Abstract
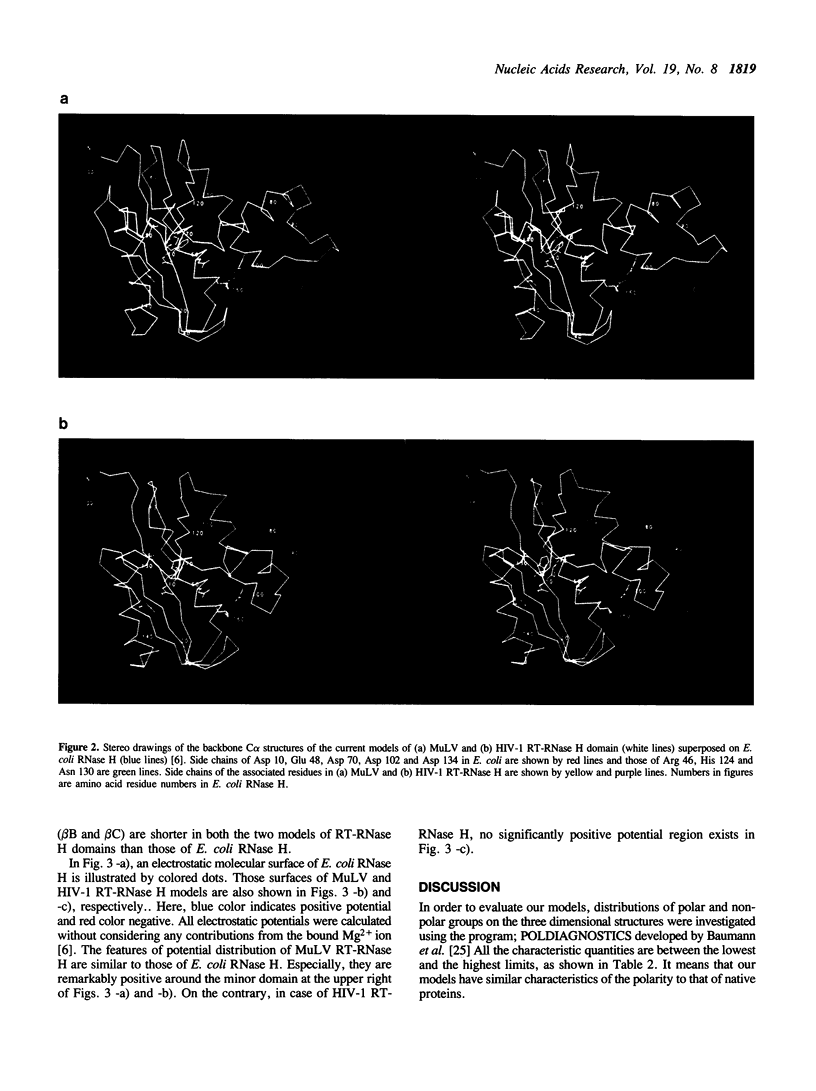
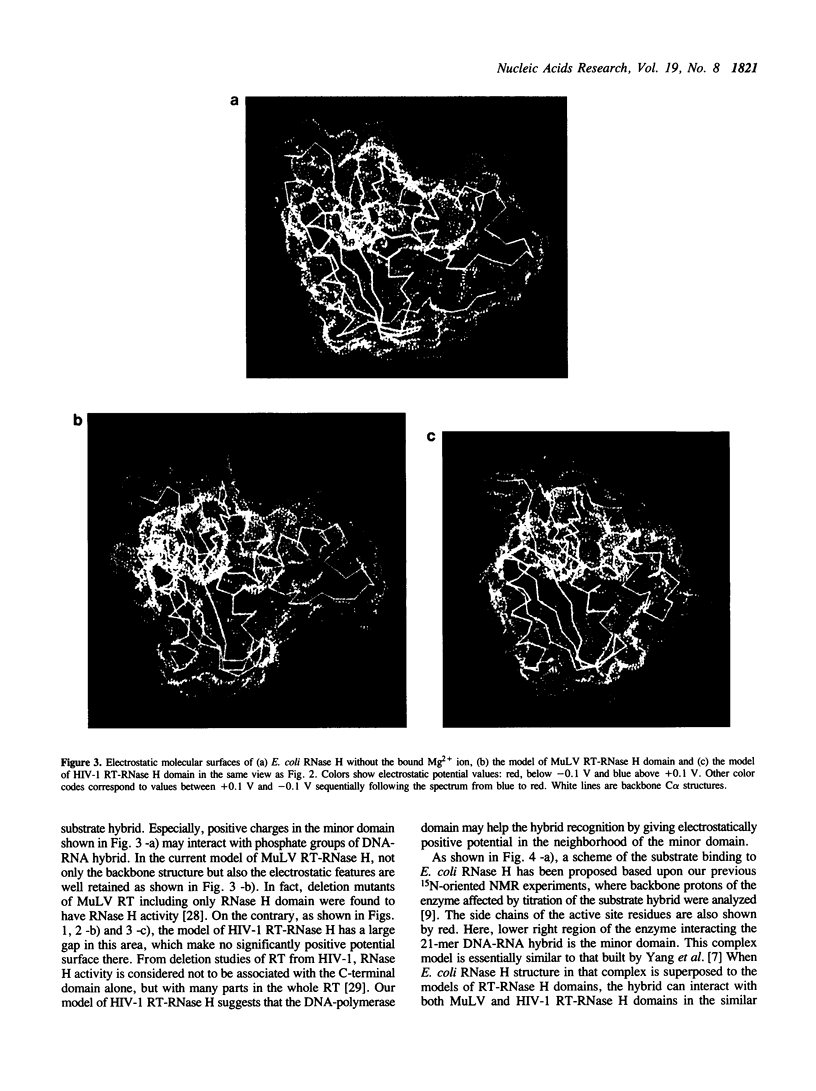
Tertiary models of ribonuclease H (RNase H) domains in reverse transcriptases (RTs) from Moloney murine leukemia virus (MuLV) and human immunodeficiency virus (HIV-1) were built based upon the X-ray structure of RNase H from Escherichia coli (E. coli RNase H). In two models of RT-RNase H domains, not only active site residues but also residues, which construct a hydrophobic core and hydrogen bonds, are located in the same positions as those of E. coli RNase H. The whole backbone structure and the electrostatic molecular surface of MuLV RT-RNase H model are similar to those of E. coli RNase H. On the contrary, HIV-1 RT-RNase H model lacks the third helix and the following loop, resulting no positive charge clusters around the hybrid recognition site. Referring the complex models of RTs with their substrate hybrid, the interaction between DNA-polymerase and RNase H domains in RTs was discussed.
Full text
PDF


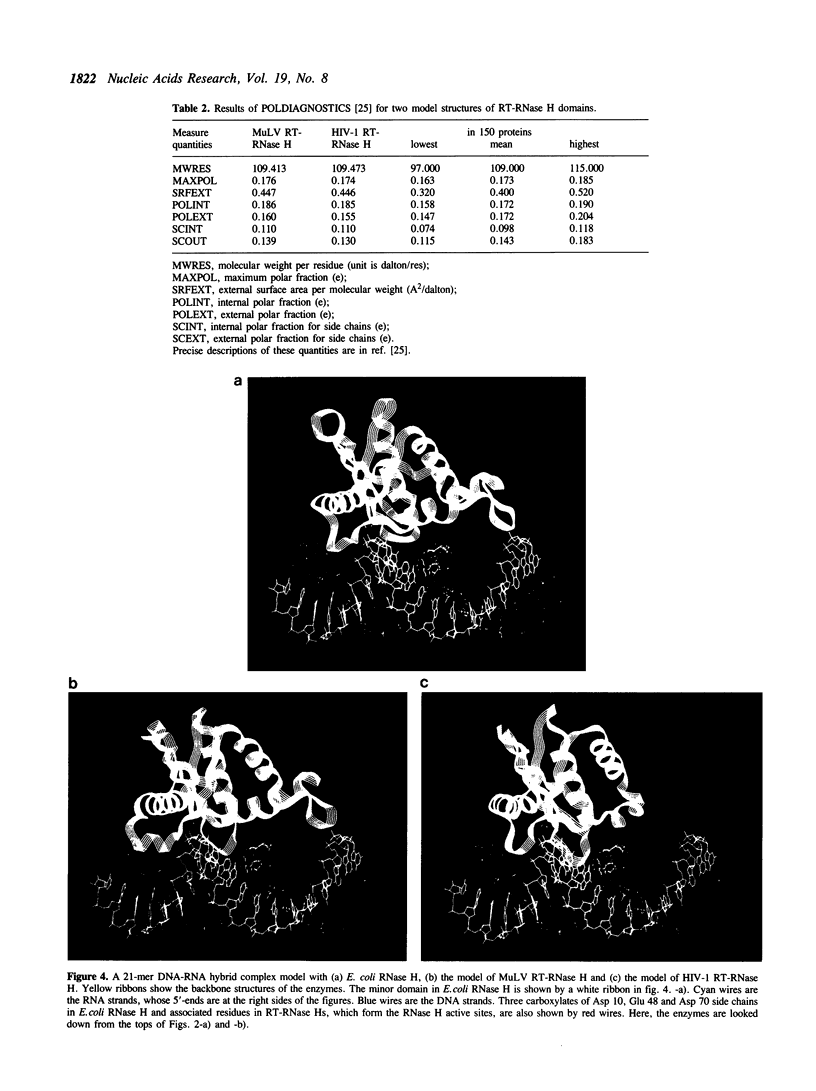

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Baltimore D. Retroviruses and retrotransposons: the role of reverse transcription in shaping the eukaryotic genome. Cell. 1985 Mar;40(3):481–482. doi: 10.1016/0092-8674(85)90190-4. [DOI] [PubMed] [Google Scholar]
- Baumann G., Frömmel C., Sander C. Polarity as a criterion in protein design. Protein Eng. 1989 Jan;2(5):329–334. doi: 10.1093/protein/2.5.329. [DOI] [PubMed] [Google Scholar]
- Bernstein F. C., Koetzle T. F., Williams G. J., Meyer E. F., Jr, Brice M. D., Rodgers J. R., Kennard O., Shimanouchi T., Tasumi M. The Protein Data Bank: a computer-based archival file for macromolecular structures. J Mol Biol. 1977 May 25;112(3):535–542. doi: 10.1016/s0022-2836(77)80200-3. [DOI] [PubMed] [Google Scholar]
