Abstract
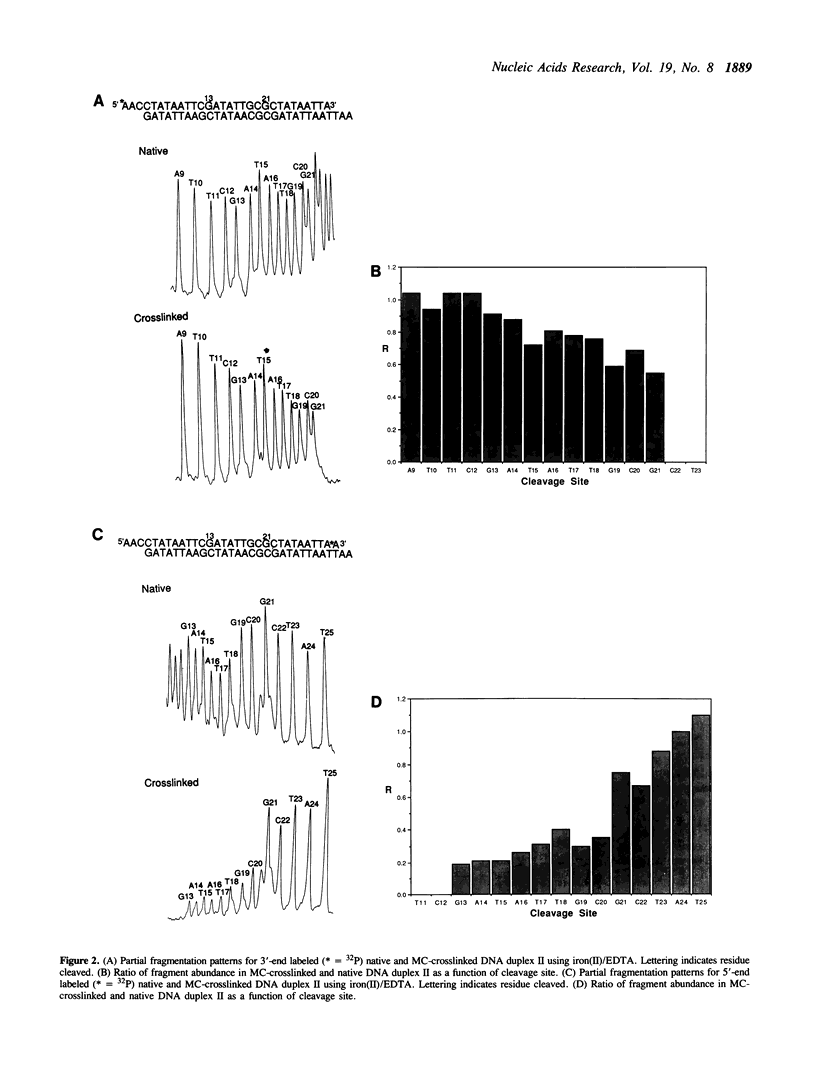
A general approach to the quantitative study of the sequence specificity of DNA interstrand crosslinking agents in synthetic duplex DNA fragments is described. In the first step, a DNA fragment previously treated with an interstrand crosslinking agent is subjected to denaturing PAGE. Not only does this distinguish crosslinked from native or monoadducted DNA, it is shown herein that isomeric crosslinked DNAs differing in position of the crosslink can in some cases be separated. In the second stage, the now fractionated crosslinked DNAs isolated from denaturing PAGE are subjected to fragmentation using iron(II)/EDTA. For those fractions which are structurally homogeneous, analysis of the resulting fragment distribution has previously been shown to reveal the crosslink position at nucleotide resolution. It is shown herein that in fractions which are structurally heterogeneous due to differences in position of crosslink, this analysis quantifies the relative extent of crosslinking at distinct sites. Using this method it is shown that reductively activated mitomycin C crosslinks the duplex sequences 5'-GCGC and 5'-TCGA with 3 +/- 1:1 relative efficiency.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Borowy-Borowski H., Lipman R., Tomasz M. Recognition between mitomycin C and specific DNA sequences for cross-link formation. Biochemistry. 1990 Mar 27;29(12):2999–3006. doi: 10.1021/bi00464a016. [DOI] [PubMed] [Google Scholar]
- Boyer V., Moustacchi E., Sage E. Sequence specificity in photoreaction of various psoralen derivatives with DNA: role in biological activity. Biochemistry. 1988 Apr 19;27(8):3011–3018. doi: 10.1021/bi00408a052. [DOI] [PubMed] [Google Scholar]
- Cech T. R. Alkaline gel electrophoresis of deoxyribonucleic acid photoreacted with trimethylpsoralen: rapid and sensitive detection of interstrand cross-links. Biochemistry. 1981 Mar 17;20(6):1431–1437. doi: 10.1021/bi00509a005. [DOI] [PubMed] [Google Scholar]
- Gamper H., Piette J., Hearst J. E. Efficient formation of a crosslinkable HMT monoadduct at the Kpn I recognition site. Photochem Photobiol. 1984 Jul;40(1):29–34. doi: 10.1111/j.1751-1097.1984.tb04549.x. [DOI] [PubMed] [Google Scholar]
