The Notch-driven transcription factor RBP-J inhibits osteoclast formation in response to TNF.
Abstract
Tumor necrosis factor (TNF) plays a key role in the pathogenesis of inflammatory bone resorption and associated morbidity in diseases such as rheumatoid arthritis and periodontitis. Mechanisms that regulate the direct osteoclastogenic properties of TNF to limit pathological bone resorption in inflammatory settings are mostly unknown. Here, we show that the transcription factor recombinant recognition sequence binding protein at the Jκ site (RBP-J) strongly suppresses TNF-induced osteoclastogenesis and inflammatory bone resorption, but has minimal effects on physiological bone remodeling. Myeloid-specific deletion of RBP-J converted TNF into a potent osteoclastogenic factor that could function independently of receptor activator of NF-κB (RANK) signaling. In the absence of RBP-J, TNF effectively induced osteoclastogenesis and bone resorption in RANK-deficient mice. Activation of RBP-J selectively in osteoclast precursors suppressed inflammatory osteoclastogenesis and arthritic bone resorption. Mechanistically, RBP-J suppressed induction of the master regulator of osteoclastogenesis (nuclear factor of activated T cells, cytoplasmic 1) by attenuating c-Fos activation and suppressing induction of B lymphocyte–induced maturation protein-1, thereby preventing the down-regulation of transcriptional repressors such as IRF-8 that block osteoclast differentiation. Thus, RBP-J regulates the balance between activating and repressive signals that regulate osteoclastogenesis. These findings identify RBP-J as a key upstream negative regulator of osteoclastogenesis that restrains excessive bone resorption in inflammatory settings.
TNF is an inflammatory cytokine important for immunity and inflammation. The resounding success of TNF blockade therapy has demonstrated a key role for TNF in the pathogenesis of autoimmune/inflammatory diseases such as rheumatoid arthritis (RA), inflammatory bowel disease, and psoriasis (Locksley et al., 2001; Sethi et al., 2009; Taylor and Feldmann, 2009). In addition to driving chronic inflammation, TNF has been implicated in pathological bone resorption (osteolysis) that accompanies inflammatory arthritis and periodontitis and represents an important component of morbidity as it contributes to pain, loss of function, and deformity (Boyce et al., 2006; Teitelbaum, 2006; Schett and Teitelbaum, 2009). An established mechanism by which TNF promotes inflammatory bone resorption is activation of osteoblasts and tissue stromal cells to express receptor activator of NF-κB (RANK) ligand (RANKL), the key factor that induces differentiation and function of osteoclasts, which are multinucleated myeloid lineage cells that are capable of efficient bone resorption. In addition, TNF can act directly on osteoclast precursors, often in synergy with RANKL, to promote osteoclastogenesis (Azuma et al., 2000; Kobayashi et al., 2000; Lam et al., 2000; Li et al., 2000; Kim et al., 2005; Boyce et al., 2006; Teitelbaum, 2006; Yao et al., 2006; Schett and Teitelbaum, 2009). Despite activating similar signaling pathways as does RANKL, TNF does not effectively induce osteoclast differentiation in the absence of RANKL; mechanisms that regulate the direct osteoclastogenic properties of TNF to limit pathological bone resorption in inflammatory settings are mostly unknown (Yao et al., 2009).
RANKL is a member of the TNF family of cytokines that acts in concert with macrophage colony-stimulating factor (M-CSF) and co-stimulatory immunoreceptor tyrosine-based activation motif (ITAM)–associated receptors and integrins to function as the major physiological inducer of osteoclastogenesis. RANKL works by inducing the expression and function of nuclear factor of activated T cells, cytoplasmic 1 (NFATc1), a transcription factor that serves as a “master regulator” of osteoclastogenesis and activates expression of genes important for osteoclast differentiation, fusion, and bone resorption. The positive signaling pathways used by the RANKL receptor RANK to activate NFATc1 are well established and include activation of canonical and noncanonical NF-κB pathways, mitogen-activated kinase (MAPK) pathways leading to activation of AP-1 and CREB transcription factors, and calcium signaling; effective calcium signaling depends on activation of co-stimulatory ITAM-associated receptors (Takayanagi, 2007; Novack and Teitelbaum, 2008). More recently, it has become clear that osteoclastogenesis is restrained by transcriptional repressors that are constitutively expressed in osteoclast precursors and inhibit expression of NFATc1 and osteoclast-related genes (Lee et al., 2006; Hu et al., 2007; Kim et al., 2007; Zhao et al., 2009; Miyauchi et al., 2010; Zhao and Ivashkiv, 2011). RANK signaling needs to overcome the barrier imposed by these transcriptional repressors in order for osteoclastogenesis to proceed. Constitutively expressed repressors of osteoclastogenesis include Eos, inhibitors of differentiation/DNA binding (Ids), v-maf musculoaponeurotic fibrosarcoma oncogene family protein B (MafB), IFN regulatory factor-8 (IRF-8), and B cell lymphoma 6 (Bcl6), and RANKL overcomes inhibition by down-regulating their expression and repressive function (Lee et al., 2006; Hu et al., 2007; Kim et al., 2007 Zhao et al., 2009; Miyauchi et al., 2010; Zhao and Ivashkiv, 2011). One mechanism by which RANK signaling down-regulates MafB, IRF-8, and Bcl6 expression is via induction of B lymphocyte–induced maturation protein-1 (Blimp1), a repressor of transcription of the genes that encode MafB, IRF-8, and Bcl6 (Miyauchi et al., 2010; Nishikawa et al., 2010). Understanding of mechanisms underlying the mutual antagonism between RANK signaling and transcriptional repressors of osteoclastogenesis is limited, and the role of transcriptional repressors in regulating inflammatory and TNF-mediated osteoclastogenesis and bone resorption has not been explored.
Recombinant recognition sequence binding protein at the Jκ site (RBP-J, also named RBP-Jκ, CSL, or CBF1) is a nuclear DNA-binding protein that can function as either a transcriptional repressor or activator depending on the partner proteins with which it interacts (Kopan and Ilagan, 2009). RBP-J is basally expressed in most cell types and is activated by interaction with other proteins. RBP-J is best known as a mediator of signaling by the canonical Notch pathway (Kopan and Ilagan, 2009), where Notch receptor cytoplasmic domains translocate to the nucleus, bind to RBP-J, and induce RBP-J transcription-activating function. Accumulating evidence shows that RBP-J is also critically involved in other signaling pathways, such as the Wnt–β-catenin (Shimizu et al., 2008) and NF-κB pathways (Plaisance et al., 1997; Izumiya et al., 2009), and is also targeted by viral proteins (Hayward, 2004; Izumiya et al., 2009) and cellular proteins of unknown function (Taniguchi et al., 1998; Beres et al., 2006). Thus, RBP-J functions as a central transcription factor that receives inputs from several signaling pathways. RBP-J regulates cell differentiation, proliferation, and survival, and plays important roles in cell fate decisions and diverse cellular functions, such as stem cell maintenance, neurogenesis, and lymphocyte development (Maillard et al., 2005; Kopan and Ilagan, 2009). In myeloid lineage cells, RBP-J has been implicated in inflammatory macrophage activation (Hu et al., 2008), DC differentiation, and maintenance of CD8− DC populations (Caton et al., 2007). Although many of these functions are related to its role in Notch signaling, RBP-J function is context-dependent, and under inflammatory conditions RBP-J plays a key role in expression of immune response genes not related to canonical Notch signaling (Hu et al., 2008). RBPJ allelic polymorphisms have recently been linked with RA (Stahl et al., 2010), but mechanisms by which RBP-J may contribute to RA pathogenesis are not known, and the role of RBP-J in osteoclastogenesis and inflammatory bone resorption has not been investigated.
RBP-J modulates signaling by at least two pathways that have been implicated in osteoclastogenesis: NF-κB and the Notch pathway, which has been shown to modestly suppress physiological RANKL-induced osteoclastogenesis (Yamada et al., 2003; Bai et al., 2008). Thus, we investigated the role of RBP-J in osteoclastogenesis and bone resorption. RBP-J modestly suppressed RANKL-induced osteoclastogenesis in vitro and had no discernable effect on physiological bone remodeling in vivo. In striking contrast, myeloid-specific deletion of RBP-J resulted in dramatically increased TNF-induced osteoclastogenesis, comparable to that induced by RANKL in control cells, and in severe bone destruction in a TNF-induced inflammatory bone resorption model. TNF was able to induce osteoclast differentiation and inflammatory bone resorption in RBP-J–deficient cells and mice even in the absence of RANK signaling; thus, the full osteoclastogenic potential of TNF leading to increased bone pathology was revealed in the absence of RBP-J. Concordant with a suppressive role in osteoclastogenesis, forced activation of RBP-J suppressed inflammatory and arthritic bone resorption. Mechanistically, RBP-J suppressed induction of NFATc1 by attenuating AP-1 activation and suppressing induction of Blimp1, thereby preventing down-regulation of repressors of osteoclastogenesis including IRF-8. Thus, RBP-J functions as a central upstream factor that controls the balance between pathways that activate and inhibit osteoclastogenesis. These findings identify a key role for RBP-J in restraining TNF-induced inflammatory osteoclastogenesis and provide insight into mechanisms that regulate the transcriptional repressor network that suppresses osteoclastogenesis. The prominent and selective role that RBP-J plays in inhibiting osteoclastogenesis in inflammatory settings suggests therapeutic targeting of RBP-J and upstream pathways as a new approach to suppressing inflammatory bone resorption.
