Abstract
Marek’s disease (MD) is a lymphoproliferative disease induced by Marek’s disease virus (MDV) infection. To augment vaccination measures in MD control, host genetic resistant to MD becomes obviously more and more important. To elucidate the mechanism of MD-resistance, most of researches were focused on the genetic differences between resistant and susceptible chickens. However, epigenetic features between MD resistant and susceptible chickens are poorly characterized. Using bisulfite pyrosequencing method, we found some candidate genes have higher promoter methylation in the MD-susceptible (L72) chickens than in the MD-resistant (L63) chickens. The hypermethylated genes, involved in cellular component organization, responding to stimulus, cell adhesion, and immune system process, may play important role in susceptibility to disease by deregulation of these genes. MDV infection induced the expression changes of all three methyltransferases genes (DNMT1, DNMT3a, and DNMT3b) in both lines of chickens. The DNMT1 was up-regulated in L72, whereas the DNMT3b was down-regulated in L63 at 21 dpi. Interestingly, a dynamic change of promoter methylation was observed during MDV life cycle. Some genes, including HDAC9, GH, STAT1, CIITA, FABP3, LATS2, and H2Ac, showed differential methylation behaviors between the two lines of chickens. In summary, the findings from this study suggested that DNA methylation heterogeneity and MDV infection induced methylation alterations differences existed between the two lines of chickens. Therefore, it is suggested that epigenetic mechanisms may be involved in modulating the resistance and/or susceptibility to MD in chickens.
Keywords: chicken, Marek’s disease, MD-resistance, MD-susceptibility, DNA methylation
Introduction
Marek’s disease (MD) is a lymphoproliferative disease of chickens caused by Marek’s disease virus (MDV) with pathological features including mononuclear cell-infiltration in the peripheral nerves, skin, and muscle (Davison and Nair, 2004). MDV is classified into the Mardivirus genus due to its genome content (Davison, 2002) and biological effect on lymphocytes like EBV (Epstein, 2001). MDV life cycle in its host can be divided into four phases, including an early cytolytic phase from 2 to 7 days post infection (dpi), a latency phase around 7–10 dpi, a late cytolytic phase starting from 18 dpi and a proliferation phase after 28 dpi (Calnek, 1986, 2001). Although MD is controlled by vaccination, the virulence of MDV has being evolved over time and resulted in more severe brain edema and acute deaths even after vaccination (Witter, 1997; Osterrieder et al., 2006). MD remains a problem in the poultry industry worldwide (Churchill et al., 1969). Since the inheritance and resistance to MD was first observed (Asmundson and Biely, 1932), MD-resistant and -susceptible chickens have been bred by those including Stone (lines 6 and 7; Bacon et al., 2000), Hutt, and Cole (lines N and P; Davison and Nair, 2004). Nowadays, the selection of genetically disease resistant chickens is especially important in MD control. A better understanding in the mechanisms of MD-resistance and -susceptibility should be of great value in developing better strategies to further prevent and control MD.
In recent years, most of the studies are focused on genetic variations between MD-resistant and susceptible chickens (Gilmour et al., 1976; Fredericksen et al., 1977; Kaiser et al., 2003; Sarson et al., 2008a). However, little is done on epigenetic differences between the two kinds of chickens. Epigenetics is the study of alterations in phenotypes that are not brought about by changes in DNA sequences, but by factors including DNA methylation, histone modifications, and so on (Allis et al., 2006). DNA methylation is known as a post-replication modification found on the 5-C position of cytosine mainly in CpG dinucleotides, generated and maintained by three methyltransferases – DNMT1, DNMT3a, and DNMT3b (Allis et al., 2006). In mammals, DNA methylation was found playing important role in development, imprinting, carcinogenesis, and other diseases (Feinberg and Tycko, 2004; Feng et al., 2010). Notably, we found two DNA mutations in DNMT3b (Yu et al., 2008a) and a higher promoter methylation level of ALVE and TVB in the spleen of MD-susceptible chickens (L72) compared to that of MD-resistant chickens (L63; Yu et al., 2008b), and the methylation level in CD4 promoter region was down-regulated in the former but not in the later at 21 dpi (Luo et al., 2011).
To advance the understanding of functional patterns of DNA methylation in disease resistance or susceptibility, we extended the scope of examination to 18 interested genes, which include STAT1, CIITA, NK-lysin, CD44, IL12, and GH1 that the expression levels of these gene are alterable upon MDV challenge (Liu et al., 2001; Abdul-Careem et al., 2006; Parcells and Burgess, 2008; Sarson et al., 2008a,b; Heidari et al., 2010; Thanthrige-Don et al., 2010). Some of the 18 genes were also chosen based on our previous temporal microarray data, which include FABP3, HDAC9, IL28RA, MON2, and THBS2 (Luo et al., 2011; Yu et al., 2011).
Materials and Methods
Animals, challenge trial, and sample collection
Specific pathogen free chickens from two highly inbred White Leghorn lines, the L63 and L72, were used. Chickens from each of the lines were divided into two groups. One group was challenged with a very virulent plus MDV (vv + MDV), 648A passage 40, intra-abdominally at day 5 post hatch at a 500 plaque-forming unit (PFU) dosage, the other was not challenged and was assigned as the control group. Fresh spleen samples were respectively collected at 5, 10, and 21 dpi from both groups, and placed in RNAlater (Qiagen, Valencia, CA, USA) immediately, and then stored at −80°C.
All of the experimental chickens were challenged and maintained in a BSL-2 facility at the Avian Disease and Oncology Laboratory (ADOL), East Lansing, Michigan. The chickens were handled closely following animal usage procedures established by the ADOL ACUC committee.
DNA extraction, bisulfite treatment, and pyrosequencing
DNA was extracted from 20 ∼ 30 mg spleen by NucleoSpin® Tissue Kits (Macherey-Nagel, Bethlehem, PA, USA). Bisulfite treatment of 1 μg DNA per chicken was performed using EZ DNA Methylation-Gold Kit™ (ZYMO Research, Irvine, CA, USA). Primers for PCR and pyrosequencing were designed with PSQ Assay Design software (Biotage, Charlotte, NC, USA; Table A1 in Appendix). For cost saving purposes, a universal primer (5′-GGGACACCGCTGATCGTTTA-3′) was used in the PCR assays (Yu et al., 2008a). PCR was carried out using Hotstar Taq DNA polymerase (Qiagen, Valencia, CA, USA) in 20 μl reactions in iCycler (Bio-Rad, Hercules, CA, USA) Detection System as follows: samples were denatured at 95°C for 15 min, followed by 50 cycles at 95°C for 30 s, 55–60°C for 30 s, 72°C for 30 s, and then extended at 72°C for 10 min. DNA methylation level analysis was performed on the Pyro Q-CpG system (PyroMark ID, Biotage, Charlotte, NC, USA) as previously described (Colella et al., 2003; Yu et al., 2008a).
RNA extraction and quantitative real-time RT-PCR
RNA from 30 ∼ 50 mg spleen was extracted using the RNeasy Mini Kit (Qiagen, USA). Reverse transcription was carried out in 20 μl with 1 μg of total RNA by using SuperScriptTM III Reverse Transcriptase (Invitrogen, Carlsbad, CA, USA) and oligo (dT)12–18 primers (Invitrogen, Carlsbad, CA, USA). Primers (Table A2 in Appendix) for quantitative real-time RT-PCR were designed using Primer3 online primer designer system1. Quantitative real-time RT-PCR was performed on the iCycler iQ PCR system (Bio-Rad, USA) in a final volume of 20 μl using QuantiTect SYBR Green PCR Kit (Qiagen) with following procedures: denatured at 95°C for 15 min, followed by 40 cycles at 95°C for 30 s, 55–60°C for 30 s, 72°C for 30 s, then extended at 72°C for 10 min. Each reaction was replicated. The housekeeping gene GAPDH (glyceraldehyde-3-phosphate dehydrogenase) was used to normalize the loading amount of cDNA.
Functional analysis and statistics
The GO Biological Process analysis of the genes was analyzed by PANTHER2. Student’s t-test was used to analyze the differences of the promoter methylation level and the gene expression before and after MDV infection.
Results
Differential methylation patterns between the L63 and L72
To determine the different methylation levels of genes between the MD-resistant L63 and the MD-susceptible L72 chickens, we analyzed the DNA methylation status of promoters for 18 genes by bisulfite pyrosequencing method. The results showed that most of the 18 genes, including LATS2, MON2, IL28RA, STAT1, CD44, H2Ac, TNFSF10, IL12, FABP3, and CIITA, were hypomethylated (methylation level <40%); few of them, ITGB5, THBS2, and HDAC9, had intermediate methylation level (between 40% and 60%), and the rest (IGF2, GH1, NK-lysin, and TGFβ3) had hypermethylation methylation level (>60%) in the control groups of both lines (Table A3 in Appendix). However, some of the CpGs of CD82 had a very low methylation level (<10%) and others had an intermediate methylation level (Table A3 in Appendix).
