High-throughput sequencing reveals stability of the intestinal IgA repertoire after plasma cell depletion and changes in repertoire diversity with age and microbial colonization.
Abstract
Intestinal immunoglobulin A (IgA) ensures host defense and symbiosis with our commensal microbiota. Yet previous studies hint at a surprisingly low diversity of intestinal IgA, and it is unknown to what extent the diverse Ig arsenal generated by somatic recombination and diversification is actually used. In this study, we analyze more than one million mouse IgA sequences to describe the shaping of the intestinal IgA repertoire, its determinants, and stability over time. We show that expanded and infrequent clones combine to form highly diverse polyclonal IgA repertoires with very little overlap between individual mice. Selective homing allows expanded clones to evenly seed the small but not large intestine. Repertoire diversity increases during aging in a dual process. On the one hand, microbiota-, T cell–, and transcription factor RORγt–dependent but Peyer’s patch–independent somatic mutations drive the diversification of expanded clones, and on the other hand, new clones are introduced into the repertoire of aged mice. An individual’s IgA repertoire is stable and recalled after plasma cell depletion, which is indicative of functional memory. These data provide a conceptual framework to understand the dynamic changes in the IgA repertoires to match environmental and intrinsic stimuli.
The intestine represents the major Ig-producing compartment in mammals (Brandtzaeg and Johansen, 2005; Macpherson et al., 2008). Most intestinal plasma cells produce IgA and express the J chain, allowing for transepithelial transport and release of secretory IgA (SIgA) into the gut lumen (Brandtzaeg, 2009). Among the many functions of SIgA are the vaccination-induced protection against cholera toxin (Lycke and Holmgren, 1986) and protection against Salmonella enterica (Wijburg et al., 2006) and influenza infection (Asahi et al., 2002). In addition to these classical immunoprotective functions, IgA plays a central role in regulating host-microbiota homeostasis. Few plasma cells are present in neonates and in germ-free mice, but plasma cell numbers rapidly increase upon colonization with commensal microbiota, and even transient exposure to live bacteria induces a stable plasma cell population (Hapfelmeier et al., 2010). Colonization of germ-free mice lacking B cells results in enhanced translocation of bacteria to gut-draining mesenteric LNs (mLNs) compared with WT mice, and SIgA deficiency increases titers of flora-binding IgG in serum (Johansen et al., 1999; Macpherson and Uhr, 2004; Sait et al., 2007). These observations show that SIgA limits the access of intestinal microbiota to systemic tissues. However, mechanistically, this process is poorly understood, and even less is known about the specificity and diversity of the IgA repertoire. Intestinal IgA binds to the commensal microflora, and such reactivity seems to be specific for distinct bacterial epitopes (Hapfelmeier et al., 2010). Moreover, screening a set of IgA hybridoma lines, no more than ∼20% of antibodies were rated polyreactive, whereas most clones did not show cross-reactivity (Benckert et al., 2011). This hints at a diverse IgA repertoire, potentially reflecting the complexity of the intestinal microbiota and antigenic load. Once induced, microbe-specific IgA levels are comparably stable over time but become attenuated when new bacterial species are introduced to the microbiota (Hapfelmeier et al., 2010). Therefore, the IgA repertoire might be dynamically shaped to mirror the actual composition of the intestinal microbiota. Because changes in microbiotal composition are rapid during infancy and early life (Dominguez-Bello et al., 2011), one might speculate that the IgA repertoire could undergo similarly profound remodeling during this period of life. Moreover, mice capable of SIgA production and secretion but with an impaired capacity to acquire somatic mutations (SMs) showed a dysbiosis of their microbiota and hyperplasia of gut-associated lymphoid tissues (GALTs; Wei et al., 2011). This indicates that not only do microbiota induce specific IgA responses, but the specificity of the IgA repertoire may in turn also affect the composition of the microbiota.
Considering the near-infinite number of potential CDR3 (complementarity determining region 3) sequences and the complexity of the antigen load, it is surprising that the study of IgA repertoire diversity by CDR3 length analysis and sequencing hints at an oligoclonal IgA repertoire of low diversity. In particular, the frequent observation of clonally related plasma cells (Dunn-Walters et al., 1997; Holtmeier et al., 2000; Stoel et al., 2005, 2008; Yuvaraj et al., 2009) was interpreted to indicate low repertoire diversity. However, previous studies used conventional sequencing technologies and had to extrapolate from the analysis of few sequences to the overall IgA repertoire. In this study, we used high-throughput sequencing to investigate the shaping of the IgA repertoire, its determinants, and stability over time, as well as differences between individuals. Analyzing more than one million VH sequences, we show that the IgA repertoire comprised both highly expanded and low frequency clones. These two components combined to make a highly polyclonal IgA repertoire. Expanded clones were evenly distributed along the small but not large intestine and dominated the repertoire in young mice. However, during aging, repertoire diversity increased by the ongoing accumulation of low frequent clones and microbiota-, T cell–, and transcription factor RORγt–dependent but Peyer’s patch (PP)–independent hypermutation. Moreover, expanded clones were rapidly recalled after plasma cell depletion, indicating that the intestinal IgA system possesses memory characteristics.
RESULTS
Expanded and low frequency clones constitute polyclonal individual IgA repertoires
To establish comprehensive IgA repertoire analysis, RNA was isolated from whole small intestine (SI) tissue and transcribed into cDNA, and VH sequences were amplified by PCR with primers annealing to FR1 (frame work region 1) and the Cα domain. Amplicons of ∼450-bp length were sequenced by 454 sequencing, and for each sample 5,000–30,000 sequences of expected length, including the flanking Cα domain primer and encoding intact Ig, were analyzed. Previous studies described clonally related sequences even when comparably few sequences were analyzed (Dunn-Walters et al., 1997, 2000; Holtmeier et al., 2000; Stoel et al., 2005; Yuvaraj et al., 2009). Indeed, in 99.7 ± 0.3%, we observed at least one identical sequence pair within a set of 25 CDR3 sequences (5,000 randomly picked sets of 25 sequences obtained from six 10–11-wk-old mice), and some clones accounted for >1% of all sequences. However, along with such highly expanded clones, we observed unique sequences. In a set of 5,000 sequences analyzed per mouse, 25–35% of all full-length VH sequences and 3.9 ± 0.8% of all CDR3 sequences were obtained only once (mean ± SD in six 10–11-wk-old C57BL/6 mice). This suggests that the IgA plasma cell pool includes highly expanded clones along with low frequency clones. Therefore, the presence of clonally related sequences observed in small sequence sets does not by itself indicate low diversity and oligoclonality of the entire repertoire.
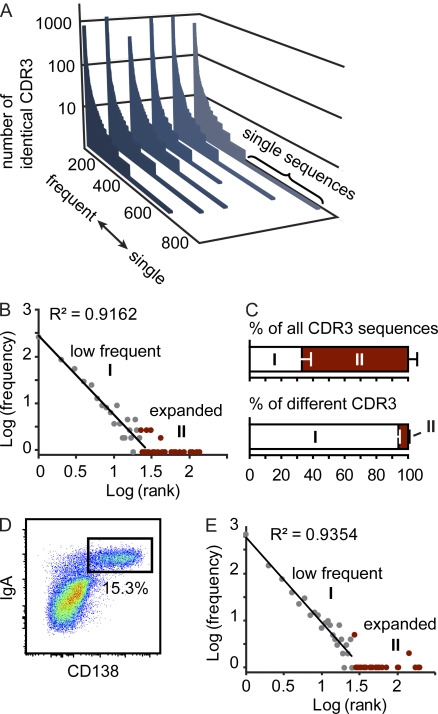
To visualize this feature of the IgA repertoire, we sorted all different CDR3 sequences in a set of 5,000 sequences from most frequent to single sequences. These sorted sequence sets were aligned along the x axis, and on the y axis we indicated how many times the respective sequence was observed (Fig. 1 A). This diagram illustrates the distribution of the IgA repertoire, which is composed of expanded and lowly frequent clones. To allow for further analysis of the data, each CDR3 sequence was assigned a rank based on its abundance. Sequences obtained only once were given rank 1, sequences obtained twice rank 2, and high abundant sequences given a rank equaling to how many times the respective sequence was observed. We next determined the frequency of each rank, e.g., among a representative set of 5,000 CDR3 sequences, we identified 294 sequences of rank 1; thus, the frequency of rank 1 was set to 294. Log transformation of rank and frequency allowed formally separating two components of the repertoire. A first component followed the power law, i.e., appeared as straight line, whereas the second component presented as a long tail in log frequency-rank diagrams (Fig. 1 B). In this respect, the IgA repertoire resembled previously described distributions of human CD8+ memory T cells (Naumov et al., 2003, 2011) and mouse regulatory T cells (Haribhai et al., 2011). Calculating the cutoff point separating both components (Naumov et al., 2003) showed that in a set of 5,000 sequences, sequences obtained less than roughly 20 times (19.4 ± 3.2; n = 6) contributed to the first component, whereas sequences obtained more frequently belonged to the second component of the repertoire. Notably, expanded clones contributed substantially to the total sequence pool but made up only a small proportion of all different CDR3 sequences (Fig. 1 C).
Figure 1.