RESULTS
RBP-J limits TNF-induced osteoclastogenesis
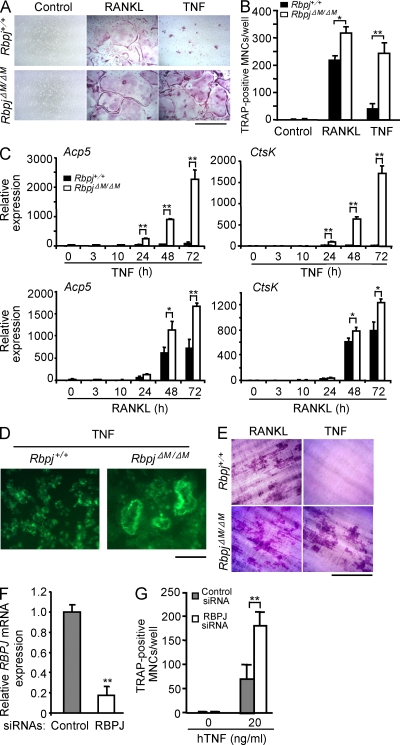
Global loss of RBP-J expression in mice leads to early embryonic lethality (Oka et al., 1995). Thus, to determine the role of RBP-J in osteoclastogenesis, we deleted Rbpj in myeloid lineage osteoclast precursors by crossing Rbpjflox/flox mice (Tanigaki et al., 2002) with LysMcre mice that express Cre under control of the myeloid-specific lysozyme M promoter. We used Rbpjflox/floxLysMcre(+) mice (hereafter referred to as RbpjΔM/ΔM) and littermate controls with a Rbpj+/+ LysMcre(+) genotype (hereafter referred to as Rbpj+/+) in most experiments. The extent of RBP-J deletion in vitro and in vivo was typically 70–80% (unpublished data). We first examined in vitro osteoclast differentiation using BM-derived macrophages (BMMs) as osteoclast precursors (Fig. 1, A and B). As expected, in Rbpj+/+ BMMs TNF induced a low number of small osteoclast-like tartrate-resistant acid phosphatase (TRAP)+ multinucleated cells (MNCs) relative to the positive control RANKL (Fig. 1, A and B). RBP-J deficiency resulted in a modest increase in RANKL-induced osteoclast differentiation. Strikingly, TNF induced a dramatically greater number of multinucleated osteoclasts in RbpjΔM/ΔM cells than in Rbpj+/+ cells, suggesting that RBP-J plays a more prominent role in limiting TNF-induced osteoclastogenesis. Moreover, the extent of osteoclastogenesis induced by TNF in RbpjΔM/ΔM cells was comparable to that induced by RANKL in control wild-type cells (Fig. 1 B), suggesting that RBP-J deficiency switches TNF signaling and function to be similar to that of RANKL in driving osteoclast differentiation. In parallel with increased generation of TRAP+ polykaryons, the expression of osteoclast marker genes Acp5 (encoding TRAP) and Ctsk (encoding cathepsin K) was dramatically enhanced in TNF-treated RbpjΔM/ΔM cells relative to control cells (Fig. 1 C). As expected, RANKL induced higher expression of these genes in control cells than did TNF, and the augmentation of RANKL-induced gene expression when RBP-J was deleted was more modest. Furthermore, the TNF-induced osteoclasts generated from RBP-J–deficient precursors formed actin rings (Fig. 1 D), and formed resorptive pits on dentin slices (Fig. 1 E) on a level comparable to that induced by RANKL in wild-type cells, indicating that TNF-induced RBP-J–deficient osteoclasts are functional and possess bone resorptive capability. Similar results were obtained using Rbpjflox/flox littermates as controls (unpublished data), or when Rbpj was deleted using Mx1-Cre (unpublished data). Knockdown of RBP-J expression in human osteoclast precursors using RNA interference resulted in enhanced TNF-induced osteoclast differentiation (Fig. 1, F and G). These results establish that RBP-J restrains osteoclastogenesis in vitro, and plays a key role in preventing osteoclastogenesis by the inflammatory cytokine TNF.
Figure 1.
TNF-induced osteoclastogenesis is restrained by RBP-J. BMMs derived from Rbpj+/+ and RbpjΔM/ΔM littermates were stimulated with RANKL or TNF in the presence of M-CSF. After 4 d, TRAP staining was performed (A), and the number of TRAP-positive MNCs per well was counted (B). TRAP-positive cells appear red in the photographs. Bar, 200 µm. *, P < 0.05; **, P < 0.01. (C) Quantitative real-time PCR analysis of mRNA expression of Acp5 (encoding TRAP) and Ctsk (encoding cathepsin K) in BMMs from Rbpj+/+ and RbpjΔM/ΔM littermates treated with TNF or RANKL in the presence of M-CSF for the indicated times. *, P < 0.05; **, P < 0.01. (D) Fluorescent images of actin ring staining from cell cultures of BMMs from Rbpj+/+ and RbpjΔM/ΔM littermates treated with TNF in the presence of M-CSF on dentin slices for 4 d. Bar, 100 µm. (E) Toluidine blue–stained dentin resorption pits formed by the osteoclasts derived from Rbpj+/+ and RbpjΔM/ΔM BMMs treated with RANKL or TNF in the presence of M-CSF for 7 d. Bar, 200 µm. (F) Quantitative real-time PCR analysis of RBPJ mRNA in human CD14-positive monocytes transfected with human RBPJ-specific siRNAs (RBPJ) or nontargeting control siRNAs (Control), and cultured for 2 d in the presence of M-CSF. **, P < 0.01. (G) Osteoclastogenesis of human osteoclast precursors transfected with RBP-J siRNAs or nontargeting control siRNAs in the presence of TNF with M-CSF for 6 d. The number of TRAP-positive MNCs per well was counted. **, P < 0.01. Data are representative of at least 20 independent experiments (A and B), or at least 3 independent experiments in (C–G).
RBP-J played a more modest role in restraining RANKL-induced osteoclastogenesis than TNF-induced osteoclastogenesis. One explanation for this difference is that RANKL stimulation resulted in a rapid decrease in Rbpj expression within 24 h, thereby decreasing the ability of RBP-J to restrain osteoclast differentiation (unpublished data). This decrease in expression after RANKL stimulation is similar to that observed for other repressors of osteoclastogenesis (Lee et al., 2006; Hu et al., 2007; Kim et al., 2007; Zhao et al., 2009; Miyauchi et al., 2010; Zhao and Ivashkiv, 2011) and functions to release osteoclast precursors from inhibitors of the osteoclast differentiation pathway. In contrast, TNF did not decrease but instead slightly enhanced RBP-J expression during a 7-d culture (unpublished data). This maintained expression of RBP-J after TNF, but not after RANKL, stimulation helps explain why RBP-J is a stronger suppressor of TNF-induced osteoclastogenesis. In addition, consistent with induction of RBP-J activity by inflammatory Toll-like receptor signaling (Hu et al., 2008), TNF induced RBP-J activity much more potently than RANKL, as assessed by induction of RBP-J–dependent genes such as Jag1 (Foldi et al., 2010). Jagged1 was more strongly induced by TNF than by RANKL (unpublished data). As Jagged1 activates the Notch signaling pathway to inhibit osteoclastogenesis (Bai et al., 2008), TNF-induced Jagged1 functions as a homeostatic feedback inhibitor to restrain osteoclastogenesis, suggesting that TNF induces feedback inhibition more effectively than does RANKL. We further found that induction of Jagged1 expression in BMMs was dependent on RBP-J (unpublished data). Thus, TNF-induced feedback inhibition is itself dependent on RBP-J, supporting a key upstream function of RBP-J. Induction of Jagged1 represents one aspect of feedback inhibition, but it is likely that TNF induces additional feedback mechanisms. Collectively, this suggests that inflammatory factors such as TNF activate RBP-J activity more effectively than the homeostatic cytokine RANKL. Feedback inhibition is an important function of RBP-J in most systems studied to date (Hu et al., 2008; Kopan and Ilagan, 2009), suggesting that activation of RBP-J by TNF induces feedback inhibition that results in a greater role for RBP-J in restraining osteoclastogenesis induced by TNF than by RANKL.
TNF-induced osteoclastogenesis and bone resorption in vivo are restrained by RBP-J
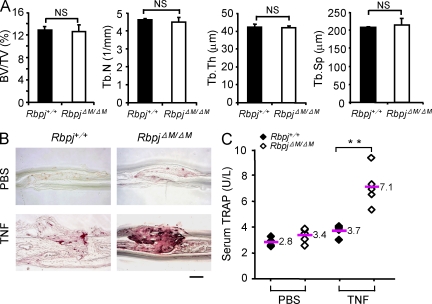
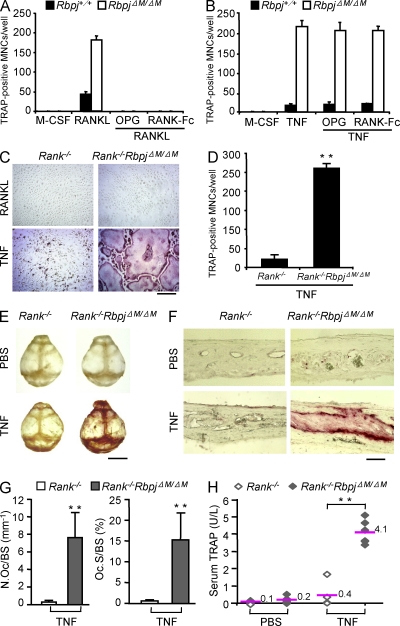
Consistent with the mild increase in osteoclast differentiation induced by RANKL in the absence of RBP-J, RbpjΔM/ΔM mice with 70–80% RBP-J deletion did not exhibit obvious defects in bone phenotype compared with Rbpj+/+ littermates (Fig. 2 A and not depicted), suggesting that RBP-J plays a minor role in osteoclastogenesis in development and under physiological conditions. We next investigated whether RBP-J restrains osteoclastogenesis and bone resorption under inflammatory conditions in vivo by using a well-established TNF-induced inflammatory bone resorption mouse model (Kitaura et al., 2005). Administration of TNF to the calvarial periosteum resulted in slightly enhanced osteoclast formation in Rbpj+/+ mice, whereas markedly more osteoclast formation and extensive bone destruction were observed in RbpjΔM/ΔM mice (Fig. 2 B). These findings were corroborated by higher TNF-induced serum levels of TRAP, a marker for osteoclasts and bone resorption, in RbpjΔM/ΔM mice (Fig. 2 C). TNF-induced osteoclastogenesis is highly dependent on synergy or pretreatment with RANKL in most in vitro systems, and does not occur in the absence of RANK signaling in vivo (Lam et al., 2000; Li, J., et al., 2000; Li, P., et al., 2004; Boyce et al., 2006; Teitelbaum, 2006; Schett and Teitelbaum, 2009; Yao et al., 2009). We wished to examine whether TNF could induce osteoclastogenesis and bone resorption independently of RANK signaling in the absence of RBP-J. As expected, RANKL-induced osteoclastogenesis in vitro was effectively suppressed by blockade of RANK signaling by osteoprotegerin (OPG), a decoy RANKL receptor, or by soluble RANK (Fig. 3 A); in contrast, TNF-induced osteoclastogenesis in RbpjΔM/ΔM cells was not affected by OPG or soluble RANK (Fig. 3 B). To more definitively exclude a role for RANK signaling in TNF-mediated effects, we took a genetic approach and crossed RbpjΔM/ΔM mice with RANK-deficient mice to generate double-knockout RbpjΔM/ΔM mice lacking the Tnfrsf11a gene that encodes RANK (hereafter, referred to as Rank−/−RbpjΔM/ΔM). As expected, RANKL did not induce osteoclast differentiation in either Rank−/− or Rank−/−RbpjΔM/ΔM cells (Fig. 3 C), and RBP-J deficiency did not compensate the basal osteopetrotic bone phenotype of Rank−/− mice (not depicted). Strikingly, TNF effectively induced osteoclast differentiation in Rank−/−RbpjΔM/ΔM cells (Fig. 3, C and D), although with slower kinetics. These results demonstrate that RBP-J deficiency allows TNF to induce osteoclast differentiation independently of RANK signaling in vitro. Consistent with previous reports that under many conditions TNF does not induce osteoclastogenesis and bone resorption in vivo in the absence of RANK signaling (Li, J., et al., 2000; Li, P., et al., 2004; Teitelbaum, 2006; Yao et al., 2009), TNF did not induce osteoclast formation and bone resorption in Rank−/− mice (Fig. 3, E–H). In contrast, TNF induced high levels of osteoclast formation, bone resorption, and serum TRAP in Rank−/−RbpjΔM/ΔM mice (Fig. 3, E–H). Thus, in the absence of RBP-J, TNF is able to induce osteoclastogenesis similar to and independent of RANK signaling. These results show that inflammatory osteoclastogenesis can proceed independent of RANK in the absence of the inhibitory function of RBP-J, and demonstrate a key role for RBP-J in restraining inflammatory bone resorption in vivo.