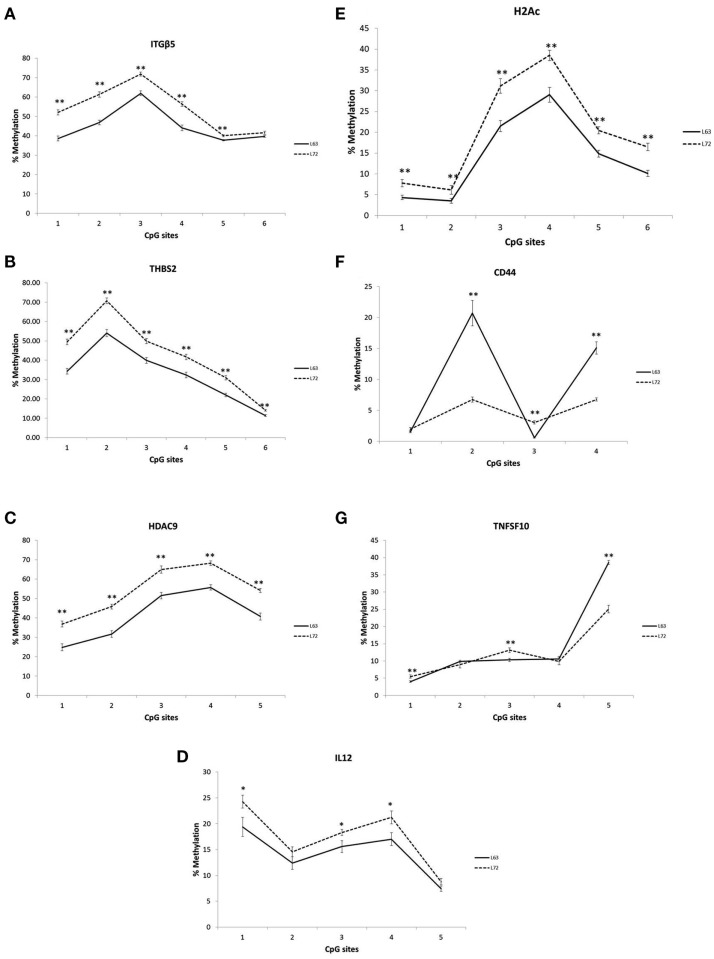
Differential promoter methylation levels were observed for ITGB5, THBS2, HDAC9, IL12, CD44, H2AC, and TNFSF10 between the L63 and L72, As showed in Figure 1, the methylation levels in all the tested CpG sites of the ITGB5, THBS2, HDAC9, IL12, H2AC were significantly higher in L72 than in L63 (P < 0.05; Figures 1A–E). However, some of the CpG sites in CD44 (CpG 2 and 4) and TNFSF10 (CpG 5) had higher level of methylation (P < 0.05), while some others (CD44 CpG 3; TNFSF10 CpG 1 and 3) had lower methylation levels in L63 than L72 (P < 0.05; Figures 1F,G).
Figure 1.
Differentiated promoter methylation levels between the L63 and L72 chickens observed in ITGβ5 (A), THBS2 (B), HDAC9 (C), IL12 (D), H2Ac (E), CD44 (F), and TNFSF10 (G). The promoter methylation levels of the genes in spleen were examined by pyrosequencing. The absolute methylation level for each CpG site from pyrosequencing result was plotted. Solid line: L63; dashed line: L72. *P < 0.05, **P < 0.01. N = 12 for each group. Error bar = SEM.
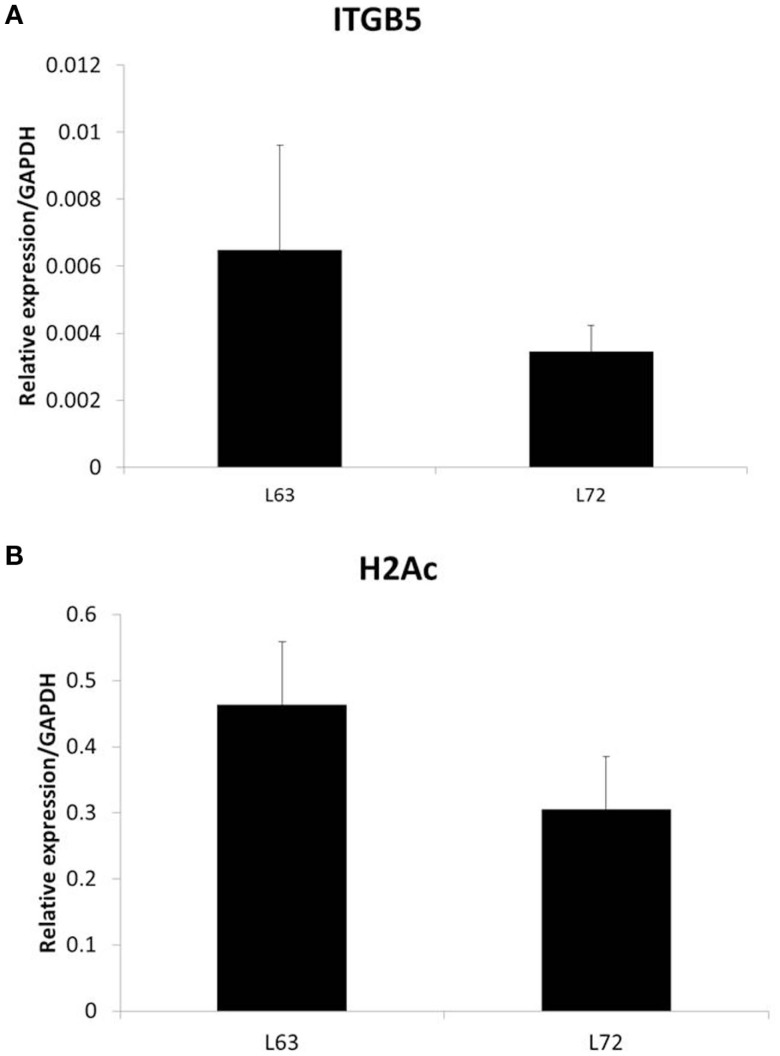
To test if the differential promoter methylation levels of these genes are related with gene expression, we randomly chose two genes, ITGB5 and H2Ac, and did quantitative RT-PCR. We found that the expression levels of the two genes, whose promoter methylation is higher in L72 chicken, is lower in these chickens (Figure 2).
Figure 2.
Expression of ITGB5 (A) and H2Ac (B) in non-infected chicken samples. N = 4 for each group. Error bar = STD.
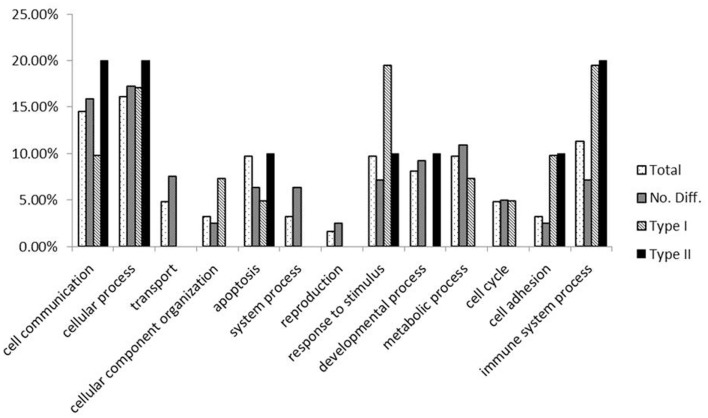
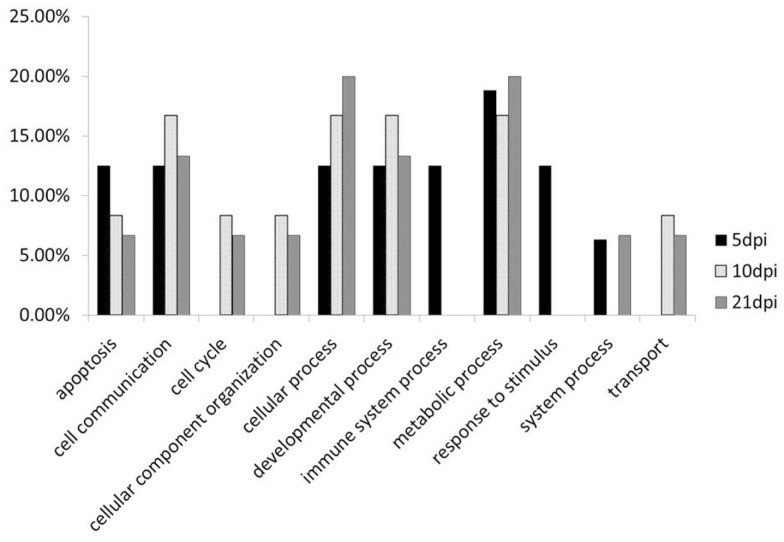
Functional analysis of the genes (Figure 3) showed that, in comparison to the whole gene set we examined in this experiment, genes with lower methylation levels in L63 are mainly enriched in cellular component organization, response to stimulus, cell adhesion, and immune system process. In contrast, an under-enrichment of these genes was shown in cell communication, transport, system process, reproduction, and developmental process. For genes with a varied methylation levels between L63 and L72, they are over-represented in functions of cell adhesion and immune system process. However, for the genes with similar methylation between the L63 and L72, no under or over-represented biological functions was identified.
Figure 3.
Functional analysis of the genes by PANTHER.