The IgA repertoire comprises highly expanded and low frequency clones. (A) In a set of 5,000 sequences, all different CDR3 sequences were listed along the x axis from frequent to single sequences, and the number of sequence reads for each different CDR3 sequence were displayed on the y axis. Each slice represents an independently analyzed mouse. (B) Log(frequency) versus log(rank) diagram for one representative mouse reveals two components of the CDR3 population. A first component (I) follows power law characteristics and comprises clones present at low frequency. A second component (II) comprises highly expanded clones and appears as a long tail in the diagram and is marked in red. (C) Bars depict the contribution of both components to all CDR3 sequences and different CDR3 sequences (mean + SD). All data are based on the analysis of six 10–11-wk-old C57BL/6 mice housed under SPF conditions. (D) Representative pseudocolor plot demonstrating IgA/CD138 staining gated on live (DAPI−) single SI lamina propria cells. (E) Log(frequency) versus log(rank) diagram of 5,000 CDR3 sequences obtained from sorted IgA+CD138+ plasma cells pooled from two 10-wk-old C57BL/6 mice. Results are representative of four independent experiments performed.
The repertoire’s two components might constitute an intrinsic feature of the plasma cell repertoire. Alternatively, the composite nature of the IgA repertoire observed in our whole tissue samples might reflect a mixture of different cell types carrying largely divergent amounts of IgA encoding messenger RNA, such as bona fide plasma cells expressing high levels of IgA transcripts and nonplasma cells expressing much lower amounts of IgA encoding messenger RNA. To distinguish between these alternatives, we analyzed IgA+CD138+ plasma cells sorted from the SI lamina propria (Fig. 1 D). Analyzing the sequence repertoire of sorted IgA+CD138+ cells, we observed, similar to our observation obtained from whole tissue, highly expanded along with low frequent sequences (Fig. 1 E). Moreover, log transformation of rank and frequency revealed two distinct components formally separated by a cutoff point similar to what we observed analyzing whole tissue samples (cutoff point for sorted plasma cell populations, 23.9 ± 3.4; n = 4; Fig. 1 E). We conclude that the two components are a fundamental characteristic of the intestinal IgA repertoire that needs to be considered when the diversity and nature of the IgA repertoire is discussed. Previous work used measures of ecosystem diversity, which considers the richness and evenness of a population, to describe the diversity of T cell receptor repertoires (Naumova et al., 2009). In the case of our data, the richness of the IgA population correlates to the number of different CDR3 sequences and positively correlates with diversity. Because lowly frequent clones account for the majority of all different CDR3 sequences, the repertoire’s nonexpanded first component is particularly relevant when the richness of the IgA repertoire is estimated. We therefore conclude that in contrast to previous suggestions (Stoel et al., 2005; Yuvaraj et al., 2009), the intestinal IgA repertoire is highly polyclonal.
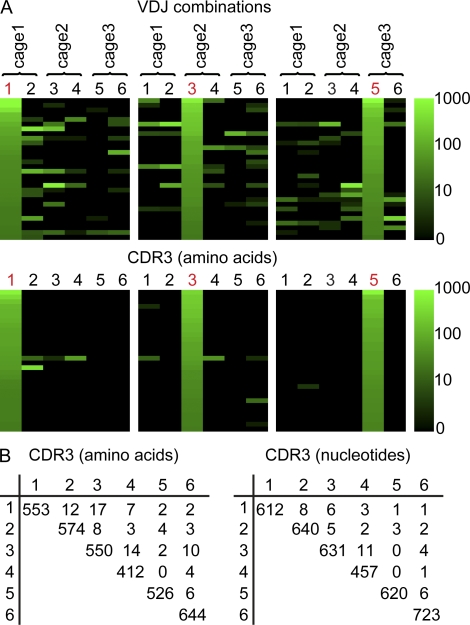
We next compared the IgA repertoire between three pairs of C57BL/6 mice housed under specified pathogen–free (SPF) conditions in three different cages. In all mice, consistently the majority of VH domain sequences belonged to the VH1 or VH5 family (48.1 ± 13.8% VH1 and 32.1 ± 14.3% VH5; n = 6). Analyzing the repertoire’s second component, i.e., highly expanded sequences, revealed that some VDJ combinations were abundantly used throughout several individual mice (Fig. 2 A). Still, when comparing CDR3 amino acid sequences in different mice, only very few CDR3 sequences expanded in one mouse were also detected in another animal (Fig. 2, A and B). Specifically, only three CDR3 sequence shared between two mice were highly expanded in both individuals, and no CDR3 amino acid sequence was observed in all mice. We quantified this observation by calculating Morisita-Horn indices (MHIs; Magurran, 1988), which score two identical populations as 1 and two completely disparate populations as 0. Comparison of CDR3 sequence repertoires of different mice consistently resulted in MHIs close to 0 (MHI 0.0013 ± 0.0020; n = 6), which is indicative of dissimilar populations. This indicates that even when exposed to the identical environment and possibly sharing a similar composition of microbiota, there is little conversion of the CDR3 sequence repertoire between different mice.
Figure 2.
Individual mice show largely nonoverlapping IgA repertoires in the intestine. (A) In a set of 5,000 sequences per sample, all VDJ combinations were enumerated, and the sequence sets were sorted from most frequent to least frequent as observed in mouse number 1, 3, or 5 (indicated in red). The number of sequence reads assigned to each VDJ combination is indicated by color code. Similarly, sequence sets were sorted according to the frequencies of CDR3 amino acid sequences. Please note that logarithmic transformation was used to generate the color code and even sequences observed only three times are nonblack. (B) CDR3 amino acid and nucleotide sequences were compared between different mice, and the number of identical sequences was enumerated. The number of different CDR3 sequences in a given animal appears in the diagonal of the table. Very few CDR3 sequences were shared between different animals. All data are based on the analysis of six 10–11-wk-old C57BL/6 mice housed under SPF conditions.
CCR9 differentially targets expanded clones to the small and large intestine
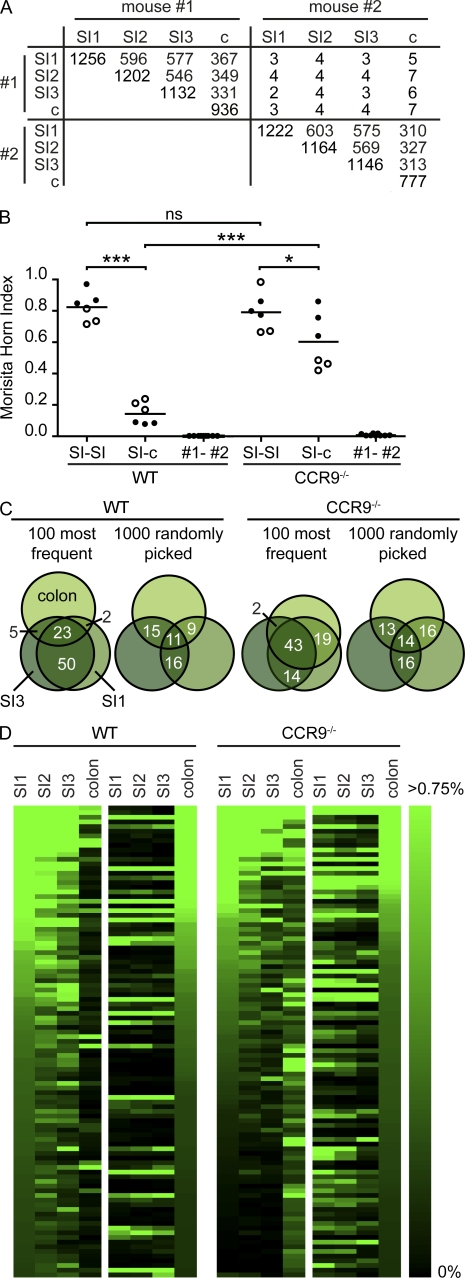
Although the sequence repertoire of different mice showed little overlap, we readily detected identical CDR3 sequences in different parts of the intestine obtained from the same animal (Fig. 3 A). CDR3 sequences from different SI fragments from the same mouse were significantly more similar than SI and colon (Fig. 3 B). Still, the sequence repertoire in SI and colon of the same mouse was more similar compared with repertoires obtained from different mice (Fig. 3 B). Notably, largely overlapping sequence repertoires between independently processed samples of the same but not different mice also ensured that expanded sequences did not result from a PCR bias and that sequence errors did not preclude conclusive data analysis. Overlap of different SI samples resulted primarily from overlapping expanded clones (Fig. 3 C), i.e., the repertoire’s second component. Sorting 10,000 CDR3 sequences obtained from the proximal jejunum from most frequent sequences to sequences observed only once, we found that the identical CDR3 sequences were similarly frequent in distal jejunum and ileum (Fig. 3 D). In contrast, many sequences abundantly present in the SI were infrequent or undetectable in the colon, and only a minority of sequences was present at high frequency in both small and large intestinal sequence sets. Sorting the sequence sets, according to their frequency in the colon, we found a comparable scenario, i.e., only a minority of the expanded sequences dominating the sequence repertoire in the colon was similarly frequent in the SI (Fig. 3 D).
Figure 3.