- Cotton F. A., Hazen E. E., Jr, Legg M. J. Staphylococcal nuclease: proposed mechanism of action based on structure of enzyme-thymidine 3',5'-bisphosphate-calcium ion complex at 1.5-A resolution. Proc Natl Acad Sci U S A. 1979 Jun;76(6):2551–2555. doi: 10.1073/pnas.76.6.2551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crouch R. J. Ribonuclease H: from discovery to 3D structure. New Biol. 1990 Sep;2(9):771–777. [PubMed] [Google Scholar]
- Doolittle R. F., Feng D. F., Johnson M. S., McClure M. A. Origins and evolutionary relationships of retroviruses. Q Rev Biol. 1989 Mar;64(1):1–30. doi: 10.1086/416128. [DOI] [PubMed] [Google Scholar]
- Gilliland G. L., Winborne E. L., Nachman J., Wlodawer A. The three-dimensional structure of recombinant bovine chymosin at 2.3 A resolution. Proteins. 1990;8(1):82–101. doi: 10.1002/prot.340080110. [DOI] [PubMed] [Google Scholar]
- Jones T. A., Thirup S. Using known substructures in protein model building and crystallography. EMBO J. 1986 Apr;5(4):819–822. doi: 10.1002/j.1460-2075.1986.tb04287.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabsch W., Sander C. Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers. 1983 Dec;22(12):2577–2637. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
- Kamphuis I. G., Kalk K. H., Swarte M. B., Drenth J. Structure of papain refined at 1.65 A resolution. J Mol Biol. 1984 Oct 25;179(2):233–256. doi: 10.1016/0022-2836(84)90467-4. [DOI] [PubMed] [Google Scholar]
