteins. Among these chaperones, heat shock protein 90 (HSP90) is unique because many of its clients are conformationally labile signal transducers with crucial roles in growth control and cell survival. HSP90 plays a key role in the conformational stabilization and maturation of mutant oncogenic signaling proteins, encompassing, for example, receptor tyrosine kinases (ErbB1 and ErbB2/HER2; Mimnaugh et al., 1996), signaling kinases (Bcr-Abl and Akt; Basso et al., 2002), NF-κB (Chen et al., 2002), c-Raf, FLT3, and steroid hormone receptors (Whitesell and Lindquist, 2005). Hsp90 is the core protein of the multicomponent machinery HSP90 that includes Hsp70, several co-chaperones, and the resident E3 ligase CHIP. Hsp90 is a dynamic ATPase, with N-terminal binding and subsequent hydrolysis of ATP which drives the conformational cycles of HSP90 chaperone activity. HSP90, a powerful antiapoptotic system, is highly up-regulated and activated specifically in cancer and is an almost ubiquitous feature of human cancers (Whitesell and Lindquist, 2005). Moreover, tumors preferentially contain Hsp90 in a higher order multi-chaperone complex with high affinity for certain small molecule inhibitors of Hsp90’s ATP-binding pocket, whereas normal tissues harbor latent, largely uncomplexed Hsp90 with low affinity for these inhibitors (Kamal et al., 2003; Moulick et al., 2011).
Pharmacological inhibition of HSP90 has been achieved by small molecules that originated from the natural ansamycin antibiotic geldanamycin (GA) and led to the clinical derivative 17AAG (17-allylamino, 17-demethoxygeldanamycin). They show potent anti-cancer activity in vitro and in vivo with a good therapeutic window and some are now in clinical trials (Taipale et al., 2010; Trepel et al., 2010). However, it is currently difficult to predict the susceptibility of individual cancers to this class of drugs. Also, there is no clear mechanistic basis to justify the combination of HSP90 inhibitors with other cancer drugs. It would therefore be highly desirable to know which HSP90 clients are critical for the anti-cancer effect of HSP90 inhibitors. At the moment, we only know a list of HSP90 clients that govern cancer cell proliferation and survival. This list is obviously incomplete. Even more importantly, the relative contribution of coexisting HSP90 clients to the anti-cancer efficacy of HSP90 inhibitors in a given tumor is currently unknown.
Macrophage migration inhibitory factor (MIF) was originally discovered as a secreted proinflammatory cytokine with a central role in innate immunity (Calandra and Roger, 2003). Recently, MIF has also been strongly implicated as tumor promoter with a central position in the inflammation–tumorigenesis axis (Bucala and Donnelly, 2007; Nemajerova et al., 2007; Conroy et al., 2010). A minor source of tumor-associated MIF is stromal and inflammatory cells secreting it into the microenvironment, which can then be taken up by tumor cells via the MIF receptor/co-receptor CD74/CD44 (Shi et al., 2006). The major source of MIF is in tumor cells themselves. Unlike other secreted cytokines that are restricted to the immune compartment in the tumor microenvironment, MIF is widely and strongly overexpressed within the cytoplasm and nucleus of malignant cells of multiple lineages. MIF overexpression in tumor cells is prominent in human cancers of breast, colon, ovary, prostate, liver, lung, pituitary, and brain (Bini et al., 1997; Meyer-Siegler, 2000; Bando et al., 2002; Tomiyasu et al., 2002; Pyle et al., 2003; Reome et al., 2004; Hagemann et al., 2005, 2007; Hira et al., 2005; He et al., 2006; Meyer-Siegler et al., 2006; Xu et al., 2008; Verjans et al., 2009; Cludts et al., 2010; Cheng et al., 2011; Zhang et al., 2011; Wang et al., 2012). Importantly, elevated intratumoral MIF levels correlate with clinical aggressiveness in cancers of the breast, lung, liver, brain, ovary, and prostate, implicating MIF in poor prognosis (Tomiyasu et al., 2002; Mitchell, 2004; Reome et al., 2004; Hagemann et al., 2005; Hira et al., 2005; Meyer-Siegler et al., 2007; Xu et al., 2008; Wang et al., 2012). Moreover, Myc- and Ras-transformed primary mouse embryo fibroblasts (MEFs) also exhibit up-regulated MIF compared with nontransformed control MEFs (Petrenko et al., 2003).
Mechanistically, MIF acts in multiple pathways to promote tumors. It increases tumor cell survival in B-CLL via secreted MIF that triggers the CD74/CD44–IL8–Bcl2 axis and the ERK pathway (Shi et al., 2006; Binsky et al., 2007). MIF also activates the Akt survival pathway (Lue et al., 2007), promotes angiogenesis via the HIF1α (Oda et al., 2008; Winner et al., 2007) or NF-κB–IL8–VEGF axes (Lin et al., 2006), and promotes invasion and migration via Rac1 activation (Rendon et al., 2007). Using MIF ablation in primary MEFs and mouse tumor models, we previously identified powerful actions of MIF within tumor cells that interfere with the two major tumor suppressor pathways, p53 and Rb-E2F, that are activated in response to oncogenic signaling. For example, we showed that primary MIF−/− embryonic fibroblasts have severe p53-dependent growth deficiencies, as well as Ras- and Myc-mediated transformation defects, which are rescued by co-deleting p53. Moreover, MIF−/− mice are more resistant than WT mice to a strong chemical carcinogen (Fingerle-Rowson et al., 2003; Petrenko et al., 2003; Nemajerova et al., 2007). Likewise, MIF deficiency in p53−/− Ras-expressing MEFs leads to reshuffling of Rb–E2F complexes and alters the DNA-binding properties of E2Fs. MIF interferes with the function of Rb and E2Fs mainly in DNA replication and does so in a transcription-independent fashion. Specifically, our data suggest that overexpressed MIF functions by directly antagonizing Rb/E2F4-mediated repression of DNA replication at ORI initiation sites (Petrenko and Moll, 2005). Consequently, overexpressed MIF strongly protects oncogene-initiated cells from apoptosis and senescence and drives their proliferation (Fingerle-Rowson et al., 2003; Petrenko et al., 2003; Petrenko and Moll, 2005; Talos et al., 2005).
In further support of MIF as an important physiological tumor promoter, genetic MIF ablation delays progression in several mouse cancer models. We reported a strong rescue effect in Myc-induced lymphomagenesis where MIF loss markedly protected Eµ-Myc transgenic mice from developing lymphomas by activating the p53 pathway (Talos et al., 2005). Moreover, MIF deletion in ApcMIN/+ mice generates fewer and smaller intestinal adenomas and decreases angiogenesis (Wilson et al., 2005). In bladder tumorigenesis induced by nitrosamine, MIF−/− mice show lower stage tumors than WT mice (Taylor et al., 2007). Finally, in response to chronic UVB exposure, MIF ablation delays skin cancer progression (Martin et al., 2009).
In sum, these data support a strong rationale for MIF as a potentially important cancer target. Targeting MIF could involve direct or indirect strategies. Within the inflammatory context, several isoxazoline-based small molecule antagonists specifically blocking the tautomerase catalytic site of MIF were developed. They inhibit MIF’s proinflammatory actions and show promising results in experimental sepsis and immuno-inflammatory diseases (Lubetsky et al., 2002). However, in cancer a unifying biochemical concept of the multiple MIF activities remains elusive, and MIF’s tautomerase activity is clearly not important (Fingerle-Rowson et al., 2009), making it difficult, if not impossible, to develop specific small molecule inhibitors that could directly bind (those undefined) critical domains of MIF to block its multiple diverse protumor activities. Alternatively, strategies to down-regulate the excess levels of MIF specific of cancer cells should also antagonize tumor growth and might be a more realistic route. This, however, would require the knowledge of a druggable mechanism that causes MIF accumulation in cancer cells.
Here, we identify HSP90 as the key mediator of MIF accumulation in cancer cells. Conversely, HSP90 inhibitors markedly suppress elevated MIF levels in vitro and in vivo. Most strikingly, this reduction of elevated MIF levels, in conjunction with reduction of the co–up-regulated HSP90 clients ErbB2 and Akt, is essential for the anti-cancer activity of the HSP90 inhibitor 17AAG in the mouse model of HER2-positive human breast cancer in vivo.
RESULTS
MIF protein is stabilized in human and mouse cancer cells
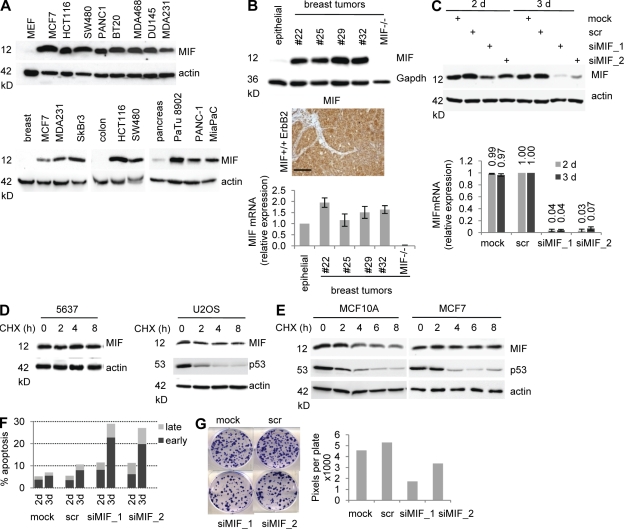
MIF silencing induces apoptosis and suppresses clonogenicity. Compared with normal cells, intracellular MIF protein in cancer cells has long been known to be highly elevated by an unknown mechanism (Bini et al., 1997; Meyer-Siegler, 2000; Bando et al., 2002; Tomiyasu et al., 2002; Pyle et al., 2003; Reome et al., 2004; Hagemann et al., 2005, 2007; Hira et al., 2005; He et al., 2006; Meyer-Siegler et al., 2006; Xu et al., 2008; Verjans et al., 2009; Cludts et al., 2010; Cheng et al., 2011; Zhang et al., 2011; Wang et al., 2012). This is illustrated by a random panel of human cancer cell lines compared with their normal tissues of origin (Fig. 1 A). Likewise, tumor cells from primary breast cancer tissues of transgenic MMTV-ErbB2 mice (Muller et al., 1988) also exhibited highly elevated levels of intracellular MIF protein (Fig. 1 B, top and middle), compared with undetectable levels in normal mammary epithelial cells isolated from fat pads of the same animals (Fig. 1 B, top, epithelial). In contrast, MIF mRNA expression in these MMTV-ErbB2 tumors increased only slightly (less than twofold) compared with normal mammary tissue (Fig. 1 B, bottom).
Figure 1.
