Abstract
Activation of the plasma membrane H+-ATPase of the yeast Saccharomyces cerevisiae by glucose is a complex process that has not yet been completely elucidated. This study aimed to shed light on the role of lipids and the lateral mobility of the enzyme complex during its activation by glucose. The significance of H+-ATPase oligomerization for the activation of H+-ATPase by glucose was shown using the strains lcb1-100 and erg6, with the disturbed synthesis of sphyngolipid and ergosterol, respectively. Experiments with GFP-fused H+-ATPase showed a decrease in fluorescence anisotropy during the course of glucose activation, suggesting structural reorganization of the molecular domains. An immunogold assay showed that the incubation with glucose results in the spatial redistribution of ATPase complexes in the plasma membrane. The data suggest that (1) to be activated by glucose, H+-ATPase is supposed to be in an oligomeric state, and (2) glucose activation is accompanied by the spatial movements of H+-ATPase clusters in the PM.
Introduction
H+-ATPase, hereafter, Pma1, of the yeast plasma membrane (PM), one of the major structural proteins of the PM, belongs to a P-type ATPase [1], [2] and is encoded by the PMA1 gene [3]. The enzyme hydrolyzes ATP and forms an electrochemical proton gradient on the PM and drives the transport of basic nutrients across the PM [4]. As has been shown previously, Pma1 consumes up to 20% of cellular ATP and, not surprisingly, its activity is under strict control [5]. The regulation of Pma1 activity was shown for the first time by Serrano, who discovered that incubation with glucose resulted in a reversible many-fold enhancement of the enzyme's activity, a decrease in Km, and an increase in Vmax [6]. The sugars utilized via the glycolytic pathway (fructose and mannose) were shown to lead to the enhancement of the enzyme's activity [6]. Sugars (D-xylose and galactose) metabolized through other pathways, as well as nonmetabolized glucose analogs (3-O-methylglucose and deoxyglucose), did not result in any enhancement.
Quite recently, it has been shown that glucose-dependent Pma1 activation is accompanied by double phosphorylation of the enzyme at the Ser-911 and Thr-912 positions [7]. Phosphorylation of these two residues is thought to eliminate the inhibitory effect of the enzyme's C-terminus on the ATP-hydrolyzing domain [7]–[9]. It is possible that the process of Pma1 phosphorylation involves protein kinase C, the activation of which is associated with the transport and phosphorylation of glucose [10].
Electron cryomicroscopy and crystallographic analysis were used previously to determine the tertiary and quaternary structure of the enzyme [11], [12]. Pma1 was shown to form detergent-resistant hexamers [13], [14], within which the monomers contact each other at the levels of both the transmembrane and cytoplasmic domains [12]. Another important finding was that the hexameric structure has internal mobility and becomes more closely packed under substrate binding [15].
As has been shown, some membrane proteins have to be associated with lipid rafts for correct incorporation into the plasma membrane [16]. To be delivered to and incorporated into the plasma membrane [17]–[20], Pma1 must also form a complex with sphingolipid. In the cells with disturbed sphingolipid synthesis, the newly synthesized native Pma1 has been shown to be routed, instead of to the PM, to the vacuole where it is degraded [17], [21]. Previous experiments with PMA1-GFP have allowed direct visualization of the raft compartmentalization of Pma1 [22].
A recently proposed model of membrane organization has suggested that all membrane proteins are contained both in the rafts and in the nonraft lipid domains and separated by vast protein-free lipid regions [23]. All protein-lipid “islands” are also thought to be bound to cytoskeleton elements [23]. This idea has aroused particular interest due to the finding of an acetylated tubulin-Pma1 complex in glucose-starved yeast and its dissociation under glucose addition [24]. Research has shown that the acetylated tubulin-Pma1 complex is dissociated very rapidly and that the glucose-induced increase in Pma1 activity occurs after its disintegration [24]. On the other hand, in S. cerevisiae, the formation and stability of Pma1-containing patches have been shown not to depend on the integrity of the actin and tubulin structures [25]. It has also been demonstrated that Pma1 molecules are relatively mobile within these patches [25].
The study of lateral mobility and oligomerization of transmembrane proteins has been based mainly on the phenomenon of fluorescence resonance energy transfer (FRET) [26]–[28]. When the donor and the acceptor carry different fluorophores, the distance between them can be assessed by changes in the fluorescence emission spectrum. If the donor and acceptor molecules carry the same fluorophore, then the intermolecular interactions can be studied by the change in fluorescence anisotropy [28], [29]. The latter method has been denoted as homo-FRET and has been widely used recently to estimate the degree of protein oligomerization [28].
Thus, it is clear that glucose activation of Pma1 is a complex process including several levels. In this work, we have attempted to assess (a) the role of sphingolipid and ergosterol in the glucose activation of Pma1 and (b) the mobility of yeast Pma1 molecules under glucose-induced activation of the enzyme.