- Kanaya S., Kohara A., Miura Y., Sekiguchi A., Iwai S., Inoue H., Ohtsuka E., Ikehara M. Identification of the amino acid residues involved in an active site of Escherichia coli ribonuclease H by site-directed mutagenesis. J Biol Chem. 1990 Mar 15;265(8):4615–4621. [PubMed] [Google Scholar]
- Karplus P. A., Schulz G. E. Refined structure of glutathione reductase at 1.54 A resolution. J Mol Biol. 1987 Jun 5;195(3):701–729. doi: 10.1016/0022-2836(87)90191-4. [DOI] [PubMed] [Google Scholar]
- Katayanagi K., Miyagawa M., Matsushima M., Ishikawa M., Kanaya S., Ikehara M., Matsuzaki T., Morikawa K. Three-dimensional structure of ribonuclease H from E. coli. Nature. 1990 Sep 20;347(6290):306–309. doi: 10.1038/347306a0. [DOI] [PubMed] [Google Scholar]
- Lindqvist Y. Refined structure of spinach glycolate oxidase at 2 A resolution. J Mol Biol. 1989 Sep 5;209(1):151–166. doi: 10.1016/0022-2836(89)90178-2. [DOI] [PubMed] [Google Scholar]
- Loebermann H., Tokuoka R., Deisenhofer J., Huber R. Human alpha 1-proteinase inhibitor. Crystal structure analysis of two crystal modifications, molecular model and preliminary analysis of the implications for function. J Mol Biol. 1984 Aug 15;177(3):531–557. [PubMed] [Google Scholar]
- Martin A. C., Cheetham J. C., Rees A. R. Modeling antibody hypervariable loops: a combined algorithm. Proc Natl Acad Sci U S A. 1989 Dec;86(23):9268–9272. doi: 10.1073/pnas.86.23.9268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oyama F., Kikuchi R., Crouch R. J., Uchida T. Intrinsic properties of reverse transcriptase in reverse transcription. Associated RNase H is essentially regarded as an endonuclease. J Biol Chem. 1989 Nov 5;264(31):18808–18817. [PubMed] [Google Scholar]