MIF protein is stabilized in human and mouse cancer cells. (A, Top) Representative immunoblot of cell lysates from the indicated human cancer cell lines compared with normal primary MEF (20 µg protein per lane). (A, Bottom) Lysates from normal human tissues (breast, colon, and pancreas) were compared with human cancer cell lines derived from the corresponding tissue types. Representative immunoblots for MIF. Actin, loading control. (B, Top) Total tissue lysates from primary breast tumors from transgenic MMTV-ErbB2 mice (each number indicates a different mouse) were compared with normal mammary epithelial cells isolated from the mammary fat pad (epithelial) by immunoblotting. MIF−/− is a control tumor from an MIF−/−ErbB2 mouse. Gapdh, loading control. (B, Middle) Immunohistochemical MIF staining of MMTV-ErbB2 tumor #25. Bar, 100 µm. Normal mouse mammary tissue contains undetectable level of MIF. (B, Bottom) Quantitative RT-PCR of MIF mRNA normalized to 36B4 mRNA in breast tumors compared with normal tissue. Relative values are given in ratio (2−ddCT). Error bars indicate the mean of two separate RT reactions of triplicates each. Epithelial and MIF−/− controls are as above. (C) Duplicate plates of U2OS cells were transfected with two different siRNAs against MIF, scrambled control siRNA (scr), or mock transfected. At 2 and 3 d after transfection, cells were harvested. Top, immunoblotting of lysates with antibodies against MIF. Bottom, total RNA was analyzed by quantitative RT-PCR. Relative values normalized to GAPDH from ratio (2−ddCT). Error bars indicate the mean of two independent experiments in triplicates each. (D and E) 5637 bladder cancer and U2OS osteosarcoma cells (D) and immortalized MCF10A and MFC7 breast cancer cells (E) were treated with 40 µg/ml CHX for the indicated times. Total cell lysates were immunoblotted for MIF. Actin, loading control. p53, positive control for translational inhibition by CHX. Representative blots from three (D) and two (E) independent experiments are shown. (F) HCT116 cells were transfected with siRNA as in Fig. 1 C. At 2 and 3 d after transfection, cells were stained with Annexin and 7-AAD to determine early and late apoptosis by flow cytometry. Each time point was determined in duplicate and the mean is plotted. (G) HCT116 cells were transfected with siRNA as in Fig. 1 C. At 3 d after transfection, equal numbers of surviving cells were seeded and cultured for 8 d. Cells were fixed, stained with crystal violet, and plates were scanned (left). Colony density was measured as total pixels per plate (right). Representative data from three independent repeats are shown.
To determine if MIF up-regulation occurs at the transcriptional or posttranslational level, we first compared the relative kinetics of down-regulation of mRNA and protein in several human cancer lines. Although MIF mRNA was already profoundly reduced after 2 d of siRNA-mediated MIF silencing, a similarly strong reduction in MIF protein occurred only after 3 d of silencing, suggesting that MIF protein stability is greatly increased in cancers with a half-life of at least 24 h (Fig. 1 C). Consistent with high MIF stability and low protein turnover, extended treatment with proteasome inhibitor MG132 for 8 h failed to further increase MIF levels (see Fig. 4, A and B, minus drug). Cycloheximide (CHX) chases verified that accumulation of MIF protein in cancer cells is a result of increased protein stability rather than increased protein synthesis. MIF protein levels in 5637 and U2OS cancer cells were completely stable over 8 h, the maximum possible length of CHX treatment as a result of cellular toxicity (Fig. 1 D). In contrast, MIF in nonmalignant MCF10A mammary epithelial cells has a half-life of <4 h, as opposed to malignant MCF7 breast cancer cells with a half-life far exceeding 8 h (Fig. 1 E). Thus, aberrant MIF up-regulation during tumorigenesis seems mainly a result of protein stabilization. Functionally, MIF silencing in tumor cells induced apoptosis (Fig. 1 F) and decreased clonogenicity (Fig. 1 G), associated with activation of p53 pathways (e.g., p21 and Noxa; not depicted) and the E2F–p73 pathway (not depicted) as previously reported (Fingerle-Rowson et al., 2003; Petrenko et al., 2003; Petrenko and Moll, 2005; Talos et al., 2005).
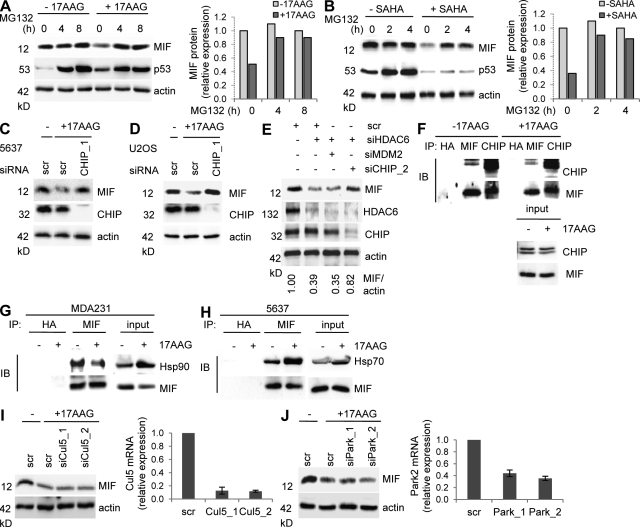
Figure 4.
CHIP ubiquitin E3 ligase is required for MIF degradation after Hsp90 inhibition in cancer cells. (A and B, Left) U2OS cancer cells were left untreated or treated with 5 µM each 17AAG (A) or SAHA (B) for 24 h with or without 10 µM of the proteasome inhibitor MG132 for the indicated final hours. Representative immunoblot analysis from three independent experiments. WT p53 serves as positive control for proteasome inhibition. Actin, loading control. (A and B, Right) Densitometric evaluations of representative immunoblots from the left. Each MIF value was normalized to its corresponding actin value. Relative values were calculated setting control cells at 0 h and without 17AAG to 1. (C and D) 5637 (C) and U2OS (D) cells were transfected with siRNA against CHIP (siCHIP_1) or control siRNA (scr). 2 d after transfection, cells were treated with 5 µM 17AAG for 24 h and MIF stability was analyzed. Representative immunoblots from two independent experiments. Actin, loading control. (E) MDA231 cells were cotransfected with siHDAC6, siMDM2, siCHIP_2, or control siRNA (scr). After 3 d, MIF levels were assessed by immunoblotting. Actin, loading control. The representative immunoblot was quantified and relative values (MIF/actin ratio) were calculated setting scr control to 1. (F) 5637 cells were treated with 5 µM 17AAG for 24 h. MG132 was added for the final 6 h. Whole cell lysates normalized for equal levels of MIF (see input) were immunoprecipitated with anti-MIF, anti-CHIP, or anti-HA control antibody (IP). MIF-bound CHIP and CHIP-bound MIF were detected by immunoblots. (G) MDA231 cells were treated with 5 µM 17AAG for 24 h. Whole cell lysates normalized for equal levels of MIF were immunoprecipitated with anti-MIF or anti-HA control antibody (IP). MIF-bound Hsp90 was detected by immunoblot. (H) 5637 cancer cells were treated as described in Fig 4 F. Whole cell lysates normalized for equal levels of MIF (see input) were immunoprecipitated with anti-MIF or anti-HA control antibody (IP). MIF-bound Hsp70 was detected by immunoblotting. (I and J) U2OS cells were transfected with two different siRNAs against Parkin (G) or Cul5 (H) or with control siRNA (scr). At 2 d after transfection, cells were cultured in parallel with 5 µM 17AAG for 24 h and MIF protein levels were analyzed by immunoblotting (left). Parkin and Cullin 5 mRNA transcripts were measured by quantitative RT-PCRs normalized to GAPDH expression (right, bar graphs). Error bars indicate the mean of two independent experiments in triplicates each.
Pharmacologic HSP90 inhibition by 17AAG or SAHA destabilizes MIF protein in cancer cells
We hypothesized that tumor-associated MIF stabilization might be a result of protection from degradation by physical association with the multi-component HSP90 chaperone complex. Up-regulation of HSP90 is tumor cell specific and accompanies malignant transformation almost ubiquitously (Whitesell and Lindquist, 2005; Taipale et al., 2010; Trepel et al., 2010). HSP90 is required for proper folding of many oncoprotein clients including HER2/ErbB2, ErbB1, Akt, c-Raf, Bcr-Abl, and FLT3 (Whitesell and Lindquist, 2005). HDAC6 is an obligate positive regulator of HSP90 by protecting the Hsp90 core protein from acetylation. Consequently, acetylation of the Hsp90 ATPase by HDAC6 knockdown or small molecule HDAC6 inhibitors inactivates HSP90 chaperone activity and triggers degradation of client proteins (Kovacs et al., 2005; Scroggins et al., 2007; Gibbs et al., 2009).
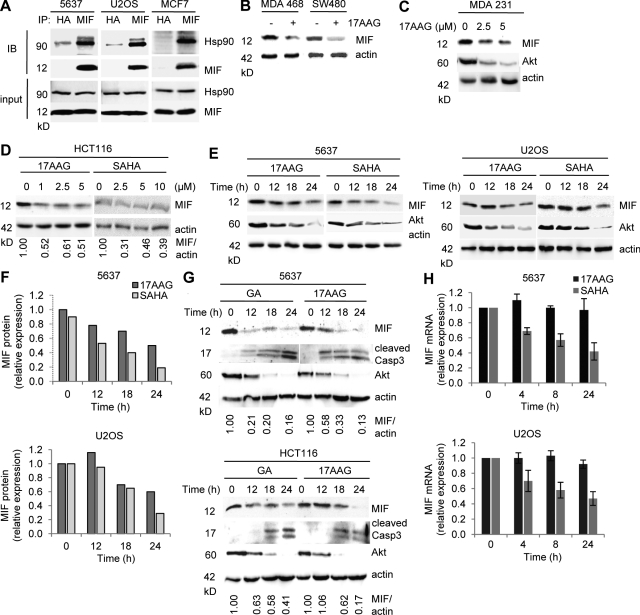
Indeed, in all analyzed cancer lines we observed a constitutive physical complex between endogenous MIF and Hsp90 (Fig. 2 A). Importantly, treatment with 17AAG, a highly specific competitive inhibitor of Hsp90 ATPase which blocks its nucleotide binding pocket and prevents client loading (Trepel et al., 2010), induced down-regulation of MIF protein in a dose- and time-dependent manner in all cancer lines tested (Fig. 2, B–G; and not depicted). Likewise, GA, another specific Hsp90 inhibitor, also induced strong down-regulation of MIF protein (Fig. 2 G and not depicted). Of note, concomitant to MIF down-regulation, 17AAG and GA induced apoptosis, indicated by cleaved caspase 3 (Fig. 2 G and not depicted). Likewise, SAHA, an inhibitor of HDACs including HDAC6, which was shown to abolish Hsp90 activity and client loading by inducing Hsp90 hyperacetylation (Bali et al., 2005; Kovacs et al., 2005), also led to MIF destabilization (Fig. 2, D–F; and not depicted). The dose- and time-dependent MIF destabilization via Hsp90 inhibition by 17AAG, GA, and SAHA was quantitated by densitometry (Fig. 2, D–G). Similarly, the prosurvival kinase Akt, a classical HSP90 client which destabilizes upon HSP90 inhibition via 17AAG, GA, or HDAC6 inhibitors (Basso et al., 2002; Bali et al., 2005), also showed destabilization upon 17AAG, GA, or SAHA treatment (Fig. 2, C, E, and G).
Figure 2.
Hsp90 inhibition by 17AAG and SAHA destabilizes MIF protein in human cancer cells. (A) Untreated 5637, U2OS, and MCF7 human cancer cells were subjected to coimmunoprecipitation with an anti-MIF antibody and immunoblotted as indicated. An anti-HA antibody served as negative precipitation control. (B–D) MDA468 and SW480 (B), MDA231 (C), and HCT116 (D) cells were treated with indicated concentrations of 17AAG or SAHA for 24 h (C and D) or with 5 µM of 17AAG for 24 h (B). Representative immunoblot analyses. Akt serves as positive control for HSP90 inhibition. Actin, loading control. Quantification of immunoblot (D) is shown as relative values (MIF/actin ratio) setting 0 h drug treatment to the value of 1. (E and F) 5637 and U2OS cells were treated with 5 µM 17AAG or SAHA for the indicated times (E). Representative immunoblots from three independent experiments are shown. Akt serves as positive control for Hsp90 inhibition. Actin, loading control. Densitometric evaluations of representative immunoblots from E are shown in F. Each MIF value was normalized to its corresponding actin value. Relative values were calculated by setting control cells at 0 h to 1. (G) 5637 (top) and HCT116 (bottom) cells were treated with 5 µM GA or 17AAG for the indicated time. Cleaved Caspase 3 indicates apoptosis. Representative immunoblots from two independent experiments are shown. Akt, positive control for Hsp90 inhibition. Actin, loading control. Quantification is as in Fig. 2 D. (H) 5637 (top) and U2OS (bottom) cells were treated with 5 µM 17AAG or SAHA for the indicated times. MIF mRNA, measured by quantitative RT-PCR, was normalized to GAPDH ratio (2−ddCT). Error bars indicate the mean of three independent experiments in triplicates each.
