Abstract
Animal and human studies of addiction indicate that the D2 dopamine receptor (DRD2) plays a critical role in the mechanism of drug reward. D2 receptor density in the brains of alcoholics has been shown to be reduced relative to controls. Previous studies of DRD2 in association with alcohol dependence using variation in the TaqI A locus were highly controversial. Recently, a synonymous mutation, C957T, in the coding region of the human DRD2 gene has been identified which appears to have functional effects including alteration in receptor availability. In order to determine if susceptibility to alcohol dependence (AD) within multiplex alcohol dependence families would be altered by the C957T in the coding region of the D2 gene, within-family association was studied in members of Caucasian multiplex alcohol dependence families. Members of control families with no personal alcohol or substance dependence history were included for case/control comparisons. Analyses performed to detect within-family association showed evidence favoring an association for the C957T polymorphism (P = 0.038). Linkage analyses of polymorphisms in this region showed that only the C957T locus remained of interest (P = 0.015). Evidence for the C957T T allele having a role in AD susceptibility at the population level using a case/control comparison was statistically marginal (P = 0.062), but was consistent with the family data results. These results support a role for DRD2 as a susceptibility gene for alcohol dependence within multiplex families at high risk for developing alcohol dependence.
Keywords: D2 receptor, alcohol dependence, multiplex families, pedigree disequilibrium test, linkage
INTRODUCTION
Twin, adoption and family studies have provided ample evidence for genetic mediation of alcohol dependence susceptibility within families [Goodwin et al., 1973; Pickens et al., 1991; Heath et al., 1999]. Twin studies tend to show greater concordance for alcohol dependence in MZ than in DZ twins [Kendler et al., 1992; McGue et al., 1992], providing estimates of heritability in the range of 0.54 to 0.58 in males [Prescott, 2001]. The search for genes that may confer increased susceptibility to alcohol dependence has included a number of genes that influence neurotransmitter regulation including the dopamine pathway.
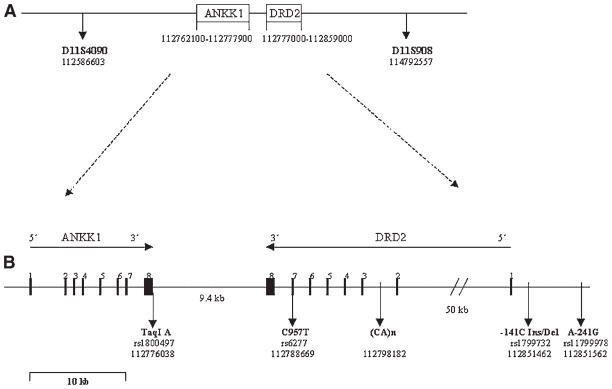
One gene that has been studied extensively in both animal and human studies of addiction is the dopamine D2 receptor system because of its apparent importance in the reward capacity of ethanol and other drugs of abuse [Volkow et al., 2002a, 2003]. DRD2 is a G-protein coupled receptor located on postsynaptic neurons, that is, centrally involved in reward-mediating mesocorticolimbic pathways. The dopamine DRD2 gene has been mapped to 11q22-23 (Fig. 1). The gene is composed of 8 exons and spans 65.8 kilobases (kb) of genomic DNA. Relative to controls, subjects with alcohol [Hietala et al., 1994; Volkow et al., 2002b], cocaine [Volkow et al., 1993], and opioid dependence [Wang et al., 1997] have been reported to have lower levels of D2 dopamine receptors.
Fig. 1.
The figure depicts a 2.21 cM region of Chromosome 11q23 that includes the human DRD2 receptor gene structure and single nucleotide polymorphism (SNP) sites included in the present analyses. The total coverage for this study is 131.4 cM. Distances are based on physical maps obtained from current NCBI databases.
One polymorphic locus that shows evidence of being in linkage disequilibrium with the DRD2 gene is a restriction fragment length polymorphism (RFLP) known as TaqI A. Much of the early work on alcohol dependence and the DRD2 gene was based on this single nucleotide polymorphism (SNP) (TaqI A) located 9.5 kb distal to the 3′ end of the gene [Grandy et al., 1989]. The A1 allele has been hypothesized to be associated with alcohol dependence. Several case/control studies have investigated allelic variation in the TaqI A polymorphism but with controversial results. Several negative results were reported in comparisons of alcohol dependent and control individuals [Bolos et al., 1990; Gelernter et al., 1991; Cook et al., 1992; Turner et al., 1992; Arinami et al., 1993; Suarez et al., 1994] though many studies did obtain positive results for alcohol dependent individuals [Amadeo et al., 1993; Noble et al., 1994; Neiswanger et al., 1995] and for polysubstance abusers [Smith et al., 1992; Lawford et al., 1997]. Averaging of A1 frequencies across 21 studies that included alcoholics and controls revealed greater frequency among alcoholics [Noble, 2003].
Because of these associations between D2 receptors and addiction, evidence that receptor density in brain might be associated with variation in the TaqI A polymorphism provides an important line of investigation. Using postmortem caudate nucleus samples from alcoholics and controls, presence of the A1 allele was reported to lower D2 receptor binding [Blum et al., 1990], a finding later confirmed in additional brain samples in which lower Bmax was found for samples with the A1 allele [Noble et al., 1991]. Using autopsy striatum samples including the caudate, Thompson et al. [1997] found a reduction in the density of D2 dopamine receptors that was associated with the presence of the A1 allele.
Investigation of possible in vivo differences in volunteers [Pohjalainen et al., 1998] has also revealed a relationship between the presence of the A1 allele and low D2 dopamine receptor binding (Bmax), affinity (Kd) and availability (Bmax/Kd) in the striatum using positron emission tomography (PET) with [11C] raclopride. A significant decrease in D2 dopamine receptor availability, reflecting a reduction in receptor density, was observed with the A1/A2 genotype in contrast to the A2/A2 genotype. These in vivo results appear to have been confirmed in another sample of healthy controls in which a relationship between A1 allele status and binding potential was found [Laruelle et al., 1998]. Finally, Jönsson et al. [1999] using [11C] raclopride and PET technology examined DRD2 polymorphisms and striatal D2 dopamine receptor density in healthy Swedish volunteers and found a positive association between the TaqI A1 allele and lower D2 dopamine receptor density. This group also found an association between the D2 promoter -141C Ins/Del and receptor density. A relationship between the A1 allele of the D2 receptor and striatal dopamine transporter (DAT) density has also been examined and found to be much higher in alcoholics with the A1/A2 genotypes than in those homozygous for the A2 allele [Laine et al., 2001].
There has been increased concern regarding the functional significance of the TaqI A polymorphism, previously reported to be associated with alcohol dependence, because it is located approximately 10 kb downstream from the DRD2 gene (GenBank database entry AF050737.1; Fig. 1). Although linkage disequilibrium (LD) has been reported to extend from the DRD2 gene to 25 kb beyond the TaqI A locus [Kidd et al., 1998], TaqI A may fall within a different coding region than the DRD2 gene. If this is correct, it would suggest that it is unlikely that the TaqI A polymorphism directly affects activity, and further suggests that the TaqI A may fall within a regulatory region downstream of the DRD2 gene.
Recently, a novel kinase gene, named ankyrin repeat kinase domain containing 1 (ANKK1) was identified that contains a single serine/threonine kinase domain and is expressed in placenta and whole spinal cord RNA [Neville et al., 2004]. The gene is a member of an extensive family of proteins involved in signal transduction pathways. Identification and characterization of the ANKK1 gene by this group suggest that the TaqI A SNP causes an amino acid substitution within the 11th ankyrin repeat of ANKK1 that may affect substrate-binding specificity. Such changes in ANKK1 activity might provide an explanation for previously observed relationships between the TaqI A locus and addictive disorders. Of some interest is the fact that ANKK1 is not expressed in brain though carriers of the A1 allele appear to show associated functional activation using fMRI in the anterior cingulate gyrus [Fosella et al., 2006]. These results suggest that while TaqI A is now known to not be within the DRD2 gene and has been mapped to the ANKK1 gene [Neville et al., 2004], TaqI A may have a role in DRD2 functioning.