Identical CDR3 sequences seed different fragments of the SI. (A) Nucleotide CDR3 sequences were compared between proximal jejunum (SI1), distal jejunum (SI2), ileum (SI3), and colon (c) of 13-wk-old C57BL/6 mice, and the number of identical sequences was enumerated. Data are based on 10,000 sequences per sample, and the number of different CDR3 sequences in each sample appears in the diagonal of the table. (B) MHIs were calculated comparing CDR3 sequence pools obtained from various gut fragments in WT and CCR9−/− mice (WT, n = 2; CCR9−/−, n = 2). Individual mice are indicated by open and closed symbols, and comparisons calculated are SI1 to SI2, SI1 to SI3, and SI2 to SI3 to describe similarity of SI fragments and SI1 to c, SI2 to c, and SI3 to c for comparison of SI- and large intestine–derived sequence pools. Comparison of CDR3 sequence pools between different mice yields MHIs close to 0 (first and second columns). Horizontal lines indicate the mean. *, P < 0.05; ***, P < 0.001. (C) Venn diagrams illustrate the overlap of CDR3 sequence pools from SI1, SI3, and colon in one WT and one CCR9−/− mouse. Numbers indicate overlap in percentages. (D) 10,000 CDR3 sequences from SI1, SI2, SI3, and colon were sorted according to their frequency in SI1 (WT, left) or colon (WT, right). Because overlap of sequence sets is driven by expanded sequences, we limited our analysis to the 100 most frequent sequences and visualized their frequency (in percentages of all sequences) by color codes. Note that upon sorting according to frequency in the proximal jejunum, more sequences with similarly high frequency could be found in CCR9−/− mice than in WT mice.
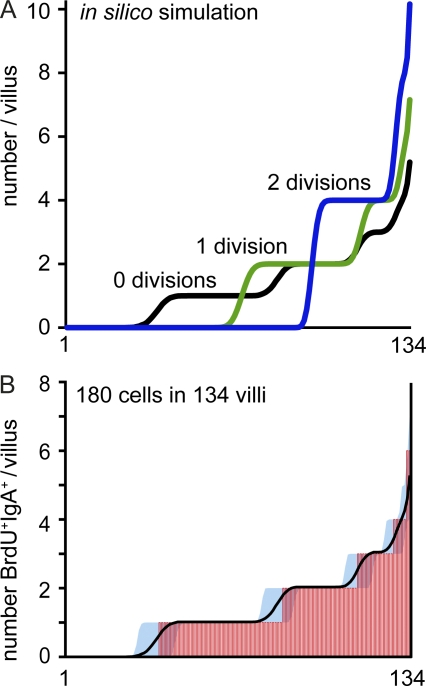
Differences in the plasma cell repertoire between SI and colon might be caused by selective homing of plasmablasts or alternatively stem from subsequent local events in the lamina propria. To study the role of plasmablast homing, we used mice lacking CCR9 (chemokine [CC motif] receptor 9), which mediates homing of plasmablasts into the SI but not colon (Pabst et al., 2004). In CCR9−/− mice, SI- and large intestine–derived sequence pools showed higher similarity (Fig. 3 B) and larger sequence overlap (Fig. 3, C and D) as compared with WT mice. Because CCR9 is down-regulated on plasma cells inside the lamina propria (Pabst et al., 2004), these data indicate that expansion of distinct specificities might occur before plasma cell homing to the lamina propria. Yet CCR9 expression is not restricted to plasmablasts, and we cannot rule out the possibility that other CCR9-expressing cells contribute to the increased repertoire overlap observed in CCR9−/− mice. To directly examine the extent of plasma cell proliferation inside the lamina propria, we compared the distribution of recently proliferated plasma cells after 7 d of continuous BrdU application with distributions predicted in silico. In a representative experiment, we observed a total of 180 BrdU+IgA+ cells in 134 villi (pooled from five mice). According to this experimental observation, we predicted distributions of 180 cells throughout 134 villi in silico assuming that cells did not proliferate, performed one division, or performed two divisions (Fig. 4 A). In silico simulations based on two divisions were distinguished by the noticeable accumulation of numerous cells within few villi, whereas 69% of all villi were devoid of any cell. Similarly, distributions obtained for single divisions inside the gut predicted that ∼49% of all villi would not contain any cell (Fig. 4 A). Both of these predicted properties did not resemble the experimentally observed spreading of BrdU+IgA+ cells throughout individual villi. In contrast, the experimental data, i.e., the observed number of BrdU+IgA+ per villus, fitted simulations that did not assume in situ proliferation (Fig. 4 B). Yet a similarly good representation of the experimental data, i.e., similarly low sum of squared errors, was obtained for models that assumed that 10% or less of all cells divided once inside the lamina propria. We therefore conclude that plasmablasts evenly homed to the SI but not colon, and in situ proliferation seems unlikely to fully explain the presence of expanded clones in the IgA repertoire.
Figure 4.
Newly generated plasma cells are evenly distributed in the gut mucosa. (A) A random distribution of 180 cells throughout 134 individual villi was predicted by in silico simulation assuming no (black), one (green), or two (blue) cell divisions. (B) Mice received BrdU continuously with the drinking water for 7 d, and the number of BrdU+IgA+ cells in individual villi was enumerated by immunofluorescence microscopy. Inspecting 134 villi (pooled from five mice), we found 180 BrdU+IgA+ cells. The x axis corresponds to individual villi sorted from villi containing no cells to villi containing multiple BrdU+IgA+ cells, and each villus is represented by a vertical red line. Mean and 95% confidence interval of the in silico simulation with no cell divisions are depicted by a black line and blue-filled area. Similar results were observed after 2 d of BrdU application.
Age, microbiota, and T cells drive repertoire diversity
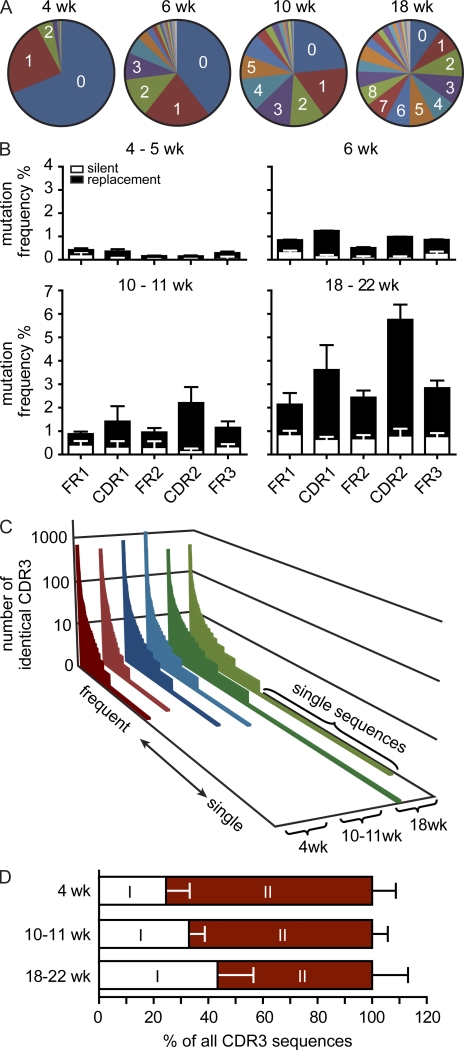
Besides CDR3 diversity, somatic hypermutation (SHM) contributes to IgA diversity, and in humans most IgA sequences are highly mutated (Barone et al., 2011; Gibbons and Spencer, 2011). Thus, we extended our analysis from CDR3 to full-length VH sequences and frequencies of SMs to investigate further determinants of repertoire diversity. In a first set of experiments, we compared the IgA repertoire in mice of various age. We did not observe significant differences with respect to VH family usage between mice of various age, but the frequency of CDR3 sequences lacking N nucleotides (N− sequences) was higher in 4-wk-old mice compared with older mice (44.6 ± 6.2 in 4-wk-old [n = 2] compared with 11.2 ± 9 in 10–11-wk-old mice [n = 6]). Lack of nontemplated N nucleotides is a characteristic feature of peritoneal B cells and B cells of fetal origin (Vale et al., 2010). Thus, we speculate that a higher frequency of N− sequences might result from B cells generated in the fetal liver prevailing in the repertoire of young mice. Yet these differences did not results in more similar IgA repertoires in young mice compared with old mice, and also comparison of CDR3 sequence repertoires of 4–5-wk-old mice resulted in an MHI close to 0 (MHI 0.00442 ± 0.00461; n = 4). Differently aged mice showed striking differences in their repertoires when we analyzed frequencies of SMs and CDR3 distributions. Whereas 4–5-wk-old mice showed few SMs, their frequencies gradually increased in 6-, 10–11-, and 18–22-wk-old mice (Fig. 5, A and B). In 18–22-wk-old WT mice housed under SPF conditions, most VH sequences showed numerous SMs. SM frequencies were highest in CDR1 and CDR2 regions, and the number of replacement mutations exceeded that of silent mutations (Fig. 5 B). This indicates that age is a critical determinant of SM frequencies. Moreover, age also affected CDR3 sequence distributions. In particular, expanded clones, i.e., the second component not following power law characteristics, were more abundant in young compared with old mice, and conversely, aged mice accumulated increasing numbers of CDR3 sequences present at low frequency (Fig. 5 C). The increase in lowly frequent clones in aged mice is also reflected in a shift of the repertoire’s two components (Fig. 5 D). This shift may indicate the ongoing recruitment of low frequent sequences into the repertoire. Yet alternatively low SM frequencies and higher frequency of N− sequences in young compared with aged mice may contribute to observed differences. Notably, expanded clones present in 18–22-wk-old mice contained as many SMs as low frequency clones (Fig. 6 A), and expanded clones of identical CDR3 showed complex phylograms, which frequently contained several major branches (Fig. 6 B). We suggest that during aging, two independent processes contribute to the diversification of the repertoire. First, on expense of expanded clones dominating the repertoire in young mice, the number of CDR3 sequences observed at low frequency increased in older mice, and second, SMs accumulated.
