Abstract
Dioxins are known to cause several human cancers through activation of the aryl hydrocarbon receptor (AhR). Harmaline and harmalol are β-carboline compounds present in several medicinal plants such as Peganum harmala. We have previously demonstrated the ability of Peganum harmala extract to inhibit TCDD-mediated induction of Cyp1a1 in murine hepatoma Hepa 1c1c7 cells. Therefore, the aim of this study is to examine the effect of harmaline and its main metabolite, harmalol, on dioxin-mediated induction of CYP1A1 in human hepatoma HepG2 cells. Our results showed that harmaline and harmalol at concentrations of (0.5–12.5 µM) significantly inhibited the dioxin-induced CYP1A1 at mRNA, protein and activity levels in a concentration-dependent manner. The role of AhR was determined by the inhibition of the TCDD-mediated induction of AhR-dependent luciferase activity and the AhR/ARNT/XRE formation by both harmaline and harmalol. In addition, harmaline significantly displaced [3H]TCDD in the competitive ligand binding assay. At posttranslational level, both harmaline and harmalol decreased the protein stability of CYP1A1, suggesting that posttranslational modifications are involved. Moreover, the posttranslational modifications of harmaline and harmalol involve ubiquitin-proteasomal pathway and direct inhibitory effects of both compounds on CYP1A1 enzyme. These data suggest that harmaline and harmalol are promising agents for preventing dioxin-mediated effects.
Keywords: Aryl hydrocarbon receptor, Carcinogenesis, CYP 1A1, Harmaline, Harmalol
1. Introduction
Environmental pollution plays a major role in chemical carcinogenesis. Numerous polycyclic aromatic hydrocarbons (PAH) and halogenated aromatic hydrocarbons (HAH) present in cigarette smoke, coal tar, automobile exhaust and charbroiled food, can lead to several types of human cancers (Mandal, 2005; Shimada and Fujii-Kuriyama, 2004). The mechanism for the carcinogenic effect of PAH and HAH involves the binding and activation of aryl hydrocarbon receptor (AhR) and its regulated gene, CYP1A1 (Denison and Nagy, 2003; Mandal, 2005). AhR is a ligand-activated transcription factor found inactive in the cytoplasm in combination with two 90-KDa heat-shock proteins (HSP90), the co-chaperone p23 and a 43-KDa protein termed hepatitis B virus X-associated protein 2 (XAP2). Activation of the AhR usually starts with binding to its ligand and the subsequent translocation to the nucleus where it heterodimerizes with AhR nuclear translocator (ARNT). The formed complex then binds to its DNA sequence called xenobiotic responsive element (XRE) that is found in the promoter region of the AhR-regulated genes including the CYP1A1 (Denison and Nagy, 2003).
CYP1A1 is a carcinogen-activating enzyme that participates in the metabolic activation of several environmental procarcinogens including PAH and HAH. CYP1A1 metabolizes several environmental pollutants to reactive compounds capable of binding to DNA and protein forming several adducts that can lead to mutagenesis and carcinogenesis (Shimada and Fujii-Kuriyama, 2004). It has been proposed previously that interfering with AhR activation and CYP1A1 can lead to prevention of several cancers. This hypothesis was substantiated by several studies that correlated the level of CYP1A1 and the development of different human cancers such as lung, colon, liver and rectal cancers (Mandal, 2005; Shah et al., 2009; Slattery et al., 2004). Therefore, the level of CYP1A1 is considered as a useful biomarker for the exposure to several carcinogens. Furthermore, the inhibition of AhR activity and its regulated gene, CYP1A1 could result in the prevention of toxic effects mediated by the AhR ligands, including carcinogenicity (Puppala et al., 2008).
β-carbolines are a large group of natural and synthetic indole alkaloids that are widely distributed in nature, including various foods, plants, marine creatures, insects, mammalians, as well as human tissues (Cao et al., 2007). β-carbolines attracted considerable attention as they possess diverse pharmacological activities such as sedative, hypnotic, anxiolytic, anticonvulsant, antitumor, antithrombotic, antiparasitic, antimicrobial, as well as antiviral activities (Cao et al., 2007). Harmaline, 4,9-dihydro-7-methoxy-1-methyl-3H-pyrido[3,4-b]indole and harmalol, 1-methyl-4,9-dihydro-3H-pyrido[3,4-b]indole-7-ol (Fig.1), are common dihydro-β-carboline compounds that are naturally found in several alcoholic beverages and medicinal plants including Peganum harmala (Zygophyllaceae) (Herraiz et al., 2010; Park et al., 2010). Harmaline and harmalol possess several pharmacological activities such as antimicrobial, antioxidant and antiprotozoal activities (Arshad et al., 2008; Moura et al., 2007). Furthermore, harmaline can interact with several enzymes and neurotransmittors including topoisomerase I, and monoamine oxidase-A (Herraiz et al., 2010; Sobhani et al., 2002).
Figure 1. Chemical structure of harmaline (4,9-Dihydro-7-methoxy-1-methyl-3H-pyrido[3,4-b]indole), and harmalol (1-Methyl-4,9-dihydro-3H-pyrido[3,4-b]indole-7-ol).
We previously demonstrated that the extract of Peganum harmala can affect AhR signaling pathway in murine hepatoma Hepa1c1c7 cells (El Gendy et al., 2010). Moreover, we showed that harmaline is one of its main active ingredients and responsible for its inhibitory effect on TCDD-mediated induction of Cyp1a1 at the catalytic activity level in Hepa 1c1c7 cells (El Gendy et al., 2010). Thus, the aim of this study is to investigate the effect of harmaline and its main metabolite, harmalol, on AhR signaling pathway and the carcinogen-activating enzyme, CYP1A1, using human hepatoma HepG2 cells. Therefore, we examined their effects on CYP1A1 at mRNA, protein, and enzyme activity levels in HepG2 cells. Furthermore, the role of the AhR signaling pathway was further studied using the XRE-luciferase activity and electrophoretic mobility shift assay (EMSA). The binding ability of harmaline and harmalol to the AhR was tested using competitive ligand binding assay. Finally, we investigated the posttranscriptional and posttranslational effects of harmaline and harmalol on CYP1A1 in HepG2 cells.
2. Material and methods
2.1. Chemicals and reagents
Cycloheximide (CHX), 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide (MTT), 7-ethoxyresorufin (7ER), fluorescamine, benzo(a)pyrene (BaP), resveratrol (Res; 99% pure) and rabbit anti-goat IgG secondary antibody were purchased from Sigma-Aldrich (St. Louis, MO). 2,3,7,8-Tetrachlorodibenzo-p-dioxin (TCDD), >99% pure, was obtained from Cambridge Isotope Laboratories (Woburn, MA). TRIzol and Lipofectamine 2000 reagents were purchased from Invitrogen (Carlsbad, CA). Primary mouse anti-human CYP1A1 antibody, primary goat anti-human glyceraldehyde-3-phosphate dehydrogenase (GAPDH) antibody and goat anti-ARNT antibody were purchased from Santa Cruz (Santa Cruz, CA). Goat anti-mouse IgG secondary antibody was obtained from R&D systems (Minneapolis, MN). High-Capacity cDNA Reverse Transcription Kit and SYBR® Green PCR Master Mix were purchased from Applied Biosystems (Foster City, CA). Harmaline hydrochloride dihydrate (>90% pure) was supplied by ACROS Organics (Morris Plains, NJ). Harmalol hydrochloride (>98% pure) was obtained from MP Biomedicals (Solon, OH). Carbobenzoxy-l-leucyl-l-leucyl-leucinal (MG-132) and actinomycin-D (Act-D) were purchased from Calbiochem (San Diego, CA). Acrylamide, N'N'-bis-methylene-acrylamide, ammonium persulphate, glycine, nitrocellulose membrane (0.45 µm), and TEMED were purchased from Bio-Rad Laboratories (Hercules, CA). Chemiluminescence Western blotting detection reagents were obtained from GE Healthcare Life Sciences (Piscataway, NJ). [γ-32P]-ATP was supplied by Perkin Elmer (Boston, MA). 2,3,7,8-Tetrachlorodibenzofuran (TCDF) and [3H]-TCDD (13 Ci/mmole) were obtained from Dr. Stephen Safe (Texas A&M University). Dual-Luciferase Reporter Assay System was obtained from Promega Corporation (Madison, WI). All other chemicals were purchased from Fisher Scientific (Toronto, ON). Real time-PCR primers were synthesized by Integrated DNA Technologies Inc. (San Diego, CA) according to previously published sequences.
2.2. Cell culture
Human hepatoma HepG2 cell line was purchased from American Type Culture Collection (Manassas, VA). Cells were maintained in Dulbecco's modified Eagle's medium, supplemented with heat-inactivated fetal bovine serum (10%, v/v), L-glutamine (2 mM), penicillin (100 IU/mL) and streptomycin (100 µg/mL). Cells were grown in 75-cm2 tissue culture flasks at 37°C in a 5% CO2 humidified incubator.
2.3. Chemical treatments
Cells were treated in serum-free medium with TCDD in presence of various concentrations of harmaline or harmalol as specified under each experiment. Harmaline, harmalol, TCDD, MG-132 and Act-D were dissolved in dimethyl sulfoxide (DMSO), whereas CHX was dissolved in sterile distilled water. In all treatments, the DMSO concentration did not exceed 0.05% (v/v).
2.4. Animals and ethics
All experimental procedures involving animals were approved by the University of Alberta Health Sciences Animal Policy and Welfare Committee. Male Hartley guinea pigs weighing 250–300 g were obtained from Charles River Canada (St. Constant, QC, Canada). All animals were exposed to 12 h light/dark cycles and were allowed free access to food and water.
2.5. Determination of cell viability
The effect of harmaline and harmalol on HepG2 cell viability was determined by measuring the capacity of reducing enzymes present in viable cells to convert MTT to formazan crystals as described previously (Mosmann, 1983).
2.6. RNA isolation and real-time polymerase chain reaction (Real-time PCR)
HepG2 cells were pre-incubated with increasing concentrations of harmaline or harmalol (0.5–12.50 µM) for 30 min before addition of TCDD (1 nM) for 6 h. Thereafter, the total RNA was isolated using TRIzol reagent, according to the manufacturer’s instructions (Invitrogen) as described previously (El Gendy and El-Kadi, 2009). Primers used in the current study were chosen from previous study (Anwar-Mohamed and El-Kadi, 2009): human CYP1A1: forward primer 5′-CGG CCC CGG CTC TCT-3′, reverse primer 5′-CGG AAG GTC TCC AGG ATG AA-3′, and human β-actin: forward primer 5′-CTG GCA CCC AGC ACA ATG-3′, reverse primer 5′-GCC GAT CCA CAC GGA GTA CT-3′. Real-time PCR reactions were performed on an ABI 7500 instrument (Applied Biosystems), using SYBR® Green PCR Master Mix as described previously (El Gendy and El-Kadi, 2009).