Although the D2 dopamine receptor system is clearly associated with addiction susceptibility, the absence of reported mutations in the human DRD2 gene had tempered interest in clinical studies of D2. Gejman et al. [1994] reported finding no structural mutations in the coding region of the D2 receptor gene in alcoholic and schizophrenic subjects. Duan et al. [2003] did identify a mutation in the human D2 receptor gene and studied six synonymous changes in the gene. One of these SNPs, the C957T polymorphism, rather than being silent, alters mRNA folding leading to decreased mRNA stability and translation, and as a consequence dramatically changes dopamine-induced up-regulation of DRD2 expression. To date, this polymorphism has been studied in schizophrenics [Lawford et al., 2005], drug dependent individuals [Xu et al., 2004; Gelernter et al., 2006] and in nicotine dependent participants [Gelernter et al., 2006; Jacobsen et al., 2006; Lerman et al., 2006]. To our knowledge, this mutation has not previously been studied in samples specifically recruited for multiplex alcohol dependent family status though alcohol dependence has been studied in the context of polysubstance abuse [Gelernter et al., 2006].
Functional polymorphisms in the 5′ promoter region of the DRD2 gene have also been identified (A-241G and -141C Ins/Del) [Arinami et al., 1997] that could affect gene regulation or expression. Elevation in D2 receptor density in postmortem brains of schizophrenics who were free of neuroleptic medication for many years [Seeman, 1992] has been reported. Additionally, a decrease in the frequency of the -141C Ins/Del allele has been reported in schizophrenics [Arinami et al., 1997]. Together these findings suggest that elevated D2 receptor density reported in schizophrenics might be the result of a reduced frequency of the -141C Ins/Del allele and further suggests a possible role of this allele in schizophrenia and other psychiatric illnesses such as alcohol dependence.
The present study was undertaken primarily to assess variation in the C957T polymorphism in multiplex alcohol dependence families by genotyping members of these multiplex families. A secondary hypothesis was that if no association between C957T and alcohol dependence were found, it might be the case that genetic variation in the 5′ promoter region of the DRD2 gene might be present. To investigate this hypothesis the A-241G and -141C Ins/Del polymorphisms were genotyped in the same set of families. Because these families had been included in a genome-wide linkage analysis [Hill et al., 2004], genotypes for microsatellites were available for further analyses. Also, previous genotyping had been done for the TaqI A and CA repeat polymorphisms from earlier work in our laboratory [Neiswanger et al., 1995; Hill et al., 1999] for a subset of individuals for whom the C957T, A-241G, and -141C Ins/Del genotyping was performed.
SUBJECTS AND METHODS
Ascertainment and Diagnostic Assessment
All members of the multiplex families and control families who participated in the study gave their written consent to do so after the nature and purpose of the study was fully explained to them. (Consent forms were approved by the University of Pittsburgh Institutional Review Board.)
Multiplex Families
Multiplex families were selected on the basis of the presence of a pair of alcohol dependent brothers or sisters. The probands were selected from among individuals in treatment for alcohol dependence in the Pittsburgh area. All proband pairs were personally interviewed using structured psychiatric interviews (Diagnostic Interview Schedule [DIS]). The DIS, though a less commonly used diagnostic instrument, has good reliability and validity [Helzer et al., 1985]. Use of this instrument made it possible to obtain diagnoses of alcohol dependence and alcohol abuse by DSM-III and IIIR criteria [American Psychiatric Association, 1982, 1987] and alcoholism by Feighner Criteria [Feighner et al., 1972]. Because the majority of individuals were assessed before the release of DSM-IV, no attempt was made to re-diagnose the sample to conform to currently prevailing nomenclature. Nevertheless, the use of more than one diagnostic system, Feighner and DSM-III, allows for comparison with other studies using these criteria in major genome searches for alcoholism susceptibility loci (e.g., Collaborative Study on the Genetics of Alcoholism). The DIS was administered to all living and cooperative participants (probands, siblings, parents). Using the DIS information, a second clinician’s information, and family history report of all other participating relatives, a best estimate diagnosis was made using Feighner criteria and DSM-III and IIIR. All symptoms were retained in computer files also allowing for use as quantitative phenotypes. The present report is based on the dichotomous phenotype in which alcoholism or alcohol dependence is coded as affected and the absence of these conditions is considered unaffected.
As noted, inclusion criteria required that a pair of same-sex adult siblings were present in the family with an alcohol dependence diagnosis. Families were excluded if the probands or any first-degree relative were considered to be primary for drug dependence (preceded alcohol dependence onset by at least 1 year), or the proband or first-degree relative met criteria for schizophrenia, or a recurrent major depressive disorder. Probands and relatives with mental retardation or physical illness precluding participation were excluded. Complete details regarding participant selection may be seen in Hill et al. [2004].
The majority of probands (80%) have three or more siblings who have contributed DNA, consented to a clinical interview, and provided family history. One or both parents have been genotyped in 86% of the families. An average of 5.1 individuals per family have been genotyped. Our rationale for having initiated the study through a double proband sampling scheme was based on the observation that restricting family ascertainment to multiplex families increases the likelihood of finding genes related to the disease of interest [Morton and Mi, 1968; Anderson et al., 1986]. This is largely due to the fact that the likelihood of finding a severe form of any particular disorder segregating within families is increased where multiple cases are found.
A total of 63 Caucasian multiplex pedigrees were available for genotyping and within-family analyses. A total of 201 males and 171 females were included. Of these participants, 226 individuals were affected and 146 were unaffected.
Case/Control Selection
Cases were chosen from among probands in the family study and supplemented with other unrelated alcohol dependent cases (spouses). No family was included more than once. A total of 63 unrelated Caucasian alcohol dependent individuals from multiplex families were chosen along with 8 additional unrelated alcohol dependent individuals. Importantly, controls were not chosen from among the multiplex families. A separate set of controls ascertained as part of a study of control families was utilized for this purpose. A total of 78 Caucasian individuals, free of alcohol dependence and other DSM-III diagnoses, served as controls.
Genotyping
STR genotyping
Blood was drawn from multiplex family members with one aliquot being used to extract DNA from whole blood and the second aliquot prepared for EBV transformation and cryopreservation. PCR conditions were as described in Hill et al. [2004]. Genotypes available for the present set of subjects included 25 STRs on Chromosome 11 that had been completed for a genome wide linkage study previously published [Hill et al., 2004]. Although significant IBD sharing had not been found in that effort, these genotypes were included with the present initiative to provide an opportunity for conducting a multipoint linkage analysis. DNA samples that had been used in this mapping effort were amplified with the ABI Linkage Marker Set Version 2 (LMSV2) primers. PCR products were analyzed on a Perkin Elmer Model 377 Automated Sequencer and electrophoresis data transferred to a Power Mac G3 and tracked as batches using GeneScan 3.1.2. This allowed for manual tracking of each gel before analysis. Each gel included two CEPH DNA (1347-02) samples to control for gel to gel allele-calling variability. Also included on each gel were allelic ladders that were created by pooling 90 DNA aliquots from the sample population. Fluorescent size markers (GeneScan 400HD Applied Biosystems, Foster City, CA) were placed in the same lanes as each sample and used to assign integer values (bins) to each peak in the allelic ladder. These bins were then used to assign allele sizes to the sample peaks.
STR allele calling was first performed using TrueAllele™ (Cybergenetics, Pittsburgh, PA) automated allele-calling software [Perlin et al., 1995], followed by checking by two experienced readers blind to family membership status. The TrueAllele™ software tracks each gel lane and measures the intensity and size of each peak profile. Using size standard and allelic ladder data, TrueAllele™ then assigns integer values to each measured peak to generate a genotype.