It was previously reported that inhibition of chromatin deacetylation by HDAC inhibitors transcriptionally represses MIF (Roger et al., 2007; Lugrin et al., 2009). In agreement, SAHA (but not 17AAG) moderately reduced MIF mRNA expression (Fig. 2 H), indicating a dual effect of SAHA in reducing MIF protein levels by inhibiting Hsp90 function via hyperacetylation and by repressing MIF transcription.
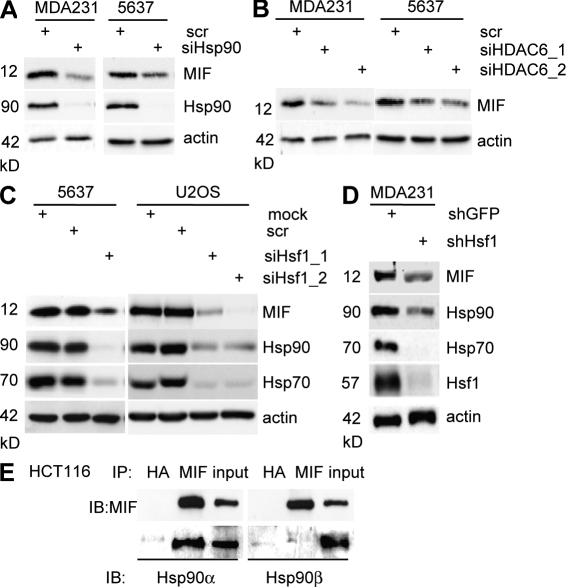
Depletion of Hsp90, HDAC6, or HSF1 all destabilize MIF protein
HDAC6 is the main cytosolic histone deacetylase and an obligate positive regulator of HSP90’s chaperone function toward client proteins (Bali et al., 2005; Kovacs et al., 2005; Scroggins et al., 2007). Toward further support of MIF as a novel HSP90 client, depletion of either Hsp90 or HDAC6 deacetylase should mimic the effect of 17AAG, GA, or SAHA seen in Fig. 2. Indeed, siRNA-mediated silencing of Hsp90 and HDAC6 strongly destabilized MIF protein in cancer cells (Fig. 3, A and B). HSF1, the master transcriptional regulator of the inducible heat shock response, controls most of the stress-inducible chaperones including Hsp90 (Xiao et al., 1999). HSF1 is frequently up-regulated in human tumors, and the HSF1-mediated stress response plays a causal, broadly supportive role in mammalian oncogenesis. Thus, as predicted, siRNA- and shRNA-mediated knockdown of HSF1 in cancer cells, which in turn down-regulates Hsp90 and Hsp70 proteins, also induced destabilization of MIF (Fig. 3, C and D; and not depicted). Of note, HSF1 primarily regulates transcription of the stress-inducible α isoform of Hsp90, whereas the β isoform is regulated by other transcription factors (Sreedhar et al., 2004). Thus, according to our model, MIF should preferentially bind to Hsp90α but not β, which is indeed the case, as confirmed by coimmunoprecipitation (Fig. 3 E). Collectively, we conclude that MIF is a novel HSP90 client in cancer cells and that it is this chaperone association that mediates MIF stabilization.
Figure 3.
Depletion of Hsp90, HDAC6, or Hsf1 all destabilize MIF protein. (A and B) MDA231 and 5637 cells were transfected with siRNA against the Hsp90 chaperone (A) or against HDAC6 (two different sequences, siHDAC6_1 and siHDAC6_2; B). After 3 d, MIF and Hsp90 protein levels were assessed by immunoblots. Representative blots from two independent experiments are shown. Actin, loading control. (C and D) 5637 and U2OS cells transfected with two different siRNAs against Hsf1 for 3 d (C), and MDA231 cells stably transfected with an shRNA against Hsf1 were immunoblotted for MIF, Hsp90, Hsp70, and Hsf1 (D). Representative blots from three independent experiments. Actin, loading control. (E) Untreated HCT116 cells were subjected to coimmunoprecipitation with anti-MIF or irrelevant anti-HA antibodies and immunoblotted with isoform-specific Hsp90 antibodies.
The E3 ubiquitin ligase CHIP and the proteasome are required for MIF degradation upon HSP90 inhibition
The rapid turnover of MIF protein after HSP90 inhibition suggests that it might be subject to proteasomal degradation under such circumstances. Indeed, the proteasome inhibitor MG132 completely blocked MIF destabilization in response to 17AAG or SAHA shown in U2OS cells (Fig. 4, A and B) and 5637 cells (not depicted). Because ubiquitination is a prerequisite for proteasomal turnover, it suggests that MIF, when no longer bound to HSP90, is modified by ubiquitin ligase. We therefore attempted to identify the E3 ligase that mediates MIF degradation.
During protein maturation in normal cells, the HSP90-associated E3 ubiquitin ligase CHIP (carboxy terminus of HSP70-interacting protein) is recruited to induce proteasomal degradation of misfolded or aggregated molecules. In cancer cells with up-regulated and activated HSP90, presentation of aberrant clients to CHIP and CHIP activity is impaired. However, inhibitors binding to the N terminus of Hsp90 can restore this function and reactivate CHIP or other E3 ligases, such as Parkin and Cullin 5, toward aberrant clients, leading to their proteasomal degradation and cellular depletion (Whitesell et al., 1994; Xu et al., 2002; Trepel et al., 2010). To test which E3 ligase plays a role in proteasomal MIF degradation that occurs after HSP90 inhibition, we silenced CHIP and then treated cells with 17AAG to inactivate Hsp90. Indeed, CHIP depletion largely prevented 17AAG-induced MIF degradation in cancer cells (Fig. 4, C and D). Likewise, CHIP depletion also partly abolished MIF degradation in cancer cells where HSP90 activity was inhibited by HDAC6 silencing (Fig. 4 E, compare lanes 2 and 4). Coimmunoprecipitations in the absence and presence of 17AAG showed that MIF was prebound in a constitutive endogenous complex with CHIP (Fig. 4 F). This is expected because in the absence of 17AAG, the stabilized HSP90 client MIF is trapped in this large chaperone complex together with the inactive Hsp70-bound CHIP ligase and multiple co-chaperones (Trepel et al., 2010). However, upon Hsp90 inhibition by 17AAG, the constitutive MIF–Hsp90 complex becomes partly disrupted (Fig. 4 G) and Hsp70 undergoes HIF1-mediated induction and activation, which in turn increases the association of Hsp70 with MIF and enhances CHIP activity toward MIF (Fig. 4 H). Other E3 ubiquitin ligases, such as MDM2, Parkin, and Cullin 5, that are also known to be involved in HSP90 client degradation (Peng et al., 2001; Morishima et al., 2008; Ehrlich et al., 2009) play no discernable role in MIF degradation. Neither silencing of MDM2 (Fig. 4 E) nor silencing of Parkin or Cullin5 (Fig. 4, I and J) could rescue 17AAG-mediated MIF destabilization. In sum, these data identify CHIP as the E3 ligase that is largely responsible for MIF degradation via proteasomes after Hsp90 inhibition in cancer cells.
17AAG-induced apoptosis and growth defects are significantly rescued by excess ectopic MIF
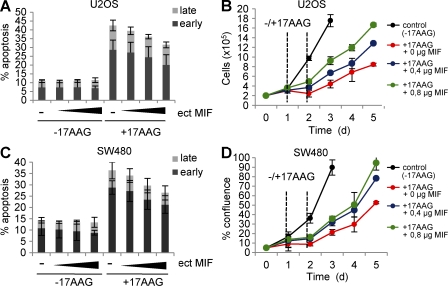
17AAG-mediated inhibition of Hsp90 in cancer cells can cause growth defects and induces apoptosis (Whitesell and Lindquist, 2005; Trepel et al., 2010), which correlates with MIF degradation (this study). Similarly, genetic knockout of MIF alone can induce growth arrest and cell death (Fingerle-Rowson et al., 2003; Petrenko et al., 2003; Petrenko and Moll, 2005; Lue et al., 2007; Nemajerova et al., 2007). To causally establish that it is specifically MIF degradation that significantly contributes to the anti-tumor effect of pharmacological Hsp90 inhibition, we used excess ectopic MIF to rescue the 17AAG-induced effects. Indeed, excess ectopic MIF that had exhausted 17AAG’s ability to degrade MIF at the concentration used (unpublished data) also partially squelched 17AAG’s ability to induce apoptosis (Fig. 5, A and C) and rescued 17AAG-induced growth defects by ∼40–50% (Fig. 5, B and D). Together, this argues that MIF degradation is a major route that mediates the cytotoxic effect of 17AAG.
Figure 5.
17AAG-induced apoptosis and growth defects are significantly rescued by excess MIF. (A and C) U2OS (A) and SW480 (C) cells were transiently transfected with increasing amounts (indicated as wedges) of MIF expression plasmid (ect MIF) or 0.8 µg empty control vector per well in 12-well plates. At day 1 after transfection, cells were treated with 5 µM 17AAG for 24 h, or left untreated, and stained with Annexin and 7-AAD to count cells in early and late apoptotic cell phases by flow cytometry. Error bars indicate the mean of three independent experiments. (B and D) U2OS (B) and SW480 (D) cells were transiently transfected with MIF expression plasmids as in A and C. At day 1 after transfection, 5 × 104 cells per 12-well plate were seeded (d0) and cultured for another 24 h. Cells were then treated with 5 µM 17AAG for 24 h (time interval indicated by vertical dashed lines) or left untreated. During subsequent culturing, cell numbers (U2OS) or cell confluence (SW480) was measured by CELIGO Cytometer using 49 squares per well. Error bars indicate the mean of two independent experiments in duplicates each. Time is in days (d).