Differential DNMT1, DNMT3a, and DNMT3b expression induced by MDV challenge
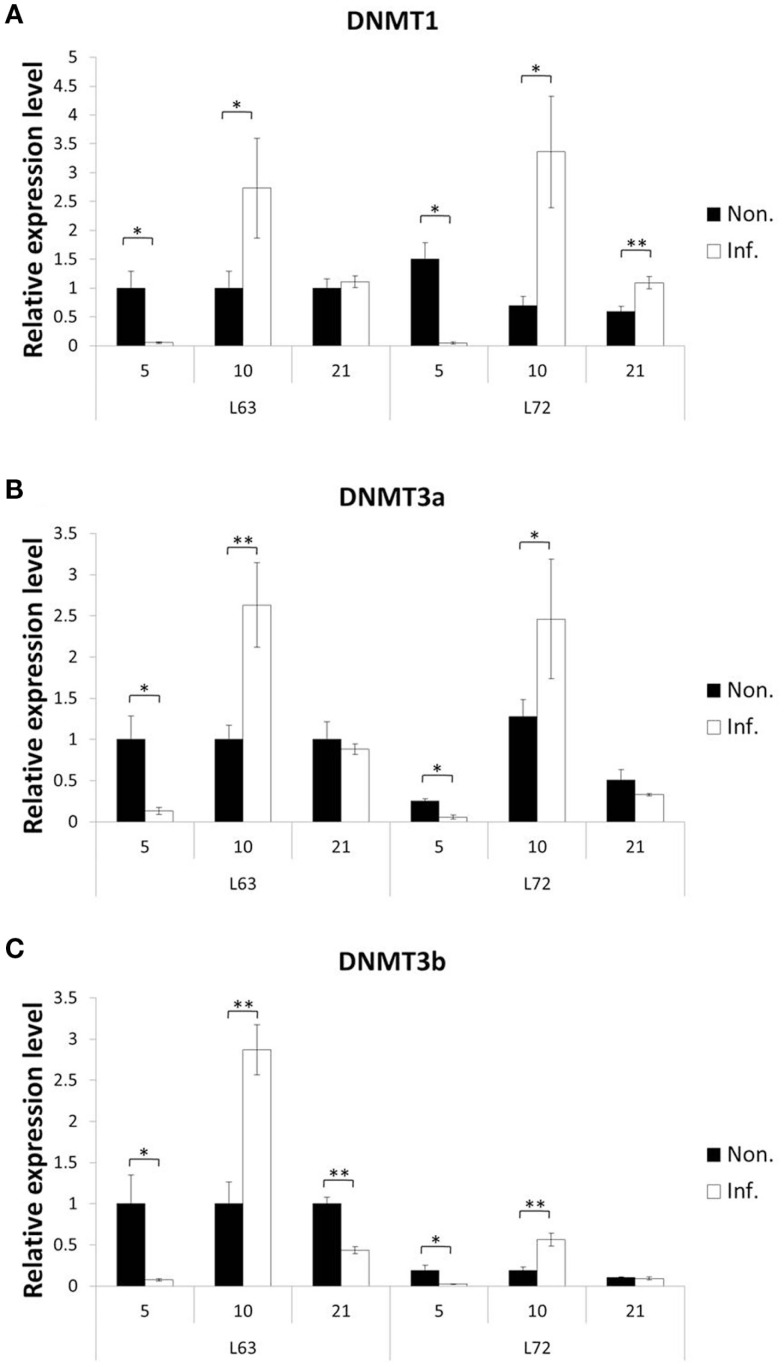
To explore how MDV challenge induces DNA methylation alteration, we first checked if the expressions of the methylation agents, three methyltransferases (DNMT1, DNMT3a, and DNMT3b), were influenced over three time points (5, 10, and 21 dpi), which represent the early cytolytic, latent, and later cytolytic phase of the virus life cycle in the host cells, respectively. Interestingly, similar trends of expression changes were observed at 5 and 10 dpi for all three DNMTs in the MDV challenged chickens of both lines (Figure 4), while at 21 dpi, the changes were much more complicated. At 21 dpi, the DNMT1 was significantly up-regulated in the infected L72 chickens compared to the L72 control group (P < 0.05). The DNMT1 was remained unchanged, however, between the infected and uninfected L63 groups (P > 0.05; Figure 4A). For DNMT3a, no expression difference was observed at 21 dpi between the infected and non-infected groups of both lines (P > 0.05; Figure 4B). However, the DNMT3b was significantly down-regulated in the infected group of L63 at 21 dpi (P < 0.05), but no differential expression was observed in L72 (Figure 4C). Overall, the expression levels of all the three DNMTs were significantly inducible by MDV infection, but with varied alteration trends and extents were found over different time points and between the infected and non-infected groups as well as between the chicken lines.
Figure 4.
Expression of DNA methyltrasferase genes after MDV infection. The expression of three DNA methyltrasferase genes, DNMT1 (A), DNMT3a (B), and DNMT3b (C), were tested by quantitative real-time RT-PCR at 5, 10, and 21 dpi in chicken spleen both resistant and susceptible to MD. The relative expression level was normalized to GAPDH gene. Black: non-infected; white: infected. *P < 0.05, **P < 0.01. N = 4 for each group. Error bar = SEM.
Aberrant methylation level induced by MDV infection
To further study DNA methylation dynamic response to MDV infection, we tested the promoter methylation of the 18 genes on 5, 10, and 21 dpi. Pairwise comparison was performed between the infected and non-infected age-matched sample groups of each chicken line for each of the CpG sites. Significant methylation level changes (P < 0.05) were detected at one or more CpG sites in all of the genes except THBS2 gene after MDV challenge. The methylation level changes of the examined genes were under 30%. The MDV-induced DNA methylation changes for CIITA, NK-lysin, FABP3, and ITGB5 were 10% above their unchallenged counterpart for each of the CpG sites. More than 10% methylation change was found in HDAC9 at 5 dpi and 7–10% changes at 21 dpi in L72. Most of the genes (12/17) had significant methylation change (P < 0.05) at more than one time point (Table A4 in Appendix; Figure 5), except for IL12, TNFSF10, and ITGB5, which were only changed at 5 dpi, and CD44, LATS2, CIITA, which were only changed at 21 dpi. In contrast between the two lines of chickens, more genes in L63 had significant methylation changes at 5 dpi, while more genes were observed with significant methylation changes in L72 at 10 and 21 dpi (Figure 6).
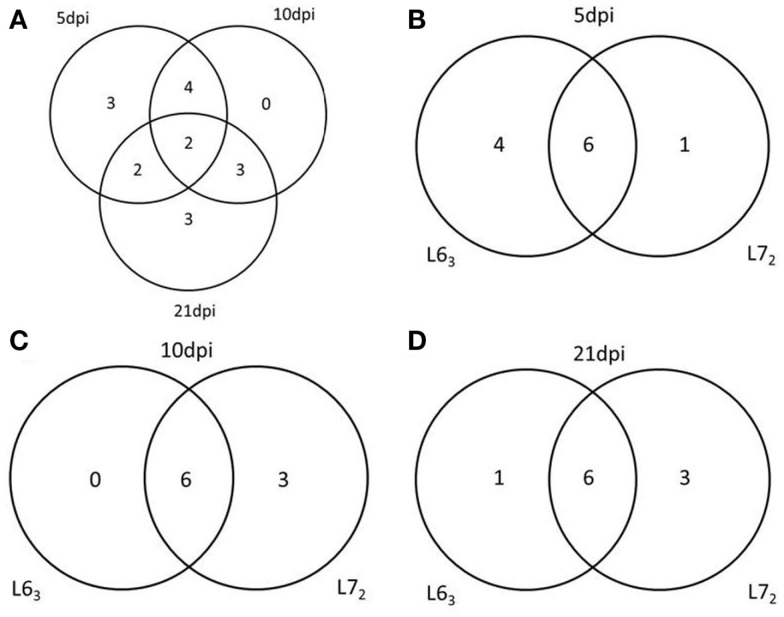
Figure 5.
Venn Diagrams of the number of genes have the methylation change at different time points and in different chicken lines. (A) The number of genes has the methylation change at 5, 10, and 21 dpi. (B–D) The number of genes has the methylation change between L63 and L72 at 5, 10, and 21 dpi respectively.
Figure 6.
Functional analysis of the genes have different methylation change between L63 and L72 at 5, 10, and 21 dpi by PANTHER.
Differentially methylation changes due to MDV challenge
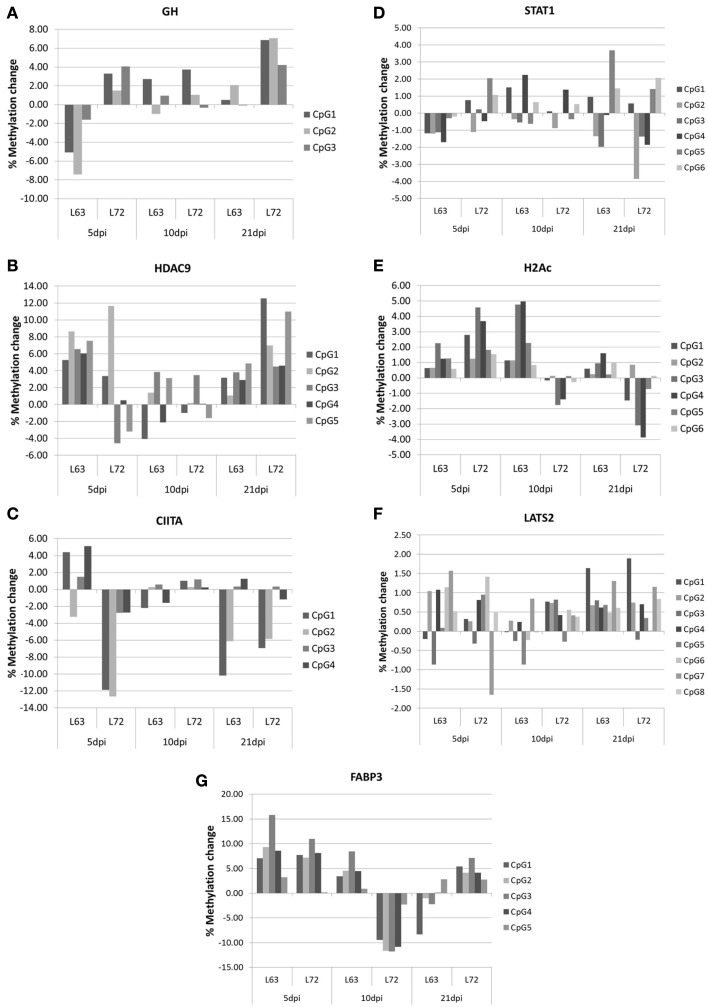
To compare the contents of the methylation change between L63 and L72, the mean methylation change of all the CpG sites was calculated for each gene. Seven out of the 18 genes (HDAC9, GH, STAT1, CIITA, FABP3, LATS2, and H2Ac) showed significant differentially averaged methylation changes (P < 0.05) between the two lines of chickens (Table 1; Figure A1 in Appendix). Functional analysis of the genes with temporal methylation changes revealed that the genes, related to apoptosis, immune system process, and response to stimulus, were over-represented at 5 dpi (Figure 6). However, genes, involved in enrichment of cell communication, were shown at 10 dpi; Genes, involved in functionality of cell cycle, cellular component organization, and transport, were over-represented at 10 and 21 dpi.
Table 1.
Differential DNA methylation change between L63 and L72 after MDV challenge.
| Time points (dpi) | Gene name | DNA methylation level change |
P value | |
|---|---|---|---|---|
| L63 | L72 | |||
| 5 | GH | −4.71 | 2.95* | 0.0146 |
| CIITA | 1.95 | −7.51* | 0.0298 | |
| STAT1 | −0.95** | 0.42 | 0.0244 | |
| H2Ac | 1.11** | 2.62** | 0.0306 | |
| 10 | FABP3 | 4.37** | −9.21** | 0.0002 |
| LATS2 | −0.01 | 0.48** | 0.0409 | |
| H2Ac | 2.53** | −0.56 | 0.0044 | |
| 21 | HDAC9 | 3.16** | 7.92** | 0.0273 |
| GH | 0.83 | 6.07** | 0.0096 | |
| FABP3 | −1.76 | 4.70** | 0.0117 | |
| H2Ac | 0.78** | −1.36 | 0.0211 | |
*P < 0.05, **P < 0.01.