SNP genotyping
SNP genotyping was performed by polymerase chain reaction (PCR) amplification of SNP containing genomic sequences by restriction fragment length polymorphism (RFLP) analysis using radiolabeled primers. For all PCR reactions, 20 μg of genomic DNA were amplified in a 7.5 μl reaction mixture containing 15 mM Tris-HCl (pH 8.4), 5 mM KCl, 2 mM MgCl2, 0.2 mM of each deoxynucleotide-5′-triphosphate, 10 pmol of each primer, and 30 units of Taq polymerase (AmpliTaq Gold, Perkin-Elmer, Waltham, MA). The reaction was performed at 95°C for 10 min followed by 10 cycles of 95°C for 15 sec, at the specific annealing temperature for 30 sec and 72°C for 1 min. The remaining 20 cycles were performed at 89°C for 15 sec, at the specific annealing temperature for 30 sec and at 72°C for 1 min. Five microliters of PCR products were digested by the appropriate restriction enzyme according to the recommendations provided by the manufacturer and electrophoresed on a 5% acrylamide gel.
A-241G polymorphism
The A-241G polymorphism was analyzed by PCR amplification of a 304 bp genomic fragment using the forward primer 5′-TGCGCGCGTGAGGCTGCCGGTTCGG-3′ and the reverse primer 5′-ACTGGCGAGCAGACGGTGAGGACCC-3′. Reaction conditions included 2.5 mM deaza-GTP and an annealing temperature of 68°C. PCR products were digested with Mae III (New England Biolabs, Ipswich, MA). The variant allele was identified by the presence of 260 and 44 bp digestion fragments. The wild-type allele was detected by the presence of a 304 bp undigested PCR product.
-141C insertion/deletion polymorphism
The -141C Ins/Del polymorphism was analyzed by PCR amplification of a 275 bp genomic fragment using the forward primer, 5′-CGGTTCGGCACTGAAGCTGGAC-3′, and the reverse primer, 5′-GACGGTGAGGACCCAGCCTGC-3′. Reaction conditions included 0.8 M betaine and an annealing temperature of 62°C for the initial 10 cycles and 60°C for the remaining 25 cycles. PCR products were digested with Bst NI (New England Biolabs). The wild-type insertion allele was detected by the presence of 133 bp and 143 bp digestion products. The variant deletion allele was identified by the presence of a 275 bp undigested DNA fragment.
C957T polymorphism
The C957T polymorphism was analyzed by amplification of a 196 bp genomic fragment using a forward primer mix 5′-ACCAYGGTCTCCACAGCACTC-3′, and the reverse primer, 5′-ATGGCGAGCATCTGAGTGGCT-3′. Reaction conditions included 10% DMSO and an annealing temperature of 62°C for the initial 10 cycles and 60°C for the remaining 25 cycles. PCR products were digested with Taq I (New England Biolabs). The variant allele was identified by the presence of 174 and 22 bp digestion products. The wild-type allele was detected by the presence of a 196 bp undigested DNA fragment.
TaqI A and DRD2-C (CA repeat)
Genotypes were available for the subjects analyzed in the present study for the TaqI A and DRD2-C CA repeat polymorphisms. Genotyping was accomplished using methods previously described [Hill et al., 1999].
STATISTICAL METHODS
Mendelian Inconsistency—PedCheck
A total of 30 markers were evaluated using PedCheck to identify Mendelian inconsistencies [O’Connell and Weeks, 1998]. Eight pedigrees had inconsistencies involving either the four SNPs or the CA repeat polymorphism. If only one child was inconsistent with their parents, the child was recoded as missing and if more than one, the entire family was dropped for the analysis of that marker.
Allele Frequency Estimation
Allele frequencies for the family data used in the linkage analyses were determined using MENDEL (version 5.0) [Lange et al., 2001], a software package that estimates population frequencies from family data using files generated through use of Mega2 [Mukhopadhyay et al., 2005]. Allele frequencies for the C957T locus were tested for Hardy–Weinberg equilibrium in controls (P = 0.637) and in cases (P = 0.377; R Genetics package, 2005; The R Foundation for Statistical Computing Version 2.1.1, 2005).
Case/Control Analyses
The first goal of the statistical analysis was to determine if evidence for association at the C957T locus could be found using independent cases and unrelated controls from an independent set of pedigrees. A contingency table for C957T in which 0, 1, or 2 T alleles were cross-tabulated by affection status (affected or unaffected) allowed for testing an additive model. A collapsed two by two contingency table allowed for testing dominant and recessive models. Both were tested using a Chi square distribution. Simulated p-values were obtained based on 10,000 replicates. In order to test the magnitude and direction of the association between the mutant genotypes and alcohol dependence, logistic regression analyses were performed. Three genetic models were again tested (dominant, recessive, and additive).
Within-Family Association
In order to determine if within-family association in the C957T polymorphism would be found in these multiplex alcohol dependence families, the Pedigree Disequilibrium Test (PDT) [Martin et al., 2000, 2001] was used. This approach was chosen because it is robust to population substructure. The allele-based sum-PDT is an extension of the TDT test [Spielman et al., 1993] that allows for testing the transmission of disease alleles from parent to offspring by including extended pedigree members, thereby increasing the power to detect association. Specifically, the PDT utilizes a composite statistic to capture differential allele transmission in case-parent trios and differential allele frequencies in discordant sib pairs [Martin et al., 2000, 2001].
A secondary hypothesis was that within-family association in the functional polymorphisms A-241G and -141C Ins/Del located in the 5′ promoter region of the dopamine D2 receptor gene might be found. Because of the close proximity of the CA repeat locus and the TaqI A polymorphisms, it was also of interest to determine if within-family association might be apparent in the CA repeat and TaqI A, markers that had previously been investigated in this sample.
Linkage Analyses
The third goal was to determine if evidence for linkage was present at the C957T locus. Genotyping was available for a total of 21 STRs that had been part of our genome-wide analysis [Hill et al., 2004] and the 4 SNPs on Chromosome 11 that were the focus of interest. In order to eliminate markers in statistically significant LD with those within the DRD2 gene, all markers were evaluated for pair-wise LD using the ldmax routine from the GOLD software package [Abecasis and Cookson, 2000]. GOLD uses the expectation-maximization algorithm to estimate the maximum likelihood of the pair-wise disequilibrium using only founders. The estimated measure linkage disequilibrium, denoted as D′, varies between 0 and 1 with larger values indicating greater disequilibrium.
Linkage analyses of the genotyping data were performed using the nonrandom sharing of alleles identical by descent (IBD) in relation to affection status as implemented in the SAGE routine SIBPAL [SAGE Version 5.0.1, 2004]. The program separately evaluates the proportion of alleles shared IBD for concordantly affected and unaffected pairs as well as discordant pairs. Two-point and multipoint IBD sharing probabilities were estimated using the GENIBD routine [SAGE Version 5.0.1, 2004]. Estimates of population allele frequencies were obtained using MENDEL (Version 5) [Lange et al., 2001]. Map locations of STRs and SNPs were obtained from the current builds of UniSTS and dbSNP, respectively (NCBI).
Two-point and multipoint LODPAL analyses were performed to determine linkage estimates. These analyses were used as a complement to the SIBPAL analyses because LODPAL allows for specification of a genetic model. LODPAL is routinely used to evaluate the contribution of affected pairs in comparison to discordant pairs. However, since greater evidence favoring increased IBD sharing occurred in the unaffected pairs, the present analysis focused on a comparison of the unaffected pairs and the discordants. We considered three modes of inheritance: additive (default option), dominant and recessive.