Materials and Methods
Strains and growth conditions
S. cerevisiae strains (Table 1) were grown on standard YPD medium (Sigma, USA) for 16–18 h (late exponential phase) and were twice washed with water and incubated in distilled water for 1 h at 28°C to completely eliminate the effect of glucose from the growth medium. The strains erg-6, lcb1-100, SEY6210, and GFP-PMA1 were a kind gift from Dr. W. Tanner (University of Regensburg, FRG); the strain BY4742 was a kind gift from Dr. J. Boek (Johns Hopkins University, Baltimore, USA); and the strain RH2874 was a kind gift from Dr. H. Riezman (University of Geneva, Switzerland).
Table 1. Yeast strains used in this study.
| Strain | Genotype | Reference |
| SEY6210 | MATα ura3-52 leu2-3,112 his3-Δ100 trp1-Δ901 lys2-801suc2-Δ9 | [30] |
| PMA1-GFP | SEY6210 except PMA1: :GFP: : kanMX4 | [22] |
| BY4742 | MATα his3-Δ1 leu2-Δ0 lys2-Δ0 ura3-Δ0 trp1-Δ901 bar1-1 | [31] |
| erg6 | BY4742 except erg6Δ: :kanMX4 | [22] |
| RH2874 | MATα leu2 ura3 lys2 trp1 bar1-1 | [32] |
| lcb1-100 | MATα lcb1-100 leu2 trp1 ura3 lys2 bar1-1 | [32] |
Incubation with sugars and cell permeabilization
The cells were resuspended in permeabilization buffer (PB) (0.5 M sorbitol, 100 mM KCl, 4 mM MgCl2, 10 mM MES, pH 6.5) at a concentration of 1 g/10 ml and 5 ml samples were poured into 15 ml test tubes. Then, 0.25 ml of 2 M glucose (deoxyglucose) (Sigma, USA) was added to the experimental samples, and 0.25 ml of water was added to the controls. The samples were incubated for 15 min at 28°C. Then, 0.25 ml of 5% Triton X-100 was added to all of the samples. Immediately after, the samples were frozen at −60°C in a deep freezer and left overnight.
Determination of Pma1 activity in situ
The permeabilized cells were thawed at 28°C, washed 3 times with 5 ml of PB, resuspended in 2 ml of the same buffer, and stored at 0°C until use. A total of 20 µl of 100 mM Mg-ATP (Boehringer, USA), 1 µl of 10 µM Bafilomycin (Sigma, USA) in DMSO (for inhibition of the vacuolar ATPase), and 50 µl of cell suspension were added to 910 µl of PB. Then, 20 µl of 5 mM VO4 was added to the controls for the inhibition of Pma1. The reaction mixture was incubated at 30°C at 1,000 rpm in a Thermomixer compact (Eppendorf, FRG) for 30 min. The cells were precipitated at 14,000 g for 3 min, and the aliquots were diluted 50–100 times with ddH2O. In a 96-well microplate, 200 µl of malachite green (Applechem, FRG) was added to 100 µl of a sample [33] and incubated at 30°C for 15 min; the content of released Pi was measured at 650 nm. Pma1 activity was calculated by the difference in the amount of inorganic Pi released in the presence and absence of 100 µM sodium orthovanadate, the specific inhibitor of Pma1 [9]. To calculate the specific enzyme activity, total cell protein was determined [34] with modifications. A total of 100 µl of the cell suspension in the PB was added to 1 ml of water and precipitated at 14,000 g. The precipitate was resuspended in 0.6 ml of water and 0.3 ml of 3 N NaOH was added and heated at 100°C for 5 min, followed by addition of 0.3 ml of 2.5% CuSO4 after cooling. Five minutes later, the mixture was centrifuged at 14,000 g, and the supernatant was measured by spectrophotometry at 555 nm. Specific Pma1 activity was expressed as nmol of inorganic Pi released from ATP per minute per mg of total cell protein.
Plasma Membrane Isolation and ATP Hydrolysis
The plasma membranes were prepared from the glucose-metabolizing yeast cells by centrifugation on a sucrose step-gradient as previously described [35]. The Pma1 assays were conducted in 96-well microplates, essentially as described [36]. The basic ATP hydrolysis assay medium consisted of 5 mM MgSO4, 4 mM ATP, 25 mM NH4Cl, and 10 mM MES-Tris, pH 6.5. The reaction was performed in 150 µl of volume with 1.0 µg of membrane protein at 30°C for 20 min. The reaction was stopped by the addition of 100 µl of a combined Stop-Color Development reagent containing 1% SDS, 0.8% ascorbic acid, 100 mM ammonium molybdate, and 0.6 M H2SO4. Pma1 activity was calculated by the difference in inorganic Pi released from ATP in the absence and presence of 10 µM sodium orthovanadate [35]. The Km was calculated using Prism software (GraphPad Software Inc., La Jolla, CA).
Determination of anisotropy
Steady-state fluorescence anisotropy measurements were performed using a Cary Eclipse spectrofluorimeter (Varian Inc. USA) equipped with an automated polarizer accessory. The cell holder temperature was kept at 28°C. An aqueous cell suspension with an absorbance about 0.3 at 600 nm was used. GFP fluorescence was excited at 480 nm, and the emission was collected at 520 nm. The excitation bandwidth was 10–20 nm, and the emission bandwidth was 20 nm. Appropriate blanks were measured using GFP-lacking cells. The difference between the cell concentrations of the main and the reference solutions was estimated from their absorbance at 600 nm. Each contribution of the reference solution was corrected for this difference using the Beer-Lambert law [37]. The blanks were subtracted from each of the fluorescence intensity values (IVV, IVH) and used to calculate the anisotropy values [38]. All of the fluorescence anisotropy values were corrected for the instrumental G factor, which was measured using a highly diluted aqueous solution of fluorescein. The values reported represent an average of three measurements with an average time of 10 s.