Figure 2.
Increased TNF-induced osteoclastogenesis and bone resorption in vivo in mice with myeloid-specific RBP-J deficiency. (A) μCT analysis of the femurs of 8-wk-old male Rbpj+/+ or RbpjΔM/ΔM littermates was performed. BV/TV, trabecular bone volume/tissue volume; Tb.N, trabecular number; Tb.Th, trabecular bone thickness; Tb.Sp, trabecular separation. n = 5 in each group. NS, no statistically significant difference. (B) TRAP staining of histological sections and (C) the concentration of serum TRAP obtained from Rbpj+/+ and RbpjΔM/ΔM littermates (8-wk-old) with or without TNF administration (75 µg/kg body weight). Bar, 200 µm in B. n = 5 per group. **, P < 0.01. Data are representative of two independent experiments (A–C).
Figure 3.
RBP-J deficiency allows TNF to induce osteoclastogenesis and bone resorption independent of RANK signaling. (A) Quantitation of TRAP-positive MNCs of the cell cultures from Rbpj+/+ and RbpjΔM/ΔM BMMs treated with RANKL, RANKL together with OPG (100 ng/ml), or RANK-Fc (5 µg/ml) in the presence of M-CSF for 4 d. (B) Quantitation of TRAP-positive MNCs of the cell cultures from Rbpj+/+ and RbpjΔM/ΔM BMMs treated with TNF, TNF together with OPG (100 ng/ml), or RANK-Fc (5 µg/ml) in the presence of M-CSF for 4 d. TRAP staining (C) and quantitation (D) of TRAP-positive MNCs in 5-d osteoclastogenic cell cultures from Rank−/− and Rank−/−RbpjΔM/ΔM BMMs. Bar, 100 µm. **, P < 0.01. (E and F) TRAP staining of mouse whole calvaria (E) and of calvarial histological sections (F) obtained from 4-wk-old Rank−/− and Rank−/−RbpjΔM/ΔM mice injected with PBS or TNF (150 µg/kg body weight). n = 5 per group. Bars: 0.5 cm (E); 100 µm (F). (G) Histomorphometric analysis of calvaria from Rank−/− and Rank−/−RbpjΔM/ΔM mice. N.Oc/BS, number of osteoclasts per bone surface; Oc.S/BS, osteoclast surface per bone surface. n = 5 per group. **, P < 0.01. (H) The concentration of serum TRAP obtained from 4-wk-old Rank−/− and Rank−/−RbpjΔM/ΔM mice injected with PBS or TNF. n = 5 for Rank−/− mice, and n = 6 for Rank−/−RbpjΔM/ΔM mice. **, P < 0.01. Data are representative of at least three (A–D) or two (E–H) independent experiments.
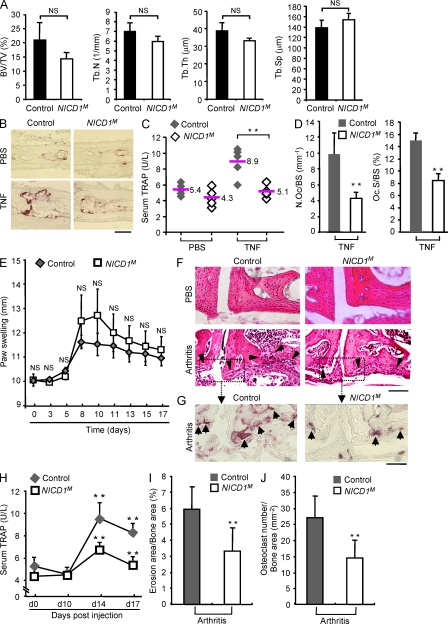
The aforementioned results suggest that activation of RBP-J in inflammatory settings functions as a feedback mechanism to limit pathological osteoclastogenesis and that further augmenting RBP-J activity would suppress bone resorption. Thus, we wished to use a gain-of-function approach to test whether boosting RBP-J activity would alleviate inflammatory bone resorption. RBP-J can be activated by several pathways (Plaisance et al., 1997; Taniguchi et al., 1998; Hayward, 2004; Maillard et al., 2005; Beres et al., 2006; Hu et al., 2008; Shimizu et al., 2008; Izumiya et al., 2009; Kopan and Ilagan, 2009); we used the best established approach of strongly activating RBP-J using forced expression of the Notch intracellular domain 1 (NICD1; Murtaugh et al., 2003; Zanotti et al., 2008; Tao et al., 2010). This approach allowed us to activate RBP-J function to supraphysiological levels and test the effects on inflammatory bone resorption (Murtaugh et al., 2003; Zanotti et al., 2008; Tao et al., 2010). We selectively activated RBP-J in myeloid osteoclast precursors in vivo using RosaNotch;LysMcre(+) mice by crossing RosaNotch mice (Murtaugh et al., 2003), where the mouse cDNA-encoding, constitutively active NICD1 was knocked into the ubiquitously expressed Rosa26 locus preceded by a STOP fragment flanked by loxP sites, with LysMcre mice (hereafter referred to as NICD1M mice). Gender- and age-matched LysMcre(+) mice were used as controls. NICD1M mice did not exhibit significant bone defects under basal conditions (Fig. 4 A); thus, both loss-of-function (Fig. 2 A) and gain-of-function (Fig. 4 A) approaches support that RBP-J plays a minor role in bone remodeling under physiological conditions. In contrast, in the TNF-induced inflammatory bone resorption mouse model (Kitaura et al., 2005), forced high-level activation of RBP-J resulted in dramatically lower amounts of TNF-induced calvarial osteoclast formation, bone resorption, and serum TRAP (Fig. 4, B–D). Thus, boosting activation of RBP-J selectively in the myeloid compartment in NICD1M mice suppressed TNF-induced osteoclastogenesis and bone resorption. We then tested whether increasing activation of RBP-J in myeloid cells would suppress bone erosion in an inflammatory arthritis model. We used the collagen antibody-induced arthritis mouse model (Terato et al., 1992; Yarilina et al., 2007) that bypasses the need to induce autoimmunity, and thus allows investigation of inflammatory bone resorption during the inflammatory effector phase of arthritis, in which TNF plays a key role. Importantly, the clinical course of inflammatory arthritis and pannus formation were not diminished in NICD1M mice (Fig. 4 E and not depicted). In striking contrast, RBP-J activation in NICD1M mice resulted in significant suppression of bone erosion (Fig. 4, F and I), osteoclast formation in resorption sites (Fig. 4, G and J), and serum TRAP amounts (Fig. 4 H). Thus, increasing RBP-J activity in the myeloid osteoclast precursor lineage using a genetic approach significantly suppressed inflammatory arthritic bone resorption, suggesting that therapeutic approaches that increase RBP-J activity may be effective in suppressing pathological osteoclastogenesis.
Figure 4.
Constitutive activation of RBP-J signaling in myeloid osteoclast precursor compartment in mice suppresses inflammatory bone resorption. (A) μCT analysis of the femurs of 8-wk-old male control or NICD1M mice was performed. BV/TV, trabecular bone volume/tissue volume; Tb.N, trabecular number; Tb.Th, trabecular bone thickness; Tb.Sp, trabecular separation. n = 5 in each group. NS, no statistically significant difference. (B) TRAP staining of histological sections and (C) the concentration of serum TRAP obtained from 6-wk-old NICD1M and control mice with or without supracalvarial TNF administration (100 µg/kg body weight). Bar, 50 µm. n = 5 per group. **, P < 0.01. (D) Histomorphometric analysis of calvaria from 6-wk-old NICD1M and control mice. N.Oc/BS, number of osteoclasts per bone surface; Oc.S/BS, osteoclast surface per bone surface. n = 5 per group. **, P < 0.01. (E) Paw swelling in arthritis induced in the 6-wk-old control and NICDM mice using anticollagen antibodies as described in Materials and methods. For each mouse, paw swelling was calculated as the sum of measurements of joint thickness of two wrists and two ankles. Paw swelling is represented as the mean ± SD for each group. n = 5 per group. NS, no statistically significant difference. (F) H&E staining of tissue sections of tarsal joints in 6-wk-old control and NICD1M mice treated with PBS or arthritis-inducing monoclonal antibody cocktail (arthritis) as described in Materials and methods. Black arrowheads indicate bone erosions. Bar, 100 µm. (G) TRAP staining of the tissue sections from the same area labeled with a dotted box in F. Black arrows show the osteoclasts (red spots) in the resorptive area. Bar, 50 µm. (H) Serum TRAP levels obtained from control and NICD1M mice at indicated times before and after induction of arthritis. n = 5 per group. **, P < 0.01. (I and J) Quantitative histomorphometric analysis of bone erosion (I) and osteoclast numbers (J) in tarsal joints. n = 5 per group. **, P < 0.01. Data are representative of two independent experiments (A–J).
RBP-J negatively regulates NFATc1 expression and function
We next explored the mechanisms by which RBP-J limits TNF-induced osteoclastogenesis and bone resorption. NFATc1 is the “master transcription factor” that controls osteoclast differentiation, and we tested whether RBP-J regulates NFATc1 expression. We found that TNF induced dramatically higher levels of NFATc1 mRNA (Fig. 5 A) and protein (Fig. 5 B) in RbpjΔM/ΔM cells than in Rbpj+/+ cells, indicating that NFATc1 expression is negatively regulated by RBP-J. The increased NFATc1 in RBP-J–deficient cells was functional because NFATc1 nuclear location was increased (not depicted), and the expression of NFATc1 target genes Itgb3 (encoding integrin β3; Fig. 5 C), and Acp5 and Ctsk (Fig. 1 C) was enhanced accordingly. Autoamplification of Nfatc1 expression through recruitment of NFATc1 to its own promoter is a determinative step for high NFATc1 induction and osteoclastogenesis. Chromatin immunoprecipitation (ChIP) assays showed that TNF-induced recruitment of NFATc1 to its own promoter, but not to control downstream sequences, was significantly enhanced in RbpjΔM/ΔM cells (Fig. 5 D and not depicted), consistent with autoamplification of expression. We next tested whether RBP-J regulated Nfatc1 transcription or mRNA stability. RBP-J deficiency did not enhance the stability of NFATc1 mRNA (not depicted). To determine whether TNF enhanced Nfatc1 transcription in RBP-J–deficient cells, we measured primary Nfatc1 transcripts using primers specific for an intronic region of the Nfatc1 gene and found that the pattern of the regulation of Nfatc1 primary transcripts by RBP-J was similar to that of steady-state mRNA (Fig. 5 E). This result was corroborated by enhanced TNF-induced recruitment of RNA polymerase II to the Nfatc1 locus in RbpjΔM/ΔM cells (Fig. 5 F). Collectively, these results suggest that the major mechanism by which RBP-J negatively regulates Nfatc1 expression is repression of transcription.
Figure 5.