Discussion
The development of disease resistance has long been a very important strategy for control of diseases in farm animals (Bishop et al., 2010; Luo et al., 2012). A better understanding on the mechanisms of disease resistance will facilitate breeding of more disease resistant animals, help to better control diseases in farm animal and also provide better models to learn disease control strategies for humans. Since the establishment of the non-MHC associated MD-resistant and -susceptible chicken lines (Line 6 and Line 7), lots of experiments have been done to elucidate the genetic mechanism of MD-resistance between the two lines of chickens (Gilmour et al., 1976; Fredericksen et al., 1977; Kaiser et al., 2003; Sarson et al., 2008a). However, not until recently, our lab started to explore their epigenetic differences between the chicken lines, which provides evidence that DNA methylation may be involved in MD-resistance or -susceptibility (Yu et al., 2008a,b; Luo et al., 2011). As we know, although the functions of DNA methylation in development, imprinting etc. were reported in mammals, it’s still unclear about its function in disease resistance. Previous study in human (Jelinek et al., 2011) and plant (Akimoto et al., 2007) showed that individuals with a higher DNA methylation level in some particular genes are susceptible to diseases or bacterial infection, which is consistent with our finding that a higher methylation level of several genes (ITGB5, THBS2, HDAC9, IL12, and H2Ac) were shown in MD-susceptible (L72) chickens. However, variable methylation level of CD44 and TNFSF10 between L63 and L72 indicated that the hypermethylation in susceptible chickens is not genome-widely. Functioning classification showed that the hypermethylated genes in susceptible chicken are showing functions of cellular component organization, response to stimulus, cell adhesion, and immune system process. Interestingly, hypermethylation of genes functioning in regulating cell adhesion was very important for the development of various cancers in human (Katto and Mahlknecht, 2011). Furthermore, expression analysis of the hypermethylated genes in the susceptible chickens showed a lower expression of these genes. The results indicated that there are specific pathways that may involve in MD-susceptibility or -resistance through hyper- or hypo-methylation of the genes included. In the future, a genome-wide DNA methylation research will be designed, which will help us explore the mechanisms further.
In previous study, the DNA methyltransferase (DNMTs) were usually found up-regulated by virus infection in human cells, like SV40 (Chuang et al., 1997) and EBV (Tsai et al., 2002). However, dynamic change of DNMTs expression was observed in vivo during MD life cycle in chicken. The DNMTs were first down-regulated at 5 dpi and then up-regulated at 10 dpi in both L63 and L72 chickens. Furthermore, different regulations of DNMTs were observed between the MD-resistant and -susceptible chickens at 21 dpi, indicating that late cytolytic phase is a critical time for DNMTs function in DNA methylation process or tumorigenesis. However, the DNMTs expression change was not necessary for the change of the methylation level change in the genes we studied. The correlation between DNMTs expression and methylation is upon chickens and time point. There are several reasons for that: First, other epigenetic mechanisms involve in the methylation change during MDV infection; second, the changed dosages of DNMTs are not efficient for the change of methylation on these genes; third, other functions of DNMTs involve. Except for establishing and maintaining the DNA methylation in cells, DNMTs also have other functions. The finding that DNMT1 was only up-regulated in MD-susceptible chicken is consistent with the observation that DNMT1 is necessary for establishing and maintaining the transformation state of cells (Bakin and Curran, 1999; Robert et al., 2003). Similarly, DNMT3B deficient mouse embryo fibroblasts were found resistant to virus induced transformation (Soejima et al., 2003), which is consistent with our finding that the down-regulation of DNMT3b was only shown in MD-resistant chicken.
Abnormal DNA methylation is a common feature of human cancer. The fact is that DNA methylation started to be changed from very early stage of transformation process and a stepwise or dynamic change was happened during carcinogenesis (Ehrlich, 2009; Novak et al., 2009). Furthermore, DNA methylation modifications at the promoter regions of genes play a critical role in the intricate host–virus interaction network (Young et al., 2000; Zheng et al., 2008). From our results, the dynamic DNA methylation change during MD progression not only indicated an interaction between MDV and host gene, but also revealed the genes with aberrant methylation level may also involve in virus induced transformation process. During MDV life cycle in chicken spleen, 5 dpi is the early cytolytic phase when B cells and some T cells were targeted by MDV (Osterrieder et al., 2006). Virus infection in this stage provokes some apoptosis, lymphoid lesion, and inflammation responses in the immune organ (Morimura et al., 1996; Baigent and Davison, 1999). Different methylation change in genes enriched in apoptosis, immune system process, and response to stimulus suggested that the expression of these genes maybe differentially regulated between the MD-resistant and -susceptible chickens, which show different response to MDV infection. Although 10 and 21 dpi represent the latency and later cytolytic or transformation stage of MDV infection, it’s very difficult to differentiate them very clearly in vivo since the latently infected cells can be mixed with the transformed cells (Davison and Nair, 2004). So we found some function enrichments like cell communication and transport are shared at 10 and 21 dpi. Genes over-represented in cell cycle and cell communications have different DNA methylation changes in L63 and L72 chicken. Since genes involve in cell cycle and cell communication play important role in carcinogenesis (Yamasaki et al., 1995; Hanahan and Weinberg, 2000, 2011), these results suggested that DNA methylation may participate in MD-resistance by disrupting pathways intriguing tumor formation.