RESULTS
Case/Control Analyses
Analysis of the three by two contingency table representing the binary disease status (affected or unaffected) and variation at the C957T locus allowed for testing an additive genetic model. A two by two contingency table was used to test dominant and recessive models. Results were evaluated using a Chi square distribution to test both allelic and genotypic frequencies among the cases and controls. Among the 81 cases the frequency of the T allele was 48% (77/162) while the frequency of the C allele was 52% (85/162). For the 78 controls the frequency of the T allele was 39% (61/156) and the frequency of the C allele 61% (95/156). Genotype frequency was used to test dominant and recessive models. Among the cases, 16 individuals carried the TT genotype, 20 were CC, and 45 were CT. For controls, 13 individuals carried the TT genotype, 30 were CC, and 35 were CT. The additive model tested whether the frequency of particular alleles covaried with affection status. These analyses showed no significant difference for the additive model (χ2 = 3.51, df = 2, P = 0.173) [simulated P = 0.181] or the recessive model (χ2 = 0.25, df = 1, P = 0.614) [simulated P = 0.690]. The dominant model showed a marginal P-value (χ2 = 3.50, df = 1, P = 0.062) [simulated P = 0.085]. Although the dominant model showed only marginal significance, it is noteworthy that the T allele was more often transmitted with alcohol dependence.
Logistic regression analyses were performed to test the magnitude and direction of the association between the mutant genotypes and alcohol dependence. As expected, evidence favoring the dominant model was again seen with an odds ratio of 1.91 (95% CI: 0.97–3.76, P = 0.06). Interestingly, the additive models indicate that the presence of the mutant allele (T) increases the odds of having alcohol dependence approximately twofold. However, relative to the CC genotype, the odds ratios were approximately equivalent for one (CT) or both (TT) copies being a mutant T allele (OR = 1.93 for heterozygotes and 1.85 for homozygous mutant).
Within-Family Association Results
Results for the A-241G, -141C Ins/Del, CA repeat and TaqI A were all nonsignificant. In contrast, results for C957T were significant (χ2 = 4.29, df = 1, P = 0.038). Parent to affected offspring transmission of a T allele, or risk allele, was 1.36 times more frequent than transmission of a C allele. Similarly, the affected member of the discordant sib pair showed an increase in the frequency of the T allele (52%) compared to that seen in the unaffected member (41%). Differing allelic transmission patterns within multiplex families at the C957T polymorphism appear to confer phenotypic variation. Because the PDT results are most powerful in the presence of linkage, the next goal was to determine if linkage might be found.
Linkage Analyses Results
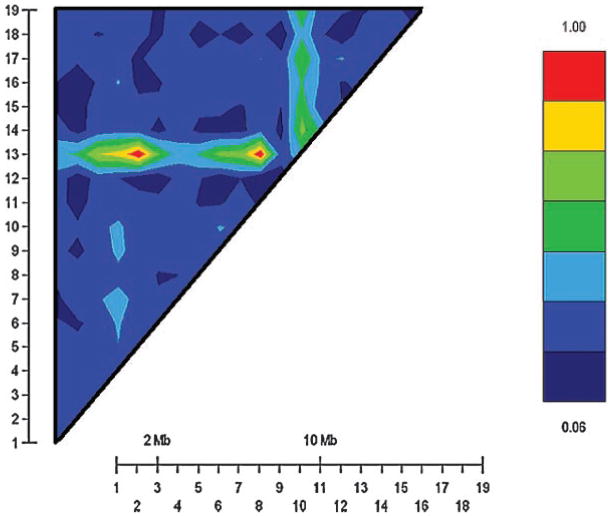
A GOLD analysis was used to estimate linkage disequilibrium for all 25 markers. An LD plot for the 19 markers used in the linkage analyses may be seen in Figure 2. The micro-satellites were designed to be approximately 9 cM apart while SNPs were chosen to be within the DRD2 gene (Fig. 1). Markers were eliminated from the set of 25 polymorphisms based on two criteria: (1) significant LD with the targeted SNP of interest, C957T; and (2) significant LD with DRD2 polymorphisms and those within 70 cM of those loci. The latter criterion was used to avoid elimination of markers that were only spuriously significant. Three STRs (D11S935, D11S901, CA repeat) and two SNPs (TaqI A and -141C Ins/Del) were excluded because of significant LD with C957T. D11S898 was eliminated because it was in significant LD with a DRD2 locus (A-241G) and was within 70 cM of the locus. For those markers in LD with C957T, we found the LD for TaqI A highly significant (D′ = 0.77, χ2 = 20.61, P = 0.00001) as was the LD between C957T and the CA repeat (D′ = 0.49, χ2 = 20.57, P = 0.0001), but less so for C957T and -141C Ins/Del (D′ = 0.74, χ2 = 4.25, P = 0.04).
Fig. 2.
Evaluation of linkage disequilibrium (LD) in the 19 markers considered in the linkage analysis. These markers are D11S4046 (1), D11S1338 (2), D11S902 (3), D11S904 (4), D11S905 (5), D11S4191 (6), D11S987 (7), D11S1314 (8), D11S937 (9), D11S4175 (10), D11S4090 (11), C957T (12), A-241G (13), D11S908 (14), D11S4127 (15), D11S925 (16), D11S4151 (17), D11S1320 (18), D11S968 (19). D11S905 (5) and A-241G (13) exhibit strong LD, but they are over 70 cM apart and the LD was not statistically significant (P = 0.27). The intense LD between D11S4090 (11) and A-241G (13) is also not statistically significant (P = 0.39).
Linkage analysis was performed on the remaining 19 markers found to not be in significant LD. The following microsatellites were analyzed: D11S4046, D11S1338, D11S902, D11S904, D11S905, D11S4191, D11S987, D11S1314, D11S937, D11S4175, D11S4090, D11S908, D11S4127, D11S925, D11S4151, D11S1320, and D11S968. The remaining SNPs, C957T, and A-241G, were analyzed along with the STRs.
The only marker to show evidence favoring linkage (two-point) was the C957T locus (Tables I and II). An adjustment for multiple comparisons was not appropriate since hypothesis testing was restricted to this locus. In the SIBPAL analysis significance was obtained for the 43 unaffected pairs with no evidence being found for the 199 affected pairs, or 168 discordant pairs. SIBPAL results were further tested by performing simulations of the data set (1,000 replicates). Simulations of the SIBPAL data confirmed the C957T results (P = 0.035 for the unaffected pairs). These SIBPAL findings showing that the 43 unaffected sib pairs shared alleles significantly more often than by chance are intriguing. Moreover, the association analysis shows that the T allele or risk allele is less frequently observed in the unaffected sibs. Also, as may be seen in Table II, two-point LOD scores for all three models show a maximum peak at C957T for the unaffected pairs, when contrasted with discordant pairs. The peak value was (LOD = 1.64) under a recessive model. Because linkage analysis of the multiplex families was performed for unaffected pairs (affected pair analysis is more typical), finding evidence for a recessive model is consistent with those obtained for the case/control regression analyses performed in which a dominant mode of inheritance was indicated as the best fitting model.
TABLE I.
Proportion of Allele Sharing IBD From SIBPAL Analysis
| Markers | P(IBD)UU | P(IBD)AU | P(IBD)AA |
|---|---|---|---|
| D11S4046 | 0.48 | 0.50 | 0.49 |
| D11S1338 | 0.49 | 0.52 | 0.49 |
| D11S902 | 0.54 | 0.50 | 0.52 |
| D11S904 | 0.54 | 0.48 | 0.50 |
| D11S905 | 0.53 | 0.53 | 0.49 |
| D11S4191 | 0.51 | 0.52 | 0.51 |
| D11S987 | 0.52 | 0.49 | 0.52 |
| D11S1314 | 0.47 | 0.49 | 0.51 |
| D11S937 | 0.55 | 0.49 | 0.51 |
| D11S4175 | 0.51 | 0.47 | 0.53 |
| D11S4090 | 0.50 | 0.49 | 0.50 |
| C957T | 0.57a | 0.49 | 0.50 |
| A-241G | 0.49 | 0.50 | 0.50 |
| D11S908 | 0.52 | 0.50 | 0.49 |
| D11S4127 | 0.53 | 0.51 | 0.48 |
| D11S925 | 0.53 | 0.49 | 0.52 |
| D11S4151 | 0.49 | 0.54 | 0.49 |
| D11S1320 | 0.46 | 0.52 | 0.50 |
| D11S968 | 0.48 | 0.54 | 0.46 |
P(IBD) scores shown in the table are estimates of the average proportion of alleles shared identical by descent by concordantly unaffected sib pairs (UU), discordant pairs (AU) pairs, and concordantly affected (AA) pairs.
