Abstract
Ribonuclease III of Escherichia coli is prominently involved in the endoribonucleolytic processing of cell and viral-encoded RNAs. Towards the goal of defining the RNA sequence and structural elements that establish specific catalytic cleavage of RNase III processing signals, this report demonstrates that a 60 nucleotide RNA (R1.1 RNA) containing the bacteriophage T7 R1.1 RNase III processing signal, can be generated by in vitro enzymatic transcription of a synthetic deoxyoligonucleotide and accurately cleaved in vitro by RNase III. Several R1.1 RNA sequence variants were prepared to contain point mutations in the internal loop which, on the basis of a hypothetical 'dsRNA mimicry' structural model of RNase III processing signals, would be predicted to inhibit cleavage by disrupting essential tertiary RNA-RNA interactions. These R1.1 sequence variants are accurately and efficiently cleaved in vitro by RNase III, indicating that the dsRNA mimicry structure, if it does exist, is not important for substrate reactivity. Also, we tested the functional importance of the strongly conserved CUU/GAA base-pair sequence by constructing R1.1 sequence variants containing base-pair changes within this element. These R1.1 variants are accurately cleaved at rates comparable to wild-type R1.1 RNA, indicating the nonessentiality of this conserved sequence element in establishing in vitro processing reactivity and selectivity.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Altman S. Ribonuclease P. Postscript. J Biol Chem. 1990 Nov 25;265(33):20053–20056. [PubMed] [Google Scholar]
- Altuvia S., Locker-Giladi H., Koby S., Ben-Nun O., Oppenheim A. B. RNase III stimulates the translation of the cIII gene of bacteriophage lambda. Proc Natl Acad Sci U S A. 1987 Sep;84(18):6511–6515. doi: 10.1073/pnas.84.18.6511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auron P. E., Weber L. D., Rich A. Comparison of transfer ribonucleic acid structures using cobra venom and S1 endonucleases. Biochemistry. 1982 Sep 14;21(19):4700–4706. doi: 10.1021/bi00262a028. [DOI] [PubMed] [Google Scholar]
- Bardwell J. C., Régnier P., Chen S. M., Nakamura Y., Grunberg-Manago M., Court D. L. Autoregulation of RNase III operon by mRNA processing. EMBO J. 1989 Nov;8(11):3401–3407. doi: 10.1002/j.1460-2075.1989.tb08504.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bram R. J., Young R. A., Steitz J. A. The ribonuclease III site flanking 23S sequences in the 30S ribosomal precursor RNA of E. coli. Cell. 1980 Feb;19(2):393–401. doi: 10.1016/0092-8674(80)90513-9. [DOI] [PubMed] [Google Scholar]