In conclusion, we found DNA methylation heterogeneity between the MD-resistant L63 and -susceptible L72 chickens. The hypermethylation of genes involved in cellular component organization, response to stimulus, cell adhesion, and immune system process may play important role in MD-susceptibility. Different from other viruses, MDV induces a dynamic expression change in DNMTs. Differential methylation changes are observed between resistant and susceptible chickens after MDV infection. All in all, the differential DNA methylation levels and DNA methylation level change induced by MDV challenge between the lines of chickens suggested that DNA methylation may play a role in host resistance and/or susceptibility to MD.
Authors’ Contributions
Juan Luo extracted DNA and RNA, performed the DNA methylation and mRNA expression experiments, analyzed the data and wrote the paper. Ying Yu extracted some of the DNA and performed some of the DNA methylation analysis. Fei Tian extracted DNA and RNA. Shuang Chang conducted the challenge trials and collected samples. Huanmin Zhang revised the paper. Jiuzhou Song designed the experiments and revised the paper.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The work was supported by USDA-NRI/NIFA 2008-35204-04660 and USDA-NRI/NIFA 2010-65205-20588.
Appendix
Table A1.
Primers for pyrosequencing analysis of promoter methylation.
| Genes | Accession No. | Primers | Sequence |
|---|---|---|---|
| ITGB5 | NM_204483 | F | 5′-GGGACACCGCTGATCGTTTA YGTGYGGAGTTYGTAGAGAT-3′ |
| R | 5′-CCCTTAAAAACTATCTCRTTCCA-3′ | ||
| Sequencing | 5′-TCTCRTTCCAATTATACAC-3′ | ||
| Assay | 5′-RACRCTACCACCCRCTACRT-3′ | ||
| CD82 | NM_001008470 | F | 5′-AGCGTTGYGAGTTTTATAGAAGTG-3′ |
| R | 5′-GGGACACCGCTGATCGTTTA AACCCTCRCTCRACTACTTTACC-3′ | ||
| Sequencing | 5′-AAGTGAGAATAATGTAATGG-3′ | ||
| Assay | 5′-TAGYGGTTAGTAGTTYGGTATTTYGTTGTTATYGTAGYGTTGTAATYGTT-3′ | ||
| HDAC9 | NM_001030981 | F | 5′-TTGGGATATGGGTTGTCGAAAT-3′ |
| R | 5′-GGGACACCGCTGATCGTTTA GCTAATACTCTCGTTCGCAACATS′ | ||
| Sequencing | 5′-TGGGTTGTCGAAATAGTT-3′ | ||
| Assay | 5′-TYGYGGGATTGTTGTGYGTGGGYGYGGTAGAAATTATGTTGCGAACGAGA-3′ | ||
| STAT1 | NM001012914 | F | 5′-TGTAAYGAAGTAAAATAGGYGAGA-3′ |
| R | 5′-GGGACACCGCTGATCGTTTA TCAACCTACACTACRCAACCTAA-3′ | ||
| Sequencing | 5′-TAAAATAGGCGAGATATAAG-3′ | ||
| Assay | 5′-TAYGYGAGTYGTTYGYGAGGTAGGGTCGTT-3′ | ||
| TGFB5 | NM_205454 | F | 5′-GYGAGGATATTTATTTGGAAGAG-3′ |
| R | 5′-GGGACACCGCTGATCGTTTA CCCAAAAAATATCACCTCCAAT-3′ | ||
| Sequencing | 5′-GAGTTTGGGTTGGGTA-3′ | ||
| Assay | 5′-TAYGTAGTATTYGGAATTTTGTTYGAAATAGGTTGGTGTTGTTTTTTTTTG YGGAGGATA YGTTAAAGYG-3′ | ||
| Nk-lysin | NM001044680 | F | 5′-GYGTTAGTTGAATTTTAGAGTTTAAAG-3′ |
| R | 5′-GGGACACCGCTGATCGTTTA TTTATAAATTTTTCTCCACTACTACTAAT-3′ | ||
| Sequencing | 5′-AATTTTAGAGTTTAAAGGGA-3′ | ||
| Assay | 5′-GYGGAGAYGGAGTATAATATTATAYGTATTATTAAYGTTAYGTAGTTTTT-3′ | ||
| IL28RA | XM 417841 | F | 5′-GGGACACCGCTGATCGTTTAAGGATGTGCGAGGTAGAATATTG-3′ |
| R | 5′-CAAACCCTACAACAACCACATAAT-3′ | ||
| Sequencing | 5′-CCCTACAACAACCACATAA-3′ | ||
| Assay | 5′-TCRCTATATACTAACCRCCACRTTCCCAACAACRCACTAACRACTACAAC0A ATATTCTACCTCGCAC-3′ | ||
| M0N2 | NM 001199605 | F | 5′-TTATTGCGGTAGGGGTTAATATT-3′ |
| R | 5′-GGGACACCGCTGATCGTTTA CAAACTAAACGCTATCCTAAACT-3′ | ||
| Sequencing | 5′-CGGTAGGGGTTAATATTTT-3′ | ||
| Assay | 5′-YGGGAGAYGTTAGYGGYGGGGATGGYGTTTTGTAGAGAGTAGTTTAGGATA-3′ | ||
| THBS2 | NM 001001755 | F | 5′-GGGACACCGCTGATCGTTTA GGGTGTATGTAGAAAAGGGAATGT-3′ |
| R | 5′-TTCAACACGATACTATTCCTACCC-3′ | ||
| Sequencing | 5′-ACATAACTACATCTCCATAT-3′ | ||
| Assay | 5′-ACRTACRCTCCCACAATAAATAAAACAAACRACRACCRCTTAAACRTACAA ACATTCCCTTTTCTACATA C-3′ | ||
| CD44 | NM_204860 | F | 5′-GTTTTTTTAAAATTTGTGTGGTTGT-3′ |
| R | 5′-GGGACACCGCTGATCGTTTA AAACTCCATCAAAAATCACACC-3′ | ||
| Sequencing | 5′-GGTTGTTTAGTTAGAATTTA-3′ | ||
| Assay | 5′-YGGTTTTTYGYGGTTTTTTTTGTTTTGTTTCGTAAT-3′ | ||
| IL12 | NM_213588 | F | 5′-GTCGATGTCGTGTTTTGTTATGT-3′ |
| R | 5′-GGGACACCGCTGATCGTTTA CCACGAAATTCCCAACTCTCA-3′ | ||
| Sequencing | 5′-TTTTGTTCGATGAAATTG-3′ | ||
| Assay | 5′-ATGYGGGATYGGTGGTTGTYGTAGGAGTTGYGTTGTTTTTATGTYGGTGG AGGAGTAGG AGTTTTTTTTT-3′ | ||
| LATS2 | XM417143 | F | 5′-GGGACACCGCTGATCGTTTATTTTGGTAGAAAGTTGGTGTGAAT-3′ |
| R | 5′-CACCATATAACACTTCCCTACCTC-3′ | ||
| Sequencing | 5′-CCTACCTCACAAAAACC-3′ | ||
| Assay | 5′-TCRCCCRTCTTACAAACRATTCACCRTCTCRCCATCTTCTCCCCCRCTCCT TCAACTCRACRAATTCACA CCAACTTTCTACCAAAA-3′ | ||
| GH | NM_204359 | F | 5′-GGGACACCGCTGATCGTTTA GATTGGTGTGGAAAGGAGGAAGA-3′ |
| R | 5′-CAAAAACAAATCGAACCCACAAC-3′ | ||
| Sequencing | 5′-CTCCTACAATTATCCATCC-3′ | ||
| Assay | 5′-CACRTTCTACCTCRTACRACTCAAAAATAAATATACTAAAACT-3′ | ||
| IGF2 | NM_001030342 | F | 5′-AAGTATAACGTGTGGTAGAAGAAGAGTT-3′ |
| R | 5′-GGGACACCGCTGATCGTTTA TCGCCCTAACTTCCTCAACTACT-3′ | ||
| Sequencing | 5′-CGTGTGGTAGAAGAAGAGT-3′ | ||
| Assay | 5′-TYGTAGYGGTTGTAGYGGGAGGTGTTAGGTATTTTGYGTGTTYGTYGGTAT YGGTGGTAGGCGGAGGGGTTGTAAGT-3′ | ||
| TNFSF10 | NM 204379 | F | 5′-GAGGGGAGGTTTAGGTTGGATATT-3′ |
| R | 5′-GGGACACCGCTGATCGTTTA ACCGCCCACATCCCTCAATA-3′ | ||
| Sequencing | 5′-GGGGTGGAGTAGTGGTATA-3′ | ||
| Assay | 5′-GTYGTTYGGGGAGYGGTGGAGTTATYGTTTTTGGAAGTGTTTAGAGTYGTGGGGATGTGGTATTGAGGGATGT-3′ | ||
| H2Ac | NM_001079475 | F | 5′-AGTGGGGGACGTGCGAAATA-3′ |
| R | 5′-GGGACACCGCTGATCGTTTA CCCCGCCCTTCCTCTTTTATAAC-3′ | ||
| Sequencing | 5′-TTATTGGGTAGATTTGGAT-3′ | ||
| Assay | 5′-TYGYGGYGTTATTGGTYGGAGYGAGTGAGAGAGTATATYGGTTAATYGGAAAGYGAGTYG GGTYGTTGYGGGAGGTTATAAAAGAGGAAG GGCG-3′ | ||
| CIITA | NC 006101.2 | F | 5′-CGGGAATTTTTACGTTAGGTTTATAGTG-3′ |
| R | 5′-GGGACACCGCTGATCGTTTAAACGCGAAACGAAAAAACTCCT-3′ | ||
| Sequencing | 5′-TTTTTACGTTAGGTTTATAG-3′ | ||
| Assay | 5′-TGTYGTYGYGGTATTTTAGTYGTTYGGTYGGGTTGYGGGGYGGTTTYGTT TTTTTTGGGGGYGGTTGTGGGAGCGGAGGAGTTTTTT-3′ | ||
| FABP3 | NM 001030889 | F | 5′-AGAGGGGGAAATTGAGGTA-3′ |
| R | 5′-GGGACACCGCTGATCGTTTA AACACACACACACGATCC-3′ | ||
| Sequencing | 5′-GGGGGAAATTGAGGTA-3′ | ||