2.7. Protein extraction and Western blot analysis
HepG2 cells were treated with increasing concentrations of harmaline or harmalol (0.5–12.5 µM) for 30 min before the addition of TCDD (1 nM) for further 24 h. Thereafter, cells were collected and the cell lysate was obtained using lysis buffer. Proteins (50 µg) in sample buffer were separated using 10% SDS-PAGE and electrophoretically transferred to a nitrocellulose membrane. The bands were visualized with the enhanced chemiluminescence method according to the manufacturer's instructions (GE Healthcare, Piscataway, NJ). The intensity of CYP1A1 protein bands was quantified relative to the signals obtained for GAPDH protein, using a densitometer (TBX, Tobias associates, Inc., PA).
2.8. Determination of CYP1A1 enzymatic activity
CYP1A1-dependent 7-ethoxyresorufin O-deethylase (EROD) activity was performed on intact, living cells using 7ER as a substrate as described previously (Sinal and Bend, 1997). The CYP1A1 enzymatic activity was normalized to cellular protein content using a modified fluorescence method (Lorenzen and Kennedy, 1993).
2.9. Transient transfection and luciferase assay
HepG2 cells (3×104 cells per well) were plated onto 12-well cell culture plates. Each well was co-transfected with XRE-driven luciferase reporter plasmid pGudLuc 6.1 (1.5 µg) and the renilla luciferase pRL-CMV vector (0.1 µg), used for normalization of transfection efficiency. The pRL-CMV vector was obtained from Promega Corporation (Madison,WI). Transfection procedure was carried out using Lipofectamine 2000 reagent according to the manufacturer's instructions (Invitrogen), the luciferase assay was performed according to the manufacturer's instructions (Promega) and luciferase activity was reported as relative light unit (RLU) of firefly luciferase to renilla luciferase (Fluc/Rluc).
2.10. Electrophoretic mobility shift assay (EMSA)
Guinea pig hepatic cytosolic extracts (2 mg) were incubated with harmaline or harmalol (250 µM) for 30 min before the addition of TCDD (20 nM) for 2 h. The ability of harmaline or harmalol to affect the transformation and DNA binding of TCDD-activated AhR was tested using a synthetic pair of post-labeled oligonucleotides for XRE binding site as described previously (Denison et al., 2002).
2.11. Competitive ligand binding assay
Hydroxyapatite (HAP) assay was performed to determine the ligand binding ability of harmaline and harmalol as described previously (Denison et al., 1986). Breifly, untreated guinea pig hepatic cytosol was diluted to 2 mg/mL in MEDG buffer (3-(N-morpholino) propanesulfonic acid (25 mM), pH 7.5, ethylenediaminetetraacetic acid (1 mM), dithiotreitol (1 mM), and glycerol (10%, v/v). Several aliquots of guinea pig cytosols (200 µL) were incubated at room temperature for 1 h with [3H]TCDD (2 nM) alone (total binding), [3H]TCDD (2 nM), and TCDF (200 nM, 100-fold excess of competitor, nonspecific binding) or [3H]TCDD (2 nM) in the presence of increasing concentrations of harmaline or harmalol (25 µM and 50 µM). All chemicals were dissolved in DMSO, in which DMSO content in reactions was adjusted to 2% (v/v) where necessary. Thereafter, hydroxyapatite suspension (250 µL) was added to the different reaction mixtures and incubated for an additional 30 min with gentle vortexing every 10 min. The reactions were washed three times with 1 mL of MEGT buffer (3-(N-morpholino) propanesulfonic acid (25 mM), pH 7.5, ethylenediaminetetraacetic acid (1 mM), glycerol (10%, v/v), and Tween 80 (0.5%, v/v). The HAP pellets were transferred to 4 mL scintillation vials, scintillation cocktail was added, and reactions were counted in a scintillation counter.
2.12. CYP1A1 mRNA stability
The half-life of CYP1A1 mRNA was analyzed by the Act-D chase assay. HepG2 cells were pre-treated with TCDD (1 nM) for 6 h. Thereafter, cells were washed and incubated with Act-D (5 µg/mL), to inhibit further RNA synthesis, immediately before treatment with harmaline (2.5 µM) or harmalol (2.5 µM). Total RNA was extracted at 0, 1, 3, 6, and 12 h after incubation with harmaline or harmalol. Thereafter, real-time PCR was carried out as described previously (El Gendy and El-Kadi, 2009). The mRNA half-life values were determined from semilog plots of mRNA amounts, expressed as percentage of treatment at t = 0, versus time.
2.13. CYP1A1 protein stability
The half-life of CYP1A1 protein was analyzed by the CHX chase assay. HepG2 cells were pre-treated with TCDD (1 nM) for 24 h. Cells were then washed three times with PBS and incubated with CHX (10 µg/mL), to inhibit further protein synthesis, immediately before treatment with harmaline (2.5 µM) or harmalol (2.5 µM). Cell homogenates were extracted at 0, 1, 3, 6, and 12 h after incubation with harmaline or harmalol. CYP1A1 protein was measured by Western blotting as described previously (El Gendy and El-Kadi, 2010). The protein half-life values were determined from semilog plots of integrated densities versus time.
2.14. Direct inhibitory study
The direct inhibitory effect of harmaline and harmalol on CYP1A1 enzyme was determined using a method similar to that described for EROD assay, with slight modifications. Briefly, HepG2 cells were incubated with 1nM TCDD for 24 h. Thereafter, media were removed from the cells, washed three times with PBS, increasing concentrations of harmaline and harmalol (0.5–12.5 µM) in assay buffer (Tris (0.05 M), NaCl (0.1 M), pH 7.8) were added to the cells for 15 min prior to the addition of 7ER (2 µM final concentration) as a substrate for EROD measurement and normalized for cellular protein content.
2.13. Statistical analysis
All results are presented as mean ± S.E.M., and statistical differences between treatment groups were determined using one way ANOVA followed by Student-Newman-Keuls post hoc test using SigmaStat 3.5 program for Windows, Systat Software Inc. (San Jose, CA).
3. Results
3.1. Effect of harmaline and harmalol on cell viability
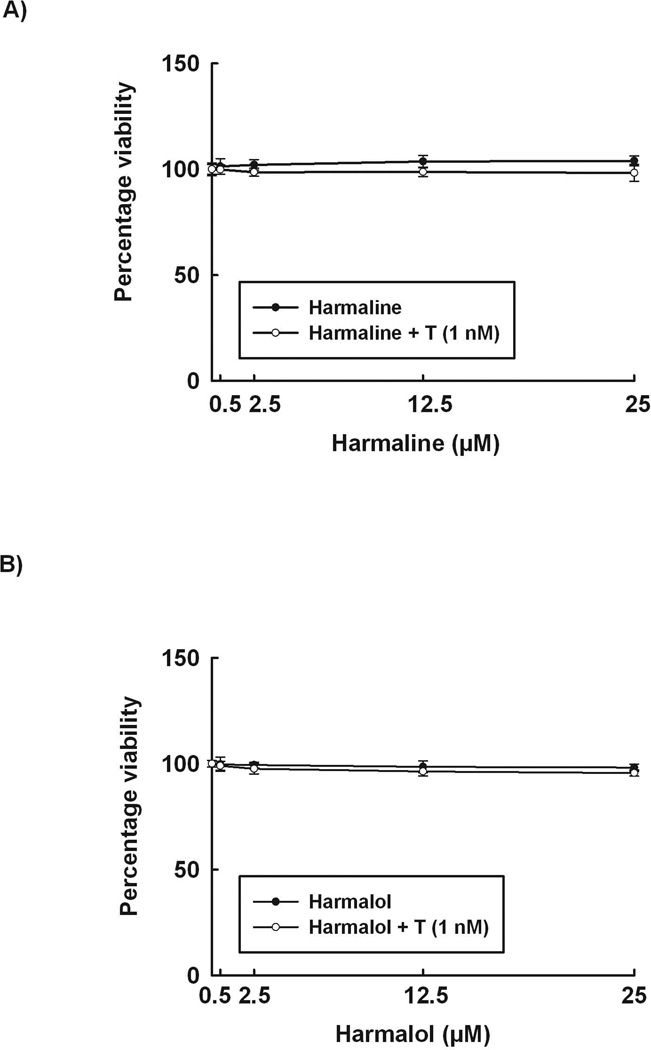
To determine the nontoxic concentrations that will be used in our study, increasing concentrations of harmaline and harmalol were incubated with HepG2 cells in presence and absence of TCDD for 24 h. Our results demonstrate that neither harmaline nor harmalol (0–25 µM) significantly affected cell viability when incubated with human hepatoma cells for 24 h either in presence or absence of TCDD (Fig. 2).
Figure 2. Effect of harmaline and harmalol on HepG2 cell viability.
The effect of different concentrations of harmaline (A) and harmalol (B) on HepG2 cell viability was tested using the MTT assay. Data are expressed as percent of control, which is set at 100%, ± S.E.M. (n = 5). (+) P < 0.05 compared with control (C), (*) P < 0.05 compared with TCDD (T).