- Chen S. M., Takiff H. E., Barber A. M., Dubois G. C., Bardwell J. C., Court D. L. Expression and characterization of RNase III and Era proteins. Products of the rnc operon of Escherichia coli. J Biol Chem. 1990 Feb 15;265(5):2888–2895. [PubMed] [Google Scholar]
- Daniels D. L., Subbarao M. N., Blattner F. R., Lozeron H. A. Q-mediated late gene transcription of bacteriophage lambda: RNA start point and RNase III processing sites in vivo. Virology. 1988 Dec;167(2):568–577. [PubMed] [Google Scholar]
- Donis-Keller H. Phy M: an RNase activity specific for U and A residues useful in RNA sequence analysis. Nucleic Acids Res. 1980 Jul 25;8(14):3133–3142. doi: 10.1093/nar/8.14.3133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Draper D. E. How do proteins recognize specific RNA sites? New clues from autogenously regulated ribosomal proteins. Trends Biochem Sci. 1989 Aug;14(8):335–338. doi: 10.1016/0968-0004(89)90167-9. [DOI] [PubMed] [Google Scholar]
- Dunn J. J. RNase III cleavage of single-stranded RNA. Effect of ionic strength on the fideltiy of cleavage. J Biol Chem. 1976 Jun 25;251(12):3807–3814. [PubMed] [Google Scholar]
- Dunn J. J., Studier F. W. Complete nucleotide sequence of bacteriophage T7 DNA and the locations of T7 genetic elements. J Mol Biol. 1983 Jun 5;166(4):477–535. doi: 10.1016/s0022-2836(83)80282-4. [DOI] [PubMed] [Google Scholar]
- Dunn J. J., Studier F. W. Effect of RNAase III, cleavage on translation of bacteriophage T7 messenger RNAs. J Mol Biol. 1975 Dec 15;99(3):487–499. doi: 10.1016/s0022-2836(75)80140-9. [DOI] [PubMed] [Google Scholar]
- Forster A. C., Symons R. H. Self-cleavage of virusoid RNA is performed by the proposed 55-nucleotide active site. Cell. 1987 Jul 3;50(1):9–16. doi: 10.1016/0092-8674(87)90657-x. [DOI] [PubMed] [Google Scholar]
- Francklyn C., Schimmel P. Aminoacylation of RNA minihelices with alanine. Nature. 1989 Feb 2;337(6206):478–481. doi: 10.1038/337478a0. [DOI] [PubMed] [Google Scholar]
- Freier S. M., Kierzek R., Jaeger J. A., Sugimoto N., Caruthers M. H., Neilson T., Turner D. H. Improved free-energy parameters for predictions of RNA duplex stability. Proc Natl Acad Sci U S A. 1986 Dec;83(24):9373–9377. doi: 10.1073/pnas.83.24.9373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gegenheimer P., Apirion D. Precursors to 16S and 23S ribosomal RNA from a ribonuclear III-strain of Escherichia coli contain intact RNase III processing sites. Nucleic Acids Res. 1980 Apr 25;8(8):1873–1891. doi: 10.1093/nar/8.8.1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gegenheimer P., Apirion D. Processing of procaryotic ribonucleic acid. Microbiol Rev. 1981 Dec;45(4):502–541. doi: 10.1128/mr.45.4.502-541.