Figure 5.
IgA repertoire diversity increases with age. (A) Diagrams depict the number of SMs observed in representative 4-, 6-, 10-, and 18-wk-old mice. (B) Bars depict mean + SD SM frequencies in FR1, FR2, FR3, CDR1, and CDR2 observed in the following number of animals: 4–5 wk, n = 4; 6 wk, n = 2; 10–11 wk, n = 6; and 18–22 wk, n = 5. (C) CDR3 sequences were sorted by rank, from high to low frequency, and the number of identical sequences was plotted (compare with Fig. 1 A). Please note that the number of unique sequences increased with age. (D) Bars depict the contribution of both components to all CDR3 sequences (mean + SD).
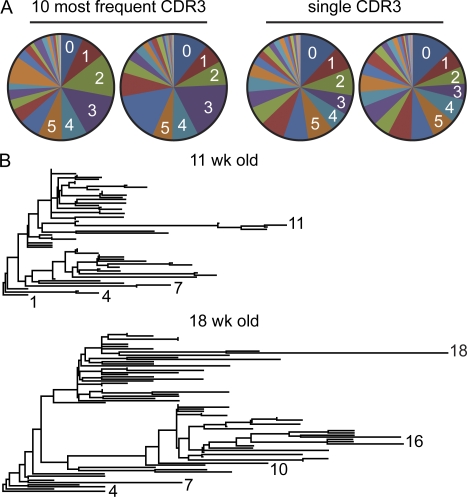
Figure 6.
Expanded clones accumulated extensive SMs. (A) Diagrams depict the frequencies of SMs in two representative 18-wk-old mice (out of five 18–22-wk-old mice analyzed) in sequences comprising the 10 most frequent CDR3 sequences or in VH sequences carrying CDR3 sequences that were present only once in a set of 17,000 (left) and 23,000 (right) sequences. (B) Phylogram demonstrating clonal relationship of the most frequent CDR3 sequences in one representative 11- and 18-wk-old mouse. Numbers indicate the number of SMs in selected sequences.
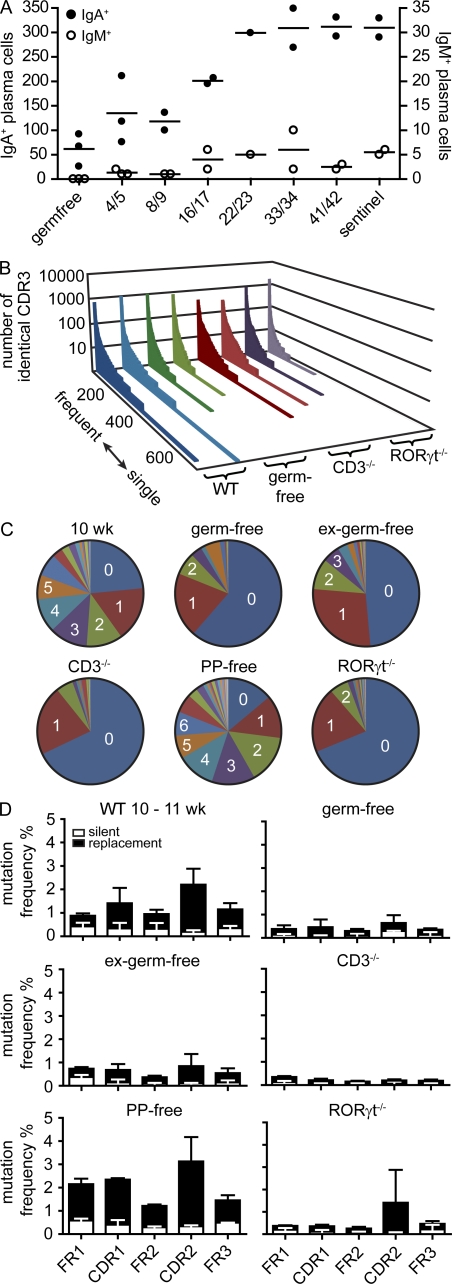
Next, we analyzed the role of microbiota in generating repertoire diversity. Whereas germ-free mice harbored only few intestinal plasma cells, cohousing of germ-free mice with SPF mice resulted in rapid induction of plasma cells in the lamina propria. After 4 wk, the number of IgA+ and IgM+ cells was indistinguishable between ex–germ-free mice and sentinels housed under SPF conditions (Fig. 7 A). Notably, IgA+ and IgM+ cell numbers increased with similar kinetics during the filling of the lamina propria in ex–germ-free mice (Fig. 7 A). This indicates that most intestinal IgA-secreting cells undergo class switch recombination before entering the lamina propria and is consistent with the lack of overt proliferating plasma cells in the lamina propria described in the previous section. The CDR3 sequence repertoire in germ-free mice showed the characteristic two components, low frequent along with highly expanded clones, also observed in SPF-reared mice (Fig. 7 B), indicating that a polyclonal IgA repertoire can be generated independent of microbial stimulation. Still, germ-free mice showed few SMs compared with age-matched SPF mice (Fig. 7, C and D). Even though after 4 wk of cohousing plasma cell numbers were indistinguishable, VH domain sequences from ex–germ-free mice still showed fewer SMs than those from age-matched SPF mice (Fig. 7, C and D), indicating that SHM was delayed compared with accumulation of plasma cells. Similarly, in T cell–deficient mice, we observed a polyclonal CDR3 population, whereas SM frequencies were rather low. Notably, mice treated in utero with lymphotoxin-β-receptor fusion protein, which blocks the development of PPs, showed similarly high SM frequencies compared with PP-sufficient mice (Fig. 7, C and D). Besides PPs, the SI harbors a large number of small-sized lymphoid aggregates, which in response to microbial stimulation can develop into B cell–rich isolated lymphoid follicles (ILFs; Pabst et al., 2006). Because, first, ILFs have been suggested to support PPs and T cell–independent IgA switch (Tsuji et al., 2008) and, second, our experimental approach of PP ablation results in increased formation of ILFs (McDonald et al., 2005), we next analyzed the IgA repertoire in RORγt−/− mice. RORγt−/− mice lack lymphoid tissue inducer cells and in consequence do not develop LNs, PPs, and ILFs (Eberl and Littman, 2004). Still, aged RORγt−/− mice generate intestinal plasma cells (Tsuji et al., 2008). In contrast to PP-deficient mice, 20-wk-old RORγt−/− mice showed very few SMs, similar to the situation in T cell–deficient mice (Fig. 7, C and D). No differences were observed in any of these mice with respect to VH family usage. In conclusion, these findings indicate that CDR3 diversity could be generated independent of microbiota, T cells, and GALTs. In contrast, diversification of the repertoire by SHM required microbiota and T cells but not PPs. Still, SMs were absent in aged RORγt−/− mice, even though these mice possess T cells. This may indicate that RORγt-dependent lymphoid tissue inducer cells might directly contribute to SHM. Alternatively, lack of SMs in RORγt−/− mice might be a consequence of impaired lymphoid organogenesis. We therefore propose that different lymphoid compartments, potentially comprising PPs, mLNs, ILFs, and spleen, might have overlapping functions in driving T cell–dependent accumulation of SMs.
Figure 7.
SHM requires T cells, microbiota, and RORγt but not PPs. (A) Germ-free mice were colonized by cohousing with sentinels raised under SPF conditions for the indicated number of days. Symbols indicate the total number of IgA+ and IgM+ cells in individual mice observed by histology counting six fields of view from at least three different sections per mouse covering different fragments of the SI. Horizontal lines indicate the mean. (B) In a set of 5,000 sequences, all different CDR3 sequences were listed along the x axis from frequent to single sequences, and the number of sequence reads for each different CDR3 sequence was displayed on the y axis. Each slice represents an independently performed experiment as indicated. (C) Diagrams depict the number of SMs observed in representative 10-wk-old WT (see Fig. 5), germ-free, ex–germ-free, CD3−/−, PP-free, and RORγt−/− mice. (D) Bars depict mean + SD SM frequencies in FR1, FR2, FR3, CDR1, and CDR2 observed in the following number of animals: 10–11-wk-old WT mice (n = 6; identical to data in Fig. 5 B and shown to improved readability), germ-free mice (9 wk; n = 2), ex–germ-free mice (11 wk; n = 2), CD3−/− mice (10 wk; n = 3), mice lacking PPs (12 wk; n = 2), and RORγt−/− mice (20 wk; n = 2). For IgA sequences from PP-free mice, RNA was isolated out of sorted IgA+CD138+ intestinal plasma cells. For each mouse, a minimum of 10,000 sequences were analyzed.
