Abstract
FlaSh-YFP, a fluorescent protein (FP) voltage sensor that is a fusion of the Shaker potassium channel with yellow fluorescent protein (YFP), is primarily expressed in the endoplasmic reticulum (ER) of mammalian cells, possibly due to misfolded monomers. In an effort to improve plasma membrane expression, the FP was split into two non-fluorescent halves. Each half was randomly inserted into Shaker monomers via a transposon reaction. Shaker subunits containing the 5' half were co-expressed with Shaker subunits containing the 3' half. Tetramerization of Shaker subunits is required for re-conjugation of the FP. The misfolded monomers trapped in ER are unlikely to tetramerize and reconstitute the beta-can structure, and thus intracellular fluorescence might be reduced. This split-can transposon approach yielded 56 fluorescent probes, 30 (54%) of which were expressed at the plasma membrane and were capable of optically reporting changes in membrane potential. The largest signal from these novel FP-sensors was a −1.4% in ΔF/F for a 100 mV depolarization, with on time constants of about 15 ms and off time constants of about 200 ms. This split-can transposon approach has the potential to improve other multimeric probes.
Keywords: Split-can, transposon, Venus, Shaker, membrane localization, FP voltage sensor, genetically encoded voltage sensor
1 Introduction
Imaging of neuronal activity can provide detailed spatial information about changes in calcium concentration (Brown et al., 1975), pH (Tolkovsky and Richards, 1987), hemodynamics (Grinvald et al., 1986; Kwong et al., 1992) and membrane potential (Davila et al., 1973). Voltage imaging can simultaneously record activity in many locations of a single cell (Canepari et al., 2008; Grinvald et al., 1981), the activities of multiple cells (Salzberg et al., 1977; Zecević et al., 1989) or population signals from many brain regions (Grinvald et al., 1982; Zochowski and Cohen, 2005).
There are two types of voltage sensitive optical probes, organic dyes and genetically-encoded probes (FP voltage sensors). While the organic dyes are widely used with their characteristics of high sensitivity, fast dynamics (Loew et al., 1985) and linear response (Gupta et al., 1981), these organic dyes lack staining specificity. Genetically-encoded probes are not as well developed, but they are able to achieve cell type specificity because their expression can be determined by a cell type specific promoter.
Thus far FP voltage sensors have been conjugates of membrane resident voltage sensors and fluorescent proteins (FPs). The voltage sensors have been either voltage-gated ion channels or voltage-sensitive phosphatases (Baker et al., 2008). The conformation change of the voltage sensor during a membrane voltage change either directly changes the FP conformation or influences the environment surrounding the chromophore, resulting in a fluorescence change.
It is essential for the FP voltage sensor to be expressed in the plasma membrane where the probe can sense the voltage change. Ideally, all of the fluorescence would come from membrane bound sensor. Intense intracellular fluorescence will increase the non-responsive background intensity and thereby decrease the voltage sensitivity (ΔF/F) and signal-to-noise ratio. Unfortunately, very little of the first generation FP voltage sensors reached the plasma membrane in mammalian cells even though they expressed well in Xenopus oocytes. The first FP voltage sensor, FlaSh (Siegel and Isacoff, 1997), did not work well in mammalian systems due to poor membrane targeting and high background fluorescence (Carter and Shieh, 2010). Three other first generation FP voltage sensors, Flare (Baker et al., 2007), SPARC (Ataka and Pieribone, 2002), and VSFP-1 (Sakai et al., 2001) all had poor plasma membrane expression but strong intracellular fluorescence in Human Embryonic Kidney (HEK) 293 cells and in primary cultured hippocampal neurons (Baker et al., 2007). The perinuclear expression pattern of the three probes suggested that they were retained in the ER or Golgi apparatus. No voltage sensitive signal was recorded from these three constructs in mammalian cells (Baker et al., 2007). The second and third generation voltage sensitive probes contain the voltage sensitive phosphatase from Ciona intestinalis (Ci-VSP), instead of ion channels. These probes are well localized to the plasma membrane in mammalian cells (Akemann et al., 2010; Tsutsui et al., 2008). We tried to find a way to improve membrane expression of probes using ion channels in mammalian cells, so that we wouldn't be limited to a single type of voltage sensor for probe design.