RBP-J deficiency results in increased TNF-induced NFATc1 expression and function. (A) Quantitative real-time PCR analysis of mRNA expression of Nfatc1 in osteoclastogenic cell cultures from Rbpj+/+ and RbpjΔM/ΔM BMMs treated with TNF for the indicated times. *, P < 0.05; **, P < 0.01. (B) Immunoblot analysis of NFATc1 expression in whole-cell lysates obtained from Rbpj+/+ and RbpjΔM/ΔM BMMs at the indicated time points after stimulation with TNF. p38 was measured as a loading control. (C) Quantitative real-time PCR analysis of mRNA expression of Itgb3 (encoding integrin β3) in osteoclastogenic cell cultures from Rbpj+/+ and RbpjΔM/ΔM BMMs treated with TNF for the indicated times. **, P < 0.01. (D) ChIP analysis of NFATc1 occupancy at the promoter of Nfatc1 in the Rbpj+/+ and RbpjΔM/ΔM BMMs treated without or with TNF for 48 h. **, P < 0.01. (E) Quantitative real-time PCR analysis of expression of primary transcripts of Nfatc1 in osteoclastogenic cell cultures from Rbpj+/+ and RbpjΔM/ΔM BMMs treated with TNF. **, P < 0.01. (F) ChIP analysis of polymerase II occupancy at the Nfatc1 locus in Rbpj+/+ and RbpjΔM/ΔM BMMs treated without or with TNF for 24 h. Data are representative of at least three independent experiments (A–F). **, P < 0.01.
We next wished to use a gain-of-function approach to corroborate that RBP-J suppresses NFATc1 expression. Activation of RBP-J transcriptional function by expressing NICD1 in osteoclast precursors suppressed RANKL-induced NFATc1 expression (Fig. 6 A). Consistent with decreased NFATc1 expression, RANKL-induced osteoclast differentiation was significantly suppressed in NICD1M cells relative to control cells (Fig. 6 B). We then used RNAi-mediated knock down of RBP-J expression (Fig. 6 C) to confirm that NICD1-induced suppression of NFATc1 and osteoclastogenesis was mediated by RBP-J. Indeed, knockdown of RBP-J expression significantly reversed NICD1-induced suppression of NFATc1 expression and osteoclastogenesis (Fig. 6, D and E). Collectively, the results indicate that activation of RBP-J suppresses NFATc1 expression and osteoclastogenesis.
Figure 6.
Constitutive activation of RBP-J signaling in myeloid osteoclast precursors suppresses NFATc1 expression and osteoclastogenesis. (A) Immunoblot analysis of NFATc1 in control and NICD1M BMMs treated with RANKL for the indicated times. p38 was measured as a loading control. (B) Quantitation of TRAP-positive MNCs in cell cultures from control and NICD1M BMMs treated with RANKL in the presence of M-CSF for 3 d. *, P < 0.05. (C) Quantitative real-time PCR analysis of Rbpj mRNA in control and NICD1M BMMs transfected with siRNA targeting Rbpj or nontargeting control siRNAs (Control) for 24 h. **, P < 0.01. (D) Immunoblot analysis of NFATc1 in control and NICD1M BMMs transfected with Rbpj or control siRNAs and stimulated with RANKL for 24 h. p38 was measured as a loading control. (E) Quantitation of TRAP-positive MNCs in cell cultures from control and NICD1M BMMs transfected with Rbpj or control siRNAs and stimulated with RANKL for 3 d. *, P < 0.05; NS, no statistically significant difference. Data are representative of at least three independent experiments (A–E).
RBP-J suppresses NFATc1 induction by attenuating AP-1 activation
Next, we sought to investigate the mechanisms by which RBP-J suppresses Nfatc1 transcription. Such repression could be a direct function of RBP-J or could occur indirectly via regulation of upstream mediators of Nfatc1 expression. We did not observe direct regulation of Nfatc1 expression by RBP-J (unpublished data), suggesting that instead RBP-J regulates TNF-induced signaling pathways and transcription factors important for Nfatc1 expression. We systematically analyzed the effect of RBP-J on upstream factors and signaling pathways that regulate Nfatc1. RBP-J deficiency did not affect expression levels of the M-CSF receptor or RANK (unpublished data) and the proliferation of osteoclast precursors (unpublished data). TNF did not induce TRAF6 activity in the presence or absence of RBP-J, and RBP-J deficiency did not affect TNF-induced activation of canonical and noncanonical NF-κB pathways, and resulted in only minimal increases in MAPK activity, c-Fos mRNA, and c-Jun protein expression that were not consistently apparent in all experiments (Fig. 7 A and not depicted). On the other hand, RBP-J deficiency resulted in substantial increases in nuclear c-Fos protein expression, especially at later time points after TNF stimulation (Fig. 7 A); these increases could not be explained by increased c-Fos mRNA and suggest regulation of c-Fos expression at the protein level. The late phase of Fos expression was dependent on ongoing TNF signaling, as it was abrogated when TNF signaling was blocked 24 h after addition of TNF (unpublished data); however, we cannot exclude that autocrine/paracrine signaling contributes to sustained Fos expression. ChIP assays showed that TNF-induced recruitment of c-Fos to its target Nfatc1 promoter but not to control downstream sequences was significantly increased in the absence of RBP-J (Fig. 7 B and not depicted), indicating that RBP-J deficiency leads to enhanced expression of c-Fos and suggesting that this c-Fos is functional in increasing transcription of Nfatc1. To further corroborate that the increased amount of c-Fos contributes to increased TNF-induced osteoclast differentiation in RbpjΔM/ΔM cells, we used RNAi to partially knock down c-Fos expression in TNF-treated RbpjΔM/ΔM cells to mimic the levels of c-Fos expression in Rbpj+/+ cells (Fig. 7 C). This partial decrease in c-Fos expression markedly decreased TNF-induced osteoclastogenesis in RbpjΔM/ΔM cells (Fig. 7, D and E). Collectively, the results show that RBP-J down-regulates Nfatc1 expression at least in part by suppressing expression of c-Fos, a direct activator of the Nfatc1 promoter.
Figure 7.
RBP-J inhibits TNF-induced c-Fos expression and function. (A) Immunoblot analysis of c-Fos and c-Jun in nuclear and cytoplasmic fractions of Rbpj+/+ and RbpjΔM/ΔM BMMs obtained at the indicated time points after stimulation with TNF. TBP and p38 were measured as loading controls for nuclear and cytoplasmic fractions, respectively. (B) ChIP analysis of c-Fos occupancy at Nfatc1 promoter in Rbpj+/+ and RbpjΔM/ΔM BMMs treated without or with TNF for 40 h. **, P < 0.01. (C) Immunoblot analysis of c-Fos in Rbpj+/+ and RbpjΔM/ΔM BMMs transfected with Fos-specific short interfering RNAs (Fos) or nontargeting control siRNAs (Control), and treated with TNF for 16 h. (D and E) Osteoclastogenesis assays of Rbpj+/+ and RbpjΔM/ΔM BMMs transfected with siRNA targeting Fos mRNA or nontargeting control siRNAs (Control) in the presence of TNF. TRAP staining is shown in D and quantitated in E. Bar, 50 µm. **, P < 0.01. Data are representative of at least three independent experiments (A–E).
RBP-J prevents down-regulation of osteoclastogenic repressor IRF-8
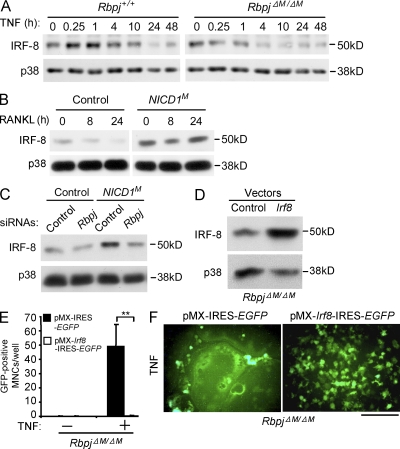
It has recently become clear that positive signaling is insufficient to induce NFATc1 and osteoclastogenesis unless the barrier imposed by transcriptional repressors is overcome. Among repressors of osteoclastogenesis, IRF-8 plays an important role in restraining osteoclast differentiation in inflammatory settings and down-regulation of IRF-8 expression is required for osteoclast differentiation (Zhao et al., 2009). Deficiency of RBP-J resulted in increased and accelerated down-regulation of IRF-8 after TNF stimulation of osteoclast precursors (Fig. 8 A); IRF8 down-regulation after RANKL stimulation was less affected, which is consistent with a more important role for RBP-J in regulating TNF responses after it is activated by TNF (not depicted). The evidence that RBP-J augments IRF8 expression was corroborated by gain-of function experiments showing that NICD1 increases IRF8 expression (Fig. 8 B), and this increase is dependent on RBP-J (Fig. 8 C). To test the functional significance of RBP-J–mediated up-regulation of IRF-8, we used retroviral transduction to reconstitute IRF-8 expression in RbpjΔM/ΔM osteoclast precursors (Fig. 8 D). Forced expression of IRF-8 in RBP-J–deficient osteoclast precursors abolished the enhanced induction of osteoclast differentiation by TNF (Fig. 8, D–F), but did not affect osteoclast precursor proliferation or survival (not depicted). Thus, the accelerated down-regulation of IRF-8 in RBP-J–deficient cells contributes to the increased osteoclastogenic phenotype. These results suggest that RBP-J suppresses osteoclastogenesis in part by maintaining expression of IRF-8, a transcriptional repressor that suppresses NFATc1 expression and function.
Figure 8.
RBP-J maintains expression of IRF-8, which suppresses osteoclastogenesis. (A) Immunoblot analysis of IRF-8 expression in Rbpj+/+ and RbpjΔM/ΔM BMMs obtained at the indicated time points after stimulation with TNF. p38 was measured as a loading control. (B) Immunoblot analysis of IRF-8 in control and NICDM BMMs stimulated with RANKL for the indicated times. (C) Immunoblot analysis of IRF-8 in control and NICD1M BMMs transfected with Rbpj or control siRNAs. p38 was measured as a loading control. (D) Immunoblot analysis of IRF-8 in RbpjΔM/ΔM BMMs transduced with the retroviral vectors pMX-IRES-EGFP (Control) or pMX-Irf8-IRES-EGFP (Irf8). p38 was measured as a loading control. (E) Osteoclastogenesis assays using RbpjΔM/ΔM BMMs transduced with the retroviral vectors pMX-IRES-EGFP or pMX-Irf8-IRES-EGFP without or with TNF stimulation. The number of GFP-positive MNCs (osteoclasts) per well was counted. **, P < 0.01. (F) Fluorescent images of the RbpjΔM/ΔM BMMs transduced with the retroviral vectors pMX-IRES-EGFP or pMX-Irf8-IRES-EGFP with TNF stimulation. The GFP-positive multinucleate cells (osteoclasts) appear as large green cells. Bar, 100 µm. Data are representative of at least three independent experiments (A–F).