- Hertzberg R. P., Dervan P. B. Cleavage of DNA with methidiumpropyl-EDTA-iron(II): reaction conditions and product analyses. Biochemistry. 1984 Aug 14;23(17):3934–3945. doi: 10.1021/bi00312a022. [DOI] [PubMed] [Google Scholar]
- Kochel T. J., Sinden R. R. Hyperreactivity of B-Z junctions to 4,5',8-trimethylpsoralen photobinding assayed by an exonuclease III/photoreversal mapping procedure. J Mol Biol. 1989 Jan 5;205(1):91–102. doi: 10.1016/0022-2836(89)90367-7. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Piette J. G., Hearst J. E. Termination sites of the in vitro nick-translation reaction on DNA that had photoreacted with psoralen. Proc Natl Acad Sci U S A. 1983 Sep;80(18):5540–5544. doi: 10.1073/pnas.80.18.5540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts J. D., Van Houten B., Qu Y., Farrell N. P. Interaction of novel bis(platinum) complexes with DNA. Nucleic Acids Res. 1989 Dec 11;17(23):9719–9733. doi: 10.1093/nar/17.23.9719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sage E., Moustacchi E. Sequence context effects on 8-methoxypsoralen photobinding to defined DNA fragments. Biochemistry. 1987 Jun 16;26(12):3307–3314. doi: 10.1021/bi00386a010. [DOI] [PubMed] [Google Scholar]
- Teng S. P., Woodson S. A., Crothers D. M. DNA sequence specificity of mitomycin cross-linking. Biochemistry. 1989 May 2;28(9):3901–3907. doi: 10.1021/bi00435a041. [DOI] [PubMed] [Google Scholar]
- Tullius T. D., Dombroski B. A., Churchill M. E., Kam L. Hydroxyl radical footprinting: a high-resolution method for mapping protein-DNA contacts. Methods Enzymol. 1987;155:537–558. doi: 10.1016/0076-6879(87)55035-2. [DOI] [PubMed] [Google Scholar]
- Weidner M. F., Sigurdsson S. T., Hopkins P. B. Sequence preferences of DNA interstrand cross-linking agents: dG-to-dG cross-linking at 5'-CG by structurally simplified analogues of mitomycin C. Biochemistry. 1990 Oct 2;29(39):9225–9233. doi: 10.1021/bi00491a017. [DOI] [PubMed] [Google Scholar]
- Zhen W. P., Buchardt O., Nielsen H., Nielsen P. E. Site specificity of psoralen-DNA interstrand cross-linking determined by nuclease Bal31 digestion. Biochemistry. 1986 Oct 21;25(21):6598–6603. doi: 10.1021/bi00369a039. [DOI] [PubMed] [Google Scholar]