Several different FPs have been used to replace wtGFP in FlaSh, including YFP (Guerrero et al., 2002). We attempted to reduce the intracellular fluorescence of FlaSh-YFP by rendering misfolded protein nonfluorescent. FlaSh-YFP, a derivative of the drosophila Shaker potassium channel, is a tetramer that needs to assemble to leave the ER (Nagaya and Papazian, 1997; Reddy and Corley, 1998; Robinson and Deutsch, 2005). Our strategy was to use split-cans and insert the two halves into separate monomers which can fluoresce only after tetramerization. When the fluorescent protein (FP) is divided properly into two fragments, neither half fluoresces (Ozawa, 2006; Wilson et al., 2004). However, the two halves can give rise to a functional chromophore when brought into proper proximity. This split-can strategy has been used to report protein-protein interactions (Jackrel et al., 2010; Ozawa, 2009) and calcium sensitive protein conjugation (Lindman et al., 2009).
For these experiments the YFP of FlaSh-YFP was replaced with venus, which has mutations to improve its maturation and brightness (Nagai et al., 2002). Venus was split into two non-fluorescent half-cans, which were fused individually with inactivated Shaker W434F α-subunit monomers. Two subunits containing the different halves of venus were co-expressed in mammalian cells with the hope that the tetramerization of the correctly folded monomers would induce the assembly of the split-can halves. This split-can strategy offers two possible improvements. Misfolded monomers would be unlikely to tetramerize and thereby remain non-fluorescent. Second, because each subunit no longer needs to accommodate a complete beta-can structure, correct folding may be more likely.
Because FlaSh-YFP’s original YFP insertion site in the Shaker potassium channel downstream of the S6 domain may not be the optimal position to allow the two half-cans to get close enough to assemble, potential insertion sites were explored for each of the two half-cans using transposon reactions to pseudo-randomly insert the venus halves at various locations in the channel protein (Mealer et al., 2008).