RBP-J negatively regulates Blimp1 expression
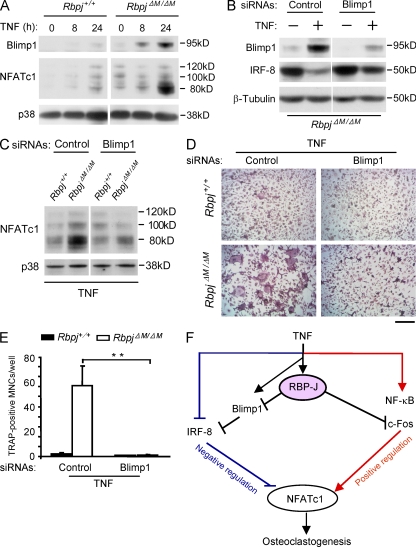
Recently, Blimp1 has been placed upstream of several repressors of osteoclastogenesis, including IRF-8, during osteoclastogenesis; an increase in Blimp1 expression after RANKL stimulation serves to down-regulate expression of repressors of osteoclastogenesis, and thus “release the brakes” on osteoclastogenesis (Miyauchi et al., 2010; Nishikawa et al., 2010). TNF induced lower levels of Blimp1 than did RANKL in control wild-type cells (not depicted), but TNF induced substantially higher Blimp1 expression in RBP-J–deficient cells than in Rbpj+/+ cells (Fig. 9 A); increases in Blimp1 protein in RBP-J–deficient cells (Fig. 9 A) were more striking than increases in Blimp1 mRNA (not depicted), suggesting a component of posttranscriptional control that is likely indirect. These results indicate that RBP-J restrains Blimp1 expression. RNAi-mediated knock down of Blimp1 (encoded by the Prdm1 gene) in TNF-stimulated RBP-J–deficient cells resulted in partial but consistent reversion of IRF-8 expression (Fig. 9 B), with a concomitant decrease in NFATc1 expression (Fig. 9 C). Furthermore, knock down of Blimp1 reversed the enhanced osteoclastogenesis induced by TNF in RBP-J–deficient cells (Fig. 9, D and E). These results place RBP-J upstream of Blimp1, which in turn regulates expression of transcriptional repressors of osteoclastogenesis, such as IRF-8 (Fig. 9 F).
Figure 9.
RBP-J prevents IRF-8 down-regulation by blocking TNF-induced Blimp1 expression. (A) Immunoblot analysis of the expression of Blimp1 and NFATc1 in Rbpj+/+ and RbpjΔM/ΔM BMMs obtained at the indicated time points after stimulation with TNF. p38 was measured as a loading control. (B) RbpjΔM/ΔM BMMs were transfected with Blimp1-specific siRNAs or nontargeting control siRNAs (Control), and treated without or with TNF for 24 h. Blimp1 and IRF-8 expression was assessed by immunoblot. β-Tubulin was measured as a loading control. (C) Rbpj+/+ and RbpjΔM/ΔM BMMs were transfected with Blimp1-specific siRNAs or nontargeting control siRNAs (Control), and treated with TNF for 24 h. NFATc1 expression was assessed by immunoblot. p38 was measured as a loading control. (D and E) Osteoclastogenesis assays of Rbpj+/+ and RbpjΔM/ΔM BMMs transfected with siRNA targeting Blimp1 mRNA or nontargeting control siRNAs (Control) in the presence of TNF for 4 d. TRAP staining is shown in D and quantitated in E. Bar, 50 µm. **, P < 0.01. (F) Model for the regulation of TNF-induced osteoclastogenesis by RBP-J. RBP-J functions as a central upstream regulator of the balance between activating and inhibitory pathways by suppressing c-Fos expression and preventing Blimp1-mediated down-regulation of IRF-8. Data are representative of at least three independent experiments (A–E).
DISCUSSION
Inflammatory bone resorption is a major clinical problem and cause of morbidity in diseases such as RA and periodontitis, but mechanisms that restrain and can prevent bone loss in inflammatory settings are not well understood. In this study, we have identified transcription factor RBP-J as a key and central negative regulator of osteoclastogenesis that plays a prominent role in suppressing TNF-induced osteoclast differentiation and limiting inflammatory bone resorption. RBP-J inhibited osteoclastogenesis by suppressing expression of the key positive regulator c-Fos, and by augmenting expression of the transcriptional repressor IRF-8, which imposes a “brake” that prevents induction of the NFATc1-mediated osteoclast differentiation program. These findings provide insight into mechanisms that control the balance between positive and negative pathways that determine the extent of osteoclastogenesis and identify a new therapeutic target for inhibition of pathological inflammatory bone resorption.
Many inflammatory cytokines, such as IL-1 and members of the TNF family, promote osteoclastogenesis in concert with RANKL (Teitelbaum, 2006; Schett and Teitelbaum, 2009). RANK is a member of the TNFR family of receptors. It is not clear why other TNFR family members that activate signaling pathways similar to those of RANK, including TNFRs, are weak inducers of osteoclastogenesis in the absence of RANK, and why RANK signaling is required for osteoclastogenesis and bone resorption in vivo under most conditions studied to date (Li, J., et al., 2000; Li, P., et al., 2004; Teitelbaum, 2006; Yao et al., 2009). A previous work (Yao et al., 2009) showed that TNF induces greater expression of the NF-κB pathway inhibitor p100 than does RANKL, leading to suppression of NF-κB signaling that is required for osteoclastogenesis. Together with our findings that TNF activates RBP-J signaling, which in turn suppresses osteoclastogenesis, the results support the idea that TNF activates feedback inhibitory mechanisms that are not effectively engaged by RANKL, and this stronger feedback inhibition helps explain why TNF is a weaker inducer of osteoclast differentiation than RANKL. Thus, differences in osteoclastogenic potential between RANKL and TNF may not be entirely explained by induction of a qualitatively unique signal by RANK, as previously supposed, but instead by stronger induction of feedback inhibition by TNF.
The suppressive functions of RBP-J on TNF-induced osteoclastogenesis are particularly potent, as, in the absence of RBP-J, TNF induced osteoclast differentiation comparably to RANKL. Furthermore, deletion of RBP-J allowed TNF to induce osteoclastogenesis independently of RANK signaling, and TNF was able to induce substantial inflammatory bone resorption in vivo in the absence of RANK. These findings support the notion that RBP-J serves as a key negative regulator that blocks osteoclast differentiation in response to inflammatory cytokines, and thus serves an important homeostatic function to prevent excessive bone resorption in inflammatory settings. An important implication of these findings is that if RBP-J signaling is not sufficiently engaged in chronic inflammatory diseases, TNF and potentially other cytokines will strongly promote osteoclastogenesis, potentially independently of RANK. RBP-J activity at sites of inflammation can potentially be attenuated by cytokines that activate Jak-STAT signaling and are pathogenic in diseases such as RA (Ivashkiv and Hu, 2003; Androutsellis-Theotokis et al., 2006; Hu et al., 2008). In addition, the homeostatic role of RBP-J in inflammatory osteolysis in RA may be compromised by RBPJ allelic variants that have been associated with RA (Stahl et al., 2010). Interestingly, allelic variants of PRDM1 (encoding Blimp1) have also been linked with RA (Raychaudhuri et al., 2009). The genetic linkage with RA of two components of the TNF–RBP-J–Blimp1 pathway that we have discovered further supports a role for this pathway in disease pathogenesis.
Our findings suggest that RBP-J potently suppresses osteoclastogenesis because it controls both positive and negative factors that regulate osteoclast differentiation (Fig. 9 F). RBP-J allows an integrated and coordinate regulation of the balance between positive and negative osteoclastogenic signaling. RBP-J-mediated regulation of the ratio of positive to negative signaling is conceptually similar to alterations in the RANKL/OPG ratio that have a more profound effect on osteoclastogenesis than changes in solely the positive or negative regulator. Positive signaling pathways and transcription factors that induce NFATc1 and promote osteoclastogenesis have been extensively studied and are well characterized (Takayanagi, 2007; Novack and Teitelbaum, 2008). Recently, the importance of negative regulation of osteoclastogenesis by transcriptional repressors, including Ids, Eos, MafB, IRF-8, and Bcl6, has been appreciated (Lee et al., 2006; Hu et al., 2007; Kim et al., 2007; Zhao et al., 2009; Miyauchi et al., 2010). These factors suppress transcription of Nfatc1 and its target genes, and their expression is down-regulated during osteoclastogenesis to allow the gene expression program associated with osteoclast differentiation to proceed. Recent work showing that MafB, IRF-8, and Bcl6 are coordinately down-regulated by the transcriptional repressor Blimp1 during osteoclastogenesis (Miyauchi et al., 2010; Nishikawa et al., 2010) suggests that a network of transcriptional repressors functions to control and fine tune the osteoclast differentiation process. Little is known about mechanisms that regulate Blimp1 expression in the early phases of osteoclastogenesis (Nishikawa et al., 2010), but our work shows that RBP-J restrains Blimp1 induction, and thus prevents subsequent down-regulation of repressors such as IRF-8 (Fig. 9 F). These results place RBP-J upstream of Blimp1 and suggest that RBP-J functions as an upstream negative regulator of osteoclast differentiation that plays a key role in preventing induction of the master positive regulator NFATc1. Although the regulation of Nfatc1 by RBP-J is most likely indirect and occurs via c-Fos and IRF-8, RBP-J may directly regulate expression of genes relevant for osteoclastogenesis, including some of the genes analyzed in this study. Our ability to address this issue has been limited by lack of RBP-J antibodies appropriate for ChIP and attempts to perform RBP-J ChIP have not been successful (unpublished data). Future study to identify direct RBP-J targets, including genome wide analysis, will provide further understanding of the inhibitory mechanisms of RBP-J.
One striking finding was that RBP-J played a more prominent and selective role in restraining inflammatory bone resorption than in physiological bone remodeling. To our knowledge, RBP-J and RelB (Vaira et al., 2008) are the only transcription factors implicated in preferentially regulating inflammatory or pathological osteoclastogenesis relative to basal osteoclastogenesis, which may offer an opportunity for selective therapeutic targeting of inflammatory bone resorption. An important question is why deletion of RBP-J resulted in a dramatic increase in TNF-induced osteoclastogenesis, but only a modest increase in RANKL-induced osteoclastogenesis. One contributing factor is that RANK signaling results in down-regulation of RBP-J expression, thus overcoming its repressive function, whereas TNFR signaling does not (unpublished data). In addition, we have found that TNF activated RBP-J much more potently than did RANKL, as RBP-J target Jagged1 (Foldi et al., 2010) was much more markedly induced by TNF than by RANKL, thus engaging an RBP-J–mediated feedback inhibitory loop that restrains osteoclastogenesis (unpublished data). These findings suggest that RBP-J signaling becomes activated by inflammatory factors as part of a feedback inhibitory loop that functions as a homeostatic mechanism. Because of its central regulation of c-Fos and the Blimp1-associated transcriptional repressor network, once RBP-J is activated it will restrain osteoclastogenesis induced by multiple factors, including RANKL. RBP-J activity will be submaximal and insufficient to prevent bone resorption under some conditions, such as physiological bone remodeling where it is weakly engaged by RANK, and moreover can be suppressed by factors such as cytokines that activate Jak-STAT signaling (Androutsellis-Theotokis et al., 2006; Hu et al., 2008; Kopan and Ilagan, 2009). Thus, further boosting of RBP-J activity using alternative means and signaling pathways, as we did in a proof-of-concept approach using NICD1 expression, may be effective in suppressing osteoclastogenesis in pathological settings.