3.2. Effect of harmaline on dioxin-induced CYP1A1 mRNA, protein, and activity levels in HepG2 cells
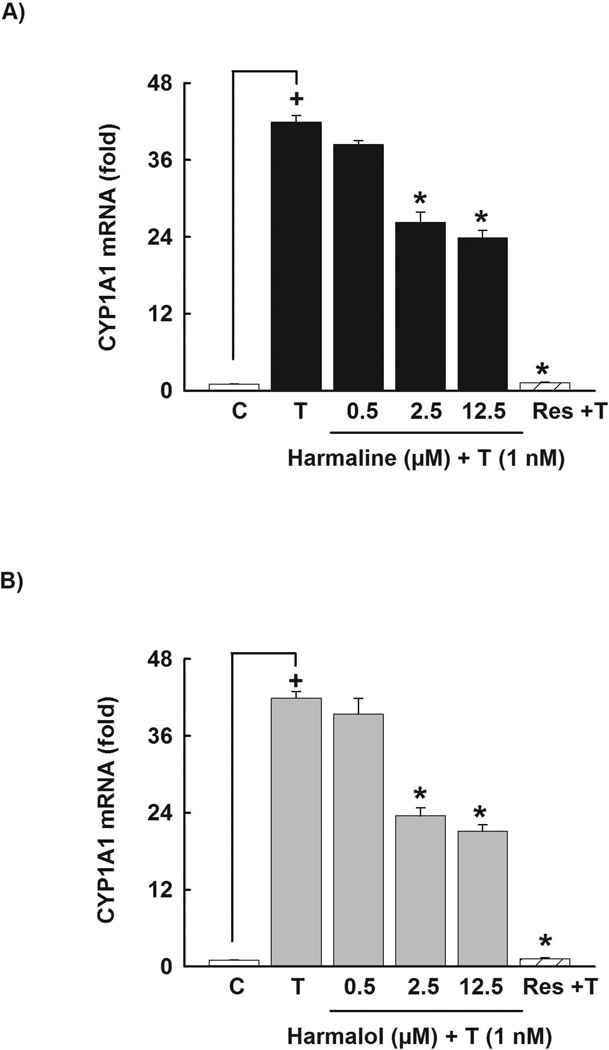
To investigate the ability of harmaline to alter the CYP1A1 mRNA level, HepG2 cells were pre-incubated with increasing concentrations of harmaline (0.5–12.50 µM) for 30 min before the addition of TCDD for 6 h. Real-time PCR was employed to quantify CYP1A1 mRNA level. As shown in Fig 3A, harmaline significantly decreased the TCDD-mediated induction of CYP1A1 mRNA in a concentration-dependent manner by 28% and 43% with harmaline concentrations of 2.5 and 12.5 µM, respectively (Fig. 3A). The positive control, resveratrol (12.5 µM) significantly decreased the TCDD-mediated induction of CYP1A1 mRNA by 97% (Fig. 3A).
Figure 3. Effect of harmaline and harmalol on CYP1A1 mRNA in HepG2 cells.
Cells were pre-incubated with increasing concentrations of harmaline (A) or harmalol (B) for 30 min before the addition of TCDD (1nM) for an additional 6 h. The amount of CYP1A1 mRNA was quantified using real-time PCR and normalized to β-actin housekeeping gene. Resveratrol (Res, 12.5 µM) was used as a positive control. Values represent the mean of fold change ± S.E.M. (n=4). (+) P < 0.05 compared with control (C), (*) P < 0.05 compared with TCDD (T).
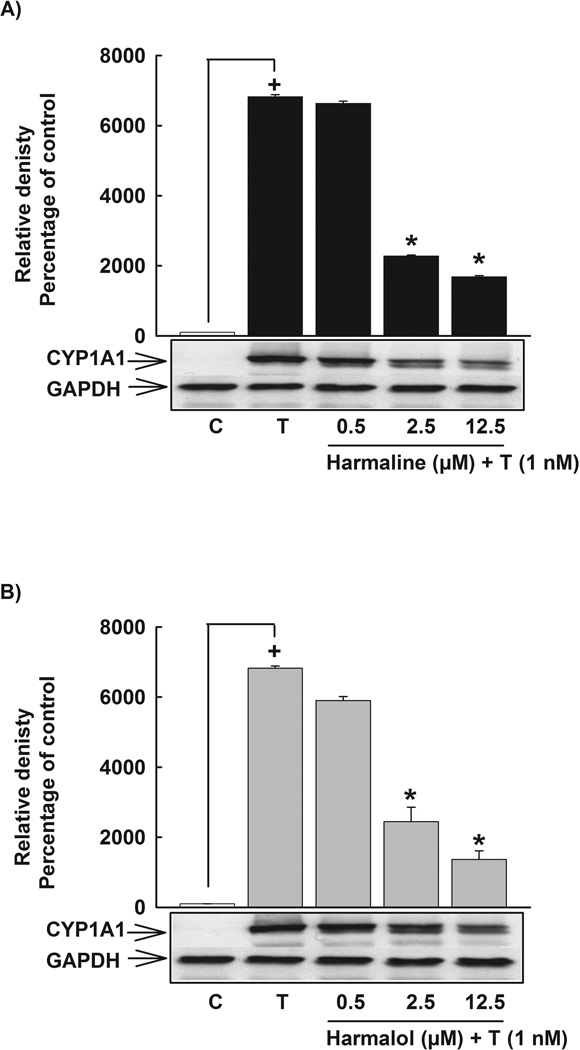
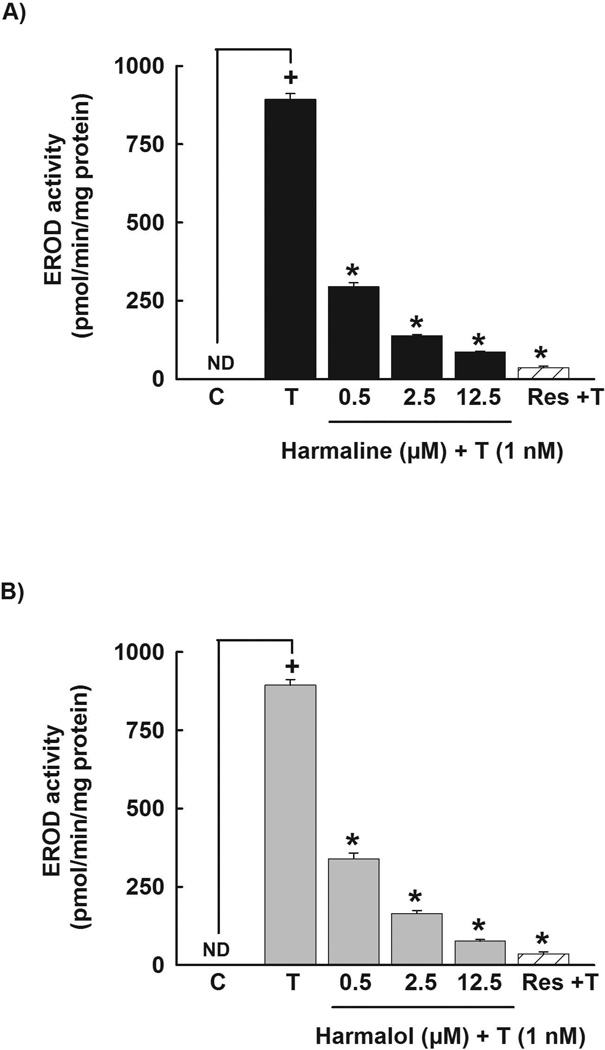
In order to assess the effect of harmaline on CYP1A1 protein level, HepG2 cells were pre-incubated with increasing concentrations of harmaline (0.5–12.5 µM) for 30 min before the incubation with TCDD (1 nM) for 24 h. Thereafter, the level of CYP1A1 protein was measured using Western blot analysis. Our results showed that harmaline significantly decreased the TCDD-mediated induction of CYP1A1 protein in a concentration-dependent manner, where it showed 66% and 76% decrease in CYP1A1 protein with harmaline concentrations of 2.5 and 12.5 µM, respectively (Fig. 4A). To test whether the effect of harmaline is translated to the CYP1A1 catalytic activity, HepG2 cells were pre-incubated with increasing concentrations of harmaline (0.5–12.5 µM) for 30 min before the incubation with TCDD (1 nM) for 24 h. Thereafter, CYP1A1 catalytic activity was determined using EROD assay. Our results showed that harmaline significantly decreased the TCDD-mediated induction of the CYP1A1 catalytic activity by 67%, 80% and 90% with harmaline concentrations of 0.5, 2.5 and 12.5 µM, respectively (Fig. 5A). Resveratrol (12.5 µM) significantly decreased the TCDD-mediated induction of CYP1A1 catalytic activity by 96% (Fig. 5A).
Figure 4. Effect of harmaline and harmalol on CYP1A1 protein in HepG2 cells.
Cells were pre-incubated with increasing concentrations of harmaline (A) or harmalol (B) for 30 min before the addition of TCDD (1nM) for an additional 24 h. Protein was separated on a 10% SDS-PAGE and CYP1A1 protein was detected using the enhanced chemiluminescence method. The intensity of bands was normalized to GAPDH signals, which was used as loading control. One of three representative experiments is shown. (+) P < 0.05 compared with control (C), (*) P < 0.05 compared with TCDD (T).
Figure 5. Effect of harmaline and harmalol on TCDD-mediated induction of CYP1A1 catalytic activity in HepG2 cells.
Cells were pre-incubated with increasing concentrations of harmaline (A) or harmalol (B) for 30 min before the addition of TCDD (1nM) for an additional 24 h. The amount of CYP1A1 activity was determined using CYP1A1-dependent EROD assay. Resveratrol (Res, 12.5 µM) was used as a positive control. Values represent mean activity ± S.E.M. (n = 8). (+) P < 0.05 compared with control (C), (*) P < 0.05 compared with TCDD (T).
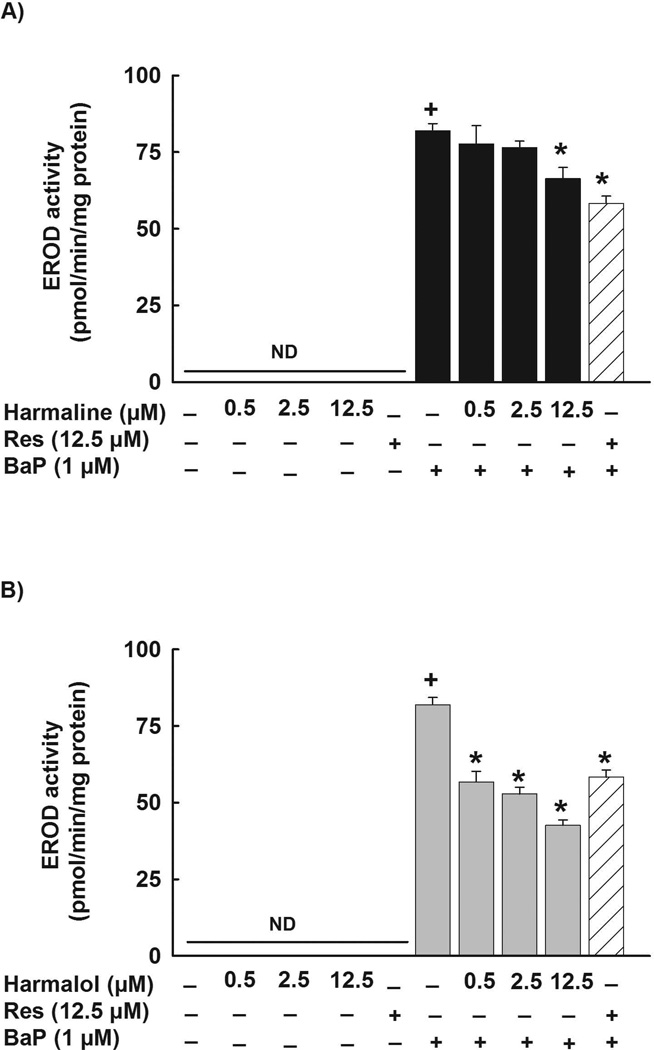
To test whether the inhibitory effects of harmaline are not AhR ligand specific, HepG2 cells were pre-incubated with increasing concentrations of harmaline before the addition of BaP (1 µM). Thereafter, CYP1A1 catalytic activity was determined using 7ER as a substrate. Fig.6A shows that harmaline decreased the BaP-mediated induction of the CYP1A1 catalytic activity by 5%, 7%, and 20% with harmaline concentrations of 0.5, 2.5 and 12.5 µM, respectively (Fig. 6A). However, the effect was only significant at the highest concentration tested, 12.5 µM (Fig. 6A). Furthermore, resveratrol (12.5 µM) significantly decreased the BaP-mediated induction of CYP1A1 catalytic activity by 29% (Fig. 6A). In addition, harmaline alone did not affect the constitutive CYP1A1 at the catalytic activity level in HepG2 cells (Fig. 6A).