Of the 120 combinations that we tested, 44 combinations had fluorescence on the plasma membrane, and 30 of these had detectable voltage sensitivity. The largest sensitivity was a −1.4% change in ΔF/F for a 100 mV depolarization. Many of the probes had a relatively fast on time constants of about 15 milliseconds, but much slower off time constants of about 200 milliseconds.
2 Material and Methods
2.1 Split-can venus
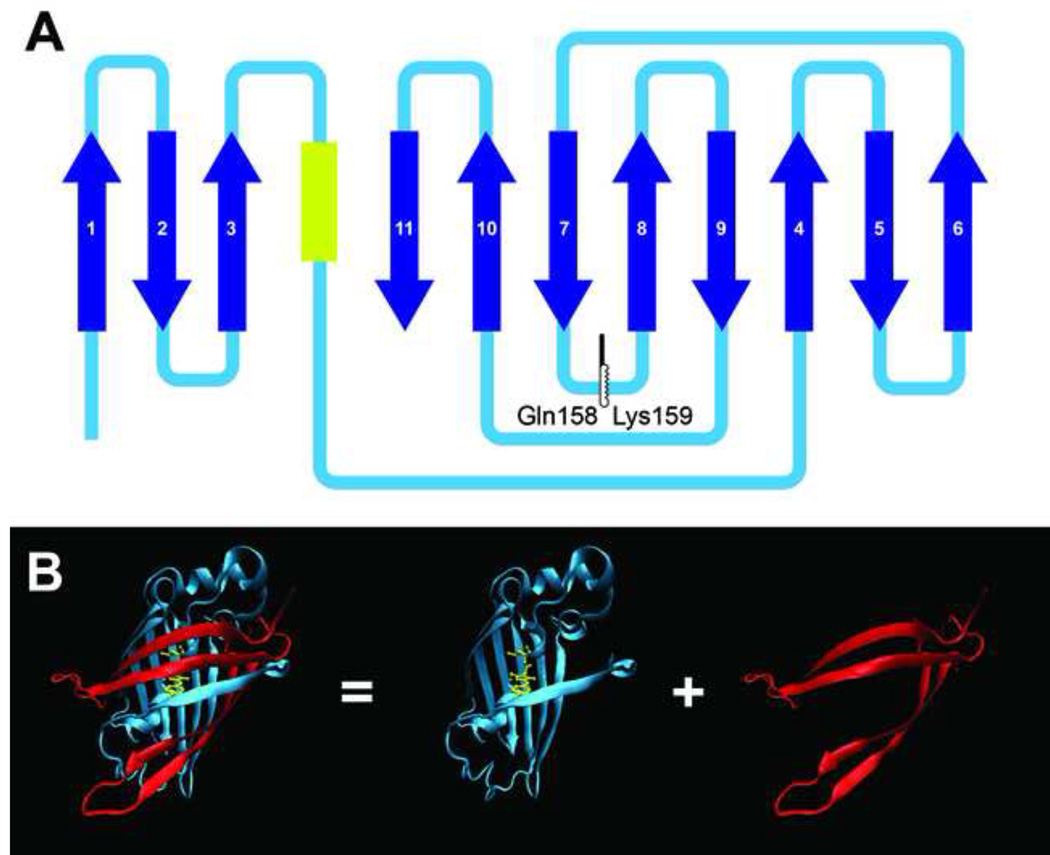
The full length venus was split between Glutamine158 and Lysine159 (figure 1A) (Mealer et al., 2006). The front half contains most of the can structure and the chromophore, and the rear half contains four β-sheets of the can structure (figure 1B).
Figure 1. Split-can venus.
A) The venus is split into two segments at Glutamine158/Lysine159. The yellow segment indicates the chromophore. The dark blue arrows indicate the β-sheets. B) The 3D structure of full length venus (PDB file 1MYW) (Rekas et al., 2002) and the two segments. The 3D structure is rendered by the Visual Molecular Dynamics (VMD) program. (Humphrey et al., 1996)
2.2 Transposon Reaction
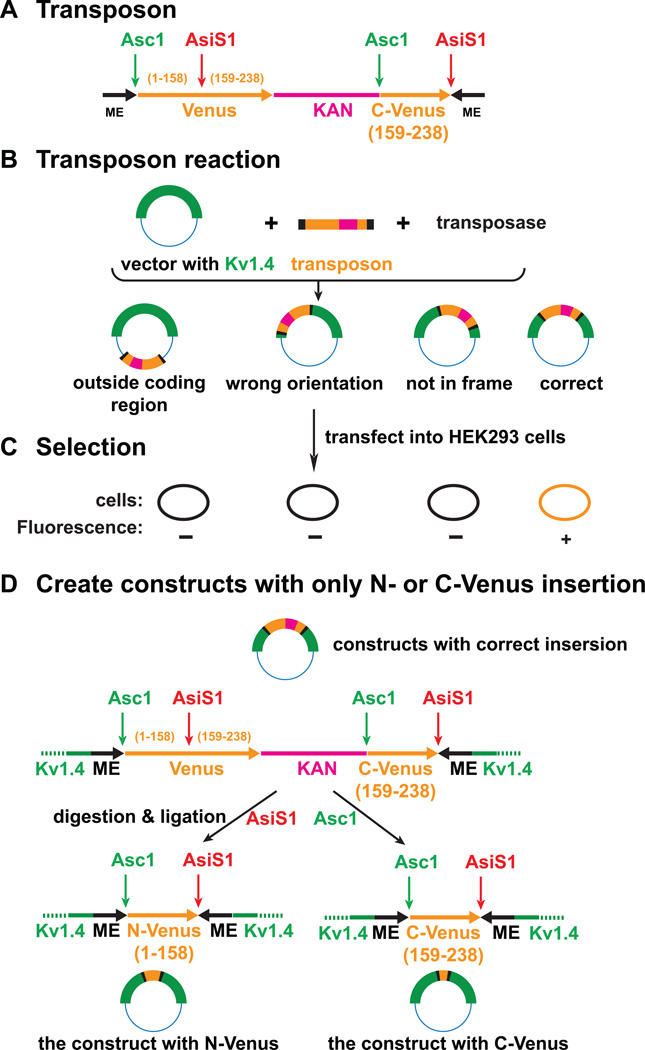
To randomly insert fluorescent protein fragments into the sequence encoding the Shaker subunit, we created a new synthetic Tn5 transposon (Pralle et al., 2006). The transposon carries two mosaic end (ME) sequences required for the transposition, a complete coding sequence for venus, a Kanamycin resistance and a C terminal fragment of venus (figure 2A). The transposase inserted this transposon into various sites of a vector encoding the Shaker W434F mutant sequence (figure 2B). The initial products then were selected by detecting fluorescence in transiently transfected HEK293 cells to identify the correct insertions, which are in the coding region, in frame, and in the correct orientation (figure 2C). AsiS1 and Asc1 restriction sites are positioned within the transposon. Restriction digestions with either AsiS1 or Asc1 and ligations produced full-length Shaker subunits with either N-terminal or C-terminal segments of venus at the original transposon insertion site (figure 2D).