Our results show that, in the myeloid osteoclast lineage, RBP-J plays a key role in suppressing inflammatory osteoclastogenesis and bone resorption. Notch signaling is the best known regulator of RBP-J function, which raises the question of the role of the Notch pathway upstream of RBP-J in contributing to the RBP-J–mediated regulation of TNF responses that we observed. However, inactivation of Notch signaling in osteoclast precursors by deleting Notch1 or ADAM10, an enzyme required for Notch receptor activation, mildly increased RANKL-induced osteoclastogenesis (consistent with previous work; Bai et al., 2008), but did not enhance TNF-induced osteoclastogenesis (unpublished data). Thus, the effects of RBP-J on TNF-induced osteoclastogenesis and inflammatory bone resorption may be related to, but are at least partially distinct from, signaling by Notch receptors. In contrast to TNF-driven inflammatory bone resorption as studied herein, a key role for Notch signaling in physiological bone remodeling in humans has been demonstrated in two recent studies linking Notch2 mutations with Hajdu-Cheney syndrome, a disorder characterized by severe and progressive bone loss (Isidor et al., 2011; Simpson et al., 2011). However, the mechanism by which Notch2 mutations (expressed in all cell types) affect bone phenotype is not clear, as Notch has context-dependent and opposing functions in both osteoblast and osteoclast lineages and can either increase or diminish bone mass (Yamada et al., 2003; Bai et al., 2008; Engin et al., 2008; Fukushima et al., 2008; Hilton et al., 2008; Zanotti et al., 2008; Kopan and Ilagan, 2009; Engin and Lee, 2010; Tao et al., 2010; Sethi et al., 2011). As RBP-J is a major mediator of Notch signaling, it likely plays a role in mediating the effects of Notch on bone phenotype, especially in the osteoblast lineage, where the Notch pathway plays a key role (Engin et al., 2008; Hilton et al., 2008; Zanotti et al., 2008; Tao et al., 2010). On the other hand, in the myeloid lineage, Notch receptors play a modest role in physiological osteoclastogenesis (Bai et al., 2008), and thus the more prominent inhibitory role of myeloid RBP-J in osteoclastogenesis under inflammatory conditions that we observed is likely explained by activation of RBP-J function by inflammatory signaling and possibly by additional upstream pathways (Plaisance et al., 1997; Taniguchi et al., 1998; Hayward, 2004; Maillard et al., 2005; Beres et al., 2006; Hu et al., 2008; Bai et al., 2008; Shimizu et al., 2008; Izumiya et al., 2009).
Our results suggest that increasing RBP-J activity during inflammation has therapeutic implications for suppressing osteoclastogenesis and associated pathological bone resorption. RBP-J is an attractive potential therapeutic target not only because of its potent suppressive functions but because it also serves as a nuclear integrator of many signaling pathways, including Notch and Wnt–GSK3–β-catenin pathways (Shimizu et al., 2008; Kopan and Ilagan, 2009). The multiple approaches that can be taken to increase RBP-J activity offer an opportunity to explore various therapeutic strategies to optimize efficacy while avoiding undesired effects. In conclusion, our work identifies RBP-J as a potent inhibitor of inflammatory bone resorption and an attractive potential therapeutic target.
MATERIALS AND METHODS
Mice and analysis of bone phenotype.
We generated mice with myeloid-specific deletion of Rbpj by crossing Rbpjflox/flox mice (Tanigaki et al., 2002) with mice with a lysozyme M promoter-driven Cre transgene on the C57/BL6 background (known as LysMcre; The Jackson Laboratory). 8-wk-old Rbpjflox/floxLysMcre(+) mice (referred to as RbpjΔM/ΔM) and their littermates with Rbpj+/+LysMcre(+) genotype as controls (referred to as Rbpj+/+) were used for experiments. In addition to myeloid-specific RBP-J knockout mice generated using Lysozyme M-cre, we generated mice with an inducible deletion of Rbpj (Rbpjf/fMx1-Cre) by crossing Rbpjflox/flox (referred to as Rbpjf/f) with mice with an Mx1 promoter-driven Cre transgene on the C57/BL6 background (The Jackson Laboratory). Littermates with Rbpjf/fMx1-Cre or Rbpj+/+Mx1-Cre genotypes were intraperitoneally injected with 200 µg per mouse of poly I:C 3 times in 5 d to induce RBP-J deletion, and BMMs from these mice were used 3 wk later for experiments. Tnfrsf11a (encoding RANK) knockout mice (referred as to Rank−/− mice) have been described previously (Li et al., 2000) and were obtained from Amgen. We generated double-knockout mice (Rank−/−RbpjΔM/ΔM mice) with both Rank deletion and myeloid-specific deletion of Rbpj by crossing Rank−/− mice with RbpjΔM/ΔM mice. 4-wk-old Rank−/− mice and Rank−/−RbpjΔM/ΔM mice were used. RosaNotch mice have been described previously (Murtaugh et al., 2003) and were obtained from The Jackson Laboratory. We generated mice with myeloid-specific constitutive expression of Notch intracellular domain 1 (NICD1) by crossing RosaNotch mice (Murtaugh et al., 2003) with LysMcre mice (referred to as NICD1M mice). 6-wk-old gender- and age-matched NICD1M mice and LysMcre mice as controls were used.
For in vivo analysis, we used an established mouse model of TNF-induced inflammatory bone resorption (Kitaura et al., 2005) with minor modifications. In brief, TNF at the doses indicated in the figure legends was administered daily to the calvarial periosteum of mice for five consecutive days, and then the mice were sacrificed, serum was collected, and calvarial bones were subjected to sectioning, TRAP staining, and histological analysis. Inflammatory arthritis was induced by injecting mice with collagen antibodies (Arthrogen-CIA Arthritis-Inducing Monoclonal Antibody Cocktail; Millipore) following the manufacturer’s protocol. The thickness of wrists and ankles was measured with a dial-type caliper (Bel-Art Products) in millimeters on the days indicated in the figure legends. Serum was collected on day 0, 10, 14, and 17, and mice were sacrificed on day 17. Paws were fixed overnight in 4% paraformaldehyde, decalcified in 10% EDTA (pH adjusted to 7.4) until the bones were pliable, trimmed, and embedded in paraffin. Serial paraffin sections (6 µm) of hind paws were stained with hematoxylin and eosin (H&E) and TRAP (Zhao et al., 2009). The bone erosion area and pannus area in tarsal joints quantified in a blinded manner in H&E-stained sections (3 per mouse) by calculating the proportions of bone erosion area and pannus area relative to the total tarsal bone area. Osteoclast numbers in the resorptive region were counted in TRAP-stained serial sections. Five mice per group were assessed. Bioquant Osteo II software was used for bone histomorphometry. All mouse experiments were approved by Institutional Animal Care and Use Committee of the Hospital for Special Surgery.
Reagents.
Mouse and human M-CSF, TNF, and sRANKL were purchased from PeproTech. Recombinant human OPG was purchased from PeproTech. Human RANK-Fc was provided by Amgen.
Cell culture.
Mouse BM cells were harvested from tibiae and femora and cultured overnight in Petri dishes (Midwest Scientific) in α-MEM medium (Invitrogen) with 10% FBS (HyClone) and 20 ng/ml of M-CSF (PeproTech). Nonadherent cells were then replated and cultured in the same medium with 20 ng/ml of M-CSF for 3 d to obtain BMMs, which are capable of differentiating into osteoclasts, and thus were used as osteoclast precursors. The attached BMMs were scraped, seeded at a density of 4.5 × 104/cm2, and cultured in α-MEM medium with 10% FBS and 20 ng/ml of M-CSF for 1 d. The cells were then treated without or with optimized concentrations of RANKL (80 ng/ml) or TNF (40 ng/ml) in the presence of M-CSF (20 ng/ml) for times indicated in the figure legends. Human osteoclast cultures were performed as described previously (Zhao et al., 2009). 4 wells in 96-well plates for each condition were used for cultures and TRAP staining. TRAP staining was performed with an acid phosphatase leukocyte diagnostic kit (Sigma-Aldrich) in accordance with the manufacturer’s instructions. For actin ring staining, the cells on dentin slices were fixed with 4% paraformaldehyde for 10 min at room temperature (RT), washed with PBS and permeabilized with 0.1% Triton X-100 in PBS for 5 min at RT, and then stained with 0.2 µM FITC-conjugated phalloidin (Sigma-Aldrich) for 60 min at 37°C.The experiments using human cells were approved by the Hospital for Special Surgery Institutional Review Board.
Pit formation assay.
BMMs were plated on dentin slices (4 mm diam, 0.2 mm thick; 5 × 104 cells/slice) in 96-well culture plates with 200 µl culture medium/well. The cells were cultured for 7 d with TNF or RANKL in the presence of M-CSF with media exchanges every 3 d. The dentin slices were washed with water, and pits formed by mature osteoclasts on the dentin slices were stained with 1% toluidine blue O (Sigma-Aldrich).
Retroviral gene transduction.
The pMX-Irf8-IRES-EGFP and pMX-IRES-EGFP retroviral vectors were previously described (Zhao et al., 2009). Retrovirus packaging was performed by transfecting the retroviral vectors into Plat-E retroviral packaging cell line (Cell Biolabs) using FuGENE HD (Roche) according to the manufacturer’s instructions and as previously described (Zhao et al., 2009). Mouse BMMs were infected with the recombinant retroviruses in the presence of 8 ng/ml M-CSF and 8 µg/ml polybrene for 8 h, and the media was changed before subsequent treatments as indicated in the figure legends.
RNA interference.
Small interfering RNAs (siRNAs) specifically targeting human RBP-J or nontargeting control siRNAs (Thermo Fisher Scientific) were transfected into primary human CD14+ monocytes with the Lonza Nucleofector device set to program Y-001 using the Human Monocyte Nucleofector kit (Lonza). Mouse siRNAs specifically targeting RBP-J, c-Fos, Blimp1, and nontargeting control siRNAs were obtained from Thermo Fisher Scientific and were transfected into murine BMMs using TransIT-TKO transfection reagent (Mirus) in accordance with the manufacturer’s instructions.
Reverse transcription and real-time PCR.
Reverse transcription and real-time PCR were performed as previously described (Zhao et al., 2009). Primary transcripts were measured with primers that amplify either exon–intron junctions or intronic sequences. The primers for real-time PCR were as follows: human RBPJ, 5′-TTCAAAAACCCCGTTGTCTC-3′ and 5′-CAAAAACCAACCAACCAACC-3′; Nfatc1, 5′-CCCGTCACATTCTGGTCCAT-3′ and 5′-CAAGTAACCGTGTAGCTCCACAA-3′; Nfatc1 (for detection of primary transcript), 5′-ACGGGTAGCAGAGCCAGAAGTTAG-3′ and 5′-ACCCCCACCTGGAGATCATTAGTT-3′; Acp5, 5′-ACGGCTACTTGCGGTTTC-3′ and 5′-TCCTTGGGAGGCTGGTC-3′; Ctsk, 5′-AAGATATTGGTGGCTTTGG-3′ and 5′-ATCGCTGCGTCCCTCT-3′; Itgb3, 5′-CCGGGGGACTTAATGAGACCACTT-3′ and 5′-ACGCCCCAAATCCCACCCATACA-3′; Csf1r, 5′-TCCACCGGGACGTAGCA-3′ and 5′-CCAGTCCAAAGTCCCCAATCT-3′; Tnfsf11a, 5′-CCAGCAGGGAAGCAAATC-3′ and 5′-GGTGAAACACTGGCTTAAACTG-3′; Rbpj, 5′-CGGCCTCCACCCAAACGACT-3′ and 5′-TCCAACCACTGCCCATAAGATACA-3′; Gapdh, 5′-ATCAAGAAGGTGGTGAAGCA-3′ and 5′-GACAACCTGGTCCTCAGTGT-3′.