In the MMTV-ErbB2 mouse model of human HER2-positive breast cancer, genetic MIF loss delays cancer progression by activating p53
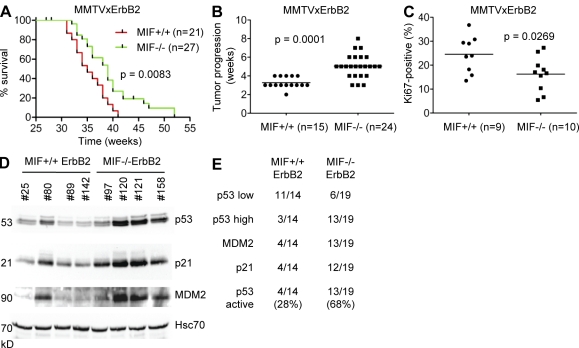
To date, a causal tumor-promoting role of aberrantly accumulated MIF in cancer cells in vivo has only been established in a few cancer types. Using MIF knockout mice, we and others showed that MIF specifically promotes B cell lymphomagenesis in transgenic EµMyc mice (Talos et al., 2005), ulcerative colitis-induced colorectal tumorigenesis (Wilson et al., 2005), nitrosamine-induced bladder cancer (Taylor et al., 2007), and UVB-induced skin cancer (Martin et al., 2009). It is currently unclear, however, what exact role MIF overexpression plays in breast cancer, the leading female cancer type (Bando et al., 2002; Xu et al., 2008; Verjans et al., 2009). Thus, we generated a genetically defined breast cancer model in mice. To this end, we used transgenic MMTV-ErbB2 mice, which exhibit 100% penetrance of spontaneously developing multifocal breast cancer by 30–40 wk of age and are an excellent model for the molecular HER2+ subtype of human breast cancer (Muller et al., 1988). Mammary tumorigenesis by ErbB2 is mediated via activation of Ras signaling and the PI3–Akt kinase pathway that inhibits proapoptotic proteins such as BAD, Forkhead, and caspase 9. MMTV-ErbB2 mice were crossed with MIF-null mice (Fingerle-Rowson et al., 2003) and female offspring were analyzed for cancer development. Both MIF+/+ and MIF−/− mice developed well differentiated mammary adenocarcinoma with identical histology (bulky tumor nodules with malignant epithelial cells delineated by scant stromal septae that contain fibroblasts with a paucity of inflammatory cells) and comparable expression of the ErbB2 transgene (unpublished data). Of note, as predicted by tumor-specific activation of the HSP90 chaperone complex, ErbB2 cancers in MIF+/+ mice exhibit marked overexpression of MIF in malignant breast epithelium compared with normal intervening stroma (Fig. 1 B, top and middle). No significant difference was seen in the time it took for tumor onset (median onset in MIF+/+ mice 32.5 wk, n = 21; and in MIF−/− mice 34 wk, n = 27; P = 0.1343) and the number of tumors developed per mouse (2.86 tumors per mouse in MIF+/+ mice, n = 15; and 2.66 tumors per mouse in MIF−/− mice, n = 24; P = 0.6544). Importantly, however, MIF−/− ErbB2 mice survived significantly longer (median survival 39 wk vs. 35 wk for MIF+/+ ErbB2 mice, P = 0.0083), with six MIF−/− ErbB2 mice surviving up to 52 wk. In contrast, 100% of MIF+/+ErbB2 mice were dead by 41 wk (Fig. 6 A). The extended survival was mainly a result of slower tumor growth in MIF−/− ErbB2 mice (5.3 wk in MIF−/− mice vs. 3.1 wk in MIF+/+ mice) to reach the allowable endpoint volume of 900 mm3 (P = 0.0001; Fig. 6 B). In turn, delayed tumor progression in MIF−/−ErbB2 mice is a result of decreased proliferation, as indicated by lower Ki67 staining in MIF−/− tumor tissues (Fig. 6 C, mean Ki67 staining of MIF−/− 16% vs. MIF+/+ 24%; P = 0.0269), whereas apoptosis was insignificant in both genotypes (unpublished data).
Figure 6.
In the MMTV-ErbB2 mouse model of breast cancer, genetic MIF loss delays cancer progression by activating p53. (A) Kaplan-Meier survival curves of MIF+/+ErbB2 (n = 21) and MIF−/−ErbB2 (n = 27) mice. Log-rank test, P = 0.0083. Note that 3 of the original 27 MIF−/− ErbB2 mice and 6 of the original 21 MIF+/+ ErbB2 mice were censored because they died from salivary cancers rather than breast cancers. (B) Speed of breast tumor growth/progression in MIF+/+ ErbB2 versus MIF−/− ErbB2 mice, calculated from the time it took from when the first of several tumors in an animal was initially palpable until it had reached the allowable endpoint volume of 900 mm3. Data are shown as scatter plot. Student’s t test, P = 0.0001. Horizontal bars indicate the mean of all values. (C) Ki67 staining of histological sections of MIF+/+ErbB2 (n = 9) and MIF−/−ErbB2 (n = 10) tumors was quantitated using a digital mask (ImageJ software). Eight random fields (20× magnification) of three standardized hematoxylin-counterstained tumor sections per mouse per genotype were counted. The number of Ki67-positive cells was calculated as percentage of total nuclei. Student’s t test, P = 0.0269. Horizontal bars indicate the mean of all values. (D) Lysates from representative tumors of MIF+/+ and MIF−/− ErbB2 mice were analyzed by immunoblot for levels of p53 and its target genes p21 and MDM2. Each number indicates a different mouse. Hcs70, loading control. (E) Summary of all MIF+/+ErbB2 (n = 14) and MIF−/−ErbB2 (n = 19) tumors analyzed by immunoblotting as in Fig. 6 D. Compared with an MIF+/+ reference tumor (tumor #25 in Fig. 6 D and Fig. 1 B), p53 low means the same p53 protein levels and p53 high means higher p53 protein levels. p53 was scored as activated if p53 levels were high and p21 and MDM2 levels were up-regulated.
MMTV-ErbB2–induced breast tumors rarely exhibit p53 mutations/deletions, nor do they undergo WT p53 accumulation indicative of p53 activation (Taneja et al., 2010). Using genetic analysis, we previously showed that MIF depletion activates the p53 pathway (Fingerle-Rowson et al., 2003; Petrenko et al., 2003; Petrenko and Moll, 2005; Talos et al., 2005). Thus, we hypothesized that p53 activation could be a major determinant responsible for the delayed tumor progression and extended survival of MIF−/−ErbB2 mice. To test this notion, all ErbB2 tumors were analyzed for p53 levels by immunoblots. Indeed, the majority of MIF−/−ErbB2 tumors (68%, 13 of 19 tumors) showed significant p53 accumulation, compared with only 21% (3 of 14 tumors) of MIF+/+ErbB2 tumors (representative examples in Fig. 6 D). Moreover, almost all tumors in this p53-activated MIF−/− group (12 or 13 of 19) showed concomitant induction of the p53 target genes p21 and MDM2 (Fig. 6 E, examples in Fig. 6 D), compared with only 28% (4 of 14) of MIF+/+ tumors. We sequence confirmed (exons 2–9) the WT status of accumulated p53 in 11 of 11 MIF−/− tumors with high p53 levels. No tumor showed Puma activation (unpublished data), consistent with the absence of apoptosis in this tumor type.
In sum, these data indicate that MIF is a major tumor promoter in ErbB2-driven breast cancer in vivo. Even more importantly, the results also predict that pharmacologic MIF suppression via HSP90 inhibition might have meaningful anti-tumor effects in the animal.
Hsp90 inhibition via systemic 17AAG treatment induces marked growth inhibition in MIF+/+ErbB2 tumors but shows little impact in MIF−/−ErbB2 tumors
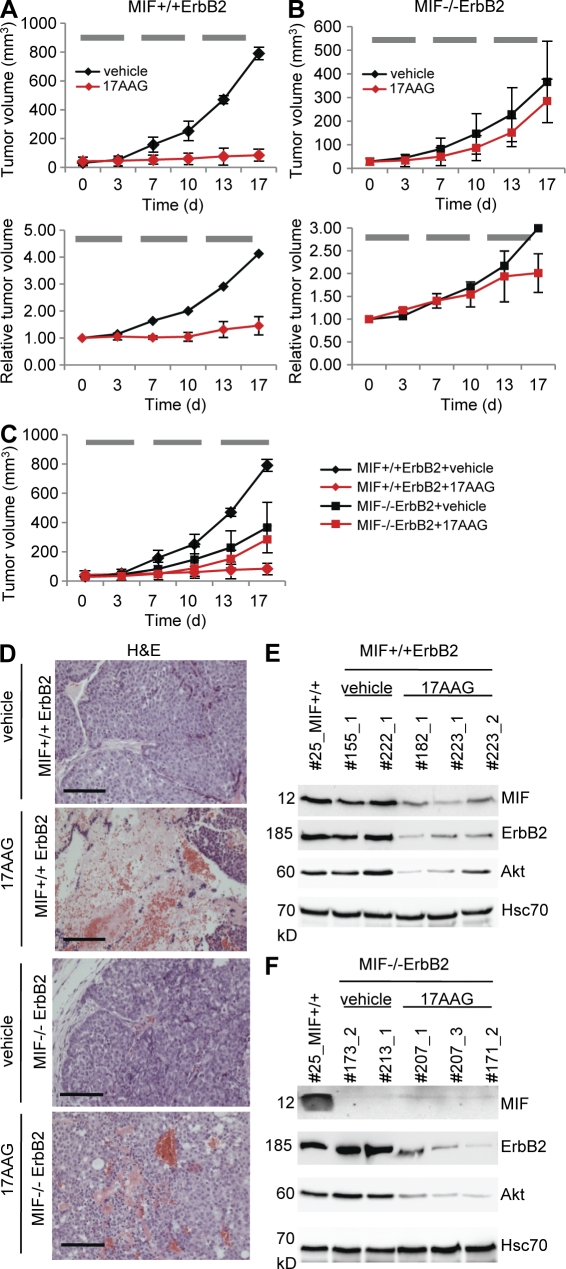
To date, 17AAG-mediated inhibition of Hsp90 function was shown to attenuate tumor progression in several human cancer xenograft models. However, although correlated with down-regulating HSP90 clients like ErbB2, Akt, and androgen receptor (Solit et al., 2002; Banerji et al., 2005; Eiseman et al., 2005; Williams et al., 2007), a causal dependence of the 17AAG-induced tumor suppression on the reduction of specific clients has not been proven. To test whether 17AAG down-regulates aberrantly stabilized MIF and consequently impairs tumor progression in our spontaneous transgenic breast cancers in vivo, we treated MIF+/+ErbB2 and MIF−/−ErbB2 mice systemically with 60 mg/kg 17AAG or vehicle by intraperitoneal injections 5 d a week for 3 wk. Indeed, rapid tumor growth in MIF+/+ErbB2 mice was brought to a complete halt in 17AAG-treated animals compared with vehicle-treated mice (Fig. 7 A, top, compare red with black line; for details see Fig. S1) and was accompanied by marked drug-induced tumor necrosis (Fig. 7 D, top). Importantly, this dramatic response in MIF+/+ErbB2 tumors was associated with destabilization of elevated MIF levels as well as the other HSP90 clients ErbB2 and Akt, as expected (Fig. 7 E).
Figure 7.
Hsp90 inhibition via systemic 17AAG treatment induces marked growth inhibition in MIF+/+ErbB2 tumors but shows no impact in MIF−/−ErbB2 tumors. (A–C) Median tumor volumes in response to treatment. Time course is in days (d). Mice with small comparably sized breast tumors (mostly <50 mm3; A and B, top) and larger tumors (mostly >200 mm3; A and B, bottom) of MIF+/+ErbB2 (A) and MIF−/−ErbB2 (B) genotypes were treated with intraperitoneal injections of 17AAG (red lines) or vehicle (EPL diluent, black lines) 5 d per week for 3 wk. Horizontal gray bars indicate the time windows of daily 17AAG treatments. During treatment, tumor sizes were monitored twice a week. Top, for small tumors, four independent experiments for treatment/genotype combinations were performed on different days in a total of 22 mice (seven mice in experiment 1; five mice in experiment 2; four mice in experiment 3; and six mice in experiment 4). MIF+/+ErbB2 + vehicle: 5 tumors in 4 mice; MIF+/+ErbB2 + 17AAG: 8 tumors in 5 mice; MIF−/−ErbB2 + vehicle: 9 tumors in 5 mice; MIF−/−ErbB2 + 17AAG: 16 tumors in 8 mice. Each independent experiment was done side by side for all treatment/genotype combinations. For clarity, data are shown separately in A (MIF+/+ErbB2) and B (MIF−/−ErbB2). Overlay of both genotypes is graphed in C. For detailed listing of all small tumors, see Fig. S1. Bottom, larger tumors (mostly > 200 mm3) were analyzed in one experiment side by side in a total of eight mice. For clarity, treatment/genotype combinations are shown separately in A (MIF+/+ErbB2) and B (MIF−/−ErbB2). The response rates of the larger tumors were normalized to their respective starting tumor volumes. As expected, vehicle-treated small and large control tumors showed a similar tumor progression (compare black lines, top and bottom). MIF+/+ErbB2 + vehicle: one tumor in one mouse. MIF+/+ErbB2 + 17AAG: three tumors in three mice. MIF−/−ErbB2 + vehicle: one tumor in one mouse. MIF−/−ErbB2 + 17AAG: three tumors in three mice. For detailed listing of all larger tumors, see Fig. S2. Error bars indicate the mean of all tumors measured per treatment/genotype combination. (D) Tumors from MIF+/+ErbB2 and MIF−/−ErbB2 mice treated with intraperitoneal 17AAG or vehicle were stained by H&E. Bars, 100 µm. (E and F) Mice were sacrificed 8 h after the final dose of intraperitoneal 17AAG or vehicle on day 17 (see A–C) and breast tumors were harvested. Lysates of MIF+/+ErbB2 (E) and MIF−/−ErbB2 tumors (F) treated with 17AAG or vehicle (EPL) were immunoblotted. Effective inhibition of Hsp90 by 17AAG was confirmed by degradation of MIF, ErbB2, and Akt. Hcs70, loading control. Each number indicates a different mouse. Tumor #25 served as reference tumor also used in Figs. 1 B and 6 D.