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottesman M., Oppenheim A., Court D. Retroregulation: control of gene expression from sites distal to the gene. Cell. 1982 Jul;29(3):727–728. doi: 10.1016/0092-8674(82)90434-2. [DOI] [PubMed] [Google Scholar]
- Grodberg J., Dunn J. J. ompT encodes the Escherichia coli outer membrane protease that cleaves T7 RNA polymerase during purification. J Bacteriol. 1988 Mar;170(3):1245–1253. doi: 10.1128/jb.170.3.1245-1253.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groebe D. R., Uhlenbeck O. C. Thermal stability of RNA hairpins containing a four-membered loop and a bulge nucleotide. Biochemistry. 1989 Jan 24;28(2):742–747. doi: 10.1021/bi00428a049. [DOI] [PubMed] [Google Scholar]
- Guarneros G., Montañez C., Hernandez T., Court D. Posttranscriptional control of bacteriophage lambda gene expression from a site distal to the gene. Proc Natl Acad Sci U S A. 1982 Jan;79(2):238–242. doi: 10.1073/pnas.79.2.238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heitman J., Model P. Mutants of the EcoRI endonuclease with promiscuous substrate specificity implicate residues involved in substrate recognition. EMBO J. 1990 Oct;9(10):3369–3378. doi: 10.1002/j.1460-2075.1990.tb07538.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heus H. A., Uhlenbeck O. C., Pardi A. Sequence-dependent structural variations of hammerhead RNA enzymes. Nucleic Acids Res. 1990 Mar 11;18(5):1103–1108. doi: 10.1093/nar/18.5.1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones M. H., Guthrie C. Unexpected flexibility in an evolutionarily conserved protein-RNA interaction: genetic analysis of the Sm binding site. EMBO J. 1990 Aug;9(8):2555–2561. doi: 10.1002/j.1460-2075.1990.tb07436.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konarska M. M., Sharp P. A. Replication of RNA by the DNA-dependent RNA polymerase of phage T7. Cell. 1989 May 5;57(3):423–431. doi: 10.1016/0092-8674(89)90917-3. [DOI] [PubMed] [Google Scholar]
- Krinke L., Wulff D. L. The cleavage specificity of RNase III. Nucleic Acids Res. 1990 Aug 25;18(16):4809–4815. doi: 10.1093/nar/18.16.4809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesser D. R., Kurpiewski M. R., Jen-Jacobson L. The energetic basis of specificity in the Eco RI endonuclease--DNA interaction. Science. 1990 Nov 9;250(4982):776–786. doi: 10.1126/science.2237428. [DOI] [PubMed] [Google Scholar]