Only C957T was significant (P = 0.015) Only Caucasian families were used in these analyses.
TABLE II.
Linkage Analyses Under Three Models—LODPAL LOD Scores
| Markers | Additive
|
Dominant
|
Recessive
|
|||
|---|---|---|---|---|---|---|
| Two-point | Multipoint | Two-point | Multipoint | Two-point | Multipoint | |
| D11S4046 | <0.01 | 0.08 | 0.01 | 0.06 | <0.01 | 0.09 |
| D11S1338 | <0.01 | 0.02 | <0.01 | 0.01 | 0.04 | 0.03 |
| D11S902 | 0.25 | 0.10 | 0.15 | 0.08 | 0.33 | 0.10 |
| D11S904 | 0.12 | 0.44 | 0.26 | 0.70 | 0.04 | 0.22 |
| D11S905 | 0.13 | 0.31 | 0.11 | 0.11 | 0.18 | 0.55 |
| D11S4191 | <0.01 | <0.01 | 0.01 | 0.03 | <0.01 | <0.01 |
| D11S987 | 0.06 | 0.05 | 0.05 | 0.05 | 0.10 | 0.05 |
| D11S1314 | 0.05 | 0.03 | 0.02 | 0.03 | 0.09 | 0.05 |
| D11S937 | 0.41 | 0.42 | 0.22 | 0.25 | 0.56 | 0.54 |
| D11S4175 | 0.38 | 0.18 | 0.43 | 0.23 | 0.30 | 0.13 |
| D11S4090 | 0.02 | 0.24 | 0.07 | 0.39 | 0.07 | 0.25 |
| C957T | 1.44 | 0.26 | 1.19 | 0.38 | 1.64 | 0.29 |
| A-241G | 0.01 | 0.25 | <0.01 | 0.34 | 0.04 | 0.30 |
| D11S908 | 0.10 | 0.32 | 0.02 | 0.16 | 0.21 | 0.46 |
| D11S4127 | 0.04 | 0.12 | 0.06 | 0.05 | 0.02 | 0.18 |
| D11S925 | 0.11 | 0.01 | 0.12 | 0.02 | 0.11 | 0.01 |
| D11S4151 | <0.01 | <0.01 | 0.09 | <0.01 | <0.01 | <0.01 |
| D11S1320 | <0.01 | <0.01 | <0.01 | <0.01 | <0.01 | <0.01 |
| D11S968 | <0.01 | <0.01 | 0.26 | <0.01 | <0.01 | <0.01 |
Only Caucasian families were used in these analyses.
DISCUSSION
In summary, the present study points to a mutation in the DRD2 gene, C957T, that confers greater likelihood of developing alcohol dependence in families selected for multiple cases of AD. The Pedigree Disequilibrium Test (PDT) was used to evaluate evidence for within-family association between AD and the C957T locus. Confirmation of our within-family association result was tested using linkage analysis of polymorphisms in the vicinity of the DRD2 gene, including those in the promoter region, but C957T remained the only polymorphism of interest. At the population level, C957T showed a trend toward differences between the cases and controls with a twofold increase in likelihood of carrying the T allele among the alcohol dependent subjects. Because these results in multiplex alcohol dependent families and in case/control comparisons are consistent with previous studies showing a reduced number of DRD2 receptors in alcohol dependent individuals, we conclude that alterations in C957T may be, at least in part, responsible for this reduction in receptors. If so, these findings may have important clinical significance in suggesting targets for pharmacological intervention in alcohol dependence.
Limitations of the present study include the rather modest SIBPAL and LODPAL linkage results, especially for the multipoint analyses. However, a pattern of results was obtained suggesting non-random variation. Without available estimates of population frequency for C957T, MENDEL was used to estimate population frequencies for the markers studied. The present results may have been biased toward the null hypothesis because multiplex families were used to estimate these population frequencies.
A further limitation was the resource available for evaluating C957T variation at the population level. A total of 159 Caucasian cases and controls were analyzed using a regression model that evaluated recessive, dominant and additive models. These analyses suggested only marginally significant results under a dominant model. Nevertheless, evidence supporting the within-family PDT results was obtained. Specifically, the T allele had a frequency of 39% in control subjects who had been screened for absence of alcohol dependence. Unselected Caucasians without screening for absence of psychiatric disorder appear to have a higher frequency (48%) (NCBI, dbSNP, build 126). This suggests that reduction in T allele frequency may be a protective factor for alcohol dependence. This observation is also consistent with our results showing that unaffected members of multiplex alcohol dependence families are less likely to carry a T allele and presumably are more likely to have greater D2 receptor availability than their affected relatives.
The greater significance of linkage findings for the unaffected pairs deserves comment. It has been our observation that in multiplex families where greater genetic susceptibility occurs, there is an accompanying increase in environmental pressures to begin drinking early and to drink to the point of intoxication in part because older siblings or parents drink excessively. This would mean that some alcohol dependent individuals might be sporadic cases from the standpoint of genetic variation. If so, both genetically based and sporadic affecteds can be expected in these families. In contrast, unaffecteds in a multiplex family environment are extreme for their family type in having escaped this environmental pressure to drink excessively. For this reason, there is less heterogeneity among unaffected individuals resulting in greater capacity to discern non-random assortment within unaffected pairs from these multiplex families when applying linkage analysis.
It may be worthwhile to speculate regarding the differing pattern of results seen in the within-family analysis and those seen in the case/control comparison. First, we recognize that our case/control comparison was probably underpowered for detection of a true difference if it exists at the population level. However, evidence for greater odds of carrying the T allele was seen in the regression models tested with almost a twofold greater likelihood in cases than in controls. The additive model showed approximately equal odds whether one or both were T alleles. This finding is in agreement with PET studies in normal volunteers showing T carriers having lower binding activity than CC volunteers and approximately equal pharmacological striatal DRD2 binding activity if the individual carried one or both T alleles [Hirvonen et al. 2004].
Variation in the C957T mutation appears to confer greater or lesser mRNA stability and translation [Duan et al., 2003]. This variation may be most salient in individuals with greater genetic loading for alcohol dependence. The behavioral importance of this genetic variation at the population level may be less because multiple environmental influences may have greater weight in individuals without a family history of alcohol dependence. Results from other clinical populations suggest the importance of DRD2 variants. Recently, Volkow et al. [2006] demonstrated that unaffected members of alcoholic families exhibit higher than normal D2 receptor availability than do unaffected individuals from control families without any alcohol dependent relatives. These results are consistent with the present findings in which the unaffected members of our multiplex alcohol dependent families were more often carriers of the C allele, the variant that is presumed to be associated with greater D2 receptor availability.
Lerman et al. [2006] evaluated response to nicotine replacement therapy (NRT), finding that individuals homozygous for the C957T T allele who presumably have reduced receptor availability exhibit a better response to NRT. In accordance with results presented by Duan et al. [2003] which had been confirmed by Hirvonen et al. [2004] and Lerman et al. [2006] interpreted their findings to indicate that those individuals homozygous for the T allele, and presumably with decreased mRNA stability and translation, exhibited a better response to NRT. Recently, Jacobsen et al. [2006] found that nicotine administration worsened verbal working memory (VBM) and processing efficiency in brain regions supporting VBM in carriers of the 957T allele.