Immunogold assay
The cells were fixed with 1.25% glutaraldehyde-1% paraformaldehyde in 0.1 M cacodylate buffer (pH 7.0). The samples were washed with the buffer, dehydrated in ethanol, infiltrated with LR white, omitting acceleration, and cured for 40 h at 50°C. The ultrathin sections obtained were mounted on copper grids covered with a formvar support. This was followed by incubation with polyclonal anti-Pma1 antibodies (a kind gift from Dr. C. Slayman, Yale University, USA), diluted (1∶100) in PBS for 2 h at 30°C, and again washed 2 times for 5 min in PBS. The grids were incubated with protein A (Serva, FRG) conjugated with 15-nm-diameter gold particles for 1 h at 30°C and washed 2 times for 5 min in PBS and finally once in water. After air drying, the samples were stained with a 3% uranyl acetate solution in 70% ethanol for 30 min and washed 2 times for 5 min in PBS. The sections were examined in a JEM100B (JEOL, Japan) electron microscope at an accelerating voltage of 80 kV.
Results and Discussion
The association of Pma1 with lipid rafts has been studied quite thoroughly [16]–[21], [39]. However, little is known about the role of lipids in glucose activation of Pma1.
To shed light on this issue, we selected the mutant erg6, which is deficient in ergosterol, and the mutant lcb1-100, which is defective in sphingoid base synthesis. The incubation of prestarved cells with 100 mM glucose or 100 mM deoxyglucose for 15 min yielded the results shown in Table 2. As expected, both of the parent strains, BY4742 and RH2874, demonstrated a glucose effect with different degrees of intensity. Unsurprisingly, the erg6 strain also showed glucose activation comparable with the parent strain. Previously, it was reported that disturbances in ergosterol synthesis in no way affected Pma1 biogenesis, secretion on the PM, or stability [19]. In addition, ergosterol was shown to occur mainly in the membrane patches containing arginine and uracyl symporters but not Pma1 [40].
Table 2. Pma1 activity in situ (nmol Pi/min/mg total cell protein, n = 3±SD) after 15-min incubation of S. cerevisiae whole cells with 100 mM glucose or 100 mM deoxyglucose. The change of activity in % of initial activity is given in parenthesis.
| Strain | Buffer | 100 mM glucose | 100 mM deoxyglucose |
| BY 4742 | 7.36±2.94 (100) | 14.64±1.16 (198.94) | 5.64±1.54 (76.6) |
| erg6 | 22.47±0.35 (100) | 44.0±0.1 (195.85) | 24.8±0.35 (110.38) |
| RH 2874 | 29.62±0.75 (100) | 38.68±1.74 (130.59) | 21.27±0.36 (71.81) |
| lcb1-100 | 38.34±0.96 (100) | 35.73±2.54 (93.2) | 36.18±0.13 (94.35) |
| SEY 6210 | 18.5±0.89 (100) | 52.36±0.85 (283.1) | 16.8±2.19 (91) |
| PMA1-GFP | 7.27±1.68 (100) | 23.19±0.22 (319.1) | 5.59±0.98 (77) |
In contrast, in strain lcb1-100, incubation with glucose did not increase the enzyme activity and even slightly reduced it. At the same time, the basal Pma1 activity in strain lcb1-100 was somewhat higher than in the parent strain RH2874. The latter result was quite unexpected, because previously it had been shown for the lcb1-100 strain that only a minor part of the newly synthesized Pma1 reaches PM at 30°C, whereas more than 90% of the enzyme is rerouted to the vacuole to be degraded [17]. However, later work suggested that this effect depended in many respects on the cultivation medium [20]. It should be noted that the growth of the lcb1-100 strain on the YPD medium was almost twice as low as that of the parent strain RH2874 (data not shown). With the exception of the erg6 strain, deoxyglucose did not result in an increase in the Pma1 activity, in agreement with the Serrano's data [6].