Figure 6. Effect of harmaline and harmalol on BaP-mediated induction of CYP1A1 catalytic activity in HepG2 cells.
Cells were incubated with increasing concentrations of harmaline (A) or harmalol (B) in the absence and presence of BaP (1 µM) for 24 h. The amount of CYP1A1 activity was determined using CYP1A1-dependent EROD assay. Resveratrol (Res, 12.5 µM) was used as a positive control. Values represent mean activity ± S.E.M. (n = 8). (+) P < 0.05 compared with control (C), (*) P < 0.05 compared with BaP.
3.3. Effect of harmalol on dioxin-induced CYP1A1 mRNA, protein, and activity levels in HepG2 cells
In an attempt to explore whether the effect of harmaline is due to its active metabolite, we examined the effect of harmalol on the CYP1A1 mRNA level in human HepG2 cells. Our results showed that harmalol significantly decreased the TCDD-mediated induction of CYP1A1 mRNA in HepG2 cells in a concentration-dependent manner by 43% and 50% with harmalol concentrations of 2.5 and 12.5 µM, respectively (Fig. 3B). Resveratrol (12.5 µM) was used as a positive control, where it showed a significant decrease of TCDD-mediated induction of CYP1A1 mRNA by 97% (Fig. 3B).
Consistent with the CYP1A1 mRNA results, harmalol significantly decreased the TCDD-mediated induction of CYP1A1 at the protein level by 64% and 80% with harmalol concentrations of 2.5 and 12.5 µM, respectively (Fig. 4B). At the CYP1A1 activity level, harmalol showed a higher effect, where it significantly decreased the induction level of CYP1A1 catalytic activity by 62%, 82% and 91% with harmalol concentrations of 0.5, 2.5 and 12.5 µM, respectively (Fig. 5B). Resveratrol (12.5 µM) significantly decreased the TCDD-mediated induction of CYP1A1 catalytic activity by 96% (Fig. 5B). Furthermore, harmalol significantly decreased the BaP-mediated induction of CYP1A1 catalytic activity in HepG2 cells in a concentration-dependent manner by 31%, 36% and 48% with harmalol concentrations of 0.5, 2.5 and 12.5 µM, respectively (Fig. 6B). Furthermore, resveratrol (12.5 µM) decreased the BaP-mediated induction of CYP1A1 by 29% (Fig. 6B). In addition, neither harmalol (0.5–12.5 µM) nor resveratrol (12.5 µM) alone significantly increased the constitutive CYP1A1 at the catalytic activity level in HepG2 cells (Fig. 6B).
3.4. Transcriptional effects of harmaline and harmalol on CYP1A1 gene
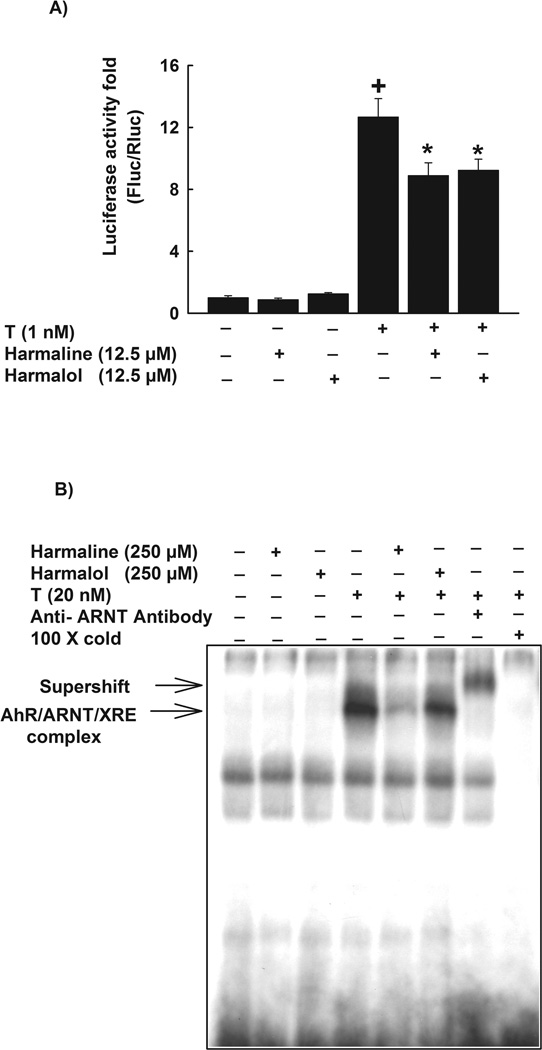
To investigate the underlying mechanisms for the effect of harmaline and harmalol on CYP1A1, we tested the role of transcriptional mechanism. For this purpose, HepG2 cells were transiently co-transfected with the XRE-driven luciferase reporter gene and renilla luciferase vector, which was used for normalization of transfection efficiency. Our results demonstrated that neither harmaline nor harmalol alone significantly affected the level of luciferase activation relative to control (Fig. 7A). On the other hand, TCDD alone significantly induced the luciferase activity by 1300% as compared with the control. Of interest, harmaline and harmalol significantly decreased the TCDD-induced luciferase activity by 30% and 27%, respectively (Fig. 7A).
Figure 7. Effect of harmaline and harmalol on XRE-luciferase activity and electrophoretic mobility shift assay (EMSA).
(A) HepG2 cells were transiently co-transfected with XRE-luciferase reporter plasmid pGudLuc 6.1. and renilla luciferase control plasmid pRL-CMV. Cells were treated with vehicle, harmaline (12.5 µM) or harmalol (12.5 µM) for 30 min before the addition of TCDD (1nM) for an additional 24 h. Cells were lysed and luciferase activity is reported as relative light unit (RLU) of firefly luciferase to renilla luciferase (Fluc/Rluc) (mean ± S.E.M., n = 4). (+) P < 0.05 compared with control (C), (*) P < 0.05 compared with TCDD (T). (B) In vitro AhR activity was measured by EMSA using guinea pig hepatic cytosolic extracts. Cytosolic extracts were pre-incubated with vehicle, harmaline (250 µM) or harmalol (250 µM) for 30 min before the addition of TCDD (20 nM) for 2 h. The mixtures were tested for binding activity to a [γ-32P]-labeled XRE consensus oligonucleotide for an additional 15 min. The products of this binding were separated on a 4% polyacrylamide gel. The specificity of the binding was confirmed by competition assays using anti-ARNT antibody or a 100 fold molar excess of unlabeled XRE. AhR-ARNT-XRE complex formed on the gel was visualized by autoradiography. One representative of three experiments is shown.
In an attempt to elucidate the direct effect of harmaline and harmalol on AhR activation and the subsequent DNA binding to XRE, EMSA was performed on untreated guinea pig hepatic cytosol incubated either with vehicle, harmaline or harmalol in the absence and presence of TCDD (20 nM) for 2 h. Similar to luciferase results, both harmaline and harmalol (250 µM) alone did not alter the AhR activity (Fig. 7B). TCDD (20 nM) alone induced the AhR activity and the formation of AhR/ARNT/XRE complex. On the other hand, pre-incubation of cytosolic extracts with harmaline or harmalol significantly inhibited the TCDD-mediated activation of AhR and the formation of AhR/ARNT/XRE complex (Fig. 7B). The specificity of the binding was confirmed by the competition assays using anti-ARNT antibody or a 100 fold molar excess of unlabeled XRE (Fig. 7B).
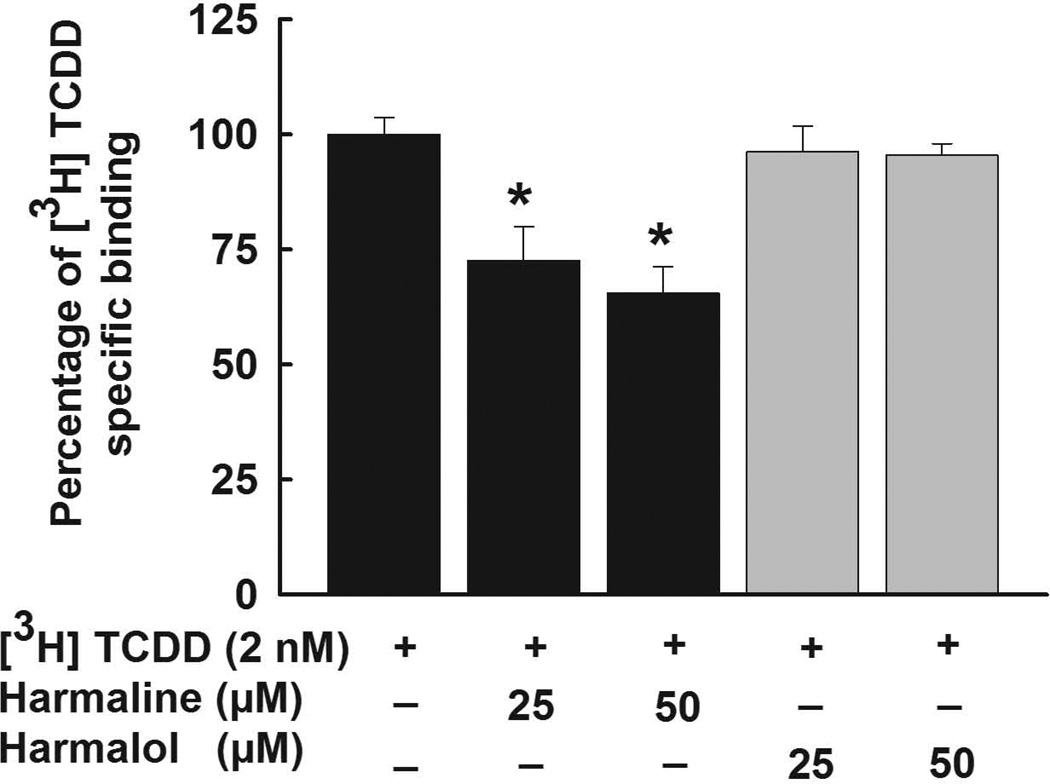
To examine whether harmaline or harmalol are direct ligands for the AhR, a ligand competition binding assay using hydroxyapatite was performed using untreated guinea pig hepatic cytosols (Fig. 8). In this assay the total binding is the overall binding of [3H]-TCDD to cytosolic AhR protein. However, to account for the non-specific binding that happens not through the AhR or not through the ligand-binding center of the AhR, reactions were conducted in the presence of 100-fold excess of the competitor. We have chosen TCDF rather than TCDD because of its higher solubility as TCDD would not be soluble at 200 nM. Thus, the specific binding of [3H]-TCDD to the AhR was calculated by subtracting the non-specific binding from the total binding. Our results showed that harmaline at concentrations of 25 µM and 50 µM was able to significantly displace [3H]-TCDD (2 nM) by 28% and 35%, respectively (Fig. 8). On the other hand, harmalol did not show a significant displacement of [3H]-TCDD (Fig. 8).