- March P. E., Gonzalez M. A. Characterization of the biochemical properties of recombinant ribonuclease III. Nucleic Acids Res. 1990 Jun 11;18(11):3293–3298. doi: 10.1093/nar/18.11.3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milligan J. F., Groebe D. R., Witherell G. W., Uhlenbeck O. C. Oligoribonucleotide synthesis using T7 RNA polymerase and synthetic DNA templates. Nucleic Acids Res. 1987 Nov 11;15(21):8783–8798. doi: 10.1093/nar/15.21.8783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholson A. W., Niebling K. R., McOsker P. L., Robertson H. D. Accurate in vitro cleavage by RNase III of phosphorothioate-substituted RNA processing signals in bacteriophage T7 early mRNA. Nucleic Acids Res. 1988 Feb 25;16(4):1577–1591. doi: 10.1093/nar/16.4.1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oakley J. L., Coleman J. E. Structure of a promoter for T7 RNA polymerase. Proc Natl Acad Sci U S A. 1977 Oct;74(10):4266–4270. doi: 10.1073/pnas.74.10.4266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panayotatos N., Truong K. Cleavage within an RNase III site can control mRNA stability and protein synthesis in vivo. Nucleic Acids Res. 1985 Apr 11;13(7):2227–2240. doi: 10.1093/nar/13.7.2227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portier C., Dondon L., Grunberg-Manago M., Régnier P. The first step in the functional inactivation of the Escherichia coli polynucleotide phosphorylase messenger is a ribonuclease III processing at the 5' end. EMBO J. 1987 Jul;6(7):2165–2170. doi: 10.1002/j.1460-2075.1987.tb02484.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pragai B., Apirion D. Processing of bacteriophage T4 tRNAs. The role of RNAase III. J Mol Biol. 1981 Dec 15;153(3):619–630. doi: 10.1016/0022-2836(81)90410-1. [DOI] [PubMed] [Google Scholar]
- Robertson H. D., Dickson E., Dunn J. J. A nucleotide sequence from a ribonuclease III processing site in bacteriophage T7 RNA. Proc Natl Acad Sci U S A. 1977 Mar;74(3):822–826. doi: 10.1073/pnas.74.3.822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson H. D. Escherichia coli ribonuclease III cleavage sites. Cell. 1982 Oct;30(3):669–672. doi: 10.1016/0092-8674(82)90270-7. [DOI] [PubMed] [Google Scholar]
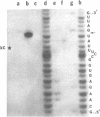
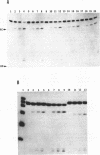
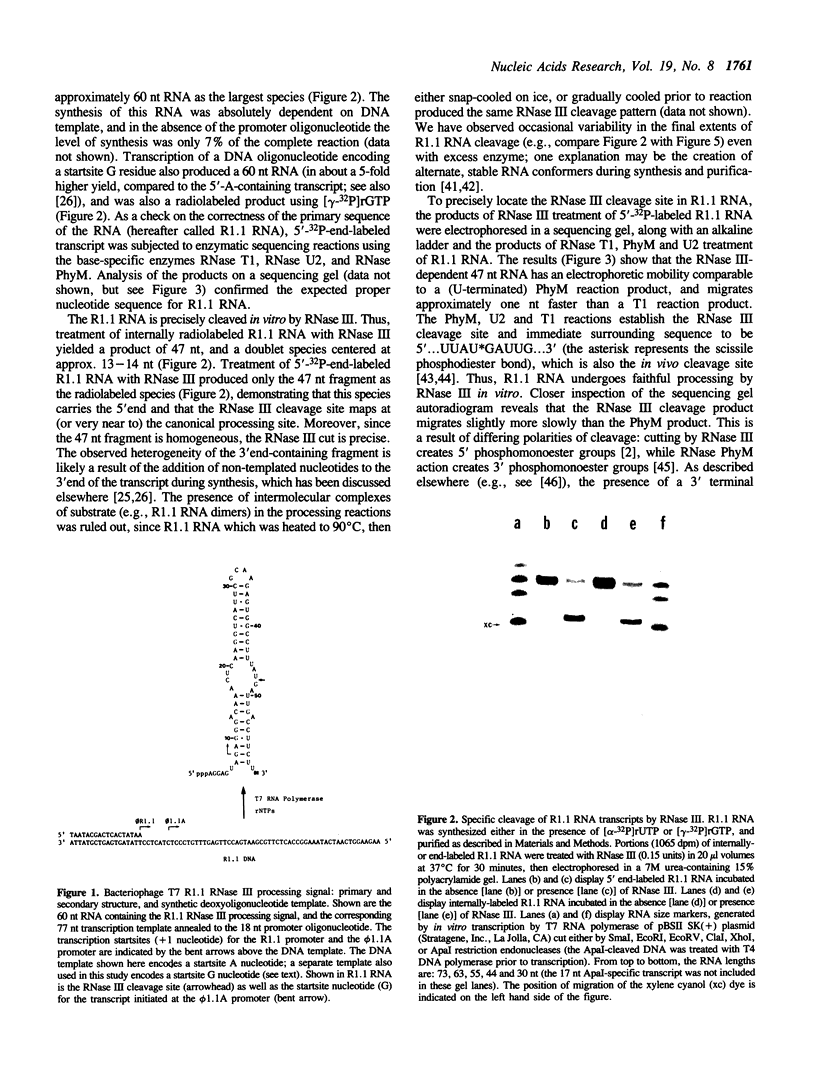
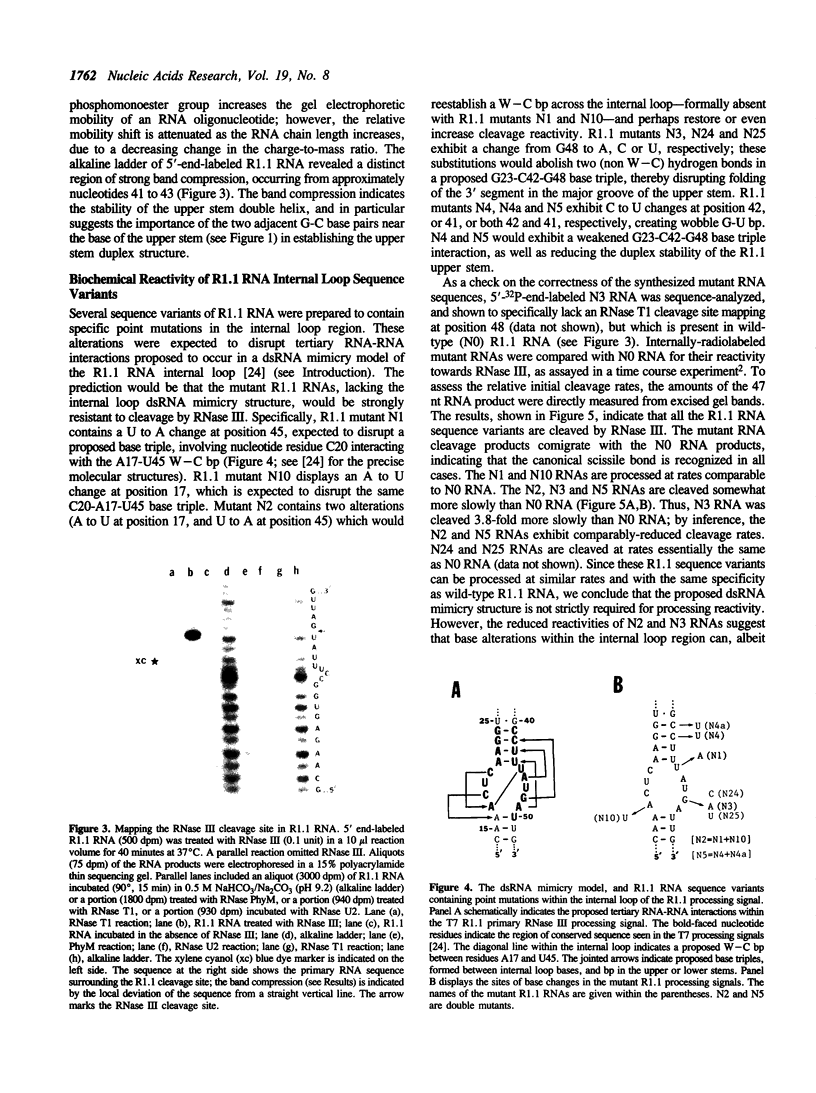