| Assay | 5′-YGGGAGYGTTYGTGGGGATAYGYGGGATCGTGTGTGTGTGTTGGGGGT-3′ |
Y stands for C/T, and R stands for G/A. Bold Y or R in the assay sequence is the CpG sites analyzed in each region.
Table A2.
Primers for quantitative real-time RT-PCR.
| Genes | Primers | Sequence |
|---|---|---|
| ITGβ5 | F | 5′-GTTTGGGGAGACCTGTGAGA-3′ |
| R | 5′-TCATCCTTGCAGTGCTTTTG-3′ | |
| H2Ac | F | 5′-CGGAAAGCAGGGCGGGAAG-3′ |
| R | 5′-GTCAGGTACTCCAGCACGG-3′ | |
| DNMT1 | F | 5′-CCACCAAAAGGAAATCAGAG-3′ |
| R | 5′-TAATCCTCTTCTCATCTTGCT-3′ | |
| DNMT3a | F | 5′-ATGAACGAGAAGGAAGACATC-3′ |
| R | 5′-GCAAAGAGGTGGCGGATCAC-3′ | |
| DNMT3b | F | 5′-CGTTACTTCTGGGGCAACCTC-3′ |
| R | 5′-ATGACAGGGATGCTCCAGGAC-3′ | |
| GAPDH | F | 5′-GAGGGTAGTGAAGGCTGCTG-3′ |
| R | 5′-ACCAGGAAACAAGCTTGACG-3′ |
Table A3.
Promoter Methylation levels of L63 and L72 not challenged with MDV.
| Genes | Lines | CpG sites |
Note | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | Hypo. | ||
| LATS2 | L63 | 0.73 ± 0.91 | 0.12 ± 0.41 | 3.60 ± 2.33 | 0.56 ± 0.84 | 1.19 ± 1.31 | 0.71 ± 1.30 | 1.13 ± 2.49 | 0.32 ± 0.64 | |
| L72 | 0.78 ± 0.97 | 0.67 ± 0.88 | 3.20 ± 1.32 | 0.78 ± 0.89 | 1.30 ± 1.22 | 0.81 ± 1.27 | 2.09 ± 2.80 | 0.56 ± 0.70 | ||
| MON2 | L63 | 1.04 ± 1.37 | 1.50 ± 1.48 | 1.59 ± 1.80 | 0.99 ± 1.80 | 0.57 ± 1010 | N/A/A | N/A/A | N/A/A | |
| L72 | 1.94 ± 1.11 | 1.34 ± 1.50 | 2.47 ± 3.25 | 1.53 ± 2.09 | 1.01 ± 1.30 | N/A/A | N/A/A | N/A/A | ||
| IL28RA | L63 | 4.77 ± 1.14 | 7.91 ± 1.85 | 2.09 ± 1.47 | 5.00 ± 1.25 | 3.50 ± 0.81 | N/A/A | N/A/A | N/A/A | |
| L72 | 3.90 ± 1.08 | 7.95 ± 1.34 | 2.49 ± 0.56 | 5.10 ± 1.75 | 3.90 ± 1.38 | N/A/A | N/A/A | N/A/A | ||
| STAT1 | L63 | 1.41 ± 1.31 | 2.22 ± 1.22 | 2.97 ± 1.48 | 2.59 ± 1.76 | 1.26 ± 1.17 | 1.98 ± 1.27 | N/A/A | N/A/A | |
| L72 | 1.06 ± 1.28 | 3.26 ± 1.93 | 2.91 ± 1.65 | 3.10 ± 0.70 | 0.71 ± 1.07 | 1.50 ± 1.47 | N/A/A | N/A/A | ||
| CD44 | L63 | 1.53 ± 0.60 | 20.72 ± 7.08 | 0.58 ± 0.16 | 15.10 ± 3.39 | N/A/A | N/A/A | N/A/A | N/A/A | |
| L72 | 1.94 ± 1.02 | 6.73 ± 1.43 | 3.04 ± 0.91 | 6.76 ± 0.93 | N/A/A | N/A/A | N/A/A | N/A/A | ||
| H2Ac | L63 | 4.33 ± 0.57 | 3.55 ± 0.59 | 21.51 ± 1.32 | 29.03 ± 1.76 | 14.83 ± 0.81 | 10.09 ± 0.74 | N/A/A | N/A/A | |
| L72 | 7.81 ± 0.91 | 6.16 ± 1.02 | 31.18 ± 1.75 | 38.50 ± 1.25 | 20.48 ± 0.87 | 16.50 ± 0.87 | N/A/A | N/A/A | ||
| TNFSF10 | L63 | 4.02 ± 0.90 | 9.92 ± 1.09 | 10.33 ± 1.67 | 10.63 ± 2.35 | 38.60 ± 2.04 | N/A/A | N/A/A | N/A/A | |
| L72 | 5.48 ± 1.58 | 8.93 ± 3.23 | 13.15 ± 2.42 | 9.91 ± 3.54 | 25.07 ± 3.75 | N/A/A | N/A/A | N/A/A | ||
| IL12 | L63 | 19.38 ± 6.42 | 12.38 ± 4.22 | 15.60 ± 3.94 | 17.02 ± 4.29 | 7.43 ± 1.76 | N/A/A | N/A/A | N/A/A | |
| L72 | 24.25 ± 4.29 | 14.57 ± 3.30 | 18.30 ± 2.01 | 21.22 ± 4.25 | 8.78 ± 2.16 | N/A/A | N/A/A | N/A/A | ||
| FABP3 | L63 | 35.99 ± 7.36 | 35.24 ± 4.20 | 48.05 ± 7.12 | 32.69 ± 4.39 | 4.16 ± 1.81 | 20.43 ± 8.12 | N/A/A | N/A/A | |
| L72 | 28.22 ± 6.60 | 30.27 ± 8.05 | 39.98 ± 12.15 | 28.38 ± 7.52 | 4.85 ± 1.46 | 17.03 ± 5.14 | N/A/A | N/A/A | ||
| CIITA | L63 | 25.57 ± 4.55 | 29.20 ± 6.72 | 4.26 ± 3.64 | 3.22 ± 2.96 | N/A/A | N/A/A | N/A/A | N/A/A | |
| L72 | 19.76 ± 7.59 | 24.70 ± 8.94 | 4.56 ± 4.72 | 4.30 ± 5.96 | N/A/A | N/A/A | N/A/A | N/A/A | ||
| ITGB5 | L63 | 38.62 ± 4.45 | 46.81 ± 4.08 | 61.94 ± 4.47 | 44.06 ± 4.88 | 37.74 ± 1.32 | 39.63 ± 1.57 | N/A/A | N/A/A | Inter. |
| L72 | 52.04 ± 4.62 | 61.32 ± 4.76 | 71.83 ± 3.54 | 56.37 ± 4.09 | 40.05 ± 1.86 | 41.53 ± 5.97 | N/A/A | N/A/A | ||
| THBS2 | L63 | 34.31 ± 5.01 | 54.07 ± 6.46 | 32.32 ± 4.90 | 22.03 ± 3.13 | 11.37 ± 1.85 | N/A/A | N/A/A | N/A/A | |
| L72 | 49.45 ± 5.16 | 70.68 ± 5.04 | 41.68 ± 4.63 | 30.91 ± 3.82 | 14.08 ± 1.45 | N/A/A | N/A/A | N/A/A | ||
| HDAC9 | L63 | 24.83 ± 6.00 | 31.65 ± 6.03 | 51.50 ± 5.82 | 55.74 ± 4.78 | 40.77 ± 6.22 | N/A/A | N/A/A | N/A/A | |
| L72 | 36.80 ± 5.36 | 45.83 ± 4.20 | 64.86 ± 6.47 | 68.25 ± 3.94 | 54.15 ± 3.48 | N/A/A | N/A/A | N/A/A | ||
| IGF2 | L63 | 89.69 ± 4.08 | 89.25 ± 2.35 | 88.05 ± 3.85 | 77.81 ± 3.72 | 79.39 ± 3.46 | 49.77 ± 3.85 | 68.19 ± 7.40 | 88.95 ± 1.92 | Hyper. |
| L72 | 91.55 ± 3.95 | 92.06 ± 2.54 | 89.78 ± 2.74 | 81.15 ± 4.68 | 82.37 ± 10.41 | 52.64 ± 5.77 | 72.41 ± 6.71 | 89.97 ± 2.02 | ||
| GH1 | L63 | 63.12 ± 2.75 | 48.64 ± 2.51 | 80.64 ± 2.54 | N/A/A | N/A/A | N/A/A | N/A/A | N/A/A | |
| L72 | 61.26 ± 2.19 | 45.13 ± 1.90 | 79.23 ± 1.91 | N/A/A | N/A/A | N/A/A | N/A/A | N/A/A | ||
| NK-lysin | L63 | 89.47 ± 2.33 | 42.98 ± 5.29 | 82.46 ± 2.20 | 78.32 ± 1.72 | 62.63 ± 2.16 | N/A/A | N/A/A | N/A/A | |
| L72 | 91.79 ± 2.63 | 40.57 ± 5.47 | 80.53 ± 2.45 | 78.14 ± 1.88 | 66.56 ± 1.52 | N/A/A | N/A/A | N/A/A | ||
| TGFB3 | L63 | 88.04 ± 1.60 | 90.98 ± 2.19 | 90.39 ± 2.28 | 80.86 ± 1.60 | 68.43 ± 2.13 | 74.39 ± 3.46 | N/A/A | N/A/A | |
| L72 | 89.88 ± 2.59 | 92.91 ± 2.58 | 93.51 ± 2.66 | 82.90 ± 2.09 | 72.94 ± 3.09 | 77.00 ± 2.68 | N/A/A | N/A/A | ||
| CD82 | L63 | 3.86 ± 1.56 | 5.24 ± 1.86 | 3.97 ± 1.61 | 3.98 ± 0.98 | 3.65 ± 2.02 | 1.88 ± 1.36 | 40.48 ± 5.54 | 52.78 ± 3.63 | Hypo + |
| L72 | 3.16 ± 0.99 | 4.41 ± 1.31 | 3.59 ± 1.12 | 4.41 ± 1.40 | 2.69 ± 1.23 | 1.85 ± 1.27 | 41.22 ± 5.29 | 51.61 ± 2.67 | Inter. | |
Methylation level shown in each cell = mean ± STD.
Hypo., hypomethylation; Hyper., hypermethylation; Inter., intermediate methylation.
N/A, data not available.
N = 12 for each group.
Table A4.
Promoter methylation level change at different CpG sites of genes after MDV challenge.
| Gene name | Time points (dpi) | Lines | % Methylation level change after MDV infection of different CpG sites |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | |||
| GH | 5 | L63 | −5.09** | −7.44** | −1.60 | |||||
| L72 | 3.29 | 1.50 | 4.07 | |||||||
| 10 | L63 | 2.73* | −1.00 | 0.95 | ||||||
| L72 | 3.75* | 1.04 | −0.33 | |||||||
| 21 | L63 | 0.50 | 2.08* | −0.09 | ||||||
| L72 | 6.87* | 7.10** | 4.23** | |||||||
| CD44 | 5 | L63 | −0.23 | 0.71 | −0.02 | 0.40 | ||||
| L72 | 0.40 | −4.25 | −1.26 | 0.36 | ||||||
| 10 | L63 | −0.56 | 2.39 | −0.13 | −1.63 | |||||
| L72 | 0.32 | 0.29 | 1.34 | 0.59 | ||||||
| 21 | L63 | 0.03 | −3.30 | 0.21 | 0.16 | |||||
| L72 | −0.77 | −3.17** | −1.57 | 1.39 | ||||||