Figure 8. AhR ligand binding ability of harmaline and harmalol.
Untreated guinea pig hepatic cytosol (2 mg/mL) was incubated with [3H]-TCDD (2 nM) alone (total binding), [3H]-TCDD (2 nM) and TCDF (200 nM, 100-fold excess of competitor, nonspecific binding), or [3H]-TCDD (2 nM) in the presence of increasing concentrations of harmaline or harmalol (25, and 50 µM) and the samples analyzed by the hydroxyapatite assay as described under Material and Methods. Values were adjusted for nonspecific binding and expressed as % specific binding relative to the absence of a competitor ligand. Values are presented as the mean ± S.E.M. (n = 9). (*) P < 0.05 compared with [3H]-TCDD.
3.5. Posttranscriptional modification of CYP1A1 mRNA by harmaline and harmalol
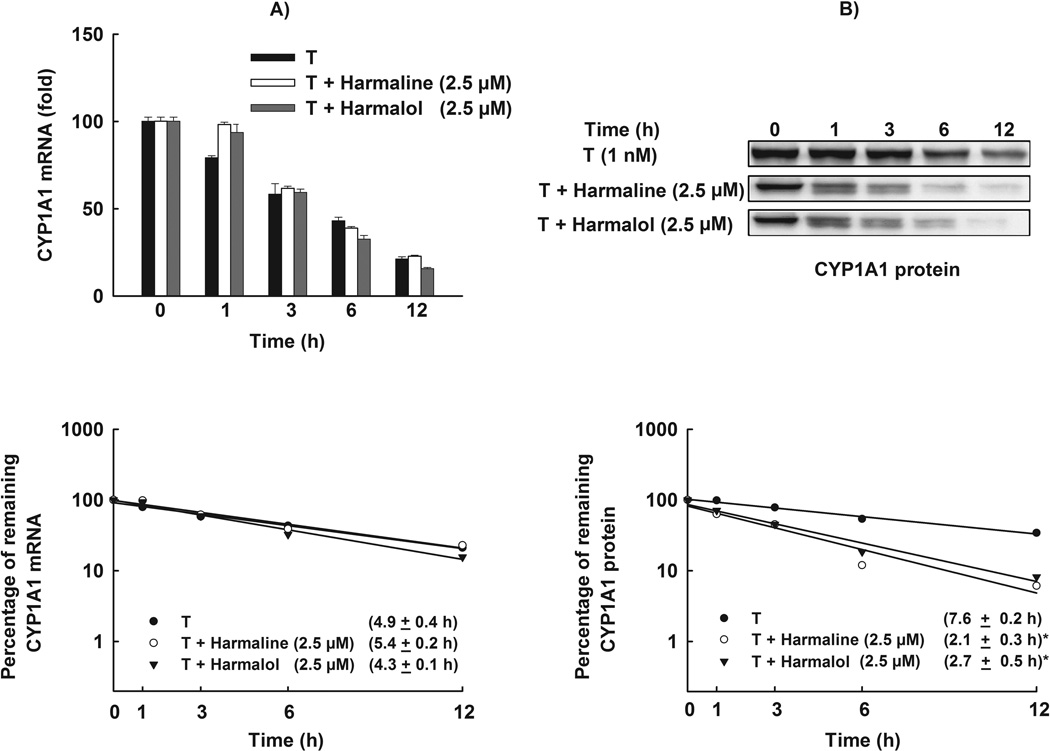
The effect of a substance on the mRNA expression is a function of both its effect on the transcription rate and the elimination rate through processing or degradation. Therefore, we tested the effect of harmaline and harmalol on the stability of CYP1A1 mRNA transcripts in HepG2 cells, using the Act-D chase experiment. If the effect of harmaline or harmalol involves stabilization of CYP1A1 mRNA, an increase in the half-life would be expected. Our results showed that TCDD-induced CYP1A1 mRNA degraded with a half-life of 4.9 ± 0.4 h (Fig. 9A). Moreover, treatment with harmaline or harmalol did not significantly alter CYP1A1 mRNA half-life which was 5.4 ± 0.2 h and 4.3 ± 0.1 h, respectively (Fig. 9A).
Figure 9. Effect of harmaline and harmalol on CYP1A1 mRNA and protein stability.
HepG2 cells were treated with TCDD (1 nM) for 6 h for mRNA stability and 24 h for protein stability assays. Thereafter, the cells were washed and incubated with fresh media containing harmaline (2.5 µM) or harmalol (2.5 µM) plus Act-D (5 µg/mL, the mRNA synthesis inhibitor) or CHX (10 µg/mL, the protein translation inhibitor). (A) Total RNA was extracted at 0, 1, 3, 6 and 12 h after incubation with harmaline or harmalol and subjected to real-time PCR. (B) Protein was separated on a 10% SDS-PAGE and CYP1A1 protein was detected using the enhanced chemiluminescence method. The intensities of CYP1A1 protein bands were normalized to GAPDH signals, which were used as loading controls (data not shown). mRNA and protein decay curves were analyzed individually, and the half-life was estimated from the slope of a straight line fitted by linear regression analysis (r2 ≥ 0.85) to a semilog plot, expressed as a percent of treatment at time = 0 h (maximum, 100%) level, versus time. The half-lives obtained from three independent experiments were then used to calculate the mean half-life (mean ± S.E.M., n = 3). (*) P < 0.05 compared with TCDD (T).
3.6. Posttranslational modification of CYP1A1 protein by harmaline and harmalol
The effect of harmaline and harmalol on the protein and the activity levels was much higher than that obtained with the mRNA level which raises the possibility of posttranslational modification. Therefore, we tested the effect of harmaline and harmalol on CYP1A1 protein stability in HepG2 cells using CHX chase experiment. Our results showed that CYP1A1 protein induced by TCDD degraded with a half-life of 7.6 ± 0.2 h (Fig. 9B). Furthermore, harmaline and harmalol significantly reduced the stability of CYP1A1 protein which degraded with half-lives of 2.1 ± 0.3 h and 2.7 ± 0.5 h, respectively (Fig. 9B).
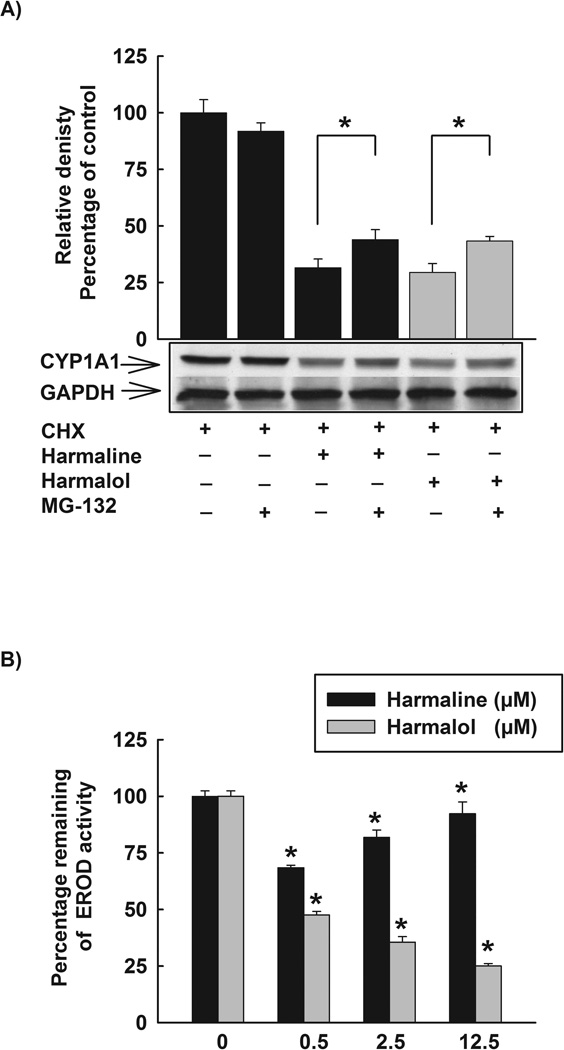
To further investigate the underlying mechanisms of the posttranslational modifications of harmaline and harmalol, we tested the role of ubiquitin-proteasomal pathway. Therefore, HepG2 cells were treated with TCDD (1 nM) for 24 h, thereafter, cells were washed three times with PBS and incubated with fresh media containing CHX (10 µg/mL) and DMSO, CHX (10 µg/mL) and harmaline (2.5 µM) or CHX (10 µg/mL) and harmalol (2.5 µM) in absence and presence of the proteasomal inhibitor, MG-132 (0.5 µM). After 6 h incubation, total protein was extracted and CYP1A1 protein was detected using Western blot analysis. Fig. 10A shows that MG-132 alone did not significantly alter CYP1A1 protein level. On the other hand, MG-132 significantly induced the level of CYP1A1 protein for both harmaline and harmalol treated cells.