Although some controversy surrounds the direction of the effects of allelic variation [Hirvonen et al., 2004, 2005], results of all studies taken together do suggest that allelic variation confers a functional effect. This effect may be most apparent in those who have already developed the phenotype of interest as in the case of smokers with known susceptibility to nicotine dependence. Similarly, multiplex alcohol dependence families may be most likely to reveal phenotypic/genotypic variation because of the selected nature of these families that results in greater susceptibility than seen in the general population.
Additionally, the role of genetic variation in the 5′ promoter region of the DRD2 gene was determined by genotyping members of these multiplex families for the A-241G and -141C Ins/Del polymorphisms. These genotypes were evaluated using PDT to determine if within-family association between alcohol dependence status and allele status might be found. A significant PDT result was not found for either of these mutations in the promoter region of the gene. Because the A-241G and -141C Ins/Del results were not statistically significant, case/control genotyping and analyses were not undertaken. The absence of a significant PDT result for these promoter region polymorphisms may have been the result of far fewer parents being heterozygous at the A-241G and -141C Ins/Del loci than was the case for the C957T locus. At any rate, the present results are not in agreement with previous reports that have used case/control comparisons. A positive result for promoter region variants has been found in a comparison of 130 Mexican-American alcoholic men and 251 nonalcoholic controls [Konishi et al., 2004]. Also, Lerman et al. [2006] found that nicotine dependent individuals homozygous for the -141C Ins/Del showed an improved treatment response to buproprion. A significant case/control association between heroin dependence and allelic variation in both -141C Ins/Del and A-241G has been reported in a Chinese sample [Xu et al., 2004] though the results were not seen in a German sample studied by this group of investigators.
Finally, it should be noted that the current analyses did not test for linkage of the controversial TaqI A locus due to the statistically significant LD between TaqI A and C957T. Previously, we reported an absence of a within-family association of the TaqI A polymorphism and alcohol dependence, though a significant case/control variation was seen [Neiswanger et al., 1995]. Of interest is the fact that Neville et al. [2004] have localized the TaqI A polymorphism to a region approximately 9.5 kb upstream from the DRD2 gene in the Ankyrin (ANKK1) gene. Two studies found evidence for linkage disequilibrium between the TaqI A and the C957T polymorphisms [Duan et al., 2003; Xu et al., 2004] as was found in the present study. This suggests those previously reported positive associations between various addictions (e.g., alcohol dependence and nicotine dependence) and the TaqI A polymorphism may have been the result of variations in the C957T polymorphism which is known to have functional effects on dopaminergic availability. The present demonstration of within-family association between variation in this mutation and likelihood of developing alcohol dependence in multiplex alcohol dependence families suggests opportunities for pharmacological intervention for alcohol dependence.
Acknowledgments
This work was supported by National Institute on Alcohol Abuse and Alcoholism awards AA 005909, AA 008082, AA 05168 to SYH and training grant support 1T32 MH20053 and Fogarty US-India 1D43 TW 006180 to DEW.
References
- Abecasis GR, Cookson WO. GOLD-graphical overview of linkage disequilibrium. Bioinformatics. 2000;16:182–183. doi: 10.1093/bioinformatics/16.2.182. [DOI] [PubMed] [Google Scholar]
- Amadeo S, Fourcade ML, Abbar M, Leroux MG, Castelnau D, Vanisse JL, Mallet J. Association between D2 receptor gene polymorphism and alcoholism. Psychiatr Genet. 1993;3:130–135. [Google Scholar]
- American Psychiatric Association. Diagnostic and statistical manual of mental disorders. 3. Washington, DC: American Psychiatric Association; 1982. [Google Scholar]
- American Psychiatric Association. Diagnostic and statistical manual of mental disorders. 3. Washington, DC: American Psychiatric Association; 1987. revised. [Google Scholar]
- Anderson VE, Hauser WA, Rich SS. Genetic heterogeneity in the epilepsies. Adv Neurol. 1986;44:59–75. [PubMed] [Google Scholar]
- Arinami T, Itokawa M, Komiyama T, Mitsushio H, Mori H, Mifune H, Hamaguchi H, Toru M. Association between severity of alcoholism and the A1 allele of the dopamine D2 receptor gene TaqI A RFLP in Japanese. Biol Psychiatry. 1993;33:108–114. doi: 10.1016/0006-3223(93)90309-2. [DOI] [PubMed] [Google Scholar]
- Arinami T, Gao M, Hamaguchi H, Toru M. A functional polymorphism in the promoter region of the dopamine D2 receptor gene is associated with schizophrenia. Hum Mol Genet. 1997;6:577–582. doi: 10.1093/hmg/6.4.577. [DOI] [PubMed] [Google Scholar]
- Blum K, Noble EP, Sheridan PJ, Montgomery A, Ritchie T, Jagadeeswaran P, Nogami H, Briggs AH, Cohn JB. Allelic association of human dopamine D2 receptor gene in alcoholism. JAMA. 1990;263:2055–2060. [PubMed] [Google Scholar]
- Bolos AM, Dean M, Lucas-Derse S, Ramsburg M, Brown GL, Goldman D. Population and pedigree studies reveal a lack of association between the dopamine D2 receptor gene and alcoholism. JAMA. 1990;264:3156–3160. [PubMed] [Google Scholar]
- Cook BL, Wang ZW, Crowe RR, Hauser R, Freimer M. Alcoholism and the D2 receptor gene. Alcohol Clin Exp Res. 1992;16:806–809. doi: 10.1111/j.1530-0277.1992.tb00683.x. [DOI] [PubMed] [Google Scholar]
- Duan J, Wainwright MS, Comeron JM, Saitou N, Sanders AR, Gelernter J, Gejman PV. Synonymous mutations in the human dopamine receptor D2 (DRD2) affect mRNA stability and synthesis of the receptor. Hum Mol Genet. 2003;12:205–216. doi: 10.1093/hmg/ddg055. [DOI] [PubMed] [Google Scholar]
- Feighner JP, Robins E, Guze SB, Woodruff RA, Jr, Winokur G, Munoz R. Diagnostic criteria for use in psychiatric research. Arch Gen Psychiatry. 1972;26:57–63. doi: 10.1001/archpsyc.1972.01750190059011. [DOI] [PubMed] [Google Scholar]
- Fosella J, Green AE, Fan J. Evaluation of a structural polymorphism in the ankyrin repeat and kinase domain containing 1 (ANKK1) gene and the activation of executive attention networks. Cogn Affect Behav Neurosci. 2006;6:71–78. doi: 10.3758/cabn.6.1.71. [DOI] [PubMed] [Google Scholar]
- Gejman PV, Ram A, Gelernter J, Friedman E, Cao Q, Pickar D, et al. No structural mutation in the dopamine D2 receptor gene in alcoholism or schizophrenia: Analysis using denaturing gradient gel electrophoresis. JAMA. 1994;271:204–208. [PubMed] [Google Scholar]