| CIITA | 5 | L63 | 4.41 | −3.23 | 1.50 | 5.11 | ||||
| L72 | −11.86* | −12.67 | −2.78 | −2.74 | ||||||
| 10 | L63 | −2.21 | 0.29 | 0.59 | −1.60 | |||||
| L72 | 1.04 | 0.28 | 1.19 | 0.24 | ||||||
| 21 | L63 | −10.17** | −6.12* | 0.34 | 1.27 | |||||
| L72 | −6.95** | −5.83* | 0.36 | −1.18 | ||||||
| NK-lysin | 5 | L63 | 4.24* | 18.21** | 6.25** | 6.72** | 1.20 | |||
| L72 | 2.69 | 29.14** | 4.23 | 4.82* | −0.40 | |||||
| 10 | L63 | 1.61 | 8.62 | 1.63 | 3.46 | 1.28 | ||||
| L72 | −1.03 | 7.60 | 1.63 | 1.33 | 2.90 | |||||
| 21 | L63 | −0.92* | −3.59 | −1.22 | −1.55 | −1.69** | ||||
| L72 | −1.69** | −2.27 | −2.06 | −2.00* | −1.99 | |||||
| THBS2 | 5 | L63 | 8.06 | 7.03 | 5.51 | 3.73 | 4.83 | |||
| L72 | −1.75 | 5.07 | 1.58 | 8.38 | 2.97 | |||||
| 10 | L63 | 5.74 | 6.32 | 0.61 | 2.03 | 0.05 | ||||
| L72 | −1.25 | 1.46 | 0.33 | −0.43 | 1.35 | |||||
| 21 | L63 | −1.69 | −1.38 | 2.49 | 1.50 | 2.07 | ||||
| L72 | 1.31 | 1.06 | −0.20 | −6.24 | −0.06 | |||||
| M0N2 | 5 | L63 | 0.63 | −1.33 | −2.87* | −1.69 | −0.10 | |||
| L72 | −1.43 | 2.22 | 0.35 | −3.06 | −0.09 | |||||
| 10 | L63 | 1.86 | −0.62 | 0.59 | 0.75 | 0.41 | ||||
| L72 | −1.17 | −0.76 | −5.96* | 0.42 | −0.52 | |||||
| 21 | L63 | −0.33 | 0.43 | 0.56 | 0.29 | −0.33 | ||||
| L72 | 0.94 | −1.18 | −0.63 | 0.64 | −0.06 | |||||
| HDAC9 | 5 | L63 | 5.28 | 8.65 | 6.56 | 6.04 | 7.54 | |||
| L72 | 3.37 | 11.67** | −4.57 | 0.48 | −3.20 | |||||
| 10 | L63 | −4.07 | 1.41 | 3.86 | −2.11 | 3.13 | ||||
| L72 | −1.01 | 0.17 | 3.47 | 0.07 | −1.60 | |||||
| 21 | L63 | 3.18 | 1.08 | 3.80 | 2.90 | 4.86 | ||||
| L72 | 12.56** | 7.00** | 4.48 | 4.59 | 10.99** | |||||
| FABP3 | 5 | L63 | 7.07 | 9.34* | 15.81* | 8.58* | 3.22* | |||
| L72 | 7.75 | 7.22 | 10.96 | 8.09 | 0.13 | |||||
| 10 | L63 | 3.43 | 4.52 | 8.48 | 4.50 | 0.92 | ||||
| L72 | −9.44* | −11.67** | −11.80** | −10.82** | −2.31** | |||||
| 21 | L63 | −8.35 | −1.06 | −2.23 | 0.03 | 2.81 | ||||
| L72 | 5.40 | 4.11 | 7.09 | 4.15 | 2.74 | |||||
| IL28RA | 5 | L63 | −1.87 | −3.97* | −0.54 | −0.92 | −0.46 | |||
| L72 | −2.75* | −0.64 | −1.12** | −1.80 | 1.50 | |||||
| 10 | L63 | −2.05** | −0.60 | −0.77 | −0.65 | 0.02 | ||||
| L72 | −1.67* | −1.01 | 0.07 | −1.15 | 0.70 | |||||
| 21 | L63 | 0.81 | 0.03 | 1.20 | 1.41 | 0.04 | ||||
| L72 | 0.11 | 0.65 | −0.58 | −0.12 | 0.21 | |||||
| IL12 | 5 | L63 | 3.19* | 4.27 | −0.42 | 0.66 | −1.46* | |||
| L72 | −0.68 | −0.68 | 0.13 | −4.91 | −1.43 | |||||
| 10 | L63 | 0.39 | −0.88 | 0.60 | −3.44 | −0.28 | ||||
| L72 | −1.07 | −4.03 | −1.05 | −5.39 | −1.00 | |||||
| 21 | L63 | −2.02 | −3.49 | −3.58 | 0.12 | −0.32 | ||||
| L72 | 3.06 | 1.85 | −0.17 | −3.05 | 1.08 | |||||
| TNFSF10 | 5 | L63 | 0.23 | −1.73* | −1.70 | −1.24 | −1.11 | |||
| L72 | −0.25 | −2.79* | −3.72* | −1.43 | −2.24 | |||||
| 10 | L63 | 1.23 | 0.48 | −0.19 | 0.81 | −2.94 | ||||
| L72 | −1.01 | 0.68 | −1.19 | 0.06 | 1.80 | |||||
| 21 | L63 | 0.38 | 0.12 | 0.19 | −1.29 | −1.54 | ||||
| L72 | −1.37 | 2.79 | −2.24 | 2.07 | 3.67 | |||||
| ITGB5 | 5 | L63 | 21.99** | 24.91** | 25.41** | 21.58** | 1.54* | 0.38 | ||
| L72 | 16.68** | 20.25** | 17.04** | 15.07** | 1.73 | −0.77 | ||||
| 10 | L63 | 3.73 | −0.14 | −3.03 | −1.15 | −0.24 | −2.14 | |||
| L72 | −0.81 | −2.01 | −0.54 | −1.81 | −1.25 | −2.76 | ||||
| 21 | L63 | 6.85 | 7.81 | 5.15 | 6.66 | 2.51 | 1.05 | |||
| L72 | 8.92 | 5.84 | 5.91 | 7.19 | 2.30 | 1.75 | ||||
| STAT1 | 5 | L63 | −1.18 | −1.20 | −1.13 | −1.69 | −0.30 | −0.19 | ||
| L72 | 0.77 | −1.10 | 0.23 | −0.48 | 2.05* | 1.07 | ||||
| 10 | L63 | 1.51* | −0.35 | −0.55 | 2.24** | −0.62 | 0.64 | |||
| L72 | 0.12 | −0.86 | 0.01 | 1.39** | −0.36 | 0.53 | ||||
| 21 | L63 | 0.95 | −1.35 | −1.98 | −0.10 | 3.69* | 1.46 | |||
| L72 | 0.58 | −3.86* | −1.37 | −1.86* | 1.42 | 2.08* | ||||
| TGFB3 | 5 | L63 | −0.44 | 4.22** | 5.32 | 0.25** | 6.90 | 5.63 | ||
| L72 | 3.62 | 2.66 | 2.97* | 2.38 | 2.15 | −1.95 | ||||
| 10 | L63 | 1.34 | 0.84 | −0.85** | 3.36** | 5.54 | 3.20 | |||
| L72 | −0.57 | −2.24 | −0.10 | 1.58 | 4.74 | 1.38 | ||||
| 21 | L63 | 2.38 | 3.58 | 3.92* | −0.33 | −1.25 | −4.07 | |||
| L72 | −0.19 | 0.69 | −0.34 | −1.03 | 0.80 | −0.13 | ||||
| H2AC | 5 | L63 | 0.63 | 0.65* | 2.26 | 1.26 | 1.26 | 0.60 | ||
| L72 | 2.79 | 1.25 | 4.58 | 3.69* | 1.83 | 1.55 | ||||
| 10 | L63 | 1.14 | 1.13 | 4.78 | 4.98 | 2.27* | 0.85** | |||
| L72 | −0.15 | 0.14 | −1.76** | −1.40 | 0.12 | −0.28 | ||||
| 21 | L63 | 0.60 | 0.25 | 0.95 | 1.61 | 0.22 | 1.04 | |||
| L72 | −1.47* | 0.87 | −3.09** | −3.88** | −0.72 | 0.13 | ||||
| CD82 | 5 | L63 | 1.85 | 2.32 | 0.33 | 0.50 | 0.18 | −0.47 | −4.79 | −2.36 |
| L72 | 2.06 | 0.04 | 2.27 | 0.39 | −0.57 | −0.15 | −5.52 | −2.09 | ||
| 10 | L63 | −1.87* | −2.07** | −1.42 | 0.92 | −2.17 | 3.88* | 1.60 | −0.24 | |
| L72 | 0.55 | −0.80 | −1.08 | 0.00 | 0.86 | 2.65 | 0.09 | 1.27 | ||
| 21 | L63 | 0.21 | 0.88* | −0.18 | 0.47 | −0.50 | 0.44 | −0.85 | −1.30* | |
| L72 | 2.01** | 0.94* | 1.19 | 3.10** | 0.06 | 1.48 | −1.77 | −0.47 | ||
| IGF2 | 5 | L63 | 0.02* | 2.83 | 0.16 | 3.50 | −0.32 | 3.14 | 6.32* | 2.45* |
| L72 | 2.19 | 1.47 | 0.82 | 3.70 | −1.45 | 3.28 | 5.75 | 0.53 | ||
| 10 | L63 | −1.36 | 1.05 | 1.93 | 2.18 | 2.69 | 3.44 | 0.49 | 0.30 | |
| L72 | −4.09 | −1.95 | −1.67 | 3.24 | 4.55* | 3.60 | −0.21 | 0.02 | ||
| 21 | L63 | 0.34 | 0.64 | −0.12 | 0.79 | 1.62 | 2.33 | −3.51 | 0.22 | |
| L72 | −1.23 | 0.10 | −1.18 | −1.32 | 4.91 | 5.46 | 2.08 | −0.60 | ||
| LATS2 | 5 | L63 | −0.21 | 1.04 | −0.87 | 1.08 | 0.09 | 1.14 | 1.57 | 0.52 |
| L72 | 0.32 | 0.25 | −0.33 | 0.81 | 0.94 | 1.42 | −1.65 | 0.48 | ||
| 10 | L63 | −0.02 | 0.27 | −0.26 | 0.24 | −0.87 | −0.23 | 0.85 | −0.03 | |
| L72 | 0.77 | 0.74 | 0.82 | 0.42 | −0.27 | 0.55 | 0.41 | 0.38 | ||
| 21 | L63 | 1.64* | 0.67 | 0.80 | 0.62 | 0.69 | 0.48 | 1.31 | 0.60 | |
| L72 | 1.90* | 0.74 | −0.22 | 0.70 | 0.35 | 0.02 | 1.15 | 0.84 | ||
*P < 0.05, **P < 0.01.
Figure A1.
Promoter methylation change during MDV life cycle in GH (A), HDAC9 (B), CIITA (C), STAT1 (D), H2Ac (E), LATS2 (F), and FABP3 (G) The methylation change within lines was the methylation changed after MDV infection compared with the promoter methylation level before MDV infection. N = 4 for each group.