Figure 10. Posttranslational modifications of CYP1A1 by harmaline and harmalol.
(A) Effect of proteasomal inhibitor, MG-132, on the reduced CYP1A1 protein stability by harmaline and harmalol. HepG2 cells were treated with TCDD (1 nM) for 24 h. Thereafter, cells were washed three times with PBS and incubated with fresh media containing CHX (10 µg/mL, the protein translation inhibitor), CHX (10 µg/mL) and harmaline (2.5 µM) or CHX (10 µg/mL) and harmalol (2.5 µM) in the absence and presence of the proteasomal inhibitor, MG-132 (0.5 µM). After 6 h incubation, total protein was extracted and CYP1A1 protein was detected using Western blot analysis. The intensities of CYP1A1 protein bands were normalized to GAPDH signals, which were used as loading controls. Values represent mean of relative densities and expressed as percentage of control (CHX and DMSO treated cells) ± S.E.M. One of three representative experiments is shown. (*) P < 0.05 compared with the relevant treatment. (B) The direct inhibitory effects of harmaline and harmalol on CYP1A1 enzyme. HepG2 cells were pre-treated with TCDD (1nM) for 24 h, thereafter media were removed, washed three times with PBS, and increasing concentrations of harmaline and harmalol (0.5–12.5 µM) in assay buffer (Tris (0.05 M), NaCl (0.1 M), pH 7.8) were added for 15 min prior to the addition of 7ER (2 µM final concentration) for the EROD measurement. Results are expressed as percentage of remaining EROD activity (mean ± S.E.M, n = 8). (*) P < 0.05 compared with control.
Furthermore, the direct effect of harmaline or harmalol on CYP1A1 enzyme was determined. In this regard, HepG2 cells were treated for 24 h with TCDD (1 nM), thereafter, the cells were washed twice with PBS and increasing concentrations of harmaline or harmalol in assay buffer were further incubated for 15 min before the addition of the substrate (7ER, 2 µM final concentration). The remaining CYP1A1 activity was detected using EROD assay. Fig. 10B shows that harmaline and harmalol possess direct inhibitory effects on CYP1A1 enzyme, where harmaline significantly inhibited the CYP1A1 activity by 32, 18, and 8% with harmaline concentrations of 0.5, 2.5, and 12.5 µM, respectively (Fig. 10B). In addition, harmalol significantly inhibited the CYP1A1 activity by 53, 65, and 75% with harmalol concentrations of 0.5, 2.5, and 12.5 µM, respectively (Fig. 10B).
4. Discussion
To our knowledge, the present study demonstrates for the first time that harmaline and its main metabolite, harmalol, significantly inhibit the dioxin-mediated induction of CYP1A1 at the transcriptional and posttranslational levels.
Most of the chemical carcinogens in the environment are chemically inert by themselves and require metabolic activation by cytochrome P450 (CYP) enzymes to more reactive metabolites in order to exhibit carcinogenicity in experimental animals and humans (Shimada and Fujii-Kuriyama, 2004). Several studies correlated the exposure to environmental pollutants with the incidence of several human cancers such as colon, lung and kidney (Czene et al., 2002; Luch, 2005). A number of PAH and HAH present as common environmental pollutants in cigarette smoke, coal tar and grilled meat can bind to and activate AhR. The activated AhR can result in several biological and toxic effects that depend on the type of AhR ligand (Bradshaw and Bell, 2009). The effect of AhR ligands on cell cycle, inflammation, and cancer cell proliferation raised the therapeutic potential for activators/inhibitors of the AhR signaling pathway (Bradshaw and Bell, 2009; Zhao et al., 2010). Dioxins are metabolically stable AhR ligands that produce a spectrum of TCDD-like AhR-dependent toxicity and carcinogenicity (Mandal, 2005). TCDD was classified as human carcinogen since 1997 by the International Agency for Research on Cancer (IARC) (Mandal, 2005). Follow-up studies of the exposed populations to TCDD revealed the increased incidence of several human cancers, including liver cancer, and their mortality both in men and women (Mandal, 2005). Accordingly, AhR has been used as a target for screening of new chemopreventative agents (Puppala et al., 2008). Numerous AhR antagonists have shown promising results against several carcinogenic agents. It has been previously reported that the genotoxicity associated with BaP in mice was inhibited by AhR antagonists such as 3'-methoxy-4'-nitroflavone and resveratrol (Dertinger et al., 2001; Revel et al., 2003). However, several AhR antagonists lack specificity and can act as partial agonists, therefore, the search for new AhR antagonist is still in progress (Puppala et al., 2008; Signorelli and Ghidoni, 2005; Zhou and Gasiewicz, 2003).
Harmaline and harmalol are common dihydro-β-carbolines that widely distributed in the environment including alcoholic beverages and several plants such as Peganum harmala (Zygophyllaceae) (Herraiz et al., 2010; Park et al., 2010). Harmaline and harmalol possess antioxidant and hydroxyl radical-scavenging properties and they are more active than their aromatic β-carboline analogues harmine and harmol (Moura et al., 2007; Tse et al., 1991). Moreover, they inhibit lipid peroxidation in microsomal hepatic preparations (Tse et al., 1991). The antioxidant activity of harmaline and harmalol depends mainly on the stabilization of the formed radical by several resonance structures (Moura et al., 2007; Tse et al., 1991). The antioxidant properties of harmaline and harmalol were postulated to be the main cause of their antimutagenic and antigenotoxic effects in yeast and mammalian cells, respectively (Moura et al., 2007). Of particular note, several AhR ligands cause the induction of CYP1A1 leading to higher level of oxidative stress. In this context, it has been previously reported that oxidative stress might participate in the mutagenic effect of several AhR ligands such as TCDD (Matsumura, 2003; Yoshida and Ogawa, 2000). Therefore, the antioxidant properties of harmaline and harmalol can participate indirectly in the protective effects of harmaline and harmalol against dioxin-induced toxicity including carcinogenicity. However, the direct effect of harmaline and harmalol on dioxin-induced CYP1A1 has not been studied before.
In the present study, we tested the effect of harmaline and its main metabolite, harmalol, on the dioxin-induced CYP1A1 enzyme in human hepatoma HepG2 cells. The use of harmaline and its main metabolite was carried out for several reasons. First, to test whether or not the effect of harmaline on dioxin-mediated induction of CYP1A1 is due to metabolic activation. Our results showed that both the drug and its metabolite possess similar inhibitory effects on CYP1A1 in a concentration-dependent manner at mRNA, protein and activity levels. The second reason for testing harmalol is the neuropharmacological side effects of harmaline (Abe and Yamada, 2009). Finally, harmaline is metabolized in the liver and extrahepatic tissues to its main metabolite, harmalol, by the CYP enzymes, mainly CYP2D6 and CYP1A2 (Fig.1) (Wu et al., 2009; Yu et al., 2003). CYP2D6 is considered one of the most polymorphic phase I drug-metabolizing enzymes that are involved in the biotransformation of 20–30% of marketed drugs and some endogenous substrates (Wu et al., 2009). Therefore, the effect, clearance and intoxication of harmaline can be affected by CYP2D6 status (Ho et al., 1971; Yu et al., 2003). In contrast, the metabolic pathway for harmalol does not involve CYP2D6, but it involves conjugation mainly with glucuronic acid and the resultant conjugate is excreted in urine and bile (Mulder and Hagedoorn, 1974). Recently, it was reported that harmaline exhibited competitive inhibition against CYP2D6 in human liver microsomes, whereas harmalol did not show similar effects (Zhao et al., 2011). Harmaline concentrations used in this study (0.5–12.5 µM) are much lower than harmaline median inhibitory concentration for CYP2D6 enzyme (26 µM) (Zhao et al., 2011).
In the current study, we have chosen human hepatoma HepG2 cells because it has been considered as a useful model to investigate the regulation of CYP1A1 expression (Beedanagari et al., 2009; Ciolino et al., 1998). In addition, we have chosen harmaline and harmalol (0.5, 2.5 and 12.5 µM) as safe nontoxic concentrations to human HepG2 cells according to the cell viability test. In the current study, we demonstrated that harmaline and harmalol inhibit TCDD-mediated induction of CYP1A1 at mRNA, protein and activity levels in human HepG2 cells. Moreover, harmaline and harmalol significantly decreased the BaP-mediated induction of CYP1A1 at the catalytic activity level suggesting that the inhibitory effect of harmaline and harmalol is not AhR ligand specific. In agreement with our results, we previously shown that the extract of Peganum harmala fruiting tops and its active ingredient harmaline decrease the TCDD-induced level of Cyp1a1 in murine hepatoma Hepa 1c1c7 cell line (El Gendy et al., 2010). In contrast to our results, other β-carbolines such as rutaecarpine (found in Rutaceae and szechuan pepper), annomontine (found in Annonaceae and marine sponges of the order Petrosiidae) and xestomanzamine A (contained in Petrosiidae sponges), have been found to induce CYP1 enzyme activity (Haarmann-Stemmann et al., 2010).
Interestingly, the effects of harmaline and harmalol on the CYP1A1 protein and the activity levels are much higher than the effects obtained at mRNA level, suggesting several mechanisms are involved in the obtained results. Therefore, to investigate the underlying mechanisms involved in the effects of harmaline and harmalol on CYP1A1, we tested whether harmaline and harmalol inhibit CYP1A1 at transcriptional level. For this purpose we examined the effect of harmaline and harmalol on AhR-dependent luciferase reporter assay and EMSA. Our results showed that harmaline and harmalol significantly inhibited the TCDD-induced AhR-dependent luciferase activity. Furthermore, both harmaline and harmalol inhibited the TCDD-mediated activation of AhR binding to the XRE using EMSA, confirming the involvement of a transcriptional mechanism. In agreement with the luciferase activity, neither harmaline nor harmalol alone significantly induced AhR activation or transformation when incubated with guinea pig hepatic cytosols using EMSA. Moreover, both harmaline and harmalol alone did not affect CYP1A1 at the activity level in human HepG2 cells. These results suggest that both harmaline and harmalol are not partial agonists for AhR at the used concentrations. Furthermore, these results are in contrast to the aromatic β-carboline compound harman that showed an induction of CYP1A1 and luciferase activity in human HepG2 cells and an induction of the AhR activation and transformation in untreated guinea pig hepatic cytosols (El Gendy and El-Kadi, 2010). These observations may suggest that both harmaline and harmalol are AhR antagonists. To confirm that harmaline and harmalol are AhR antagonists, we performed the ligand competition binding assay. Our data showed that harmaline but not harmalol possess the ability to displace [3H]-TCDD from the binding site on AhR. These data suggest that the underlying mechanism of harmalol inhibitory effect is due to other cascade in the process of AhR activation rather than a competing for the ligand-binding center of the AhR. However, we postulate two responsible mechanisms for the inhibitory effect of harmalol on AhR. First, harmalol may interact with a second binding site on the AhR rather than the TCDD-binding site (Ciolino et al., 1998). This AhR binding site has been proposed for carotenoids and primaquine AhR-dependent induction of CYP1A1. This was substantiated by the fact that these compounds did not show a significant competition with [3H]TCDD binding site on the AhR (Delescluse et al., 2000; Fontaine et al., 1999; Gradelet et al., 1997). Second, harmalol may indirectly affect AhR (Ciolino et al., 1998). There are several proteins participating in the activation process of AhR, such as heat shock protein HSP90 (Palermo et al., 2005; Pratt, 1997), c-SRC (Enan and Matsumura, 1996), the AhR-interacting protein (Ma and Whitlock, 1997), protein kinase C (Long et al., 1998), and the proteins responsible for the recruitment of AhR or ARNT to the XRE of the corresponding genes (Beedanagari et al., 2009). Harmalol may inhibit TCDD-mediated activation of AhR by affecting one or more of these target proteins without affecting the ligand binding site of the AhR.