- Prasad V. R., Goff S. P. Linker insertion mutagenesis of the human immunodeficiency virus reverse transcriptase expressed in bacteria: definition of the minimal polymerase domain. Proc Natl Acad Sci U S A. 1989 May;86(9):3104–3108. doi: 10.1073/pnas.86.9.3104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schatz O., Mous J., Le Grice S. F. HIV-1 RT-associated ribonuclease H displays both endonuclease and 3'----5' exonuclease activity. EMBO J. 1990 Apr;9(4):1171–1176. doi: 10.1002/j.1460-2075.1990.tb08224.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scouloudi H., Baker E. N. X-ray crystallographic studies of seal myoglobin. The molecule at 2.5 A resolution. J Mol Biol. 1978 Dec 25;126(4):637–660. doi: 10.1016/0022-2836(78)90013-x. [DOI] [PubMed] [Google Scholar]
- Sutcliffe M. J., Haneef I., Carney D., Blundell T. L. Knowledge based modelling of homologous proteins, Part I: Three-dimensional frameworks derived from the simultaneous superposition of multiple structures. Protein Eng. 1987 Oct-Nov;1(5):377–384. doi: 10.1093/protein/1.5.377. [DOI] [PubMed] [Google Scholar]
- Tanese N., Goff S. P. Domain structure of the Moloney murine leukemia virus reverse transcriptase: mutational analysis and separate expression of the DNA polymerase and RNase H activities. Proc Natl Acad Sci U S A. 1988 Mar;85(6):1777–1781. doi: 10.1073/pnas.85.6.1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W., Hendrickson W. A., Crouch R. J., Satow Y. Structure of ribonuclease H phased at 2 A resolution by MAD analysis of the selenomethionyl protein. Science. 1990 Sep 21;249(4975):1398–1405. doi: 10.1126/science.2169648. [DOI] [PubMed] [Google Scholar]