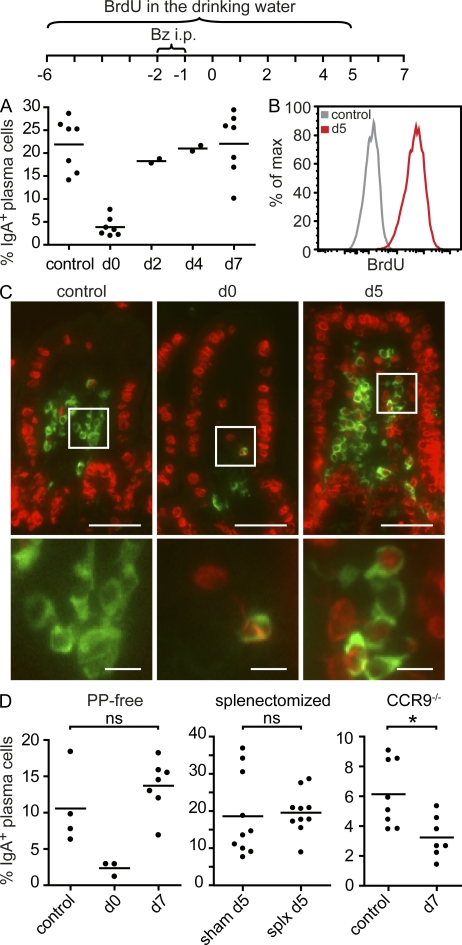
The IgA system recalls previously selected specificities after plasma cell depletion
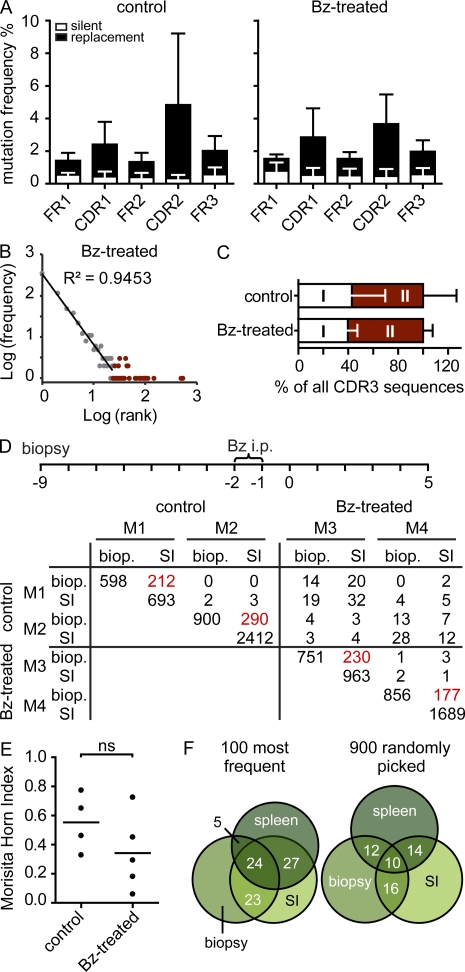
To investigate the stability of individual repertoires, we established an experimental setup that allows on the one hand to deplete intestinal plasma cells and on the other hand to follow an individual’s repertoire over time. To deplete intestinal plasma cells, we used the proteasome inhibitor Bortezomib, which is used for the treatment of myeloma patients. Bortezomib has been shown to deplete plasma cells in bone marrow and spleen (Neubert et al., 2008) and has less severe effects on other immune cells. We observed that two consecutive injections of Bortezomib depleted most intestinal IgA-secreting cells (Fig. 8, A and C). Yet numbers of IgA-secreting cells rapidly recovered after depletion, and after 5–7 d, plasma cell numbers in the SI had returned to normal values (Fig. 8, A and C). Virtually all of these plasma cells were newly generated (Fig. 8, B and C). Replenishment of plasma cell numbers after depletion showed similar kinetics in PP-deficient and splenectomized mice compared with WT mice (Fig. 8 D). However, plasma cell repopulation was delayed in CCR9−/− mice (Fig. 8 D). This suggests that the plasma cell population installed after depletion was derived from circulating progenitors that required CCR9 to enter the lamina propria. Furthermore, the plasma cell repertoire reinstalled after depletion showed SM frequencies similar to nondepleted age-matched controls (Fig. 9 A). This indicates that the plasma cell pool was reestablished from a previously selected and mutated cell population. Moreover, the reinstalled IgA repertoire showed the characteristic two components (Fig. 9 B) without major changes in their contribution (Fig. 9 C). To compare the plasma cell repertoire before depletion and after plasma cell replenishment within the same animal, we next obtained SI biopsies before Bortezomib injection. Interestingly, the repertoire observed in biopsies was similar to the repertoire recalled after plasma cell depletion (Fig. 9, D and E). Moreover, we noted a substantial overlap of the IgA repertoire in the SI and spleen of the same animal (Fig. 9 F). This indicates that after plasma cell depletion, the IgA system recalled previously selected specificities. In sum, our results suggest that an individual’s IgA repertoire undergoes ongoing modulation, by recruitment of new nonexpanded clones into the population and on the basis of already established expanded clones. Diversification of expanded clones might require the repeated entry and selection of circulating precursors in germinal centers, which might also underlie the striking stability of an individual’s IgA repertoire.
Figure 8.
Intestinal IgA-secreting plasma cell numbers rapidly recover after depletion. (A) Mice were injected i.p. on two consecutive days with Bortezomib (Bz), lamina propria cells were isolated, and IgA-secreting plasma cells were quantified by ELISPOT on days 0, 2, 4, and 7 as indicated. Circles represent individual mice analyzed and pooled from two or more experiments, and horizontal lines indicate the mean. (B) Mice received BrdU with the drinking water starting 6 d before Bortezomib injection as indicated. Lamina propria cells were isolated, and the frequency of BrdU+ cells among CD138+IgA+ cells was determined by flow cytometry. Control mice did not receive BrdU. (C) SI tissue was embedded in paraffin, sectioned, and immunostained for IgA (green) and BrdU (red). Images are representative of at least five mice analyzed in two independent experiments. Boxed areas are shown at higher magnification in the bottom panels. Bars: (top) 50 µm; (bottom) 10 µm. (D) Plasma cell numbers rapidly recover after depletion in PP-deficient mice and splenectomized mice but not in CCR9−/− mice. Mice received two consecutive injections of Bortezomib, and plasma cell numbers were quantified by ELISPOT in nondepleted mice and at days 0, 5, or 7 after depletion as depicted. Symbols represent individual mice pooled from at least two experiments. Horizontal lines indicate the mean. *, P < 0.05.
Figure 9.
Identical specificities were recalled after plasma cell depletion. (A) Bars depict mean + SD SM frequencies in FR1, FR2, FR3, CDR1, and CDR2 present in mice after plasma cell depletion or without depletion (four to five mice per group). (B) Representative log(frequency) versus log(rank) diagram of 5,000 CDR3 sequences. (C) Bars depict the contribution of both components to all CDR3 sequences (mean + SD). (D) 10,000 CDR3 sequences were compared between SI biopsies (biop) and SI of two control and two Bortezomib (Bz)-treated mice, and the number of identical CDR3 sequences was enumerated. Overlap of biopsy and SI from the same mouse is highlighted by red numbers. (E) MHIs were calculated comparing CDR3 sequences from the biopsies with the corresponding SI. Mice were 11–15 wk old at the time of sacrifice. Horizontal lines indicate the mean. (F) Venn diagrams illustrate percent overlap of the 100 most frequent CDR3 sequences and 900 randomly picked unique sequences from the SI, biopsy, and spleen of one untreated mouse.
DISCUSSION
The intestinal mucosa is constantly exposed to microbial and dietary antigens, all of which are considered to induce IgA-secreting plasma cells. Thus, the limited IgA repertoire diversity predicted in previous studies (Dunn-Walters et al., 1997, 2000; Holtmeier et al., 2000; Stoel et al., 2005; Yuvaraj et al., 2009) is difficult to reconcile with the broad range of intestinal antigens. In this study, we show that in fact each individual harbors a private polyclonal and highly diverse IgA repertoire. Because we analyzed switched Ig sequences, repertoire analyses performed in this study did not require sorting of plasma cells. Instead, RNA was isolated directly from intestinal tissue, and IgA-encoding transcripts were enriched during reverse transcription. This approach was equivalent to our results obtained when RNA was isolated out of sorted plasma cells. The IgA repertoire showed characteristics reminiscent of the T cell receptor repertoire of CD8+ memory cells (Naumov et al., 2003), i.e., the IgA repertoire comprised two components which both contributed to the overall IgA repertoire diversity. A first component contained clones present at low frequency, whereas a second component encompassed highly expanded clones. Previous studies (Dunn-Walters et al., 1997; Holtmeier et al., 2000; Stoel et al., 2005; Yuvaraj et al., 2009), which lacked the coverage to distinguish these components or wrongly assumed that expanded clones are representative of the entire plasma cell population, inevitably underestimated the number of B cell precursors giving rise to the overall plasma cell population. Expanded clones were readily detected in previous studies, and clonal expansion has been suggested to occur in the periphery (Dunn-Walters et al., 1997, 2000; Holtmeier et al., 2000) and/or locally in the lamina propria (Husband and Gowans, 1978; Yuvaraj et al., 2009). Several observations in this study support the former hypothesis. First, expanded clones were evenly distributed along the SI but not large intestine. Second, SI- and large intestine–specific sorting of specificities required chemokine receptor CCR9, and third, proliferated plasma cells were randomly distributed throughout SI villi. All of these observations suggest that clonal expansion is likely to occur predominantly before plasmablast entry into the lamina propria.
Comparing the IgA sequence repertoire of different mice, we observed very little overlap. Antibody heavy chain sequences are generated in a series of recombination events using multiple V, D, and J segments and further sequence diversification at their junctions (Jung et al., 2006). As a consequence, the number of possible antibody sequences exceeds the number of plasma cells. Still, a comprehensive analysis of the B cell repertoire in zebrafish revealed a highly stereotyped usage of VDJ combinations in young fish (Jiang et al., 2011). Moreover, adult zebrafish also showed signs of conversion of their antibody repertoires; i.e., different individuals made the same antibodies (Weinstein et al., 2009; Jiang et al., 2011). In fact, we also observed some CDR3 sequences shared between different mice, and three of these CDR3 sequences were highly expanded. At present, we cannot reliably calculate the probability to observe such overlap in nonconvergent repertoires. Still, most CDR3 sequences were unique and not shared between mice. Moreover, the similarity of the sequence repertoire was not any different comparing mice housed in the same or different cages. This observation is reminiscent of the observation made in zebrafish that in older animals, expansion of distinct clones confounded similarities of the original, nonexpanded antibody repertoire. Thus, effectively, each mouse harbored an individual IgA repertoire.