Also, we investigated the role of posttranscriptional and posttranslational mechanisms using Act-D- and CHX-chase experiments, respectively. Our results demonstrated that harmaline and harmalol did not alter the stability of CYP1A1 mRNA in HepG2 cells. However, both compounds significantly decreased the CYP1A1 protein stability in HepG2 cells. We postulate that, the presence of methoxyl or hydroxyl group in the uncompleted aromatic structures of harmaline and harmalol, respectively, plays a role in the stability of CYP1A1 protein by both compounds. This conclusion is substantiated by the fact that harman, an aromatic β-carboline, which lacks those functioning groups, did not alter the stability of CYP1A1 protein in HepG2 cells (El Gendy and El-Kadi, 2010).
Several mechanisms are introduced to explain the protein degradation including ubiquitin-proteasomal, autophagy-lysosome and calpain pathways (Taguchi et al., 2011). However, ubiquitin-proteasomal pathway possess a key role in CYP1A regulation (Pollenz, 2007; Wiseman and Vijayan, 2007). Therefore, we tested the effect of MG-132, a proteasomal inhibitor, on the reduced CYP1A1 protein stability caused by harmaline and harmalol. Our results demonstrated that inhibition of ubiquitin-proteasomal pathway significantly induced the CYP1A1 protein level of harmaline and harmalol treated cells, implying the involvement of the ubiquitin-proteasomal pathway in the posttranslational modifications of harmaline and harmalol. In addition, we tested the direct inhibitory effect of harmaline and harmalol on CYP1A1 enzyme. Incubation of harmaline and harmalol for 15 min with HepG2 cells pre-treated with TCDD for 24 h significantly reduced the level of CYP1A1 enzyme activity as measured by EROD assay, suggesting that both compounds possess a direct inhibitory effect on CYP1A1 enzyme that participate in their posttranslational modifications.
In conclusion, harmaline and harmalol inhibit the dioxin-induced CYP1A1 at transcriptional and posttranslational levels. Furthermore, these mechanisms may participate, at least in part, in the protective effects of both harmaline and its main metabolite, harmalol, against dioxin-mediated toxicity.
Acknowledgments
This work was supported by the Natural Sciences and Engineering Council of Canada (NSERC) grant RGPIN 250139 to A.O.S.E., and the National Institutes of Environmental Health Sciences research grant R01ES07685 to M.S.D. M.A.M.E. is the recipient of the Egyptian government scholarship. We are grateful to Dr. Loren Kline (University of Alberta, AB) for providing us with guinea pig livers.
Abbreviations
- AhR
aryl hydrocarbon receptor
- Act-D
actinomycin D
- BaP
benzo(a)pyrene
- CHX
cycloheximide
- CYP
cytochrome P450
- EMSA
electrophoretic mobility shift assay
- 7ER
7-ethoxyresorufin
- EROD
7-ethoxyresorufin O-deethylase
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
- HAH
halogenated aromatic hydrocarbons
- MG-132
carbobenzoxy-l-leucyl-l-leucyl-leucinal
- MTT
(3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide)
- PAH
Polycyclic aromatic hydrocarbons
- Res
resveratrol
- TCDD
2,3,7,8-tetrachlorodibenzo-p-dioxin
- TCDF
2,3,7,8-tetrachlorodibenzofuran
- XRE
xenobiotic responsive element
Footnotes
Conflict of Interest Statement
The authors have declared no conflict of interests.
References
- Abe A, Yamada H. Harmol induces apoptosis by caspase-8 activation independently of Fas/Fas ligand interaction in human lung carcinoma H596 cells. Anticancer Drugs. 2009;20:373–381. doi: 10.1097/CAD.0b013e32832a2dd9. [DOI] [PubMed] [Google Scholar]
- Anwar-Mohamed A, El-Kadi AO. Sulforaphane induces CYP1A1 mRNA, protein, and catalytic activity levels via an AhR-dependent pathway in murine hepatoma Hepa 1c1c7 and human HepG2 cells. Cancer Lett. 2009;275:93–101. doi: 10.1016/j.canlet.2008.10.003. [DOI] [PubMed] [Google Scholar]
- Arshad N, Zitterl-Eglseer K, Hasnain S, Hess M. Effect of Peganum harmala or its beta-carboline alkaloids on certain antibiotic resistant strains of bacteria and protozoa from poultry. Phytother Res. 2008;22:1533–1538. doi: 10.1002/ptr.2528. [DOI] [PubMed] [Google Scholar]
- Beedanagari SR, Bebenek I, Bui P, Hankinson O. Resveratrol inhibits dioxin-induced expression of human CYP1A1 and CYP1B1 by inhibiting recruitment of the aryl hydrocarbon receptor complex and RNA polymerase II to the regulatory regions of the corresponding genes. Toxicol Sci. 2009;110:61–67. doi: 10.1093/toxsci/kfp079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradshaw TD, Bell DR. Relevance of the aryl hydrocarbon receptor (AhR) for clinical toxicology. Clin Toxicol (Phila) 2009;47:632–642. doi: 10.1080/15563650903140423. [DOI] [PubMed] [Google Scholar]
- Cao R, Peng W, Wang Z, Xu A. beta-Carboline alkaloids: biochemical and pharmacological functions. Curr Med Chem. 2007;14:479–500. doi: 10.2174/092986707779940998. [DOI] [PubMed] [Google Scholar]
- Ciolino HP, Daschner PJ, Yeh GC. Resveratrol inhibits transcription of CYP1A1 in vitro by preventing activation of the aryl hydrocarbon receptor. Cancer Res. 1998;58:5707–5712. [PubMed] [Google Scholar]
- Czene K, Lichtenstein P, Hemminki K. Environmental and heritable causes of cancer among 9.6 million individuals in the Swedish Family-Cancer Database. Int J Cancer. 2002;99:260–266. doi: 10.1002/ijc.10332. [DOI] [PubMed] [Google Scholar]
- Delescluse C, Lemaire G, de Sousa G, Rahmani R. Is CYP1A1 induction always related to AHR signaling pathway? Toxicology. 2000;153:73–82. doi: 10.1016/s0300-483x(00)00305-x. [DOI] [PubMed] [Google Scholar]
- Denison MS, Harper PA, Okey AB. Ah receptor for 2,3,7,8-tetrachlorodibenzo-p-dioxin. Codistribution of unoccupied receptor with cytosolic marker enzymes during fractionation of mouse liver, rat liver and cultured Hepa-1c1 cells. Eur J Biochem. 1986;155:223–229. doi: 10.1111/j.1432-1033.1986.tb09480.x. [DOI] [PubMed] [Google Scholar]
- Denison MS, Nagy SR. Activation of the aryl hydrocarbon receptor by structurally diverse exogenous and endogenous chemicals. Annu Rev Pharmacol Toxicol. 2003;43:309–334. doi: 10.1146/annurev.pharmtox.43.100901.135828. [DOI] [PubMed] [Google Scholar]
- Denison MS, Pandini A, Nagy SR, Baldwin EP, Bonati L. Ligand binding and activation of the Ah receptor. Chem Biol Interact. 2002;141:3–24. doi: 10.1016/s0009-2797(02)00063-7. [DOI] [PubMed] [Google Scholar]
- Dertinger SD, Nazarenko DA, Silverstone AE, Gasiewicz TA. Aryl hydrocarbon receptor signaling plays a significant role in mediating benzo[a]pyrene- and cigarette smoke condensate-induced cytogenetic damage in vivo. Carcinogenesis. 2001;22:171–177. doi: 10.1093/carcin/22.1.171. [DOI] [PubMed] [Google Scholar]
- El Gendy MA, El-Kadi AO. Peganum harmala L. differentially modulates cytochrome P450 gene expression in human hepatoma HepG2 cells. Drug Metab Lett. 2009;3:212–216. doi: 10.2174/187231209790218163. [DOI] [PubMed] [Google Scholar]
- El Gendy MA, El-Kadi AO. Harman induces CYP1A1 enzyme through an aryl hydrocarbon receptor mechanism. Toxicol Appl Pharmacol. 2010;249:55–64. doi: 10.1016/j.taap.2010.08.014. [DOI] [PubMed] [Google Scholar]
- El Gendy MA, Somayaji V, El-Kadi AO. Peganum harmala L. is a candidate herbal plant for preventing dioxin mediated effects. Planta Med. 2010;76:671–677. doi: 10.1055/s-0029-1240633. [DOI] [PubMed] [Google Scholar]
- Enan E, Matsumura F. Identification of c-Src as the integral component of the cytosolic Ah receptor complex, transducing the signal of 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD) through the protein phosphorylation pathway. Biochem Pharmacol. 1996;52:1599–1612. doi: 10.1016/s0006-2952(96)00566-7. [DOI] [PubMed] [Google Scholar]
- Fontaine F, Delescluse C, de Sousa G, Lesca P, Rahmani R. Cytochrome 1A1 induction by primaquine in human hepatocytes and HepG2 cells: absence of binding to the aryl hydrocarbon receptor. Biochem Pharmacol. 1999;57:255–262. doi: 10.1016/s0006-2952(98)00304-9. [DOI] [PubMed] [Google Scholar]
- Gradelet S, Astorg P, Pineau T, Canivenc MC, Siess MH, Leclerc J, Lesca P. Ah receptor-dependent CYP1A induction by two carotenoids, canthaxanthin and beta-apo-8'-carotenal, with no affinity for the TCDD binding site. Biochem Pharmacol. 1997;54:307–315. doi: 10.1016/s0006-2952(97)00176-7. [DOI] [PubMed] [Google Scholar]