In contrast and as expected, vehicle-treated MIF−/−ErbB2 tumors grew more slowly as a result of lack of MIF (Fig. 7 B, top black line; see also Fig. 6). Importantly, though, and in contrast to the strong effect seen in MIF+/+ tumors, 17AAG treatment essentially failed to inhibited growth in MIF−/−ErbB2 tumors (Fig. 7 B, top red line, no significant difference to black line; Fig. 7 C superimposes data from both genotypes), despite the fact that ErbB2 and Akt were equally reduced by 17AAG in these tumors (compare Fig. 7, E and F). We repeated the 17AAG treatment experiments on additional mice starting with larger tumors (mostly >200 mm3) and preliminary results suggest that irrespective of tumor size, MIF is a critical factor in drug response (Fig. 7, A and B, bottom; for details see Fig. S2). In contrast to MIF+/+ tumors, larger MIF−/− tumors again were only slightly responsive to 17AAG treatment and became so only toward the very end of treatment, similar to what we saw for smaller tumors (Fig. 7 B, top). Thus, the intrinsically slower tumor growth of MIF−/− tumors does not mask or somehow distort the observed 17AAG effects. In aggregate, the loss or reduction of 17AAG-induced anti-tumor efficacy specifically in MIF−/−ErbB2, but not in MIF+/+ErbB2, tumors indicates that a critical in vivo target of 17AAG is, surprisingly, the tumor-promoting client MIF, in conjunction with the coexpressed ErbB2 and Akt clients. Conversely, the dramatic anti-tumor effect of 17AAG treatment in MIF+/+ErbB2 mice is also the result of MIF degradation. In sum, these data further support the notion that MIF is a pathologically important HSP90 client involved in cancer progression and that tumor-associated MIF accumulation sensitizes to a 17AAG-induced anti-tumor response.
DISCUSSION
Here, we identify MIF as a novel client of the tumor-activated HSP90 chaperone machinery and show that HSP90 is responsible for the aberrant MIF accumulation that characterizes many established human cancers. Furthermore, we show that MIF overexpression in tumor tissues is an important factor in tumor progression because mice with MIF-deficient ErbB2-driven breast cancer exhibit delayed tumor progression and prolonged survival. Together, these findings render MIF as a druggable anti-tumor target. Most importantly, our genetic MIF-ErbB2 analysis indicates that induced degradation of MIF, in addition to induced degradation of HSP90 clients from the ErbB2-Akt and other signal transduction pathways, is a critical determinant in the growth suppressive anti-tumor response to pharmacological HSP90 inhibitors in vivo.
Research during the previous decade established that aberrantly stabilized MIF is an important tumor promoter with pleiotropic actions in multiple pathways. Hence, varying degrees of increased MIF levels are found in a majority of human malignancies (Bini et al., 1997; Meyer-Siegler, 2000, 2006; Bando et al., 2002; Tomiyasu et al., 2002; Pyle et al., 2003; Reome et al., 2004; Hagemann et al., 2005, 2007; Hira et al., 2005; He et al., 2006; Xu et al., 2008; Verjans et al., 2009; Cludts et al., 2010; Cheng et al., 2011; Zhang et al., 2011; Wang et al., 2012), making MIF an attractive drug target for anti-cancer therapy. However, our current knowledge of functional interactions of MIF in cancer remains sketchy. MIF’s tautomerase activity is not important (Fingerle-Rowson et al., 2009), and more importantly a unifying concept of a biochemical mechanism of MIF activities in tumors remains elusive. This makes it difficult, if not impossible, to develop specific small molecule inhibitors that would bind critical domains of MIF to block its many diverse activities. Our results now point to a simple and effective indirect way to pharmacologically target MIF. Using 17AAG as proof of principle for this drug class, HSP90 inhibitors effectively destabilize MIF and hence diminish the tumor promoting activities of MIF in cultured human cancer cells and in ErbB2 oncogene-driven breast cancer in mice. We find that HSP90 inhibitors are effective MIF inhibitors that attain significant anti-tumor responses in vivo.
17AAG has previously been found to reduce solid tumor progression in preclinical mouse models. However, two shortcomings characterized these studies. First, all but one study were limited to tumor xenografts, questioning their predictive relevance for human cancers (Solit et al., 2002; Banerji et al., 2005; Eiseman et al., 2005; Kang et al., 2006; Williams et al., 2007; Park et al., 2008; Watanabe et al., 2009; Sun et al., 2010). In contrast, we show here that systemically administered 17AAG displays strong anti-tumor efficacy in spontaneously occurring cancers of transgenic mice that closely model HER2/ErbB2-positive breast cancer, one of the most frequent cancer subtypes in humans. Second, these earlier studies could not clearly assign the anti-tumor effect of HSP90 inhibitors to specific clients. Using genetically defined MIF-proficient and deficient versions of ErbB2 breast cancers, our study now identifies that one important determinant of the anti-cancer activity of 17AAG is its ability to specifically induce efficient degradation of MIF (Fig. 7).
Given the plethora of known HSP90 clients in tumors (Whitesell and Lindquist, 2005; Taipale et al., 2010; Trepel et al., 2010), it is surprising that MIF turns out to be so important for 17AAG-mediated inhibition of tumor growth. In this model, other HSP90 clients are also causally involved in tumor formation, notably Erbb2, the driving oncoprotein for this tumor type, which signals to PI3K/Akt. At least in this experimental setting, they appear less critical for the anti-tumor response to HSP90 interference because ErbB2 and Akt were similarly degraded by 17AAG in both MIF+/+ and MIF−/− tumors (Fig. 7, E and F) and, thus, did not correlate with drug sensitivity (Fig. 7, A and B). Collectively, although other molecular tumor types might have a different profile of dependence on HSP90-regulated oncoproteins, MIF was a critical HSP90 client in this important tumor type.
Aside from MIF overexpression shown here, the transcription factors ID1 and ID3, implicated in regulating tumor angiogenesis, represent another determinant of how transgenic ErbB2 mammary tumors respond to 17AAG. Tumors that were poorly vascularized as a result of genetic ID1/3 ablation responded better to 17AAG (deCandia et al., 2003). It remains to be determined whether MIF reduction in tumors also results in increased responsiveness to hypoxia. However, because both MIF loss and hypoxia induce a p53 response, it is conceivable that synergistic p53 activation might underlie the improved 17AAG responsiveness of poorly vascularized ID1/3-deficient tumors (deCandia et al., 2003). Even more strikingly, previous studies reported induction of MIF transcription by HIF1α (hypoxia-inducible factor 1α; Welford et al., 2006) and, conversely, HIF1α protein levels being stabilized by MIF (Winner et al., 2007; Oda et al., 2008). This raises the intriguing possibility that tumors lacking sufficient angiogenesis and/or suffering from hypoxia increase MIF and depend on MIF overexpression and, therefore, should be exquisitely sensitive to HSP90 inhibition.
Although not yet FDA approved, the clinical development of HSP90 inhibitors is making steady progress by improving formulations, oral bioavailability, further lowering the already acceptable toxicity, and adding >10 new chemically distinct molecules to the prototype 17AAG. There are currently 23 active oncology trials involving HSP90 inhibitors. 17AAG (Tanespimycin) is the most advanced and currently in phase II and III clinical trails. Of note, promising results were reported in a phase II trial of progressive HER2-positive metastatic breast cancer patients that had progressed (failed) under trastuzumab treatment. Weekly treatments with 17AAG plus trastuzumab yielded an overall response rate in 22% and an overall clinical benefit including stable disease in 59% of patients (Modi et al., 2011). Two similar trials are currently still ongoing (Trepel et al., 2010). Elevated intratumoral MIF levels have previously been shown to correlate with tumor aggressiveness and poor prognosis in conventional chemotherapy regimens. Our results suggest that the degree of MIF overexpression, and possibly a WT p53 status, represent potential predictive markers for tumor responsiveness toward HSP90 inhibitors. Whether MIF levels provide a translatable strategy for how to better use 17AAG could be tested in future clinical studies.
Combined with conventional anti-cancer drugs (DNA damaging agents, S-phase inhibitors, and antimitotics), HSP90 inhibition by 17AAG-type drugs (a direct Hsp90 core protein ATPase inhibitor) and by SAHA (an HSP90 inhibitor via HDAC6 blockade) is increasingly emerging as a promising concept for tumor therapy precisely because their effect is broad range. This is because this concept is based on targeting a central molecular hub of tumor state maintenance and because it generates a large therapeutic window to normal tissues that lack constitutive HSP90 up-regulation and activation. In the case of SAHA (Vorinostat), which is the first FDA-approved HDAC inhibitor (since 2006), the combination of Hsp90 inhibition and HDAC(6) inhibition should further enhance MIF degradation and target an even broader spectrum of tumor regulatory pathways. HDAC inhibition by SAHA contributes to MIF reduction transcriptionally (Roger et al., 2007; Lugrin et al., 2009) and, as we showed here, to MIF protein degradation by inhibiting the HDAC6-HSP90 axis (Fig. 2). Overall, our results further support the notion that in addition to targeted cancer therapeutics, such broad-range tumor drugs are also clinically useful. MIF appears at the center of such signaling pathways and serves as a major target for HSP90 inhibitors in cancer.
MATERIALS AND METHODS
Mouse models.