In young mice, the IgA repertoire was dominated by expanded clones carrying few SMs. We therefore propose that early in life, each individual selects and expands a set of founder B cells, which best fit the actual intestinal antigen load to set up the initial pool of intestinal plasma cells. Intriguingly, the clones selected at this early time might also serve as basis for the further modulation of the IgA repertoire. Frequencies of SMs progressively increased during aging, and the IgA repertoire installed after plasma cell depletion was largely similar to the repertoire before depletion. Because plasma cells are terminally differentiated cells, both observations could be explained by the existence of circulating IgA-positive B cells. Such cells might be established along with the first wave of plasmablasts seeding the SI and considered IgA memory B cells. Subsequently, their repeated reentry into germinal centers could allow for the progressive accumulation of SMs and result in the complex clonal populations of plasma cell we observed in aged mice. At the same time, an IgA memory–like population might enable the rapid recall of previously selected specificities, including SMs, after plasma cell depletion. Alternatively, the recall of identical specificities after plasma cell depletion might be viewed as amplification of ongoing immune responses. In fact, intestinal bacteria are permanently taken up into PPs and carried by gut dendritic cells into the draining mLNs (Macpherson and Uhr, 2004). Thus, the intestinal immune system is under constant stimulation, and GALTs might permanently accommodate activated B cells of the respective specificities. Indeed, recent work has shown that the IgA system lacks prime-boost characteristics (Hapfelmeier et al., 2010), an important hallmark of memory responses. In any case, our observations demonstrate that functionally the IgA system can recall previously selected specificities and that an individual’s private IgA repertoire is fixed in the sense that a limited set of expanded clones is maintained and vitally contributes to the overall plasma cell pool. Nevertheless, depletion of plasma cells has been shown to result in loss of self-reactive Ig and to improve the clinical outcome in experimental autoimmune models (Neubert et al., 2008), and further repertoire studies will be needed to see to what extent plasma cell depletion can permanently eradicate disease promoting self-reactive specificities also in the intestine.
SHM was triggered by microbiota, which on the one hand stimulate the maturation of GALTs and germinal centers and on the other hand provide a complex array of antigens. Both of these functions might contribute to the increase of SM frequencies observed after colonization of germ-free mice. Moreover, SHM was strictly dependent on T cells and the presence of RORγt. Besides T cell–dependent germinal center reactions, in mice, several alternative pathways of plasma cell generation, including peritoneal B1 cells, T cell–independent processes, and class switch recombination in the lamina propria, have been proposed (Suzuki et al., 2010; Cerutti et al., 2011). Our data do not provide strong arguments to support or dismiss the relevance of such pathways in young mice. However, in aged mice, similar to human IgA sequences (Barone et al., 2011; Gibbons and Spencer, 2011), most VH sequences carried numerous SMs. Thus, in aged mice, most IgA-secreting plasma cells seem to be generated with the help of T cells. Even though we cannot easily compare the age of humans and mice, one might speculate that the comparison of young mice, which still undergo substantial modulation of their IgA repertoire, with humans might overemphasize the presumed differences between both species. Another prerequisite of SHM seems to be the transcription factor RORγt, and RORγt−/− mice did not acquire SMs even though these mice possess a polyclonal T cell pool. One possible explanation of this observation might emphasize the function of GALTs and germinal centers for SHMs. Yet we cannot rule out the possibility that the absence of lymphoid tissue inducer cells in RORγt−/− mice or other defects in these mice might also have more direct effects on some pathways of IgA generation. Thus, further experiments will be needed to dissect the contribution of germinal center–dependent and –independent pathways of IgA induction in young and aged mice as well as in humans.
In sum, we propose two processes that simultaneously shape the mouse intestinal IgA repertoire. On the one hand, new clones can be recruited into the intestine. This process might reflect the cumulative encounter of IgA-inducing stimuli throughout life, which the system cannot appropriately account for on the basis of expanded clones already represented in the repertoire. On the other hand, each individual harbors a private set of expanded clones that undergo continued T cell–dependent maturation. This process coincides with the establishment of stable intestinal communities, and changes in the microbiota might be a major factor driving the mutation of expanded clones. However, the host immune system also reciprocally affects the composition of the microbiota (Fagarasan et al., 2002; Wei et al., 2011), and such a process might rather be regarded as reciprocal shaping of the host’s IgA repertoire along with its intestinal microbiota. Together, these processes account for the dynamic changes in the IgA repertoire described herein, offer an explanation for the repertoire’s unexpected stability after plasma cell depletion, and provide a framework to further dissect the generation and function of intestinal IgA.
MATERIALS AND METHODS
Mice.
C57BL/6, CCR9−/−, CD3−/−, and RORγt−/− mice (all backcrossed to C57BL/6 background for at least eight generations) were bred under SPF conditions or germ-free conditions at the central animal facility of the Hannover Medical School. Data displayed in Fig. 7 A are based on the analysis of ex–germ-free mice previously reported in Pabst et al. (2006). PP-free mice were generated by injecting pregnant C57BL/6 mice on gestational day 14 i.p. with 2 mg lymphotoxin receptor β–Ig fusion protein (Krege et al., 2009). Mice were carefully inspected to lack PPs before inclusion in the study. To label proliferating cells, mice were injected i.p. with 3 mg BrdU and thereafter continuously received 80 mg/100 ml BrdU with the drinking water. For plasma cell depletion, mice were injected i.p. on two consecutive days with 20 µg Bortezomib (Velcade). All animal experiments were performed in accordance with institutional guidelines and have been approved by the review board of the Hannover Medical School and the Niedersaechsische Landesamt für Verbraucherschutz und Lebensmittelsicherheit.
Mice surgery.
Mice were anesthetized using ketamine and xylazine. The abdominal cavity was opened, and an SI fragment was exposed. Approximately 1 × 2 mm2 of intestinal tissue was cut from the antimesenteric site, and the intestine was closed with suture. The SI was reintroduced into the abdominal cavity. For splenectomy, the peritoneum was opened, the spleen was located, vessels and ligaments were heat cauterized, and the spleen was cut out. The peritoneum was closed with suture, and skin was stapled.
Cell isolation, flow cytometry, and cell sorting.
For isolation of intestinal lamina propria cells, gut content and PPs were removed, and intestines were opened longitudinally and washed with cold PBS. Intestines were incubated three times for 15 min in 15 ml HBSS containing 10% FCS and 2 mM EDTA at 37°C to remove epithelial cells. After each incubation step, tubes were shaken vigorously for 10 s, and media containing epithelial cells and debris were discarded. The remaining tissue was incubated for 45 min in RPMI 1640 with 10% FCS, 0.24 mg/ml collagenase A (Roche), and 40 U/ml DNase I (Roche), and tubes were shaken vigorously for 10 s. Cell suspensions were filtered through a nylon mesh and purified by density gradient centrifugation with 40–70% Percoll (GE Healthcare). For flow cytometric analysis, cells were stained with IgA-FITC (Invitrogen) and CD138-PE (BD). BrdU staining was performed with the APC-BrdU Flow kit (BD) according to the manufacturer’s instructions. Plasma cells were sorted as DAPI−IgA+CD138+ single cells to >95% purity.
ELISPOT assay.
ELISPOT was performed to detect IgA-secreting cells. ELISPOT plates (MultiScreen HTS IP 0.45 µm; Millipore) were coated overnight at 4°C with affinity-purified anti–mouse IgA (5 µg/ml in PBS; eBioscience). The next day, plates were washed twice with RPMI 1640 and blocked with 10% FCS in RPMI 1640 at 37°C for 2 h. Isolated lamina propria cells were added to the wells and incubated overnight at 37°C and 5% CO2. The next day, cells were discarded, and plates were washed twice with deionized H2O and three times with 0.1% Tween in PBS. Detection of IgA spots was performed with biotinylated anti-IgA antibody (AbD Serotec) followed by peroxidase-labeled streptavidin (Jackson ImmunoResearch Laboratories, Inc.) and 3-amino-9-ethylcarbazole (Sigma-Aldrich) as substrate. Color development was stopped by washing several times with water. Plates were scanned, and spots were counted.
Histology.
For preparation of paraffin sections of the SI, a distal piece of the SI was washed with PBS and fixed with Bouin’s solution (Sigma-Aldrich) for 5 h at 4°C. After washing three times for 15 min with cold PBS, the SI was dehydrated with ascending concentrations of ethanol followed by xylene and finally embedded in paraffin. 6-µm sections were prepared and stained according to the manufacturer’s instructions with the BrdU Immunohistochemistry System (EMD) based on Cyanine3-labeled tyramide (PerkinElmer) as substrate in combination with IgA staining by an FITC-labeled antibody (Invitrogen). Staining was documented using an epifluorescence microscope (BX61; Olympus) and AnalysisD software (Soft Imaging System).
454 sequencing.