- Gelernter J, O’Malley S, Risch N, Kranzler HR, Krystal J, Merikangas K, et al. No association between an allele at the D2 dopamine receptor gene (DRD2) and alcoholism. JAMA. 1991;266:1801–1807. [PubMed] [Google Scholar]
- Gelernter J, Yu Y, Weiss R, Brady K, Panhuysen C, Yang B-Z, Kranzler HR, Farrer L. Haplotype spanning TTC12 and ANKK1, flanked by the DRD2 and NCAMI loci, is strongly associated to nicotine dependence in two distinct American populations. Hum Mol Genet. 2006;15:3498–3507. doi: 10.1093/hmg/ddl426. [DOI] [PubMed] [Google Scholar]
- Goodwin DW, Schulsinger F, Hermansen L, Guze SB, Winokur G. Alcohol problems in adoptees raised apart from alcoholic biological parents. Arch Gen Psychiatry. 1973;28:238–243. doi: 10.1001/archpsyc.1973.01750320068011. [DOI] [PubMed] [Google Scholar]
- Grandy DK, Litt M, Allen L, Bunzow JR, Marchionni M, Makam H, et al. The human dopamine D2 receptor gene is located on chromosome 11 at q22-q23 and identifies a Taq1 RFLP. Am J Hum Genet. 1989;45:778–785. [PMC free article] [PubMed] [Google Scholar]
- Heath AC, Madden PA, Bucholz KK, Dinwiddie SH, Slutske WS, Bierut LJ, Rohrbaugh JW, Statham DJ, Dunne MP, Whitfield JB, Martin NG. Genetic differences in alcohol sensitivity and the inheritance of alcoholism risk. Psychol Med. 1999;29:1069–1081. doi: 10.1017/s0033291799008909. [DOI] [PubMed] [Google Scholar]
- Helzer JE, Robins LN, McEvoy LT, Spitznagel EL, Stoltzman RK, Farmer A, Brockington IF. A comparison of clinical and diagnostic interview schedule diagnoses. Physician reexamination of lay-interviewed cases in the general population. Arch Gen Psychiatry. 1985;42(7):657–666. doi: 10.1001/archpsyc.1985.01790300019003. [DOI] [PubMed] [Google Scholar]
- Hietala J, West C, Syvalahti E, Nagren K, Lehikoinen P, Sonninen P, Ruotsalainen U. Striatal D2 dopamine receptor binding characteristics in vivo in patients with alcohol dependence. Psychopharmacology. 1994;116:285–290. doi: 10.1007/BF02245330. [DOI] [PubMed] [Google Scholar]
- Hill SY, Zezza N, Wipprecht G, Locke J, Neiswanger K. Personality traits and dopamine receptors (D2 and D4): Linkage studies in families of alcoholics. Am J Med Genet Neuropsychiatr Genet. 1999;88:634–641. doi: 10.1002/(sici)1096-8628(19991215)88:6<634::aid-ajmg11>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- Hill SY, Shen S, Zezza N, Hoffman EK, Perlin M, Allan W. A genome wide search for alcoholism susceptibility genes. Am J Med Genet Part B. 2004;128B:102–113. doi: 10.1002/ajmg.b.30013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirvonen M, Laakso A, Nagren K, Rinne JO, Pohjalinen T, Hietala J. C957T polymorphism of the dopamine D2 receptor (DRD2) gene affects striatal DRD2 availability in vivo. Mol Psychiatry. 2004;9:1060–1061. doi: 10.1038/sj.mp.4001561. [DOI] [PubMed] [Google Scholar]
- Hirvonen M, Laakso A, Nagren K, Rinne JO, Pohjalinen T, Hietala J. C957T polymorphism of the dopamine D2 receptor (DRD2) gene affects striatal DRD2 availability in vivo. Mol Psychiatry. 2005;10:889. doi: 10.1038/sj.mp.4001561. [DOI] [PubMed] [Google Scholar]
- Jacobsen LK, Pugh KR, Menel WE, Gelernter J. C957T polymorphisms of the dopamine D2 receptor gene modulates the effect of nictoine on working memory performance and cortical processing efficiency. Psychopharmacology. 2006;188:530–540. doi: 10.1007/s00213-006-0469-1. [DOI] [PubMed] [Google Scholar]
- Jönsson EG, Nöthen MM, Grünhage F, Farde L, Nakashima Y, Propping P, Sedvall GC. Polymorphisms in the dopamine D2 receptor gene and their relationships to striatal dopamine receptor density of healthy volunteers. Mol Psychiatry. 1999;4:290–296. doi: 10.1038/sj.mp.4000532. [DOI] [PubMed] [Google Scholar]
- Kendler KS, Heath AC, Neale MC, Kessler RC, Eaves LJ. A population-based twin study of alcoholism in women. JAMA. 1992;268:1877–1882. [PubMed] [Google Scholar]
- Kidd KK, Morar B, Castiglione CM, Zhao H, Pakstis AJ, Speed WC, Bonne-Tamir B, Lu RB, Goldman D, Lee C, Nam YS, Grandy DK, Jenkins T, Kidd JR. A global survey of haplotype frequencies and linkage disequilibrium at the DRD2 locus. Human Genet. 1998;103:211–227. doi: 10.1007/s004390050809. [DOI] [PubMed] [Google Scholar]
- Konishi T, Calvillo M, Leng A-S, Lin K-M, Wan Y-JY. Polymorphisms of the dopamine D2 receptor, serotonin transporter, and GABAA receptor B3 subunit genes and alcoholism in Mexican-Americans. Alcohol. 2004;32:45–52. doi: 10.1016/j.alcohol.2003.11.002. [DOI] [PubMed] [Google Scholar]
- Laine TPJ, Ahonen A, Rasanen P, Pohjalainen T, Tiihonen J, Hietala J. The A1 allele of the D2 dopamine receptor gene is associated with high dopamine transporter density in detoxified alcoholics. Alcohol Alcohol. 2001;36:262–265. doi: 10.1093/alcalc/36.3.262. [DOI] [PubMed] [Google Scholar]
- Lange K, Cantor R, Horvath S, Perola M, Sabatti C, Sinsheimer J, Sobel E. Mendel version 4.0: A complete package for the exact genetic analysis of discrete traits in pedigree and population data sets. Am J Hum Genet. 2001;69(Suppl):A1886. [Google Scholar]
- Laruelle M, Gelernter J, Innis RB. D2 receptors binding potential is not affected by TaqI polymorphism at the D2 receptor gene. Mol Psychiatry. 1998;3:261–265. doi: 10.1038/sj.mp.4000343. [DOI] [PubMed] [Google Scholar]
- Lawford BR, Young RM, Rowell JA, Gibson JN, Feeney GFX, Richie TL, Sydulko K, Noble EP. Association of the D2 dopamine receptor A1 allele with alcoholism: Medical severity of alcoholism and type of controls. Biol Psychiatry. 1997;41:386–393. doi: 10.1016/S0006-3223(96)00478-7. [DOI] [PubMed] [Google Scholar]
- Lawford BR, Young RM, Swagell CD, Barnes M, Burton SC, Ward W, et al. The C/C genotype of the C957T polymorphism of the dopamine D2 receptor is associated with schizophrenia. Schizophr Res. 2005;73:31–37. doi: 10.1016/j.schres.2004.08.020. [DOI] [PubMed] [Google Scholar]
- Lerman C, Jepson C, Wileyto EP, Epstein L, Rukstalis M, Patterson F, Kaufmann V, Restine S, Hawk L, Niaura R, Berfettini W. Role of functional genetic varation in the dopamine D2 receptor (DRD2) in response to buproprion and nicotine replace therapy for tobacco dependence: Results of two randomized clinical trials. Neuropsychopharmacology. 2006;31:231–242. doi: 10.1038/sj.npp.1300861. [DOI] [PubMed] [Google Scholar]
- Martin ER, Monks SA, Warren LL, Kaplan NL. A test for linkage and association in general pedigrees. Am J Hum Genet. 2000;67:146–154. doi: 10.1086/302957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin ER, Bass MP, Kaplan NL. Correcting for potential bias in the pedigree disequilibrium test. Am J Hum Genet. 2001;68:259–266. doi: 10.1086/319525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGue M, Pickens RW, Svikis DS. Sex and age effects on the inheritance of alcohol problems: A twin study. J Abnormal Psych. 1992;101:3–17. doi: 10.1037//0021-843x.101.1.3. [DOI] [PubMed] [Google Scholar]