Thus, we have shown that sphingolipid but not ergosterol is important for glucose activation of Pma1. This fact can be explained as follows: One of the consequences of sphingolipid synthesis disturbance in the lcb1-100 strain is inefficient or completely blocked Pma1 oligomerization [17], [18], [20], which probably results in the elimination of glucose activation. The difference in glucose effects on Pma1 activity in the erg6 and lcb1-100 strains may, therefore, be attributed to the sphingolipid associating with the protein at the very initial stages of biosynthesis of the enzyme and determining its oligomeric structure [18], [20]. Ergosterol, the other component of the lipid raft, appears not to participate directly in the formation of the oligomeric Pma1 complex and have no particular effect on the functioning of the protein. The idea that oligomerization of Pma1 is necessary for the glucose activation of Pma1 was indirectly confirmed in the earlier work [12]. Using electron crystallography, researchers showed that the cytoplasmic part of Pma1 in a ligand-free form consists of four domains [12]. Domain two of one Pma1 molecule directly contacts domain three of the neighboring molecule. Unfortunately, the authors of this work did not link these structural domains with the functional (ATP-binding, phosphorylation, C-terminal) domains. However, it may be hypothesized that in the absence of glucose, the nucleotide-binding domain of the Pma1 molecule is locked by the C-domain of the neighboring Pma1 molecule. In this case, glucose activation of the enzyme results in successive phosphorylation of Ser-911 and Thr-912, followed by the release of the C-tail from the nucleotide-binding domain, as demonstrated previously [7], [9]. Taking into account the intermolecular character of the described event, it may be supposed that Pma1 oligomerization is necessary for the activation of Pma1 by glucose.
Since the modern concept of glucose activation of Pma1 presupposes the movement of its C-tail [7], [9], this process could be traced using the strain PMA1-GFP, the Pma1 molecule of which carries a GFP domain at the C-terminus [22]. To elucidate the effect of this spectral marker on glucose activation of the enzyme, the influence of the 15-min incubation of whole cells with 100 mM glucose or 100 mM deoxyglucose on Pma1 activity was investigated (Table 2). Both the parent strain SEY6210 and the strain PMA1-GFP demonstrated a marked glucose effect that exceeded that of the BY4742 and RH2874 strains. Although the presence of the GFP domain resulted in a considerable decrease in basal Pma1 activity (18.5 and 7.3 nmol Pi/min/mg total cell protein for SEY6210 and PMA1-GFP, respectively), it had little effect on the Km of the glucose-activated enzyme. The Km values of Pma1 determined in the membrane fraction from glucose-activated cells of the SEY6210 and PMA1-GFP strains were 0.22 mM and 0.41 mM, respectively. It is known that the Km for glucose-activated Pma1 is 0.3–0.8 mM, in contrast to 2–4 mM for Pma1 in glucose-deprived cells [6], [36]. Thus, the PMA1-GFP strain affords an opportunity to register the glucose activation process using modern spectral methods.
Fluorescence anisotropy was used to observe the structural rearrangements in the Pma1 molecules. The value of fluorescence anisotropy r for the GFP monomer in the absence of substantial rotation of the fluorophore molecule during the fluorescence lifetime is close to the maximum value (0.4) [41]–[43]. The radiationless transfer of energy between the fluorophore molecules (homo-FRET) as a result of oligomerization of the GFP molecules has been shown to result in a decrease in the observed effective value r [44]. Table 3 shows the r values of PMA1-GFP whole cells in the absence and presence of (deoxy)glucose. Since the procedure for measuring the value of r involves subtraction of the contribution of cellular autofluorescence to the total fluorescence, the observed r value corresponds to that of the GFP molecules. As can be seen, the experimental r values (0.165–0.202) were much lower than 0.38, the value r for the monomeric and dimeric form of GFP expressed in different samples [44]. The decrease in r was typical of the homo-FRET phenomenon observed for the clusters of fluorophores approaching each other until the distance between them was less than 10 nm.
Table 3. Fluorescence depolarization (anisotropy) r of PMA1-GFP in whole cells after 15 min incubation with 100 mM glucose or deoxyglucose.
| Additions | r |
| None | 0.192±0.005 |
| 100 mM glucose | 0.165±0.005 |
| 100 mM deoxyglucose | 0.202±0.003 |
Since it has been shown previously that the supramolecular Pma1complex in the PM consists of six units [11]–[13], the value r in the absence of (deoxy)glucose (equal to 0.192) characterizes the anisotropy of the PMA1-GFP hexameric complex. The incubation of the PMA1-GFP cells with 100 mM glucose for 15 min resulted in a marked decrease in the r value by 0.026±0.005. The same incubation with deoxyglucose produced an insignificant opposite effect: an increase in the r value by 0.011±0.006. Thus, there is a correlation between the values of Pma1 activity (Table 2) and fluorescence anisotropy (Table 3). This finding implies that glucose activation of the enzyme is accompanied by certain structural rearrangements of the Pma1 molecules at the level of clusters. The following variants and their combinations are possible [44], [45]: a change in the number of PMA1-GFP molecules in a cluster or association of clusters with each other, a change in the average distance between the Pma1 molecules in a cluster, or regrouping/reorientation of the GFP domains without a change in the distance between the Pma1 molecules. The most probable explanation for the decrease in the r value in our study seems to that since the activated enzyme intensively hydrolyzed ATP, the enzyme remained in the ligand-bound conformation for some of the time. Previously, it has been shown that the cytoplasmic Pma1 domain is more closely packed in this conformation [15]. It has also been proposed that phosphorylation of Ser-911 and Thr-912 during glucose activation results in conformational changes in the Pma1 molecule in which the C-domain is released from the nucleotide-binding domain [7], [9]. In our model system, the C-tail of Pma1 carried a GFP domain. The decrease in the r value may point to approaching of GFP domains within a hexamer complex during glucose activation of Pma1. However, in the case of a high fluorophore concentration, which often occurs in the case of membrane-bound proteins, the phenomenon of homo-FRET theoretically may be not a consequence of fluorophore oligomerization. Instead, it may be the result of the close approach of monomeric proteins (the so-called concentration depolarization effect) [28].