Immunoblot analysis.
Total cell extracts were obtained using lysis buffer containing 20 mM Hepes, pH 7.0, 300 mM NaCl, 10 mM KCl, 1 mM MgCl2, 0.1% Triton X-100, 0.5 mM DTT, 20% glycerol, and 1× proteinase inhibitor cocktail (Roche). The cell membrane–permeable protease inhibitor Pefablock (1 mM) was added immediately before harvesting cells. Nuclear extracts were isolated as described previously (Hu et al., 2002). The protein concentration of extracts was quantitated using the Bradford assay (Bio-Rad Laboratories). Cell lysates (10 µg/sample) were fractionated on 7.5% SDS-PAGE, transferred to Immobilon-P membranes (Millipore), and incubated with specific antibodies. Western Lightning plus-ECL (PerkinElmer) was used for detection. c-Jun antibody was obtained from Cell Signaling Technology. c-Fos, IRF-8, Blimp1, p38, and TBP antibodies were purchased from Santa Cruz Biotechnology, Inc. NFATc1 antibody was purchased from BD, and β-tubulin antibody was obtained from Abcam.
ChIP assay.
ChIP assays were performed as previously described (Rogatsky et al., 2001). In brief, 5–10 × 106 mouse BMMs (2.5 × 106 cells/100 mm dish) per condition were treated as indicated in figure legends, and 50% of chromatin from each condition was used per immunoprecipitation. Cell fixation, lysis, and nuclear extraction were performed as previously described (Rogatsky et al., 2001). A Branson Sonifier 250 with a microtip at power output of 5 (equivalent to 12 w) was used. Samples were sonicated for 6 cycles at 20 s each separated by 1 min incubations on ice. Mean DNA fragment size of 200–500 bp was confirmed by agarose gel electrophoresis. For immunoprecipitations, 2 µg per sample of antibodies against RNA polymerase II (Millipore), c-Fos (Santa Cruz Biotechnology, Inc.), or control rabbit IgG (Santa Cruz Biotechnology, Inc.), and 4 µg per immunoprecipitation sample of NFATc1 (BD) or control mouse IgG (Santa Cruz Biotechnology, Inc.) were used. Immunoprecipitated DNA was analyzed by quantitative real-time PCR and normalized relative to 28S rRNA-encoding gene segments that reflect amounts of nonspecific background DNA precipitation in each reaction. The primers used to amplify the Nfatc1 promoter region bound by NFATc1 (−764/−550 relative to the transcription start site) are: 5′-CCGGGACGCCCATGCAATCTGTTAGTAATT-3′ and 5′-GCGGGTGCCCTGAGAAAGCTACTCTCCCTT-3′. Primers used to detect polymerase II occupancy at the Nfatc1 gene are: 5′-ACGGGTAGCAGAGCCAGAAGTTAG-3′ and 5′-ACCCCCACCTGGAGATCATTAGTT-3′. The primers that amplify 28S rRNA encoding gene segments are: 5′-GATCCTTCGATGTCGGCTCTTCCTATC-3′ and 5′-AGGGTAAAACTAACCTGTCTCACG-3′.
Statistical analysis.
Statistical analysis was performed using the Student t test (P < 0.05 was taken as statistically significant), and all data are presented as the mean ± SD.
Acknowledgments
We thank Drs. Stephen Doty, Anna Yarilina, and Nikolaus Binder for assistance with analysis of the histological sections and histomorphometry and the arthritis model. We thank Dr. Masamichi Takami for kind instruction on actin ring staining. We thank Dr. Tasuku Honjo for providing Rbpj floxed mice and Dr. Rachael Wagman at Amgen (Thousand Oaks, CA) for providing RANK-deficient mice.
This work was supported by grants from the Arthritis Foundation (B. Zhao) and the National Institutes of Health (L.B. Ivashkiv).
The authors have no conflicting financial interests.
Footnotes
Abbreviations used:
- Blimp1
- B lymphocyte–induced maturation protein-1
- BMM
- BM-derived macrophage
- ChIP
- chromatin immunoprecipitation
- IRF-8
- IFN regulatory factor-8
- M-CSF
- macrophage colony-stimulating factor
- MNC
- multinucleated cell
- NFATc1
- nuclear factor of activated T cells, cytoplasmic 1
- OPG
- osteoprotegerin
- RA
- rheumatoid arthritis
- RANK
- receptor activator of NF-κB
- RANKL
- RANK ligand
- RBP-J
- recombinant recognition sequence binding protein at the Jκ site
- siRNA
- small interfering RNA
- TRAP
- tartrate-resistant acid phosphatase
References
- Androutsellis-Theotokis A., Leker R.R., Soldner F., Hoeppner D.J., Ravin R., Poser S.W., Rueger M.A., Bae S.K., Kittappa R., McKay R.D. 2006. Notch signalling regulates stem cell numbers in vitro and in vivo. Nature. 442:823–826 10.1038/nature04940 [DOI] [PubMed] [Google Scholar]
- Azuma Y., Kaji K., Katogi R., Takeshita S., Kudo A. 2000. Tumor necrosis factor-alpha induces differentiation of and bone resorption by osteoclasts. J. Biol. Chem. 275:4858–4864 10.1074/jbc.275.7.4858 [DOI] [PubMed] [Google Scholar]
- Bai S., Kopan R., Zou W., Hilton M.J., Ong C.T., Long F., Ross F.P., Teitelbaum S.L. 2008. NOTCH1 regulates osteoclastogenesis directly in osteoclast precursors and indirectly via osteoblast lineage cells. J. Biol. Chem. 283:6509–6518 10.1074/jbc.M707000200 [DOI] [PubMed] [Google Scholar]
- Beres T.M., Masui T., Swift G.H., Shi L., Henke R.M., MacDonald R.J. 2006. PTF1 is an organ-specific and Notch-independent basic helix-loop-helix complex containing the mammalian Suppressor of Hairless (RBP-J) or its paralogue, RBP-L. Mol. Cell. Biol. 26:117–130 10.1128/MCB.26.1.117-130.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyce B.F., Schwarz E.M., Xing L. 2006. Osteoclast precursors: cytokine-stimulated immunomodulators of inflammatory bone disease. Curr. Opin. Rheumatol. 18:427–432 10.1097/01.bor.0000231913.32364.32 [DOI] [PubMed] [Google Scholar]
- Caton M.L., Smith-Raska M.R., Reizis B. 2007. Notch-RBP-J signaling controls the homeostasis of CD8− dendritic cells in the spleen. J. Exp. Med. 204:1653–1664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engin F., Lee B. 2010. NOTCHing the bone: insights into multi-functionality. Bone. 46:274–280 10.1016/j.bone.2009.05.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engin F., Yao Z., Yang T., Zhou G., Bertin T., Jiang M.M., Chen Y., Wang L., Zheng H., Sutton R.E., et al. 2008. Dimorphic effects of Notch signaling in bone homeostasis. Nat. Med. 14:299–305 10.1038/nm1712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foldi J., Chung A.Y., Xu H., Zhu J., Outtz H.H., Kitajewski J., Li Y., Hu X., Ivashkiv L.B. 2010. Autoamplification of Notch signaling in macrophages by TLR-induced and RBP-J-dependent induction of Jagged1. J. Immunol. 185:5023–5031 10.4049/jimmunol.1001544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukushima H., Nakao A., Okamoto F., Shin M., Kajiya H., Sakano S., Bigas A., Jimi E., Okabe K. 2008. The association of Notch2 and NF-kappaB accelerates RANKL-induced osteoclastogenesis. Mol. Cell. Biol. 28:6402–6412 10.1128/MCB.00299-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayward S.D. 2004. Viral interactions with the Notch pathway. Semin. Cancer Biol. 14:387–396 10.1016/j.semcancer.2004.04.018 [DOI] [PubMed] [Google Scholar]
- Hilton M.J., Tu X., Wu X., Bai S., Zhao H., Kobayashi T., Kronenberg H.M., Teitelbaum S.L., Ross F.P., Kopan R., Long F. 2008. Notch signaling maintains bone marrow mesenchymal progenitors by suppressing osteoblast differentiation. Nat. Med. 14:306–314 10.1038/nm1716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu X., Herrero C., Li W.P., Antoniv T.T., Falck-Pedersen E., Koch A.E., Woods J.M., Haines G.K., Ivashkiv L.B. 2002. Sensitization of IFN-gamma Jak-STAT signaling during macrophage activation. Nat. Immunol. 3:859–866 10.1038/ni828 [DOI] [PubMed] [Google Scholar]
- Hu R., Sharma S.M., Bronisz A., Srinivasan R., Sankar U., Ostrowski M.C. 2007. Eos, MITF, and PU.1 recruit corepressors to osteoclast-specific genes in committed myeloid progenitors. Mol. Cell. Biol. 27:4018–4027 10.1128/MCB.01839-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu X., Chung A.Y., Wu I., Foldi J., Chen J., Ji J.D., Tateya T., Kang Y.J., Han J., Gessler M., et al. 2008. Integrated regulation of Toll-like receptor responses by Notch and interferon-gamma pathways. Immunity. 29:691–703 10.1016/j.immuni.2008.08.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isidor B., Lindenbaum P., Pichon O., Bézieau S., Dina C., Jacquemont S., Martin-Coignard D., Thauvin-Robinet C., Le Merrer M., Mandel J.L., et al. 2011. Truncating mutations in the last exon of NOTCH2 cause a rare skeletal disorder with osteoporosis. Nat. Genet. 43:306–308 10.1038/ng.778 [DOI] [PubMed] [Google Scholar]
- Ivashkiv L.B., Hu X. 2003. The JAK/STAT pathway in rheumatoid arthritis: pathogenic or protective? Arthritis Rheum. 48:2092–2096 10.1002/art.11095 [DOI] [PubMed] [Google Scholar]
- Izumiya Y., Izumiya C., Hsia D., Ellison T.J., Luciw P.A., Kung H.J. 2009. NF-kappaB serves as a cellular sensor of Kaposi’s sarcoma-associated herpesvirus latency and negatively regulates K-Rta by antagonizing the RBP-Jkappa coactivator. J. Virol. 83:4435–4446 10.1128/JVI.01999-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim N., Kadono Y., Takami M., Lee J., Lee S.H., Okada F., Kim J.H., Kobayashi T., Odgren P.R., Nakano H., et al. 2005. Osteoclast differentiation independent of the TRANCE-RANK-TRAF6 axis. J. Exp. Med. 202:589–595 10.1084/jem.20050978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K., Kim J.H., Lee J., Jin H.M., Kook H., Kim K.K., Lee S.Y., Kim N. 2007. MafB negatively regulates RANKL-mediated osteoclast differentiation. Blood. 109:3253–3259 10.1182/blood-2006-09-048249 [DOI] [PubMed] [Google Scholar]