Footnotes
References
- Abdul-Careem M. F., Hunter B. D., Sarson A. J., Mayameei A., Zhou H., Sharif S. (2006). Marek’s disease virus-induced transient paralysis is associated with cytokine gene expression in the nervous system. Viral Immunol. 19, 167–176 10.1089/vim.2006.19.167 [DOI] [PubMed] [Google Scholar]
- Akimoto K., Katakami H., Kim H. J., Ogawa E., Sano C. M., Wada Y., Sano H. (2007). Epigenetic inheritance in rice plants. Ann. Bot. 100, 205–217 10.1093/aob/mcm110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allis C. D., Jenuwein T., Reinberg D., Caparros M.-L. (2006). Epigenetics. Cold Spring Harbor: Laboratory Press [Google Scholar]
- Asmundson V. S., Biely J. (1932). Inheritance and resistance to fowl paralysis (neuro-lymphomatosis gallinarum). Can. J. Res. 6, 171–176 10.1139/cjr32-012 [DOI] [Google Scholar]
- Bacon L. D., Hunt H. D., Cheng H. H. (2000). A review of the development of chicken lines to resolve genes determining resistance to diseases. Poult. Sci. 79, 1082–1093 [DOI] [PubMed] [Google Scholar]
- Baigent S. J., Davison T. F. (1999). Development and composition of lymphoid lesions in the spleens of Marek’s disease virus-infected chickens: association with virus spread and the pathogenesis of Marek’s disease. Avian Pathol. 28, 4. 10.1080/03079459994786 [DOI] [PubMed] [Google Scholar]
- Bakin A. V., Curran T. (1999). Role of DNA 5-methylcytosine transferase in cell transformation by fos. Science 283, 387–390 10.1126/science.283.5400.387 [DOI] [PubMed] [Google Scholar]
- Bishop S. C., Axford R. F. E., Nicholas F. W., Owen J. B. (eds). (2010). Breeding for Disease Resistance in Farm Animals, 3rd Edn Oxfordshire: CABI [Google Scholar]
- Calnek B. W. (1986). Marek’s disease – a model for herpesvirus oncology. Crit. Rev. Microbiol. 12, 293–320 10.3109/10408418509104432 [DOI] [PubMed] [Google Scholar]
- Calnek B. W. (2001). Pathogenesis of Marek’s disease virus infection. Curr. Top. Microbiol. Immunol. 255, 25–55 10.1007/978-3-642-56863-3_2 [DOI] [PubMed] [Google Scholar]
- Chuang L. S., Ian H. I., Koh T. W., Ng H. H., Xu G., Li B. F. (1997). Human DNA-(cytosine-5) methyltransferase-PCNA complex as a target for p21WAF1. Science 277, 1996–2000 10.1126/science.277.5334.1996 [DOI] [PubMed] [Google Scholar]
- Churchill A. E., Payne L. N., Chubb R. C. (1969). Immunization against Marek’s disease using a live attenuated virus. Nature 221, 744–747 10.1038/221744a0 [DOI] [PubMed] [Google Scholar]
- Colella S., Shen L., Baggerly K. A., Issa J. P., Krahe R. (2003). Sensitive and quantitative universal Pyrosequencing methylation analysis of CpG sites. Biotechniques 35, 146–150 [DOI] [PubMed] [Google Scholar]
- Davison A. J. (2002). Evolution of the herpesviruses. Vet. Microbiol. 86, 69–88 10.1016/S0378-1135(01)00492-8 [DOI] [PubMed] [Google Scholar]
- Davison F., Nair V. (2004). Marek’s Disease: An Evolving Problem. Oxford: Elsevier Academic Press [Google Scholar]
- Ehrlich M. (2009). DNA hypomethylation in cancer cells. Epigenomics 1, 239–259 10.2217/epi.09.33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epstein M. A. (2001). Historical background. Philos. Trans. R. Soc. Lond. B Biol. Sci. 356, 413–420 10.1098/rstb.2000.0774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg A. P., Tycko B. (2004). The history of cancer epigenetics. Nat. Rev. Cancer 4, 143–153 10.1038/nrc1279 [DOI] [PubMed] [Google Scholar]
- Feng S., Jacobsen S. E., Reik W. (2010). Epigenetic reprogramming in plant and animal development. Science 330, 622–627 10.1126/science.1190614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fredericksen T. L., Longenecker B. M., Pazderka F., Gilmour D. G., Ruth R. F. (1977). A T-cell antigen system of chickens: Ly-4 and Marek’s disease. Immunogenetics 5, 535–552 10.1007/BF01570512 [DOI] [Google Scholar]
- Gilmour D. G., Brand A., Donnelly N., Stone H. A. (1976). Bu-1 and Th-1, two loci determining surface antigens of B or T lymphocytes in the chicken. Immunogenetics 3, 549–563 10.1007/BF01576985 [DOI] [Google Scholar]
- Hanahan D., Weinberg R. A. (2000) , The hallmarks of cancer. Cell 100, 57–70 10.1016/S0092-8674(00)81683-9 [DOI] [PubMed] [Google Scholar]
- Hanahan D., Weinberg R. A. (2011). Hallmarks of cancer: the next generation. Cell 144, 646–674 10.1016/j.cell.2011.02.013 [DOI] [PubMed] [Google Scholar]
- Heidari M., Sarson A. J., Huebner M., Sharif S., Kireev D., Zhou H. (2010). Marek’s disease virus-induced immunosuppression: array analysis of chicken immune response gene expression profiling. Viral Immunol. 23, 309–319 10.1089/vim.2009.0079 [DOI] [PubMed] [Google Scholar]
- Jelinek J., Gharibyan V., Estecio M. R., Kondo K., He R., Chung W., Lu Y., Zhang N., Liang S., Kantarjian H. M., Cortes J. E., Issa J. P. (2011). Aberrant DNA methylation is associated with disease progression, resistance to imatinib and shortened survival in chronic myelogenous leukemia. PLoS ONE 6, e22110. 10.1371/journal.pone.0022110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser P., Underwood G., Davison F. (2003). Differential cytokine responses following Marek’s disease virus infection of chickens differing in resistance to Marek’s disease. J. Virol. 77, 762–768 10.1128/JVI.77.1.762-768.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katto J., Mahlknecht U. (2011). Epigenetic regulation of cellular adhesion in cancer. Carcinogenesis 32, 1414–1418 10.1093/carcin/bgr120 [DOI] [PubMed] [Google Scholar]
- Liu H. C., Cheng H. H., Tirunagaru V., Sofer L., Burnside J. (2001). A strategy to identify positional candidate genes conferring Marek’s disease resistance by integrating DNA microarrays and genetic mapping. Anim. Genet. 32, 351–359 10.1046/j.1365-2052.2001.00798.x [DOI] [PubMed] [Google Scholar]
- Luo J., Yu Y., Song J. (2012). “Epigenetics and animal health,” in Livestock Epigenetics, ed. Khatib H. (Oxford: Wiley-Blackwell; ), 10.1002/9781119949930.ch8 [DOI] [Google Scholar]
- Luo J., Yu Y., Zhang H., Tian F., Chang S., Cheng H. H., Song J. (2011). Down-regulation of promoter methylation level of CD4 gene after MDV infection in MD-susceptible chicken line. BMC Proc. 5(Suppl. 4), S7. 10.1186/1753-6561-5-S9-S7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morimura T., Ohashi K., Kon Y., Hattori M., Sugimoto C., Onuma M. (1996). Apoptosis and CD8-down-regulation in the thymus of chickens infected with Marek’s disease virus. Arch. Virol. 141, 2243–2249 10.1007/BF01718230 [DOI] [PubMed] [Google Scholar]
- Novak P., Jensen T. J., Garbe J. C., Stampfer M. R., Futscher B. W. (2009). Stepwise DNA methylation changes are linked to escape from defined proliferation barriers and mammary epithelial cell immortalization. Cancer Res. 69, 5251–5258 10.1158/0008-5472.CAN-08-4977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osterrieder N., Kamil J. P., Schumacher D., Tischer B. K., Trapp S. (2006). Marek’s disease virus: from miasma to model. Nat. Rev. Microbiol. 4, 283–294 10.1038/nrmicro1382 [DOI] [PubMed] [Google Scholar]
- Parcells M. S., Burgess S. C. (2008). Immunological aspects of Marek’s disease virus (MDV)-induced lymphoma progression. Cancer Growth Progression 11, 169 [Google Scholar]
- Robert M. F., Morin S., Beaulieu N., Gauthier F., Chute I. C., Barsalou A., MacLeod A. R. (2003). DNMT1 is required to maintain CpG methylation and aberrant gene silencing in human cancer cells. Nat. Genet. 33, 61–65 10.1038/ng1068 [DOI] [PubMed] [Google Scholar]
- Sarson A. J., Parvizi P., Lepp D., Quinton M., Sharif S. (2008a). Transcriptional analysis of host responses to Marek’s disease virus infection in genetically resistant and susceptible chickens. Anim. Genet. 39, 232–240 10.1111/j.1365-2052.2008.01710.x [DOI] [PubMed] [Google Scholar]
- Sarson A. J., Abdul-Careem M. F., Read L. R., Brisbin J. T., Sharif S. (2008b). Expression of cytotoxicity-associated genes in Marek’s disease virus-infected chickens. Viral Immunol. 21, 267–272 10.1089/vim.2007.0094 [DOI] [PubMed] [Google Scholar]
- Soejima K., Fang W., Rollins B. J. (2003). DNA methyltransferase 3b contributes to oncogenic transformation induced by SV40T antigen and activated Ras. Oncogene 22, 4723–4733 10.1038/sj.onc.1206510 [DOI] [PubMed] [Google Scholar]
- Thanthrige-Don N., Read L. R., Abdul-Careem M. F., Mohammadi H., Mallick A. I., Sharif S. (2010). Marek’s disease virus influences the expression of genes associated with IFN-gamma-inducible MHC class II expression. Viral Immunol. 23, 227–232 10.1089/vim.2009.0092 [DOI] [PubMed] [Google Scholar]
- Tsai C. N., Tsai C. L., Tse K. P., Chang H. Y., Chang Y. S. (2002). The Epstein-Barr virus oncogene product, latent membrane protein 1, induces the downregulation of E-cadherin gene expression via activation of DNA methyltransferases. Proc. Natl. Acad. Sci. U.S.A. 99, 10084–10089 10.1073/pnas.262499599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witter R. L. (1997). Increased virulence of Marek’s disease virus field isolates. Avian Dis. 41, 149–163 10.2307/1592455 [DOI] [PubMed] [Google Scholar]
- Yamasaki H., Mesnil M., Omori Y., Mironov N., Krutovskikh V. (1995). Intercellular communication and carcinogenesis. Mutat. Res. 333, 181–188 10.1016/0027-5107(95)00144-1 [DOI] [PubMed] [Google Scholar]
- Young W. B., Lindberg G. L., Link C. J., Jr. (2000). DNA methylation of helper virus increases genetic instability of retroviral vector producer cells. J. Virol. 74, 3177–3187 10.1128/JVI.74.11.5242-5249.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y., Luo J., Mitra A., Chang S., Tian F., Zhang H., Yuan P., Zhou H., Song J. (2011). Temporal transcriptome changes induced by MDV in marek’s disease-resistant and -susceptible inbred chickens. BMC Genomics 12:501. 10.1186/1471-2164-12-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y., Zhang H., Tian F., Zhang W., Fang H., Song J. (2008a). An integrated epigenetic and genetic analysis of DNA methyltransferase genes (DNMTs) in tumor resistant and susceptible chicken lines. PLoS ONE 3, e2672. 10.1371/journal.pone.0002672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y., Zhang H., Tian F., Bacon L., Zhang Y., Zhang W., Song J. (2008b). Quantitative evaluation of DNA methylation patterns for ALVE and TVB genes in a neoplastic disease susceptible and resistant chicken model. PLoS ONE 3, e1731. 10.1371/journal.pone.0001731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng D. L., Zhang L., Cheng N., Xu X., Deng Q., Teng X. M., Wang K. S., Zhang X., Huang J., Han Z. G. (2008). Epigenetic modification induced by hepatitis B virus X protein via interaction with de novo DNA methyltransferase DNMT3A. J. Hepatol. 50, 377–387 10.1016/j.jhep.2008.10.019 [DOI] [PubMed] [Google Scholar]