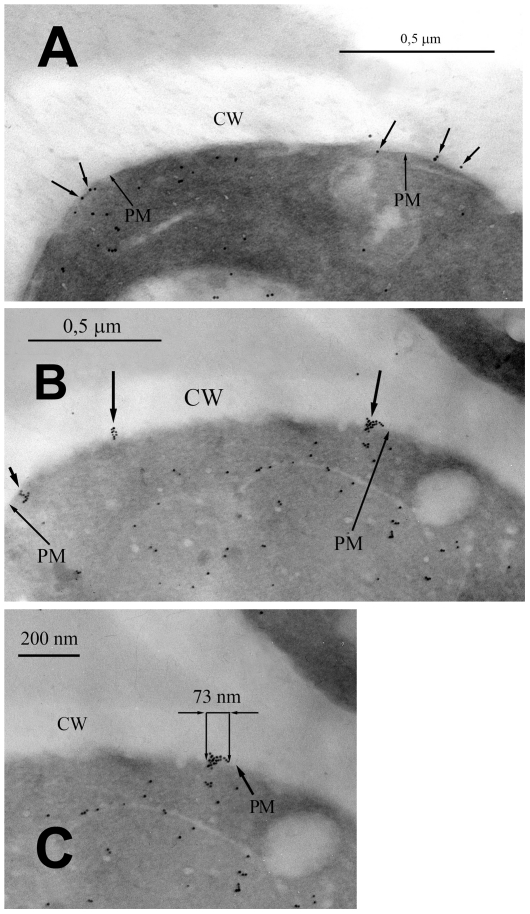
The change in the spatial distribution of the clusters containing Pma1 molecules during glucose metabolism was visualized using immunogold labeling. As Figure 1 shows, the distribution of the clusters of Pma1 molecules in the PM underwent substantial changes. In the glucose-starved cells of the parent strain SEY6210, the distribution of the clusters of Pma1 molecules was relatively uniform, and the average distance between them exceeded 10 nm (Fig. 1A). In the cells that had metabolized glucose, the clusters of Pma1 molecules aggregated in large groups of closely adjacent clusters (Fig. 1B). Since immunogold labeling was performed on a cell cross-section but not on the inner leaflet of the PM obtained by freeze-fractioning, the size of the groups cannot be analyzed using Ripley's K-function. It is interesting that the approximate sizes of these groups of clusters of Pma1 molecules (about 70 nm) (Fig. 1C) are similar to the sizes of the individual lipid rafts (25–70 nm) [46], [47]. The distance between the Pma1 clusters in such a group was comparable to the distances at which the homo-FRET effect was exhibited (10 nm); therefore, the observed decrease in fluorescence anisotropy in response to glucose addition can be at least partially explained by this phenomenon. While intracellular labeling can be observed in all of the images, we assume that it is the labeling of the newly synthesized Pma1 molecules during their delivery to PM.
Figure 1. Immunogold labeling of Pma1 in the plasma membrane of S. cerevisiae SEY6210.
(A) – glucose-starved cells, Pma1 was distributed in the membrane as single structures; (B) – cells that had metabolized glucose for 15 min, Pma1 formed large bunch-like complexes; (C) – enlarged fragment of photograph (B) CW = cell wall; PM = plasma membrane.
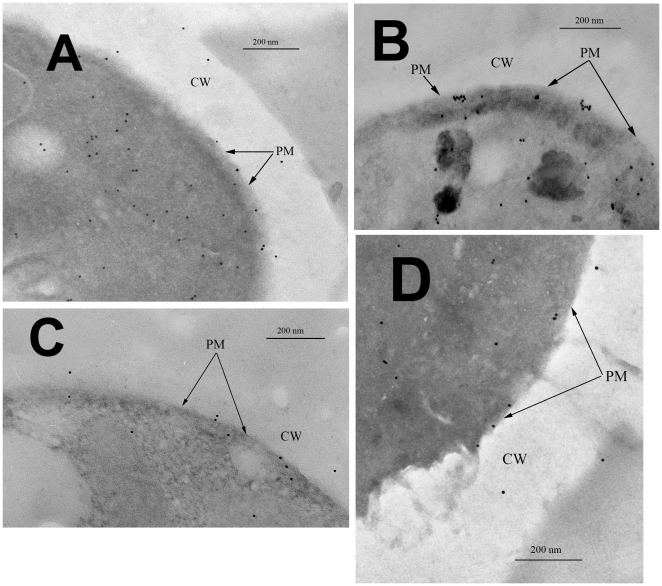
Figure 2 shows the effect of incubation with glucose on the spatial distribution of the Pma1 molecules in the PM of the erg6 and lcb1-100 strains. In the erg6 strain (Fig. 2A, B), where glucose activated Pma1, the distribution of the clusters of enzyme molecules before and after the incubation with glucose was similar to the distribution of Pma1 in the SEY6210 strain. In the absence of glucose, Pma1 in the PM was distributed uniformly as separate particles. Following the incubation with glucose, Pma1 formed groups. In contrast to the SEY6210 and erg6 strains, the incubation with glucose did not change the distribution of Pma1 in the PM of the lcb1-100 strain (Fig. 2C, D). The distribution of the Pma1 clusters remained uniform, without the formation of groups. Moreover, glucose did not exert an effect on the lcb1-100 strain (Table 2). Thus, it may be supposed that Pma1 activation by glucose is accompanied by the formation of groups of Pma1 clusters.