The activated ErbB2 transgenic mouse FVBN-Tg(MMTV-ErbB2)NK1Mul/J (The Jackson Laboratory) is one of the most commonly used spontaneous breast cancer models because of its clear phenotype and molecular mimicry of the human disease (Barrington et al., 1998; Cardiff et al., 2000). They express the activated ErbB2 (c-neu) oncogene carrying a Val664 to Glu664 mutation, driven off the MMTV promoter (Muller et al., 1988). Random transgene expression occurs in mammary gland epithelium from hemizygous mice. Tumor formation is multifocal, stochastic, and matches the transgene expression. Homozygous ErbB2 mice were crossed with homozygous MIF−/− mice (129SV background; Fingerle-Rowson et al., 2003). Heterozygous F1 offspring were crossed with MIF+/+ or MIF−/− mice generating MIF−/−ErbB2 or MIF+/+ErbB2 animals heterozygous for the MMTV-ErbB2 transgene. This F2 generation had a mixed strain background of 75% 129SV/25% FVBN. Mice were palpated for tumors twice a week. As expected, they developed breast tumors starting from 25 wk of age. After reaching a tumor size of 12 × 12 × 12 mm, mice were euthanized and tumors dissected. Tumor-free fat pads were used as control tissue. For tumor treatments, 17AAG (provided by Bristol-Myers Squibb and the National Cancer Institute at National Institutes of Health) was pre-dissolved in DMSO and further diluted in EPL egg yolk emulsion diluent (10% DMSO/EPL, provided by the National Cancer Institute). Mice with small, comparably sized palpable tumors (mostly <50 mm3) or larger tumors (mostly >200 mm3) were treated by intraperitoneal injection with 60 mg/kg 17AAG or vehicle alone for 5 d per week for 3 wk. During treatment, tumor sizes were monitored twice a week using a caliper and tumor volumes were calculated as ellipsoid (V = abc × 4/3π). At day 17, mice were euthanized and tumors dissected for analysis. All experiments were performed in full agreement with the Göttingen University Animal Care Committee and the Institutional Guidelines for Humane Use of Animals in Research.
Cell culture, reagents, siRNAs, and plasmids.
Human cancer cells H1299 (lung), U2OS (osteosarcoma) and PaTu8902, PANC-1, MiaPac (pancreas), BT20 (breast), and DU145 (prostate) were cultured in DME/10% FBS. HCT116 colon cancer cells were cultured in McCoys/10% FBS. Human cancer cells 5637 (bladder), SW480 (colon), MCF7, MDA-MB231, and MDA-MB468 (all breast) were cultured in RPMI/10%FBS. Immortalized human MCF10A mammary epithelial cells were cultured in DME/F12 media containing 5% horse serum and PS (Invitrogen), 100 µg/ml EGF, 1 mg/ml hydrocortisone, 1 mg/ml cholera toxin, and 10 mg/ml insulin (all Sigma-Aldrich). MEFs were cultured in DME/10% FBS. Freshly prepared 17AAG (diluted in methanol; EMD) and SAHA (diluted in DMSO; Enzo Life Sciences) was used as indicated. CHX and MG132 (EMD) was stored in DMSO and used as indicated. All siRNAs were purchased from Invitrogen (validated or predesigned Silencer select siRNAs). The MIF expression plasmid was generated in pcDNA3.1 (Invitrogen). siRNAs and plasmids were transfected with Lipofectamine 2000 (Invitrogen). Lysates from normal human tissues were ordered by Acris Antibodies GmbH.
Quantitative RT-PCR.
Total RNA from cells and tumor tissue was isolated using Trizol reagent (Invitrogen). Equal amounts of RNA were reverse transcribed with M-MuLV Reverse transcription (New England Biolabs, Inc.). Real-time PCR analysis was performed using PCR Master-Mix (75 mM Tris-HCl, pH 8.8, 20 mM (NH4)2SO4, 0.01% Tween-20, 3 mM MgCl2, SYBR Green 1:80,000, 0.2 mM dNTPs, 20 U/ml Taq polymerase, 0.25% Triton X-100, 0.3 M Trehalose, and 0.3 µM primers). The following primers were used: human MIF, 5′-AGCAGCTGGCGCAGGCCAC-3′ and 5′-CTCGCTGGAGCCGCCGAAGG-3′; mouse MIF, 5′-TCCGTGCCAGAGGGGTTTCTGT-3′ and 5′-ACGTTGGCAGCGTTCATGTCG-3′; mouse 36B4, 5′-GCAGATCGGGTACCCAACTGTTG-3′ and 5′-CAGCAGCCGCAAATGCAGATG-3′; and Gapdh, 5′-TGAAGGTCGGAGTCAACGGATTTG-3′ and 5′-GCAGAGATGATGACCCTTTTGGCTC-5′. Primers were used in a two-step protocol (2 min at 95°C preheating; 40 cycles at 95°C for 15 s, followed by 60°C for 1 min).
Coimmunoprecipitation and immunoblotting.
For coimmunoprecipitation, cells were lysed in NP-40 buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl2, 1% Nonident P40, 5 mM EDTA, and complete protease inhibitor mix; Roche), followed by sonification. After centrifugation, samples were precleared with protein G-Sepharose (GE Healthcare) and equal amounts of total protein were immunoprecipitated with antibodies to MIF, Hsp90, HA (all Santa Cruz Biotechnology, Inc.), or CHIP (Calbiochem). An aliquot of each lysate was used as input control. Precipitates were analyzed by immunoblotting. In brief, whole cell lysates were made with RIPA buffer (1% Triton X-100, 1% Desoxycholate, 0,1% SDS, 150 mM NaCl, 10 mM EDTA, 20 mM Tris-HCl, pH 7.5, and complete protease inhibitor mix). For tumor tissues, tissues were finely minced and lysed with RIPA buffer followed by sonication. After centrifugation, protein amounts were measured with a BCA protein assay (Thermo Fisher Scientific). For immunoblotting, equal amounts of protein lysates or precipitates were separated by SDS-gel electrophoresis, transferred onto nitrocellulose membrane (Millipore), blocked, and probed with the following antibodies: MIF (FL-115, human and mouse), p53 (DO-1), Hsp70 (W-27), pan-Hsp90 (H-114), Hsp70 (all Santa Cruz Biotechnology, Inc.), polyclonal Hsp90 α and polyclonal Hsp90 β (Enzo Life Sciences), p21 (human, OP-64; EMD), Akt (9271, human and mouse) and cleaved Caspase3 (both Cell Signaling Technology), CHIP (PC711; EMD), Gapdh, and Actin (8245 and 8227; Abcam). Antibodies used for mouse tissues were p21 (F-5), MDM2 (SMP-14), Hsc70 (B-6; all Santa Cruz Biotechnology, Inc.), p53 (Ab-1; EMD), and ErbB2 (29D8; Cell Signaling Technology).
Immunohistochemistry.
Tumors were bisected into two parts, one for protein and/or RNA extraction and the other for immunohistochemistry. Tumors were fixed overnight in 3.7% paraformaldehyde (Sigma-Aldrich) at room temperature. After repeated washing in PBS for at least 3 h, tumors were dehydrated in an alcohol series, embedded in paraffin, sectioned (4 µm), and processed for immunohistochemistry as previously described (Holembowski et al., 2011). Antibodies were Ki67 (TEC-3 and M7249; Dako), MIF (FL-115), and ErbB2 (29D8; Cell Signaling Technology).
Apoptosis assay.
Cells were trypsinized, stained with Annexin and 7-AAD, and counted by flow cytometry for early and late apoptotic phases (GUAVA Nexin; Millipore).
Survival and clonogenic assays.
For cell survival, equal numbers of treated or transfected cells were plated into 12-well plates. Cell confluence and cell numbers were measured over the indicated time periods using a Celigo Cytometer (Cyntellect). For clonogenic assays, equal numbers of transfected cells were plated and cultured. Colonies were fixed in methanol and stained with 0.5% crystal violet in 25% MeOH. For quantification, plates were scanned side by side and colony density was measured as total pixels per plate using Photoshop (Adobe).
Quantitative image analysis, statistical analysis, and densitometric evaluations.
For quantitative image analysis of histological Ki67 staining, comparable images of tumor sections from both genotypes stained with Ki67 and hematoxylin were counted using a digital mask of custom-made scripts written for ImageJ software (National Institutes of Health). The percentage of Ki67-positive nuclei relative to total cell number (hematoxylin staining) was analyzed for statistical significance, using the statistical software PRISM (GraphPad Software). Densitometric measurements for quantification of protein bands were done with the gel analysis software Laboratory Image 1D (Intas Science Imaging GmbH) and normalized for loading control.
Online supplemental material.
Fig. S1 shows an overview of small tumors measured in mice. Fig. S2 shows an overview of large tumors measured in mice. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20111117/DC1.
Acknowledgments
We thank Marie Reinert and Dun Li for technical assistance and Andreas Scheel for writing ImageJ scripts for image quantification.
This work was supported by the German Research Foundation (Deutsche Forschungsgemeinschaft), the Wilhelm Sander-Stiftung für Krebsforschung, and the Carol Baldwin Breast Cancer Research Fund. 17AAG was generously provided by Bristol-Myers Squibb and the National Cancer Institute at National Institutes of Health.
The authors have no conflicting financial interests.