Three pieces of 5-cm length each were taken from the proximal, middle, and distal part of the SI. Gut content and PPs were removed before pieces were opened longitudinally and washed with cold PBS. Intestines were homogenized in TRIZOL reagent (Invitrogen) with an Ultra Turrax (all three parts together in 5 ml or each part in 3 ml; IKA). From 1 ml of the lysate, RNA was isolated according to the manufacturer’s instructions. Biopsy samples were lysed in 0.5 ml TRIZOL, and the whole lysate was used for RNA isolation. For sequence analysis from sorted cells, lamina propria cells were isolated, IgA+CD138+ live (DAPI−) plasma cells were sorted, and RNA was isolated with the RNeasy Mini kit (QIAGEN). cDNA synthesis was performed with SuperScript III (Invitrogen) based on a mix of three gene-specific primers for the IgA locus (5′-ATCAGGCAGCCGATTATCAC-3′, 5′-TCTCCTTCTGGGCACTCG-3′, and 5′-TGAATGATGCGCCACTGT-3′). To generate template libraries of rearranged IgA sequences from IgA+ plasma cells, PCR with a primer binding in the constant Cα region 5′-CGTATCGCCTCCCTCGCGCCATCAG(MID)GAGCTCGTGGGAGTGTCAGTG-3′ in combination with a promiscuous VH primer 5′-CTATGCGCCTTGCCAGCCCGCTCAGGAGGTGCAGCTGCAGGAGTCTGG-3′ (binding to all VH genes) was performed. MID (multiple identifier) specifies a 4-nt sequence used in six different variations to identify samples within single lanes. PCR conditions were as follows: 95°C, 4 min; 25 × (94°C, 30 s; 62°C, 30 s; 72°C, 35 s); and 72°C, 10 min. Amplicons were purified by gel extraction (QIAquick Gel Extraction kit; QIAGEN) and quantified by Quant-iT dsDNA HS Assay kit (Invitrogen) measured with the Qubit fluorometer (Invitrogen). Amplicons were prepared with the GS FLX Titanium SV emPCR kit (Lib-A) for ultra-deep 454 pyrosequencing on the Genome Sequencer FLX system (Roche) as described by the manufacturer.
Sequence analysis.
Sequences were assigned to individual samples according to their MID. Sequences lacking one or both primer sequences or shorter than 320 bp were excluded. Sequences were further analyzed with ImMunoGeneTics (IMGT) HighV-QUEST (http://www.imgt.org/; Brochet et al., 2008; Lefranc et al., 2009), a web portal allowing for the analysis of high numbers of sequences. All sequences were compared against reference sequences from the IMGT database. Results obtained from IMGT were further analyzed with Excel (Microsoft) and VBA (Visual Basic for Applications), and only productive sequences were used. Mutation frequencies were calculated as the number of mutations divided by the number of all nucleotides of the given frame work regions and CDRs. Venn diagrams were generated with VennMaster 0.37.4 (2010-04-06). For phylogenetic analysis, sequences were aligned with ClustalW 2.0.12 Multiple Sequence Alignment tool (Larkin et al., 2007; Goujon et al., 2010). Based on the alignment, phylogenetic trees were calculated with ClustalW2 and displayed with dendroscope 2.7.4 (Huson et al., 2007).
Repertoire enumeration and analysis.
CDR3 sequences were sorted by frequency, and sequences with the same frequency were grouped into ranks ranging from single sequences with rank 1 and high abundant sequences given a rank number according to their frequency. To display the power law–like distribution, a log-log transformation of a frequency versus rank plot was performed. As described in the first section of Results, low frequency sequences follow the power law and lay on a straight line, whereas high frequency sequences do not. These two components of the repertoire can be divided at a critical point, which was calculated as Xc = exp((log N + log a)/−b), where N is the number of different CDR3 sequences, and a and b are the intercept and slope of the log-log plot. Of note, all ranks belonging to the power law component must have a smaller rank than Xc.
Statistical analysis.
Statistical analysis was performed with Prism software (GraphPad Software). All significant values were determined using one-way analysis of variance with Tukey’s post-hoc test, for comparing more than two groups, or an unpaired two-tailed Student’s t test, for comparison of two groups. Data are mean + SD. P-values are indicated as follows: *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Acknowledgments
We thank Jo Spencer, Per Brandtzaeg, Hedda Wardemann, Rudolf Manz, Reinhold Förster, Olga Schulz, Heike Herbrand, and Andreas Krueger for discussion of the manuscript and Michaela Friedrichsen and Sabrina Woltemate for excellent technical assistance.
This work was supported by Deutsche Forschungsgemeinschaft grant SFB900-Z1 to S. Suerbaum and grant SFB621-TPA11 to O. Pabst.
The authors declare no competing financial interests.
Footnotes
Abbreviations used:
- GALT
- gut-associated lymphoid tissue
- ILF
- isolated lymphoid follicle
- MHI
- Morisita-Horn index
- mLN
- mesenteric LN
- PP
- Peyer’s patch
- SHM
- somatic hypermutation
- SI
- small intestine
- SIgA
- secretory IgA
- SM
- somatic mutation
- SPF
- specified pathogen free
References
- Asahi Y., Yoshikawa T., Watanabe I., Iwasaki T., Hasegawa H., Sato Y., Shimada S., Nanno M., Matsuoka Y., Ohwaki M., et al. 2002. Protection against influenza virus infection in polymeric Ig receptor knockout mice immunized intranasally with adjuvant-combined vaccines. J. Immunol. 168:2930–2938 [DOI] [PubMed] [Google Scholar]
- Barone F., Vossenkamper A., Boursier L., Su W., Watson A., John S., Dunn-Walters D.K., Fields P., Wijetilleka S., Edgeworth J.D., Spencer J. 2011. IgA-producing plasma cells originate from germinal centers that are induced by B-cell receptor engagement in humans. Gastroenterology. 140:947–956 10.1053/j.gastro.2010.12.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benckert J., Schmolka N., Kreschel C., Zoller M.J., Sturm A., Wiedenmann B., Wardemann H. 2011. The majority of intestinal IgA+ and IgG+ plasmablasts in the human gut are antigen-specific. J. Clin. Invest. 121:1946–1955 10.1172/JCI44447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandtzaeg P. 2009. Mucosal immunity: Induction, dissemination, and effector functions. Scand. J. Immunol. 70:505–515 10.1111/j.1365-3083.2009.02319.x [DOI] [PubMed] [Google Scholar]
- Brandtzaeg P., Johansen F.E. 2005. Mucosal B cells: Phenotypic characteristics, transcriptional regulation, and homing properties. Immunol. Rev. 206:32–63 10.1111/j.0105-2896.2005.00283.x [DOI] [PubMed] [Google Scholar]
- Brochet X., Lefranc M.P., Giudicelli V. 2008. IMGT/V-QUEST: the highly customized and integrated system for IG and TR standardized V-J and V-D-J sequence analysis. Nucleic Acids Res. 36:W503–W508 10.1093/nar/gkn316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerutti A., Chen K., Chorny A. 2011. Immunoglobulin responses at the mucosal interface. Annu. Rev. Immunol. 29:273–293 10.1146/annurev-immunol-031210-101317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez-Bello M.G., Blaser M.J., Ley R.E., Knight R. 2011. Development of the human gastrointestinal microbiota and insights from high-throughput sequencing. Gastroenterology. 140:1713–1719 10.1053/j.gastro.2011.02.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn-Walters D.K., Boursier L., Spencer J. 1997. Hypermutation, diversity and dissemination of human intestinal lamina propria plasma cells. Eur. J. Immunol. 27:2959–2964 10.1002/eji.1830271131 [DOI] [PubMed] [Google Scholar]
- Dunn-Walters D.K., Hackett M., Boursier L., Ciclitira P.J., Morgan P., Challacombe S.J., Spencer J. 2000. Characteristics of human IgA and IgM genes used by plasma cells in the salivary gland resemble those used in duodenum but not those used in the spleen. J. Immunol. 164:1595–1601 [DOI] [PubMed] [Google Scholar]
- Eberl G., Littman D.R. 2004. Thymic origin of intestinal alphabeta T cells revealed by fate mapping of RORgammat+ cells. Science. 305:248–251 10.1126/science.1096472 [DOI] [PubMed] [Google Scholar]
- Fagarasan S., Muramatsu M., Suzuki K., Nagaoka H., Hiai H., Honjo T. 2002. Critical roles of activation-induced cytidine deaminase in the homeostasis of gut flora. Science. 298:1424–1427 10.1126/science.1077336 [DOI] [PubMed] [Google Scholar]
- Gibbons D.L., Spencer J. 2011. Mouse and human intestinal immunity: Same ballpark, different players; different rules, same score. Mucosal Immunol. 4:148–157 10.1038/mi.2010.85 [DOI] [PubMed] [Google Scholar]