Figure 2. Immunogold labeling of Pma1 in the plasma membrane of S. cerevisiae erg6 and lcb1-100.
(A) – glucose-starved cells of the erg6 strain, Pma1 was distributed in the membrane as single structures; (B) – erg6 cells that had metabolized glucose for 15 min, Pma1 formed complexes; (C) - glucose-starved cells of the lcb1-100 strain, Pma1 was distributed in the membrane as single structures; (D) – lcb1-100 cells that had metabolized glucose for 15 min, Pma1 was distributed in the membrane as single structures. CW = cell wall; PM = plasma membrane.
The revealed glucose-dependent movement of the clusters of the Pma1 molecules is noteworthy because it is a rather quick process (about 15 min). It is unclear what cellular mechanism underlies this reorganization of the Pma1 clusters, but one of the most probable reasons is the previously reported association of Pma1 with acetylated tubulin [24]. In this study, the authors showed that Pma1 in the cells that did not metabolize glucose was associated with acetylated tubulin. On glucose addition, this complex very quickly degraded. These findings, together with our data presented above, suggest that acetylated tubulin in the absence of glucose fixes Pma1 clusters on the membrane and leads to a uniform distribution. Upon glucose addition, this complex disintegrates, allowing the clusters of Pma1 molecules to move freely in the membrane plane and combine into large groups.
These findings lead to the following two conclusions: (1) for glucose activation of Pma1 to take place, the enzyme is supposed to be in the oligomeric state, and (2) glucose activation is accompanied by the spatial movement of Pma1 clusters in the PM.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was funded by the Russian Foundation for Basic Research grant #10-04-01248; Program of the Russian Academy of Sciences “Molecular and Cell Biology.” The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Palmgren MG, Nissen P. P-type ATPases. Annual review of biophysics. 2011;40:243–266. doi: 10.1146/annurev.biophys.093008.131331. [DOI] [PubMed] [Google Scholar]
- 2.Lutsenko S, Kaplan JH. Organization of P-type ATPases: significance of structural diversity. Biochemistry. 1995;34:15607–15613. doi: 10.1021/bi00048a001. [DOI] [PubMed] [Google Scholar]
- 3.Serrano R, Kielland-Brandt MC, Fink GR. Yeast plasma membrane ATPase is essential for growth and has homology with (Na++K+), K+− and Ca2+−ATPases. Nature. 1986;319:689–693. doi: 10.1038/319689a0. [DOI] [PubMed] [Google Scholar]
- 4.Rao R, Nakamoto RK, Verjovski-Almeida S, Slayman CW. Structure and function of the yeast plasma-membrane H(+)-ATPase. Annals of the New York Academy of Sciences. 1992;671:195–203. doi: 10.1111/j.1749-6632.1992.tb43796.x. [DOI] [PubMed] [Google Scholar]
- 5.Morsomme P, Slayman CW, Goffeau A. Mutagenic study of the structure, function and biogenesis of the yeast plasma membrane H(+)-ATPase. Biochimica et biophysica acta. 2000;1469:133–157. doi: 10.1016/s0304-4157(00)00015-0. [DOI] [PubMed] [Google Scholar]
- 6.Serrano R. In vivo glucose activation of the yeast plasma membrane ATPase. FEBS letters. 1983;156:11–14. doi: 10.1016/0014-5793(83)80237-3. [DOI] [PubMed] [Google Scholar]
- 7.Lecchi S, Nelson CJ, Allen KE, Swaney DL, Thompson KL, et al. Tandem phosphorylation of Ser-911 and Thr-912 at the C terminus of yeast plasma membrane H+-ATPase leads to glucose-dependent activation. The Journal of biological chemistry. 2007;282:35471–35481. doi: 10.1074/jbc.M706094200. [DOI] [PubMed] [Google Scholar]
- 8.Eraso P, Portillo F. Molecular mechanism of regulation of yeast plasma membrane H(+)-ATPase by glucose. Interaction between domains and identification of new regulatory sites. The Journal of biological chemistry. 1994;269:10393–10399. [PubMed] [Google Scholar]
- 9.Lecchi S, Allen KE, Pardo JP, Mason AB, Slayman CW. Conformational changes of yeast plasma membrane H(+)-ATPase during activation by glucose: role of threonine-912 in the carboxy-terminal tail. Biochemistry. 2005;44:16624–16632. doi: 10.1021/bi051555f. [DOI] [PubMed] [Google Scholar]
- 10.Souza MA, Tropia MJ, Brandao RL. New aspects of the glucose activation of the H(+)-ATPase in the yeast Saccharomyces cerevisiae. Microbiology. 2001;147:2849–2855. doi: 10.1099/00221287-147-10-2849. [DOI] [PubMed] [Google Scholar]
- 11.Cyrklaff M, Auer M, Kuhlbrandt W, Scarborough GA. 2-D structure of the Neurospora crassa plasma membrane ATPase as determined by electron cryomicroscopy. The EMBO journal. 1995;14:1854–1857. doi: 10.1002/j.1460-2075.1995.tb07177.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Auer M, Scarborough GA, Kuhlbrandt W. Three-dimensional map of the plasma membrane H+-ATPase in the open conformation. Nature. 1998;392:840–843. doi: 10.1038/33967. [DOI] [PubMed] [Google Scholar]