Footnotes
Abbreviations used:
- CHX
- cycloheximide
- HSP90
- heat shock protein 90
- GA
- geldanamycin
- MEF
- mouse embryo fibroblast
- MIF
- macrophage migration inhibitory factor
References
- Bali P., Pranpat M., Bradner J., Balasis M., Fiskus W., Guo F., Rocha K., Kumaraswamy S., Boyapalle S., Atadja P., et al. 2005. Inhibition of histone deacetylase 6 acetylates and disrupts the chaperone function of heat shock protein 90: a novel basis for antileukemia activity of histone deacetylase inhibitors. J. Biol. Chem. 280:26729–26734 10.1074/jbc.C500186200 [DOI] [PubMed] [Google Scholar]
- Bando H., Matsumoto G., Bando M., Muta M., Ogawa T., Funata N., Nishihira J., Koike M., Toi M. 2002. Expression of macrophage migration inhibitory factor in human breast cancer: association with nodal spread. Jpn. J. Cancer Res. 93:389–396 10.1111/j.1349-7006.2002.tb01269.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerji U., Walton M., Raynaud F., Grimshaw R., Kelland L., Valenti M., Judson I., Workman P. 2005. Pharmacokinetic-pharmacodynamic relationships for the heat shock protein 90 molecular chaperone inhibitor 17-allylamino, 17-demethoxygeldanamycin in human ovarian cancer xenograft models. Clin. Cancer Res. 11:7023–7032 10.1158/1078-0432.CCR-05-0518 [DOI] [PubMed] [Google Scholar]
- Barrington R.E., Subler M.A., Rands E., Omer C.A., Miller P.J., Hundley J.E., Koester S.K., Troyer D.A., Bearss D.J., Conner M.W., et al. 1998. A farnesyltransferase inhibitor induces tumor regression in transgenic mice harboring multiple oncogenic mutations by mediating alterations in both cell cycle control and apoptosis. Mol. Cell. Biol. 18:85–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basso A.D., Solit D.B., Chiosis G., Giri B., Tsichlis P., Rosen N. 2002. Akt forms an intracellular complex with heat shock protein 90 (Hsp90) and Cdc37 and is destabilized by inhibitors of Hsp90 function. J. Biol. Chem. 277:39858–39866 10.1074/jbc.M206322200 [DOI] [PubMed] [Google Scholar]
- Bini L., Magi B., Marzocchi B., Arcuri F., Tripodi S., Cintorino M., Sanchez J.C., Frutiger S., Hughes G., Pallini V., et al. 1997. Protein expression profiles in human breast ductal carcinoma and histologically normal tissue. Electrophoresis. 18:2832–2841 10.1002/elps.1150181519 [DOI] [PubMed] [Google Scholar]
- Binsky I., Haran M., Starlets D., Gore Y., Lantner F., Harpaz N., Leng L., Goldenberg D.M., Shvidel L., Berrebi A., et al. 2007. IL-8 secreted in a macrophage migration-inhibitory factor- and CD74-dependent manner regulates B cell chronic lymphocytic leukemia survival. Proc. Natl. Acad. Sci. USA. 104:13408–13413 10.1073/pnas.0701553104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bucala R., Donnelly S.C. 2007. Macrophage migration inhibitory factor: a probable link between inflammation and cancer. Immunity. 26:281–285 10.1016/j.immuni.2007.03.005 [DOI] [PubMed] [Google Scholar]
- Calandra T., Roger T. 2003. Macrophage migration inhibitory factor: a regulator of innate immunity. Nat. Rev. Immunol. 3:791–800 10.1038/nri1200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardiff R.D., Anver M.R., Gusterson B.A., Hennighausen L., Jensen R.A., Merino M.J., Rehm S., Russo J., Tavassoli F.A., Wakefield L.M., et al. 2000. The mammary pathology of genetically engineered mice: the consensus report and recommendations from the Annapolis meeting. Oncogene. 19:968–988 10.1038/sj.onc.1203277 [DOI] [PubMed] [Google Scholar]
- Chen G., Cao P., Goeddel D.V. 2002. TNF-induced recruitment and activation of the IKK complex require Cdc37 and Hsp90. Mol. Cell. 9:401–410 10.1016/S1097-2765(02)00450-1 [DOI] [PubMed] [Google Scholar]
- Cheng R.J., Deng W.G., Niu C.B., Li Y.Y., Fu Y. 2011. Expression of macrophage migration inhibitory factor and CD74 in cervical squamous cell carcinoma. Int. J. Gynecol. Cancer. 21:1004–1012 10.1097/IGC.0b013e31821c45b7 [DOI] [PubMed] [Google Scholar]
- Cludts S., Decaestecker C., Johnson B., Lechien J., Leroy X., Kindt N., Kaltner H., André S., Gabius H.J., Saussez S. 2010. Increased expression of macrophage migration inhibitory factor during progression to hypopharyngeal squamous cell carcinoma. Anticancer Res. 30:3313–3319 [PubMed] [Google Scholar]
- Conroy H., Mawhinney L., Donnelly S.C. 2010. Inflammation and cancer: macrophage migration inhibitory factor (MIF)—the potential missing link. QJM. 103:831–836 10.1093/qjmed/hcq148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Candia P., Solit D.B., Giri D., Brogi E., Siegel P.M., Olshen A.B., Muller W.J., Rosen N., Benezra R. 2003. Angiogenesis impairment in Id-deficient mice cooperates with an Hsp90 inhibitor to completely suppress HER2/neu-dependent breast tumors. Proc. Natl. Acad. Sci. USA. 100:12337–12342 10.1073/pnas.2031337100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrlich E.S., Wang T., Luo K., Xiao Z., Niewiadomska A.M., Martinez T., Xu W., Neckers L., Yu X.-F. 2009. Regulation of Hsp90 client proteins by a Cullin5-RING E3 ubiquitin ligase. Proc. Natl. Acad. Sci. USA. 106:20330–20335 10.1073/pnas.0810571106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eiseman J.L., Lan J., Lagattuta T.F., Hamburger D.R., Joseph E., Covey J.M., Egorin M.J. 2005. Pharmacokinetics and pharmacodynamics of 17-demethoxy 17-[[(2-dimethylamino)ethyl]amino]geldanamycin (17DMAG, NSC 707545) in C.B-17 SCID mice bearing MDA-MB-231 human breast cancer xenografts. Cancer Chemother. Pharmacol. 55:21–32 10.1007/s00280-004-0865-3 [DOI] [PubMed] [Google Scholar]
- Fingerle-Rowson G., Petrenko O., Metz C.N., Forsthuber T.G., Mitchell R.A., Huss R., Moll U., Müller W., Bucala R. 2003. The p53-dependent effects of macrophage migration inhibitory factor revealed by gene targeting. Proc. Natl. Acad. Sci. USA. 100:9354–9359 10.1073/pnas.1533295100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fingerle-Rowson G., Kaleswarapu D.R., Schlander C., Kabgani N., Brocks T., Reinart N., Busch R., Schütz A., Lue H., Du X., et al. 2009. A tautomerase-null macrophage migration-inhibitory factor (MIF) gene knock-in mouse model reveals that protein interactions and not enzymatic activity mediate MIF-dependent growth regulation. Mol. Cell. Biol. 29:1922–1932 10.1128/MCB.01907-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs A., Schwartzman J., Deng V., Alumkal J. 2009. Sulforaphane destabilizes the androgen receptor in prostate cancer cells by inactivating histone deacetylase 6. Proc. Natl. Acad. Sci. USA. 106:16663–16668 10.1073/pnas.0908908106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagemann T., Wilson J., Kulbe H., Li N.F., Leinster D.A., Charles K., Klemm F., Pukrop T., Binder C., Balkwill F.R. 2005. Macrophages induce invasiveness of epithelial cancer cells via NF-kappa B and JNK. J. Immunol. 175:1197–1205 [DOI] [PubMed] [Google Scholar]
- Hagemann T., Robinson S.C., Thompson R.G., Charles K., Kulbe H., Balkwill F.R. 2007. Ovarian cancer cell-derived migration inhibitory factor enhances tumor growth, progression, and angiogenesis. Mol. Cancer Ther. 6:1993–2002 10.1158/1535-7163.MCT-07-0118 [DOI] [PubMed] [Google Scholar]
- He X.X., Yang J., Ding Y.W., Liu W., Shen Q.Y., Xia H.H.X. 2006. Increased epithelial and serum expression of macrophage migration inhibitory factor (MIF) in gastric cancer: potential role of MIF in gastric carcinogenesis. Gut. 55:797–802 10.1136/gut.2005.078113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hira E., Ono T., Dhar D.K., El-Assal O.N., Hishikawa Y., Yamanoi A., Nagasue N. 2005. Overexpression of macrophage migration inhibitory factor induces angiogenesis and deteriorates prognosis after radical resection for hepatocellular carcinoma. Cancer. 103:588–598 10.1002/cncr.20818 [DOI] [PubMed] [Google Scholar]
- Holembowski L., Schulz R., Talos F., Scheel A., Wolff S., Dobbelstein M., Moll U.M. 2011. While p73 is essential, p63 is completely dispensable for the development of the central nervous system. Cell Cycle. 10:680–689 10.4161/cc.10.4.14859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamal A., Thao L., Sensintaffar J., Zhang L., Boehm M.F., Fritz L.C., Burrows F.J. 2003. A high-affinity conformation of Hsp90 confers tumour selectivity on Hsp90 inhibitors. Nature. 425:407–410 10.1038/nature01913 [DOI] [PubMed] [Google Scholar]
- Kang J., Kamal A., Burrows F.J., Evers B.M., Chung D.H. 2006. Inhibition of neuroblastoma xenograft growth by Hsp90 inhibitors. Anticancer Res. 26(3A):1903–1908 [PMC free article] [PubMed] [Google Scholar]
- Kovacs J.J., Murphy P.J.M., Gaillard S., Zhao X., Wu J.-T., Nicchitta C.V., Yoshida M., Toft D.O., Pratt W.B., Yao T.-P. 2005. HDAC6 regulates Hsp90 acetylation and chaperone-dependent activation of glucocorticoid receptor. Mol. Cell. 18:601–607 10.1016/j.molcel.2005.04.021 [DOI] [PubMed] [Google Scholar]
- Lin E.Y., Li J.F., Gnatovskiy L., Deng Y., Zhu L., Grzesik D.A., Qian H., Xue X.N., Pollard J.W. 2006. Macrophages regulate the angiogenic switch in a mouse model of breast cancer. Cancer Res. 66:11238–11246 10.1158/0008-5472.CAN-06-1278 [DOI] [PubMed] [Google Scholar]
- Lubetsky J.B., Dios A., Han J., Aljabari B., Ruzsicska B., Mitchell R., Lolis E., Al-Abed Y. 2002. The tautomerase active site of macrophage migration inhibitory factor is a potential target for discovery of novel anti-inflammatory agents. J. Biol. Chem. 277:24976–24982 10.1074/jbc.M203220200 [DOI] [PubMed] [Google Scholar]
- Lue H., Thiele M., Franz J., Dahl E., Speckgens S., Leng L., Fingerle-Rowson G., Bucala R., Lüscher B., Bernhagen J. 2007. Macrophage migration inhibitory factor (MIF) promotes cell survival by activation of the Akt pathway and role for CSN5/JAB1 in the control of autocrine MIF activity. Oncogene. 26:5046–5059 10.1038/sj.onc.1210318 [DOI] [PubMed] [Google Scholar]
- Lugrin J., Ding X.C., Le Roy D., Chanson A.-L., Sweep F.C.G.J., Calandra T., Roger T. 2009. Histone deacetylase inhibitors repress macrophage migration inhibitory factor (MIF) expression by targeting MIF gene transcription through a local chromatin deacetylation. Biochim. Biophys. Acta. 1793:1749–1758 10.1016/j.bbamcr.2009.09.007 [DOI] [PubMed] [Google Scholar]
- Martin J., Duncan F.J., Keiser T., Shin S., Kusewitt D.F., Oberyszyn T., Satoskar A.R., VanBuskirk A.M. 2009. Macrophage migration inhibitory factor (MIF) plays a critical role in pathogenesis of ultraviolet-B (UVB) -induced nonmelanoma skin cancer (NMSC). FASEB J. 23:720–730 10.1096/fj.08-119628 [DOI] [PubMed] [Google Scholar]
- Meyer-Siegler K.L. 2000. Increased stability of macrophage migration inhibitory factor (MIF) in DU-145 prostate cancer cells. J. Interferon Cytokine Res. 20:769–778 10.1089/10799900050151030 [DOI] [PubMed] [Google Scholar]
- Meyer-Siegler K.L., Iczkowski K.A., Leng L., Bucala R., Vera P.L. 2006. Inhibition of macrophage migration inhibitory factor or its receptor (CD74) attenuates growth and invasion of DU-145 prostate cancer cells. J. Immunol. 177:8730–8739 [DOI] [PubMed] [Google Scholar]
- Meyer-Siegler K.L., Vera P.L., Iczkowski K.A., Bifulco C., Lee A., Gregersen P.K., Leng L., Bucala R. 2007. Macrophage migration inhibitory factor (MIF) gene polymorphisms are associated with increased prostate cancer incidence. Genes Immun. 8:646–652 10.1038/sj.gene.6364427 [DOI] [PubMed] [Google Scholar]