- Kitaura H., Zhou P., Kim H.J., Novack D.V., Ross F.P., Teitelbaum S.L. 2005. M-CSF mediates TNF-induced inflammatory osteolysis. J. Clin. Invest. 115:3418–3427 10.1172/JCI26132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi K., Takahashi N., Jimi E., Udagawa N., Takami M., Kotake S., Nakagawa N., Kinosaki M., Yamaguchi K., Shima N., et al. 2000. Tumor necrosis factor alpha stimulates osteoclast differentiation by a mechanism independent of the ODF/RANKL-RANK interaction. J. Exp. Med. 191:275–286 10.1084/jem.191.2.275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopan R., Ilagan M.X. 2009. The canonical Notch signaling pathway: unfolding the activation mechanism. Cell. 137:216–233 10.1016/j.cell.2009.03.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam J., Takeshita S., Barker J.E., Kanagawa O., Ross F.P., Teitelbaum S.L. 2000. TNF-alpha induces osteoclastogenesis by direct stimulation of macrophages exposed to permissive levels of RANK ligand. J. Clin. Invest. 106:1481–1488 10.1172/JCI11176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J., Kim K., Kim J.H., Jin H.M., Choi H.K., Lee S.H., Kook H., Kim K.K., Yokota Y., Lee S.Y., et al. 2006. Id helix-loop-helix proteins negatively regulate TRANCE-mediated osteoclast differentiation. Blood. 107:2686–2693 10.1182/blood-2005-07-2798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Sarosi I., Yan X.Q., Morony S., Capparelli C., Tan H.L., McCabe S., Elliott R., Scully S., Van G., et al. 2000. RANK is the intrinsic hematopoietic cell surface receptor that controls osteoclastogenesis and regulation of bone mass and calcium metabolism. Proc. Natl. Acad. Sci. USA. 97:1566–1571 10.1073/pnas.97.4.1566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P., Schwarz E.M., O’Keefe R.J., Ma L., Boyce B.F., Xing L. 2004. RANK signaling is not required for TNFalpha-mediated increase in CD11(hi) osteoclast precursors but is essential for mature osteoclast formation in TNFalpha-mediated inflammatory arthritis. J. Bone Miner. Res. 19:207–213 10.1359/JBMR.0301233 [DOI] [PubMed] [Google Scholar]
- Locksley R.M., Killeen N., Lenardo M.J. 2001. The TNF and TNF receptor superfamilies: integrating mammalian biology. Cell. 104:487–501 10.1016/S0092-8674(01)00237-9 [DOI] [PubMed] [Google Scholar]
- Maillard I., Fang T., Pear W.S. 2005. Regulation of lymphoid development, differentiation, and function by the Notch pathway. Annu. Rev. Immunol. 23:945–974 10.1146/annurev.immunol.23.021704.115747 [DOI] [PubMed] [Google Scholar]
- Miyauchi Y., Ninomiya K., Miyamoto H., Sakamoto A., Iwasaki R., Hoshi H., Miyamoto K., Hao W., Yoshida S., Morioka H., et al. 2010. The Blimp1-Bcl6 axis is critical to regulate osteoclast differentiation and bone homeostasis. J. Exp. Med. 207:751–762 10.1084/jem.20091957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murtaugh L.C., Stanger B.Z., Kwan K.M., Melton D.A. 2003. Notch signaling controls multiple steps of pancreatic differentiation. Proc. Natl. Acad. Sci. USA. 100:14920–14925 10.1073/pnas.2436557100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishikawa K., Nakashima T., Hayashi M., Fukunaga T., Kato S., Kodama T., Takahashi S., Calame K., Takayanagi H. 2010. Blimp1-mediated repression of negative regulators is required for osteoclast differentiation. Proc. Natl. Acad. Sci. USA. 107:3117–3122 10.1073/pnas.0912779107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novack D.V., Teitelbaum S.L. 2008. The osteoclast: friend or foe? Annu. Rev. Pathol. 3:457–484 10.1146/annurev.pathmechdis.3.121806.151431 [DOI] [PubMed] [Google Scholar]
- Oka C., Nakano T., Wakeham A., de la Pompa J.L., Mori C., Sakai T., Okazaki S., Kawaichi M., Shiota K., Mak T.W., Honjo T. 1995. Disruption of the mouse RBP-J kappa gene results in early embryonic death. Development. 121:3291–3301 [DOI] [PubMed] [Google Scholar]
- Plaisance S., Vanden Berghe W., Boone E., Fiers W., Haegeman G. 1997. Recombination signal sequence binding protein Jkappa is constitutively bound to the NF-kappaB site of the interleukin-6 promoter and acts as a negative regulatory factor. Mol. Cell. Biol. 17:3733–3743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raychaudhuri S., Thomson B.P., Remmers E.F., Eyre S., Hinks A., Guiducci C., Catanese J.J., Xie G., Stahl E.A., Chen R., et al. ; BIRAC Consortium; YEAR Consortium 2009. Genetic variants at CD28, PRDM1 and CD2/CD58 are associated with rheumatoid arthritis risk. Nat. Genet. 41:1313–1318 10.1038/ng.479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogatsky I., Zarember K.A., Yamamoto K.R. 2001. Factor recruitment and TIF2/GRIP1 corepressor activity at a collagenase-3 response element that mediates regulation by phorbol esters and hormones. EMBO J. 20:6071–6083 10.1093/emboj/20.21.6071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schett G., Teitelbaum S.L. 2009. Osteoclasts and arthritis. J. Bone Miner. Res. 24:1142–1146 10.1359/jbmr.090533 [DOI] [PubMed] [Google Scholar]
- Sethi G., Sung B., Kunnumakkara A.B., Aggarwal B.B. 2009. Targeting TNF for Treatment of Cancer and Autoimmunity. Adv. Exp. Med. Biol. 647:37–51 10.1007/978-0-387-89520-8_3 [DOI] [PubMed] [Google Scholar]
- Sethi N., Dai X., Winter C.G., Kang Y. 2011. Tumor-derived JAGGED1 promotes osteolytic bone metastasis of breast cancer by engaging notch signaling in bone cells. Cancer Cell. 19:192–205 10.1016/j.ccr.2010.12.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu T., Kagawa T., Inoue T., Nonaka A., Takada S., Aburatani H., Taga T. 2008. Stabilized beta-catenin functions through TCF/LEF proteins and the Notch/RBP-Jkappa complex to promote proliferation and suppress differentiation of neural precursor cells. Mol. Cell. Biol. 28:7427–7441 10.1128/MCB.01962-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson M.A., Irving M.D., Asilmaz E., Gray M.J., Dafou D., Elmslie F.V., Mansour S., Holder S.E., Brain C.E., Burton B.K., et al. 2011. Mutations in NOTCH2 cause Hajdu-Cheney syndrome, a disorder of severe and progressive bone loss. Nat. Genet. 43:303–305 10.1038/ng.779 [DOI] [PubMed] [Google Scholar]
- Stahl E.A., Raychaudhuri S., Remmers E.F., Xie G., Eyre S., Thomson B.P., Li Y., Kurreeman F.A., Zhernakova A., Hinks A., et al. ; BIRAC Consortium; YEAR Consortium 2010. Genome-wide association study meta-analysis identifies seven new rheumatoid arthritis risk loci. Nat. Genet. 42:508–514 10.1038/ng.582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takayanagi H. 2007. Osteoimmunology: shared mechanisms and crosstalk between the immune and bone systems. Nat. Rev. Immunol. 7:292–304 10.1038/nri2062 [DOI] [PubMed] [Google Scholar]
- Tanigaki K., Han H., Yamamoto N., Tashiro K., Ikegawa M., Kuroda K., Suzuki A., Nakano T., Honjo T. 2002. Notch-RBP-J signaling is involved in cell fate determination of marginal zone B cells. Nat. Immunol. 3:443–450 10.1038/ni793 [DOI] [PubMed] [Google Scholar]
- Taniguchi Y., Furukawa T., Tun T., Han H., Honjo T. 1998. LIM protein KyoT2 negatively regulates transcription by association with the RBP-J DNA-binding protein. Mol. Cell. Biol. 18:644–654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao J., Chen S., Yang T., Dawson B., Munivez E., Bertin T., Lee B. 2010. Osteosclerosis owing to Notch gain of function is solely Rbpj-dependent. J. Bone Miner. Res. 25:2175–2183 10.1002/jbmr.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor P.C., Feldmann M. 2009. Anti-TNF biologic agents: still the therapy of choice for rheumatoid arthritis. Nat Rev Rheumatol. 5:578–582 10.1038/nrrheum.2009.181 [DOI] [PubMed] [Google Scholar]
- Teitelbaum S.L. 2006. Osteoclasts; culprits in inflammatory osteolysis. Arthritis Res. Ther. 8:201 10.1186/ar1857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terato K., Hasty K.A., Reife R.A., Cremer M.A., Kang A.H., Stuart J.M. 1992. Induction of arthritis with monoclonal antibodies to collagen. J. Immunol. 148:2103–2108 [PubMed] [Google Scholar]
- Vaira S., Johnson T., Hirbe A.C., Alhawagri M., Anwisye I., Sammut B., O’Neal J., Zou W., Weilbaecher K.N., Faccio R., Novack D.V. 2008. RelB is the NF-kappaB subunit downstream of NIK responsible for osteoclast differentiation. Proc. Natl. Acad. Sci. USA. 105:3897–3902 10.1073/pnas.0708576105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada T., Yamazaki H., Yamane T., Yoshino M., Okuyama H., Tsuneto M., Kurino T., Hayashi S., Sakano S. 2003. Regulation of osteoclast development by Notch signaling directed to osteoclast precursors and through stromal cells. Blood. 101:2227–2234 10.1182/blood-2002-06-1740 [DOI] [PubMed] [Google Scholar]
- Yao Z., Li P., Zhang Q., Schwarz E.M., Keng P., Arbini A., Boyce B.F., Xing L. 2006. Tumor necrosis factor-alpha increases circulating osteoclast precursor numbers by promoting their proliferation and differentiation in the bone marrow through up-regulation of c-Fms expression. J. Biol. Chem. 281:11846–11855 10.1074/jbc.M512624200 [DOI] [PubMed] [Google Scholar]
- Yao Z., Xing L., Boyce B.F. 2009. NF-kappaB p100 limits TNF-induced bone resorption in mice by a TRAF3-dependent mechanism. J. Clin. Invest. 119:3024–3034 10.1172/JCI38716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarilina A., DiCarlo E., Ivashkiv L.B. 2007. Suppression of the effector phase of inflammatory arthritis by double-stranded RNA is mediated by type I IFNs. J. Immunol. 178:2204–2211 [DOI] [PubMed] [Google Scholar]
- Zanotti S., Smerdel-Ramoya A., Stadmeyer L., Durant D., Radtke F., Canalis E. 2008. Notch inhibits osteoblast differentiation and causes osteopenia. Endocrinology. 149:3890–3899 10.1210/en.2008-0140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B., Ivashkiv L.B. 2011. Negative regulation of osteoclastogenesis and bone resorption by cytokines and transcriptional repressors. Arthritis Res. Ther. 13:234–243 10.1186/ar3379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao B., Takami M., Yamada A., Wang X., Koga T., Hu X., Tamura T., Ozato K., Choi Y., Ivashkiv L.B., et al. 2009. Interferon regulatory factor-8 regulates bone metabolism by suppressing osteoclastogenesis. Nat. Med. 15:1066–1071 10.1038/nm.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]