- 13.Chadwick CC, Goormaghtigh E, Scarborough GA. A hexameric form of the Neurospora crassa plasma membrane H+-ATPase. Archives of biochemistry and biophysics. 1987;252:348–356. doi: 10.1016/0003-9861(87)90041-5. [DOI] [PubMed] [Google Scholar]
- 14.Serrano R. The molecular and cellular biology of the yeast Saccharomyces, genome dynamics, protein sinthesis and energetics. In: Broach JR, Pringle JR, Jones EW, editors. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1991. [Google Scholar]
- 15.Rhee KH, Scarborough GA, Henderson R. Domain movements of plasma membrane H(+)-ATPase: 3D structures of two states by electron cryo-microscopy. The EMBO journal. 2002;21:3582–3589. doi: 10.1093/emboj/cdf385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bagnat M, Keranen S, Shevchenko A, Simons K. Lipid rafts function in biosynthetic delivery of proteins to the cell surface in yeast. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:3254–3259. doi: 10.1073/pnas.060034697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bagnat M, Chang A, Simons K. Plasma membrane proton ATPase Pma1p requires raft association for surface delivery in yeast. Molecular biology of the cell. 2001;12:4129–4138. doi: 10.1091/mbc.12.12.4129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee MC, Hamamoto S, Schekman R. Ceramide biosynthesis is required for the formation of the oligomeric H+-ATPase Pma1p in the yeast endoplasmic reticulum. The Journal of biological chemistry. 2002;277:22395–22401. doi: 10.1074/jbc.M200450200. [DOI] [PubMed] [Google Scholar]
- 19.Gaigg B, Timischl B, Corbino L, Schneiter R. Synthesis of sphingolipids with very long chain fatty acids but not ergosterol is required for routing of newly synthesized plasma membrane ATPase to the cell surface of yeast. The Journal of biological chemistry. 2005;280:22515–22522. doi: 10.1074/jbc.M413472200. [DOI] [PubMed] [Google Scholar]
- 20.Wang Q, Chang A. Sphingoid base synthesis is required for oligomerization and cell surface stability of the yeast plasma membrane ATPase, Pma1. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:12853–12858. doi: 10.1073/pnas.202115499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Toulmay A, Schneiter R. Lipid-dependent surface transport of the proton pumping ATPase: a model to study plasma membrane biogenesis in yeast. Biochimie. 2007;89:249–254. doi: 10.1016/j.biochi.2006.07.020. [DOI] [PubMed] [Google Scholar]
- 22.Malinska K, Malinsky J, Opekarova M, Tanner W. Visualization of protein compartmentation within the plasma membrane of living yeast cells. Molecular biology of the cell. 2003;14:4427–4436. doi: 10.1091/mbc.E03-04-0221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lillemeier BF, Pfeiffer JR, Surviladze Z, Wilson BS, Davis MM. Plasma membrane-associated proteins are clustered into islands attached to the cytoskeleton. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:18992–18997. doi: 10.1073/pnas.0609009103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Campetelli AN, Previtali G, Arce CA, Barra HS, Casale CH. Activation of the plasma membrane H-ATPase of Saccharomyces cerevisiae by glucose is mediated by dissociation of the H(+)-ATPase-acetylated tubulin complex. The FEBS journal. 2005;272:5742–5752. doi: 10.1111/j.1742-4658.2005.04959.x. [DOI] [PubMed] [Google Scholar]
- 25.Malinska K, Malinsky J, Opekarova M, Tanner W. Distribution of Can1p into stable domains reflects lateral protein segregation within the plasma membrane of living S. cerevisiae cells. Journal of cell science. 2004;117:6031–6041. doi: 10.1242/jcs.01493. [DOI] [PubMed] [Google Scholar]
- 26.Bader AN, Hofman EG, van Bergen En Henegouwen PM, Gerritsen HC. Imaging of protein cluster sizes by means of confocal time-gated fluorescence anisotropy microscopy. Optics express. 2007;15:6934–6945. doi: 10.1364/oe.15.006934. [DOI] [PubMed] [Google Scholar]