- Mimnaugh E.G., Chavany C., Neckers L. 1996. Polyubiquitination and proteasomal degradation of the p185c-erbB-2 receptor protein-tyrosine kinase induced by geldanamycin. J. Biol. Chem. 271:22796–22801 10.1074/jbc.271.37.22796 [DOI] [PubMed] [Google Scholar]
- Mitchell R.A. 2004. Mechanisms and effectors of MIF-dependent promotion of tumourigenesis. Cell. Signal. 16:13–19 10.1016/j.cellsig.2003.07.002 [DOI] [PubMed] [Google Scholar]
- Modi S., Stopeck A., Linden H., Solit D., Chandarlapaty S., Rosen N., D’Andrea G., Dickler M., Moynahan M.E., Sugarman S., et al. 2011. HSP90 inhibition is effective in breast cancer: a phase II trial of tanespimycin (17-AAG) plus trastuzumab in patients with HER2-positive metastatic breast cancer progressing on trastuzumab. Clin. Cancer Res. 17:5132–5139 10.1158/1078-0432.CCR-11-0072 [DOI] [PubMed] [Google Scholar]
- Morishima Y., Wang A.M., Yu Z., Pratt W.B., Osawa Y., Lieberman A.P. 2008. CHIP deletion reveals functional redundancy of E3 ligases in promoting degradation of both signaling proteins and expanded glutamine proteins. Hum. Mol. Genet. 17:3942–3952 10.1093/hmg/ddn296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moulick K., Ahn J.H., Zong H., Rodina A., Cerchietti L., Gomes DaGama E.M., Caldas-Lopes E., Beebe K., Perna F., Hatzi K., et al. 2011. Affinity-based proteomics reveal cancer-specific networks coordinated by Hsp90. Nat. Chem. Biol. 7:818–826 10.1038/nchembio.670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller W.J., Sinn E., Pattengale P.K., Wallace R., Leder P. 1988. Single-step induction of mammary adenocarcinoma in transgenic mice bearing the activated c-neu oncogene. Cell. 54:105–115 10.1016/0092-8674(88)90184-5 [DOI] [PubMed] [Google Scholar]
- Nemajerova A., Moll U.M., Petrenko O., Fingerle-Rowson G. 2007. Macrophage migration inhibitory factor coordinates DNA damage response with the proteasomal control of the cell cycle. Cell Cycle. 6:1030–1034 10.4161/cc.6.9.4163 [DOI] [PubMed] [Google Scholar]
- Oda S., Oda T., Nishi K., Takabuchi S., Wakamatsu T., Tanaka T., Adachi T., Fukuda K., Semenza G.L., Hirota K. 2008. Macrophage migration inhibitory factor activates hypoxia-inducible factor in a p53-dependent manner. PLoS ONE. 3:e2215 10.1371/journal.pone.0002215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park M.A., Zhang G., Mitchell C., Rahmani M., Hamed H., Hagan M.P., Yacoub A., Curiel D.T., Fisher P.B., Grant S., Dent P. 2008. Mitogen-activated protein kinase kinase 1/2 inhibitors and 17-allylamino-17-demethoxygeldanamycin synergize to kill human gastrointestinal tumor cells in vitro via suppression of c-FLIP-s levels and activation of CD95. Mol. Cancer Ther. 7:2633–2648 10.1158/1535-7163.MCT-08-0400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Y., Chen L., Li C., Lu W., Chen J. 2001. Inhibition of MDM2 by hsp90 contributes to mutant p53 stabilization. J. Biol. Chem. 276:40583–40590 10.1074/jbc.M102817200 [DOI] [PubMed] [Google Scholar]
- Petrenko O., Moll U.M. 2005. Macrophage migration inhibitory factor MIF interferes with the Rb-E2F pathway. Mol. Cell. 17:225–236 10.1016/j.molcel.2004.11.052 [DOI] [PubMed] [Google Scholar]
- Petrenko O., Fingerle-Rowson G., Peng T., Mitchell R.A., Metz C.N. 2003. Macrophage migration inhibitory factor deficiency is associated with altered cell growth and reduced susceptibility to Ras-mediated transformation. J. Biol. Chem. 278:11078–11085 10.1074/jbc.M211985200 [DOI] [PubMed] [Google Scholar]
- Pyle M.E., Korbonits M., Gueorguiev M., Jordan S., Kola B., Morris D.G., Meinhardt A., Powell M.P., Claret F.X., Zhang Q., et al. 2003. Macrophage migration inhibitory factor expression is increased in pituitary adenoma cell nuclei. J. Endocrinol. 176:103–110 10.1677/joe.0.1760103 [DOI] [PubMed] [Google Scholar]
- Rendon B.E., Roger T., Teneng I., Zhao M., Al-Abed Y., Calandra T., Mitchell R.A. 2007. Regulation of human lung adenocarcinoma cell migration and invasion by macrophage migration inhibitory factor. J. Biol. Chem. 282:29910–29918 10.1074/jbc.M704898200 [DOI] [PubMed] [Google Scholar]
- Reome J.B., Hylind J.C., Dutton R.W., Dobrzanski M.J. 2004. Type 1 and type 2 tumor infiltrating effector cell subpopulations in progressive breast cancer. Clin. Immunol. 111:69–81 10.1016/j.clim.2003.11.013 [DOI] [PubMed] [Google Scholar]
- Roger T., Ding X.C., Chanson A.-L., Renner P., Calandra T. 2007. Regulation of constitutive and microbial pathogen-induced human macrophage migration inhibitory factor (MIF) gene expression. Eur. J. Immunol. 37:3509–3521 10.1002/eji.200737357 [DOI] [PubMed] [Google Scholar]
- Scroggins B.T., Robzyk K., Wang D., Marcu M.G., Tsutsumi S., Beebe K., Cotter R.J., Felts S., Toft D., Karnitz L., et al. 2007. An acetylation site in the middle domain of Hsp90 regulates chaperone function. Mol. Cell. 25:151–159 10.1016/j.molcel.2006.12.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi X., Leng L., Wang T., Wang W., Du X., Li J., McDonald C., Chen Z., Murphy J.W., Lolis E., et al. 2006. CD44 is the signaling component of the macrophage migration inhibitory factor-CD74 receptor complex. Immunity. 25:595–606 10.1016/j.immuni.2006.08.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solit D.B., Zheng F.F., Drobnjak M., Münster P.N., Higgins B., Verbel D., Heller G., Tong W., Cordon-Cardo C., Agus D.B., et al. 2002. 17-Allylamino-17-demethoxygeldanamycin induces the degradation of androgen receptor and HER-2/neu and inhibits the growth of prostate cancer xenografts. Clin. Cancer Res. 8:986–993 [PubMed] [Google Scholar]
- Sreedhar A.S., Kalmár E., Csermely P., Shen Y.-F. 2004. Hsp90 isoforms: functions, expression and clinical importance. FEBS Lett. 562:11–15 10.1016/S0014-5793(04)00229-7 [DOI] [PubMed] [Google Scholar]
- Sun J., Liu L., Jiang X., Chen D., Huang Y. 2010. Therapeutic effects of radiolabeled 17-allylamino-17-demethoxygeldanamycin on human H460 nonsmall-cell lung carcinoma xenografts in mice. Cancer Biother. Radiopharm. 25:155–164 10.1089/cbr.2009.0664 [DOI] [PubMed] [Google Scholar]
- Taipale M., Jarosz D.F., Lindquist S. 2010. HSP90 at the hub of protein homeostasis: emerging mechanistic insights. Nat. Rev. Mol. Cell Biol. 11:515–528 10.1038/nrm2918 [DOI] [PubMed] [Google Scholar]
- Talos F., Mena P., Fingerle-Rowson G., Moll U.M., Petrenko O. 2005. MIF loss impairs Myc-induced lymphomagenesis. Cell Death Differ. 12:1319–1328 10.1038/sj.cdd.4401653 [DOI] [PubMed] [Google Scholar]
- Taneja P., Maglic D., Kai F., Sugiyama T., Kendig R.D., Frazier D.P., Willingham M.C., Inoue K. 2010. Critical roles of DMP1 in human epidermal growth factor receptor 2/neu-Arf-p53 signaling and breast cancer development. Cancer Res. 70:9084–9094 10.1158/0008-5472.CAN-10-0159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor J.A., III, Kuchel G.A., Hegde P., Voznesensky O.S., Claffey K., Tsimikas J., Leng L., Bucala R., Pilbeam C. 2007. Null mutation for macrophage migration inhibitory factor (MIF) is associated with less aggressive bladder cancer in mice. BMC Cancer. 7:135 10.1186/1471-2407-7-135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomiyasu M., Yoshino I., Suemitsu R., Okamoto T., Sugimachi K. 2002. Quantification of macrophage migration inhibitory factor mRNA expression in non-small cell lung cancer tissues and its clinical significance. Clin. Cancer Res. 8:3755–3760 [PubMed] [Google Scholar]
- Trepel J., Mollapour M., Giaccone G., Neckers L. 2010. Targeting the dynamic HSP90 complex in cancer. Nat. Rev. Cancer. 10:537–549 10.1038/nrc2887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verjans E., Noetzel E., Bektas N., Schütz A.K., Lue H., Lennartz B., Hartmann A., Dahl E., Bernhagen J. 2009. Dual role of macrophage migration inhibitory factor (MIF) in human breast cancer. BMC Cancer. 9:230 10.1186/1471-2407-9-230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X.B., Tian X.Y., Li Y., Li B., Li Z. 2012. Elevated expression of macrophage migration inhibitory factor correlates with tumor recurrence and poor prognosis of patients with gliomas. J. Neurooncol. 106:43–51 10.1007/s11060-011-0640-3 [DOI] [PubMed] [Google Scholar]
- Watanabe G., Behrns K.E., Kim J.-S., Kim R.D. 2009. Heat shock protein 90 inhibition abrogates hepatocellular cancer growth through cdc2-mediated G2/M cell cycle arrest and apoptosis. Cancer Chemother. Pharmacol. 64:433–443 10.1007/s00280-008-0888-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welford S.M., Bedogni B., Gradin K., Poellinger L., Broome Powell M., Giaccia A.J. 2006. HIF1alpha delays premature senescence through the activation of MIF. Genes Dev. 20:3366–3371 10.1101/gad.1471106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitesell L., Lindquist S.L. 2005. HSP90 and the chaperoning of cancer. Nat. Rev. Cancer. 5:761–772 10.1038/nrc1716 [DOI] [PubMed] [Google Scholar]
- Whitesell L., Mimnaugh E.G., De Costa B., Myers C.E., Neckers L.M. 1994. Inhibition of heat shock protein HSP90-pp60v-src heteroprotein complex formation by benzoquinone ansamycins: essential role for stress proteins in oncogenic transformation. Proc. Natl. Acad. Sci. USA. 91:8324–8328 10.1073/pnas.91.18.8324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams C.R., Tabios R., Linehan W.M., Neckers L. 2007. Intratumor injection of the Hsp90 inhibitor 17AAG decreases tumor growth and induces apoptosis in a prostate cancer xenograft model. J. Urol. 178:1528–1532 10.1016/j.juro.2007.05.120 [DOI] [PubMed] [Google Scholar]
- Wilson J.M., Coletta P.L., Cuthbert R.J., Scott N., MacLennan K., Hawcroft G., Leng L., Lubetsky J.B., Jin K.K., Lolis E., et al. 2005. Macrophage migration inhibitory factor promotes intestinal tumorigenesis. Gastroenterology. 129:1485–1503 10.1053/j.gastro.2005.07.061 [DOI] [PubMed] [Google Scholar]
- Winner M., Koong A.C., Rendon B.E., Zundel W., Mitchell R.A. 2007. Amplification of tumor hypoxic responses by macrophage migration inhibitory factor-dependent hypoxia-inducible factor stabilization. Cancer Res. 67:186–193 10.1158/0008-5472.CAN-06-3292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao X., Zuo X., Davis A.A., McMillan D.R., Curry B.B., Richardson J.A., Benjamin I.J. 1999. HSF1 is required for extra-embryonic development, postnatal growth and protection during inflammatory responses in mice. EMBO J. 18:5943–5952 10.1093/emboj/18.21.5943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W., Marcu M., Yuan X., Mimnaugh E., Patterson C., Neckers L. 2002. Chaperone-dependent E3 ubiquitin ligase CHIP mediates a degradative pathway for c-ErbB2/Neu. Proc. Natl. Acad. Sci. USA. 99:12847–12852 10.1073/pnas.202365899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X., Wang B., Ye C., Yao C., Lin Y., Huang X., Zhang Y., Wang S. 2008. Overexpression of macrophage migration inhibitory factor induces angiogenesis in human breast cancer. Cancer Lett. 261:147–157 10.1016/j.canlet.2007.11.028 [DOI] [PubMed] [Google Scholar]
- Zhang C., Liang T., Song J., Jiang S., Qu L., Hou G. 2011. Evaluation of macrophage migration inhibitory factor as an imaging marker for hepatocellular carcinoma in murine models. Scand. J. Gastroenterol. 46:720–726 10.3109/00365521.2011.568517 [DOI] [PMC free article] [PubMed] [Google Scholar]