- Morton NE, Mi MP. Multiplex families with two or more probands. Am J Hum Genet. 1968;20:361–367. [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay N, Almasy L, Schroeder M, Mulvihill WP, Weeks DE. Mega2 data-handling for facilitating genetic linkage and association studies. Bioinformatics. 2005;21:2556–2557. doi: 10.1093/bioinformatics/bti364. [DOI] [PubMed] [Google Scholar]
- Neiswanger K, Hill SY, Kaplan BR. Association and linkage studies of the TAQI A1 allele at the dopamine D2 receptor gene in samples of female and male alcoholics. Am J Med Genet (Neuropsychiatr Genet) 1995;60:267–271. doi: 10.1002/ajmg.1320600402. [DOI] [PubMed] [Google Scholar]
- Neville MJ, Johnstone EC, Walton RT. Identification and characterization of ANK K1: A novel kinase gene closely linked to DRD2 on chromosome band 11q23.1. Hum Mutat. 2004;23:540–545. doi: 10.1002/humu.20039. [DOI] [PubMed] [Google Scholar]
- Noble E. D2 dopamine receptor gene in psychiatric and neurologic disorders and its phenotypes. Am J Med Genet Part B. 2003;116B:103–125. doi: 10.1002/ajmg.b.10005. [DOI] [PubMed] [Google Scholar]
- Noble EP, Blum K, Ritchie T, Montgomery A, Sheridan PJ. Allelic association of the D2 dopamine receptor gene with receptor binding characteristics in alcoholism. Arch Gen Psychiatry. 1991;48:648–654. doi: 10.1001/archpsyc.1991.01810310066012. [DOI] [PubMed] [Google Scholar]
- Noble EP, Syndulko K, Fitch RJ, Ritchie T, Bohlman MC, Guth P, et al. D2 dopamine receptor TaqI A alleles in medically ill alcoholic and nonalcoholic patients. Alcohol Alcohol. 1994;29:729–744. [PubMed] [Google Scholar]
- O’Connell JR, Weeks DE. PedCheck: A program for identifying genotype incompatibilities in linkage analysis. Am J Hum Genet. 1998;63:259–266. doi: 10.1086/301904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perlin MW, Lankier G, Ng SK. Toward fully automated genotyping: Genotyping micro satellite markers by disconsolation. Am J Hum Genet. 1995;57:1199–1210. [PMC free article] [PubMed] [Google Scholar]
- Pickens RW, Svikis DS, McGue M, Liken DT, Hesston LL, Clayton PJ. Heterogeneity in the inheritance of alcoholism: A study of male and female twins. Arch Gen Psychiatry. 1991;48:19–28. doi: 10.1001/archpsyc.1991.01810250021002. [DOI] [PubMed] [Google Scholar]
- Pohjalainen T, Rinne JO, Nagger K, Lehikoinen P, Attila K, Syvalahti EKG, Hietala J. The A1 allele of the human D2 dopamine receptor gene predicts low D2 receptor availability in healthy volunteers. Mol Psychiatry. 1998;3:256–260. doi: 10.1038/sj.mp.4000350. [DOI] [PubMed] [Google Scholar]
- Prescott CA. The genetic epidemiologist of alcoholism: Sex differences and future directions. In: Agawam DP, Seitz HK, editors. Alcohol in health and disease. Marcel Decker, Inc; New York and Basel: 2001. pp. 125–149. [Google Scholar]
- SAGE. Statistical analysis for genetic epidemiologist. Statistical Solutions Ltd; Cork, Ireland: 2004. [Google Scholar]
- Seeman P. Dopamine receptor sequences. Therapeutic levels of narcoleptics occupy D2 receptors, cocaine occupies D4. Neuropharmacology. 1992;7:261–284. [PubMed] [Google Scholar]
- Smith SS, O’Hara BF, Persico AM, Gorelick DA, Newlin DB, Vlahov D, Solomon L, Pickens R, Uhl GR. Genetic vulnerability to drug abuse: The D2 dopamine receptor TaqI B1 restriction fragment length polymorphism appears more frequently in polysubtance abusers. Arch Gen Psychiatry. 1992;49:723–727. doi: 10.1001/archpsyc.1992.01820090051009. [DOI] [PubMed] [Google Scholar]
- Spielman RS, McGinnis RE, Ewens WJ. Transmission test for linkage disequilibrium: The insulin gene region and Insulin-dependent Diabetes Mellitus (IDDM) Am J Hum Genet. 1993;52:506–516. [PMC free article] [PubMed] [Google Scholar]
- Suarez BK, Parsian A, Hampe CL, Todd RD, Reich T, Cloninger CL. Linkage disequilibria at the D2 dopamine receptor locus (DRD2) in alcoholics and controls. Genomics. 1994;19:12–20. doi: 10.1006/geno.1994.1005. [DOI] [PubMed] [Google Scholar]
- The R Foundation for Statistical Computing Version 2.1.1, 2005.
- Thompson J, Thomas N, Singleton A, Piggott M, Lloyd S, Perry EK, Morris CM, Perry RH, Ferrier IN, Court JA. D2 dopamine receptor gene (DRD2) TaqI A polymorphism: Reduced dopamine D2 receptor binding in the human striatum associated with the A1 allele. Pharmacogenetics. 1997;7:479–484. doi: 10.1097/00008571-199712000-00006. [DOI] [PubMed] [Google Scholar]
- Turner E, Ewing J, Shilling P, Smith TL, Irwin M, Schuckit M, Kelsoe JR. Lack of association between an RFLP near the D2 dopamine receptor gene and severe alcoholism. Biol Psychiatry. 1992;31:285–290. doi: 10.1016/0006-3223(92)90052-2. [DOI] [PubMed] [Google Scholar]
- Volkow ND, Fowler JS, Wang GJ, Hitzemann R, Logan J, Schlyer DJ, Dewey SL, Wolf AP. Decreased dopamine D2 receptor availability is associated with reduced frontal metabolism in cocaine abusers. Synapse. 1993;14:169–177. doi: 10.1002/syn.890140210. [DOI] [PubMed] [Google Scholar]
- Volkow ND, Fowler JS, Wang G-J. Role of dopamine in drug reinforcement and addiction in humans: Results from imaging studies. Behav Pharmacol. 2002a;13:355–366. doi: 10.1097/00008877-200209000-00008. [DOI] [PubMed] [Google Scholar]
- Volkow ND, Wang GJ, Maynard L, Fowler JS, Jayne B, Telang F, Logan J, Ding YS, Gatley SJ, Hitzemann R, Wong C, Pappas N. Effects of alcohol detoxification on dopamine D2 receptors in alcoholics: A preliminary study. Psychiatry Res. 2002b;116:163–172. doi: 10.1016/s0925-4927(02)00087-2. [DOI] [PubMed] [Google Scholar]
- Volkow ND, Fowler JS, Wang G-J. The addicted human brain: Insights from imaging studies. J Clin Invest. 2003;111:1444–1451. doi: 10.1172/JCI18533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volkow ND, Wang G-J, Begleiter H, Porjesz B, Fowler J, Telang F, Wong C, Ma Y, Logan J, Goldstein R, Alexoff D, Thanos PK. High levels of dopamine D2 receptors in unaffected members of alcoholic families. Arch Gen Psychiatry. 2006;63:999–1008. doi: 10.1001/archpsyc.63.9.999. [DOI] [PubMed] [Google Scholar]
- Wang GJ, Volkow ND, Fowler JS, Logan J, Abumrad NN, Hitzemann RJ, Pappas NS, Pascani K. Dopamine D2 receptor availability in opiate-dependent subjects before and after naloxone-precipitated withdrawal. Neuropsychopharmacology. 1997;16:174–182. doi: 10.1016/S0893-133X(96)00184-4. [DOI] [PubMed] [Google Scholar]
- Xu K, Lichtermann D, Lipsky RH, Franke P, Liu X, Hu Y, Cao L, Schwab SG, Wildenauer DB, Bau CHD, Ferro E, Astor W, Finch T, Terry J, Taubman J, Maier W, Goldman D. Association of specific haplotypes of D2 dopamine receptor gene with vulnerability to heroin dependence in 2 distinct populations. Arch Gen Psychiatry. 2004;61:597–606. doi: 10.1001/archpsyc.61.6.597. [DOI] [PubMed] [Google Scholar]