- Haarmann-Stemmann T, Sendker J, Gotz C, Krug N, Bothe H, Fritsche E, Proksch P, Abel J. Regulation of dioxin receptor function by different beta-carboline alkaloids. Arch Toxicol. 2010;84:619–629. doi: 10.1007/s00204-010-0548-2. [DOI] [PubMed] [Google Scholar]
- Herraiz T, Gonzalez D, Ancin-Azpilicueta C, Aran VJ, Guillen H. beta-Carboline alkaloids in Peganum harmala and inhibition of human monoamine oxidase (MAO) Food Chem Toxicol. 2010;48:839–845. doi: 10.1016/j.fct.2009.12.019. [DOI] [PubMed] [Google Scholar]
- Ho BT, Estevez V, Fritchie GE, Tansey LW, Idanpaan-Heikkila J, McIsaac WM. Metabolism of harmaline in rats. Biochem Pharmacol. 1971;20:1313–1319. doi: 10.1016/0006-2952(71)90363-7. [DOI] [PubMed] [Google Scholar]
- Long WP, Pray-Grant M, Tsai JC, Perdew GH. Protein kinase C activity is required for aryl hydrocarbon receptor pathway-mediated signal transduction. Mol Pharmacol. 1998;53:691–700. doi: 10.1124/mol.53.4.691. [DOI] [PubMed] [Google Scholar]
- Lorenzen A, Kennedy SW. A fluorescence-based protein assay for use with a microplate reader. Anal Biochem. 1993;214:346–348. doi: 10.1006/abio.1993.1504. [DOI] [PubMed] [Google Scholar]
- Luch A. Nature and nurture - lessons from chemical carcinogenesis. Nat Rev Cancer. 2005;5:113–125. doi: 10.1038/nrc1546. [DOI] [PubMed] [Google Scholar]
- Ma Q, Whitlock JP., Jr A novel cytoplasmic protein that interacts with the Ah receptor, contains tetratricopeptide repeat motifs, and augments the transcriptional response to 2,3,7,8-tetrachlorodibenzo-p-dioxin. J Biol Chem. 1997;272:8878–8884. [PubMed] [Google Scholar]
- Mandal PK. Dioxin: a review of its environmental effects and its aryl hydrocarbon receptor biology. J Comp Physiol B. 2005;175:221–230. doi: 10.1007/s00360-005-0483-3. [DOI] [PubMed] [Google Scholar]
- Matsumura F. On the significance of the role of cellular stress response reactions in the toxic actions of dioxin. Biochem Pharmacol. 2003;66:527–540. doi: 10.1016/s0006-2952(03)00157-6. [DOI] [PubMed] [Google Scholar]
- Mosmann T. Rapid colorimetric assay for cellular growth and survival: application to proliferation and cytotoxicity assays. J Immunol Methods. 1983;65:55–63. doi: 10.1016/0022-1759(83)90303-4. [DOI] [PubMed] [Google Scholar]
- Moura DJ, Richter MF, Boeira JM, Pegas Henriques JA, Saffi J. Antioxidant properties of beta-carboline alkaloids are related to their antimutagenic and antigenotoxic activities. Mutagenesis. 2007;22:293–302. doi: 10.1093/mutage/gem016. [DOI] [PubMed] [Google Scholar]
- Mulder GJ, Hagedoorn AH. UDP glucuronyltransferase and phenolsulfotransferase in vivo and in vitro. Conjugation of harmol and harmalol. Biochem Pharmacol. 1974;23:2101–2109. doi: 10.1016/0006-2952(74)90575-9. [DOI] [PubMed] [Google Scholar]
- Palermo CM, Westlake CA, Gasiewicz TA. Epigallocatechin gallate inhibits aryl hydrocarbon receptor gene transcription through an indirect mechanism involving binding to a 90 kDa heat shock protein. Biochemistry. 2005;44:5041–5052. doi: 10.1021/bi047433p. [DOI] [PubMed] [Google Scholar]
- Park SY, Kim YH, Kim YH, Park G, Lee SJ. Beta-carboline alkaloids harmaline and harmalol induce melanogenesis through p38 mitogen-activated protein kinase in B16F10 mouse melanoma cells. BMB Rep. 2010;43:824–829. doi: 10.5483/BMBRep.2010.43.12.824. [DOI] [PubMed] [Google Scholar]
- Pollenz RS. Specific blockage of ligand-induced degradation of the Ah receptor by proteasome but not calpain inhibitors in cell culture lines from different species. Biochem Pharmacol. 2007;74:131–143. doi: 10.1016/j.bcp.2007.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratt WB. The role of the hsp90-based chaperone system in signal transduction by nuclear receptors and receptors signaling via MAP kinase. Annu Rev Pharmacol Toxicol. 1997;37:297–326. doi: 10.1146/annurev.pharmtox.37.1.297. [DOI] [PubMed] [Google Scholar]
- Puppala D, Lee H, Kim KB, Swanson HI. Development of an aryl hydrocarbon receptor antagonist using the proteolysis-targeting chimeric molecules approach: a potential tool for chemoprevention. Mol Pharmacol. 2008;73:1064–1071. doi: 10.1124/mol.107.040840. [DOI] [PubMed] [Google Scholar]
- Revel A, Raanani H, Younglai E, Xu J, Rogers I, Han R, Savouret JF, Casper RF. Resveratrol, a natural aryl hydrocarbon receptor antagonist, protects lung from DNA damage and apoptosis caused by benzo[a]pyrene. J Appl Toxicol. 2003;23:255–261. doi: 10.1002/jat.916. [DOI] [PubMed] [Google Scholar]
- Shah PP, Saurabh K, Pant MC, Mathur N, Parmar D. Evidence for increased cytochrome P450 1A1 expression in blood lymphocytes of lung cancer patients. Mutat Res. 2009;670:74–78. doi: 10.1016/j.mrfmmm.2009.07.006. [DOI] [PubMed] [Google Scholar]
- Shimada T, Fujii-Kuriyama Y. Metabolic activation of polycyclic aromatic hydrocarbons to carcinogens by cytochromes P450 1A1 and 1B1. Cancer Sci. 2004;95:1–6. doi: 10.1111/j.1349-7006.2004.tb03162.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Signorelli P, Ghidoni R. Resveratrol as an anticancer nutrient: molecular basis, open questions and promises. J Nutr Biochem. 2005;16:449–466. doi: 10.1016/j.jnutbio.2005.01.017. [DOI] [PubMed] [Google Scholar]
- Sinal CJ, Bend JR. Aryl hydrocarbon receptor-dependent induction of cyp1a1 by bilirubin in mouse hepatoma hepa 1c1c7 cells. Mol Pharmacol. 1997;52:590–599. doi: 10.1124/mol.52.4.590. [DOI] [PubMed] [Google Scholar]
- Slattery ML, Samowtiz W, Ma K, Murtaugh M, Sweeney C, Levin TR, Neuhausen S. CYP1A1, cigarette smoking, and colon and rectal cancer. Am J Epidemiol. 2004;160:842–852. doi: 10.1093/aje/kwh298. [DOI] [PubMed] [Google Scholar]
- Sobhani AM, Ebrahimi SA, Mahmoudian M. An in vitro evaluation of human DNA topoisomerase I inhibition by Peganum harmala L. seeds extract and its beta-carboline alkaloids. J Pharm Pharm Sci. 2002;5:19–23. [PubMed] [Google Scholar]
- Taguchi K, Motohashi H, Yamamoto M. Molecular mechanisms of the Keap1-Nrf2 pathway in stress response and cancer evolution. Genes Cells. 2011;16:123–140. doi: 10.1111/j.1365-2443.2010.01473.x. [DOI] [PubMed] [Google Scholar]
- Tse SY, Mak IT, Dickens BF. Antioxidative properties of harmane and beta-carboline alkaloids. Biochem Pharmacol. 1991;42:459–464. doi: 10.1016/0006-2952(91)90305-o. [DOI] [PubMed] [Google Scholar]
- Wiseman SB, Vijayan MM. Aryl hydrocarbon receptor signaling in rainbow trout hepatocytes: role of hsp90 and the proteasome. Comp Biochem Physiol C Toxicol Pharmacol. 2007;146:484–491. doi: 10.1016/j.cbpc.2007.05.006. [DOI] [PubMed] [Google Scholar]
- Wu C, Jiang XL, Shen HW, Yu AM. Effects of CYP2D6 status on harmaline metabolism, pharmacokinetics and pharmacodynamics, and a pharmacogenetics-based pharmacokinetic model. Biochem Pharmacol. 2009;78:617–624. doi: 10.1016/j.bcp.2009.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida R, Ogawa Y. Oxidative stress induced by 2,3,7,8-tetrachlorodibenzo-p-dioxin: an application of oxidative stress markers to cancer risk assessment of dioxins. Ind Health. 2000;38:5–14. doi: 10.2486/indhealth.38.5. [DOI] [PubMed] [Google Scholar]
- Yu AM, Idle JR, Krausz KW, Kupfer A, Gonzalez FJ. Contribution of individual cytochrome P450 isozymes to the O-demethylation of the psychotropic beta-carboline alkaloids harmaline and harmine. J Pharmacol Exp Ther. 2003;305:315–322. doi: 10.1124/jpet.102.047050. [DOI] [PubMed] [Google Scholar]
- Zhao B, Degroot DE, Hayashi A, He G, Denison MS. CH223191 is a ligand-selective antagonist of the Ah (Dioxin) receptor. Toxicol Sci. 2010;117:393–403. doi: 10.1093/toxsci/kfq217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao T, He YQ, Wang J, Ding KM, Wang CH, Wang ZT. Inhibition of Human Cytochrome P450 Enzymes 3A4 and 2D6 by beta-Carboline Alkaloids, Harmine Derivatives. Phytother Res. 2011 doi: 10.1002/ptr.3458. In press. [DOI] [PubMed] [Google Scholar]
- Zhou J, Gasiewicz TA. 3'-methoxy-4'-nitroflavone, a reported aryl hydrocarbon receptor antagonist, enhances Cyp1a1 transcription by a dioxin responsive element-dependent mechanism. Arch Biochem Biophys. 2003;416:68–80. doi: 10.1016/s0003-9861(03)00274-1. [DOI] [PubMed] [Google Scholar]