- 27.Bader AN, Hofman EG, Voortman J, en Henegouwen PM, Gerritsen HC. Homo-FRET imaging enables quantification of protein cluster sizes with subcellular resolution. Biophysical journal. 2009;97:2613–2622. doi: 10.1016/j.bpj.2009.07.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yeow EK, Clayton AH. Enumeration of oligomerization states of membrane proteins in living cells by homo-FRET spectroscopy and microscopy: theory and application. Biophysical journal. 2007;92:3098–3104. doi: 10.1529/biophysj.106.099424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lakowicz JR, Gryczynski I, Gryczynski Z, Dattelbaum JD. Anisotropy-based sensing with reference fluorophores. Analytical biochemistry. 1999;267:397–405. doi: 10.1006/abio.1998.3029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Robinson JS, Klionsky DJ, Banta LM, Emr SD. Protein sorting in Saccharomyces cerevisiae: isolation of mutants defective in the delivery and processing of multiple vacuolar hydrolases. Molecular and cellular biology. 1988;8:4936–4948. doi: 10.1128/mcb.8.11.4936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brachmann CB, Davies A, Cost GJ, Caputo E, Li J, et al. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast. 1998;14:115–132. doi: 10.1002/(SICI)1097-0061(19980130)14:2<115::AID-YEA204>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 32.Zanolari B, Friant S, Funato K, Sutterlin C, Stevenson BJ, et al. Sphingoid base synthesis requirement for endocytosis in Saccharomyces cerevisiae. The EMBO journal. 2000;19:2824–2833. doi: 10.1093/emboj/19.12.2824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stewart DJ. Sensitive automated methods for phosphate and (Na+plus K+)-ATPase. Analytical biochemistry. 1974;62:349–364. doi: 10.1016/0003-2697(74)90167-5. [DOI] [PubMed] [Google Scholar]
- 34.Herbert D, Phipps PJ, Strange RE. Chemical Analysis of Microbial Cells. In: Norris JR, Ribbons DW, editors. Methods in microbiology. N.-Y.: Academ Press; 1971. pp. 210–344. [Google Scholar]
- 35.Seto-Young D, Monk BC, Perlin DS. Assessing hydrophobic regions of the plasma membrane H(+)-ATPase from Saccharomyces cerevisiae. Biochimica et biophysica acta. 1992;1102:213–219. doi: 10.1016/0005-2728(92)90102-8. [DOI] [PubMed] [Google Scholar]
- 36.Monk BC, Kurtz MB, Marrinan JA, Perlin DS. Cloning and characterization of the plasma membrane H(+)-ATPase from Candida albicans. Journal of bacteriology. 1991;173:6826–6836. doi: 10.1128/jb.173.21.6826-6836.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Permyakov EA. Luminescent spectroscopy of proteins. Ann Arbor, London, Tokyo: CRC Press Inc; 1993. [Google Scholar]
- 38.Lacowicz JR. Principles of fluorescence spectroskopy. New York: Springer; 2006. [Google Scholar]
- 39.Gaigg B, Toulmay A, Schneiter R. Very long-chain fatty acid-containing lipids rather than sphingolipids per se are required for raft association and stable surface transport of newly synthesized plasma membrane ATPase in yeast. The Journal of biological chemistry. 2006;281:34135–34145. doi: 10.1074/jbc.M603791200. [DOI] [PubMed] [Google Scholar]
- 40.Grossmann G, Opekarova M, Malinsky J, Weig-Meckl I, Tanner W. Membrane potential governs lateral segregation of plasma membrane proteins and lipids in yeast. The EMBO journal. 2007;26:1–8. doi: 10.1038/sj.emboj.7601466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Partikian A, Olveczky B, Swaminathan R, Li Y, Verkman AS. Rapid diffusion of green fluorescent protein in the mitochondrial matrix. The Journal of cell biology. 1998;140:821–829. doi: 10.1083/jcb.140.4.821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hink MA, Griep RA, Borst JW, van Hoek A, Eppink MH, et al. Structural dynamics of green fluorescent protein alone and fused with a single chain Fv protein. The Journal of biological chemistry. 2000;275:17556–17560. doi: 10.1074/jbc.M001348200. [DOI] [PubMed] [Google Scholar]
- 43.Volkmer A, Subramaniam V, Birch DJ, Jovin TM. One- and two-photon excited fluorescence lifetimes and anisotropy decays of green fluorescent proteins. Biophysical journal. 2000;78:1589–1598. doi: 10.1016/S0006-3495(00)76711-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bader AN, Hoetzl S, Hofman EG, Voortman J, van Bergen En Henegouwen PM, et al. Homo-FRET Imaging as a Tool to Quantify Protein and Lipid Clustering. Chemphyschem: a European journal of chemical physics and physical chemistry. 2011;12:475–483. doi: 10.1002/cphc.201000801. [DOI] [PubMed] [Google Scholar]
- 45.Runnels LW, Scarlata SF. Theory and application of fluorescence homotransfer to melittin oligomerization. Biophysical journal. 1995;69:1569–1583. doi: 10.1016/S0006-3495(95)80030-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pralle A, Keller P, Florin EL, Simons K, Horber JK. Sphingolipid-cholesterol rafts diffuse as small entities in the plasma membrane of mammalian cells. The Journal of cell biology. 2000;148:997–1008. doi: 10.1083/jcb.148.5.997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Varma R, Mayor S. GPI-anchored proteins are organized in submicron domains at the cell surface. Nature. 1998;394:798–801. doi: 10.1038/29563. [DOI] [PubMed] [Google Scholar]