- Robertson H. D., Webster R. E., Zinder N. D. Purification and properties of ribonuclease III from Escherichia coli. J Biol Chem. 1968 Jan 10;243(1):82–91. [PubMed] [Google Scholar]
- Rosenberg M., Kramer R. A., Steitz J. A. The specificity of RNase III cleavage of bacteriophage T7 early messenger RNAs. Brookhaven Symp Biol. 1975 Jul;(26):277–285. [PubMed] [Google Scholar]
- Régnier P., Grunberg-Manago M. Cleavage by RNase III in the transcripts of the met Y-nus-A-infB operon of Escherichia coli releases the tRNA and initiates the decay of the downstream mRNA. J Mol Biol. 1989 Nov 20;210(2):293–302. doi: 10.1016/0022-2836(89)90331-8. [DOI] [PubMed] [Google Scholar]
- Saito H., Richardson C. C. Processing of mRNA by ribonuclease III regulates expression of gene 1.2 of bacteriophage T7. Cell. 1981 Dec;27(3 Pt 2):533–542. doi: 10.1016/0092-8674(81)90395-0. [DOI] [PubMed] [Google Scholar]
- Sampson J. R., Uhlenbeck O. C. Biochemical and physical characterization of an unmodified yeast phenylalanine transfer RNA transcribed in vitro. Proc Natl Acad Sci U S A. 1988 Feb;85(4):1033–1037. doi: 10.1073/pnas.85.4.1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmeissner U., McKenney K., Rosenberg M., Court D. Removal of a terminator structure by RNA processing regulates int gene expression. J Mol Biol. 1984 Jun 15;176(1):39–53. doi: 10.1016/0022-2836(84)90381-4. [DOI] [PubMed] [Google Scholar]
- Steege D. A., Cone K. C., Queen C., Rosenberg M. Bacteriophage lambda N gene leader RNA. RNA processing and translational initiation signals. J Biol Chem. 1987 Dec 25;262(36):17651–17658. [PubMed] [Google Scholar]
- Studier F. W., Rosenberg A. H., Dunn J. J., Dubendorff J. W. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 1990;185:60–89. doi: 10.1016/0076-6879(90)85008-c. [DOI] [PubMed] [Google Scholar]
- Tang R. S., Draper D. E. Bulge loops used to measure the helical twist of RNA in solution. Biochemistry. 1990 Jun 5;29(22):5232–5237. doi: 10.1021/bi00474a003. [DOI] [PubMed] [Google Scholar]
- Uhlenbeck O. C. A small catalytic oligoribonucleotide. Nature. 1987 Aug 13;328(6131):596–600. doi: 10.1038/328596a0. [DOI] [PubMed] [Google Scholar]
- Walstrum S. A., Uhlenbeck O. C. The self-splicing RNA of Tetrahymena is trapped in a less active conformation by gel purification. Biochemistry. 1990 Nov 20;29(46):10573–10576. doi: 10.1021/bi00498a022. [DOI] [PubMed] [Google Scholar]
- Witherell G. W., Uhlenbeck O. C. Specific RNA binding by Q beta coat protein. Biochemistry. 1989 Jan 10;28(1):71–76. doi: 10.1021/bi00427a011. [DOI] [PubMed] [Google Scholar]