- Goujon M., McWilliam H., Li W., Valentin F., Squizzato S., Paern J., Lopez R. 2010. A new bioinformatics analysis tools framework at EMBL-EBI. Nucleic Acids Res. 38:W695–W699 10.1093/nar/gkq313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hapfelmeier S., Lawson M.A., Slack E., Kirundi J.K., Stoel M., Heikenwalder M., Cahenzli J., Velykoredko Y., Balmer M.L., Endt K., et al. 2010. Reversible microbial colonization of germ-free mice reveals the dynamics of IgA immune responses. Science. 328:1705–1709 10.1126/science.1188454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haribhai D., Williams J.B., Jia S., Nickerson D., Schmitt E.G., Edwards B., Ziegelbauer J., Yassai M., Li S.H., Relland L.M., et al. 2011. A requisite role for induced regulatory T cells in tolerance based on expanding antigen receptor diversity. Immunity. 35:109–122 10.1016/j.immuni.2011.03.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holtmeier W., Hennemann A., Caspary W.F. 2000. IgA and IgM V(H) repertoires in human colon: evidence for clonally expanded B cells that are widely disseminated. Gastroenterology. 119:1253–1266 10.1053/gast.2000.20219 [DOI] [PubMed] [Google Scholar]
- Husband A.J., Gowans J.L. 1978. The origin and antigen-dependent distribution of IgA-containing cells in the intestine. J. Exp. Med. 148:1146–1160 10.1084/jem.148.5.1146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huson D.H., Richter D.C., Rausch C., Dezulian T., Franz M., Rupp R. 2007. Dendroscope: An interactive viewer for large phylogenetic trees. BMC Bioinformatics. 8:460 10.1186/1471-2105-8-460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang N., Weinstein J.A., Penland L., White R.A., III, Fisher D.S., Quake S.R. 2011. Determinism and stochasticity during maturation of the zebrafish antibody repertoire. Proc. Natl. Acad. Sci. USA. 108:5348–5353 10.1073/pnas.1014277108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansen F.E., Pekna M., Norderhaug I.N., Haneberg B., Hietala M.A., Krajci P., Betsholtz C., Brandtzaeg P. 1999. Absence of epithelial immunoglobulin A transport, with increased mucosal leakiness, in polymeric immunoglobulin receptor/secretory component-deficient mice. J. Exp. Med. 190:915–922 10.1084/jem.190.7.915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung D., Giallourakis C., Mostoslavsky R., Alt F.W. 2006. Mechanism and control of V(D)J recombination at the immunoglobulin heavy chain locus. Annu. Rev. Immunol. 24:541–570 10.1146/annurev.immunol.23.021704.115830 [DOI] [PubMed] [Google Scholar]
- Krege J., Seth S., Hardtke S., Davalos-Misslitz A.C., Förster R. 2009. Antigen-dependent rescue of nose-associated lymphoid tissue (NALT) development independent of LTbetaR and CXCR5 signaling. Eur. J. Immunol. 39:2765–2778 10.1002/eji.200939422 [DOI] [PubMed] [Google Scholar]
- Larkin M.A., Blackshields G., Brown N.P., Chenna R., McGettigan P.A., McWilliam H., Valentin F., Wallace I.M., Wilm A., Lopez R., et al. 2007. Clustal W and Clustal X version 2.0. Bioinformatics. 23:2947–2948 10.1093/bioinformatics/btm404 [DOI] [PubMed] [Google Scholar]
- Lefranc M.P., Giudicelli V., Ginestoux C., Jabado-Michaloud J., Folch G., Bellahcene F., Wu Y., Gemrot E., Brochet X., Lane J., et al. 2009. IMGT, the international ImMunoGeneTics information system. Nucleic Acids Res. 37:D1006–D1012 10.1093/nar/gkn838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lycke N., Holmgren J. 1986. Intestinal mucosal memory and presence of memory cells in lamina propria and Peyer’s patches in mice 2 years after oral immunization with cholera toxin. Scand. J. Immunol. 23:611–616 10.1111/j.1365-3083.1986.tb01995.x [DOI] [PubMed] [Google Scholar]
- Macpherson A.J., Uhr T. 2004. Induction of protective IgA by intestinal dendritic cells carrying commensal bacteria. Science. 303:1662–1665 10.1126/science.1091334 [DOI] [PubMed] [Google Scholar]
- Macpherson A.J., McCoy K.D., Johansen F.E., Brandtzaeg P. 2008. The immune geography of IgA induction and function. Mucosal Immunol. 1:11–22 10.1038/mi.2007.6 [DOI] [PubMed] [Google Scholar]
- Magurran, A.E. 1988. Ecological Diversity and Its Measurement. Princeton University Press, Princeton, NJ. 192 pp.
- McDonald K.G., McDonough J.S., Newberry R.D. 2005. Adaptive immune responses are dispensable for isolated lymphoid follicle formation: antigen-naive, lymphotoxin-sufficient B lymphocytes drive the formation of mature isolated lymphoid follicles. J. Immunol. 174:5720–5728 [DOI] [PubMed] [Google Scholar]
- Naumov Y.N., Naumova E.N., Hogan K.T., Selin L.K., Gorski J. 2003. A fractal clonotype distribution in the CD8+ memory T cell repertoire could optimize potential for immune responses. J. Immunol. 170:3994–4001 [DOI] [PubMed] [Google Scholar]
- Naumov Y.N., Naumova E.N., Yassai M.B., Gorski J. 2011. Selective T cell expansion during aging of CD8 memory repertoires to influenza revealed by modeling. J. Immunol. 186:6617–6624 10.4049/jimmunol.1100091 [DOI] [PubMed] [Google Scholar]
- Naumova E.N., Gorski J., Naumov Y.N. 2009. Two compensatory pathways maintain long-term stability and diversity in CD8 T cell memory repertoires. J. Immunol. 183:2851–2858 10.4049/jimmunol.0900162 [DOI] [PubMed] [Google Scholar]
- Neubert K., Meister S., Moser K., Weisel F., Maseda D., Amann K., Wiethe C., Winkler T.H., Kalden J.R., Manz R.A., Voll R.E. 2008. The proteasome inhibitor bortezomib depletes plasma cells and protects mice with lupus-like disease from nephritis. Nat. Med. 14:748–755 10.1038/nm1763 [DOI] [PubMed] [Google Scholar]
- Pabst O., Ohl L., Wendland M., Wurbel M.A., Kremmer E., Malissen B., Förster R. 2004. Chemokine receptor CCR9 contributes to the localization of plasma cells to the small intestine. J. Exp. Med. 199:411–416 10.1084/jem.20030996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pabst O., Herbrand H., Friedrichsen M., Velaga S., Dorsch M., Berhardt G., Worbs T., Macpherson A.J., Förster R. 2006. Adaptation of solitary intestinal lymphoid tissue in response to microbiota and chemokine receptor CCR7 signaling. J. Immunol. 177:6824–6832 [DOI] [PubMed] [Google Scholar]
- Sait L.C., Galic M., Price J.D., Simpfendorfer K.R., Diavatopoulos D.A., Uren T.K., Janssen P.H., Wijburg O.L., Strugnell R.A. 2007. Secretory antibodies reduce systemic antibody responses against the gastrointestinal commensal flora. Int. Immunol. 19:257–265 10.1093/intimm/dxl142 [DOI] [PubMed] [Google Scholar]
- Stoel M., Jiang H.Q., van Diemen C.C., Bun J.C., Dammers P.M., Thurnheer M.C., Kroese F.G., Cebra J.J., Bos N.A. 2005. Restricted IgA repertoire in both B-1 and B-2 cell-derived gut plasmablasts. J. Immunol. 174:1046–1054 [DOI] [PubMed] [Google Scholar]
- Stoel M., Evenhuis W.N., Kroese F.G., Bos N.A. 2008. Rat salivary gland reveals a more restricted IgA repertoire than ileum. Mol. Immunol. 45:719–727 10.1016/j.molimm.2007.07.001 [DOI] [PubMed] [Google Scholar]
- Suzuki K., Maruya M., Kawamoto S., Fagarasan S. 2010. Roles of B-1 and B-2 cells in innate and acquired IgA-mediated immunity. Immunol. Rev. 237:180–190 10.1111/j.1600-065X.2010.00941.x [DOI] [PubMed] [Google Scholar]
- Tsuji M., Suzuki K., Kitamura H., Maruya M., Kinoshita K., Ivanov I.I., Itoh K., Littman D.R., Fagarasan S. 2008. Requirement for lymphoid tissue-inducer cells in isolated follicle formation and T cell-independent immunoglobulin A generation in the gut. Immunity. 29:261–271 10.1016/j.immuni.2008.05.014 [DOI] [PubMed] [Google Scholar]
- Vale A.M., Tanner J.M., Schelonka R.L., Zhuang Y., Zemlin M., Gartland G.L., Schroeder H.W., Jr 2010. The peritoneal cavity B-2 antibody repertoire appears to reflect many of the same selective pressures that shape the B-1a and B-1b repertoires. J. Immunol. 185:6085–6095 10.4049/jimmunol.1001423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei M., Shinkura R., Doi Y., Maruya M., Fagarasan S., Honjo T. 2011. Mice carrying a knock-in mutation of Aicda resulting in a defect in somatic hypermutation have impaired gut homeostasis and compromised mucosal defense. Nat. Immunol. 12:264–270 10.1038/ni.1991 [DOI] [PubMed] [Google Scholar]
- Weinstein J.A., Jiang N., White R.A., III, Fisher D.S., Quake S.R. 2009. High-throughput sequencing of the zebrafish antibody repertoire. Science. 324:807–810 10.1126/science.1170020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wijburg O.L., Uren T.K., Simpfendorfer K., Johansen F.E., Brandtzaeg P., Strugnell R.A. 2006. Innate secretory antibodies protect against natural Salmonella typhimurium infection. J. Exp. Med. 203:21–26 10.1084/jem.20052093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuvaraj S., Dijkstra G., Burgerhof J.G., Dammers P.M., Stoel M., Visser A., Kroese F.G., Bos N.A. 2009. Evidence for local expansion of IgA plasma cell precursors in human ileum. J. Immunol. 183:4871–4878 10.4049/jimmunol.0901315 [DOI] [PubMed] [Google Scholar]