Figure 2. Preparation of the DNA constructs.
A) Structure of the Tn5 transposon: The black segments: 19-bp mosaic end (ME) sequences required for transposition. The orange segment on left: a full length sequence for venus. The pink segment: the sequence for Kanamycin resistance. The orange segment on the right: C-terminal sequence of venus. B) Transposon reaction: The transposase inserts the transposon into the targeted vector at various locations. C) The products of the transposon reaction are tested by looking for fluorescence in transfected HEK293 cells. The ones with wrong orientation, out-of-frame or out-of-coding-region do not produce venus. D) The constructs with correct insertions were cut by either AsiS1 or AsC1 and then ligated. The deletions generated constructs containing either the N-terminal segment or the C-terminal segment of the venus.
2.3 Transfection
HEK293 cells or NIE115 neuroblastoma cells were plated on coverslips coated with poly-L-lysine in 24-well plates. The cells were co-transfected with two fusion protein constructs (0.4 µg of each), one containing the front half and the other containing the rear half of venus, using 2 µl (of) lipofectamine 2000 (Invitrogen). All combinations listed in table 2 were tested in HEK293 cells. Only a portion of the combinations were tested in NIE115 cells.
Table 2.
ΔF/F and Tau-on of the voltage sensitive combinations in HEK293 cells. The signal sizes and the Tau-on values were measured from the response to 100 mv depolarization steps from a resting potential of −70 mv.
| A1f | E7f | A8f | H6f | B6f | E3f | G2f | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| % | ms | % | ms | % | ms | % | ms | % | ms | % | ms | % | ms | |
| C3r | −0.08 | 19 | ||||||||||||
| A1r | −0.19 | 22 | −0.12 | 17 | ||||||||||
| H5r | −0.26 | 11 | −0.28 | 12 | −0.11 | 19 | ||||||||
| F7r | 0.9 | 24 | −0.14 | 8 | −0.13 | 18 | ||||||||
| B5r | −1.35 | 15 | −0.07 | 12 | ||||||||||
| A8r | −0.05 | 26 | ||||||||||||
| D2r | −0.15 | 17 | −0.03 | 32 | ||||||||||
| H1r | −0.14 | 11 | −0.07 | 25 | −0.01 | 13 | ||||||||
| H6r | −0.08 | 16 | −0.10 | 33 | −0.04 | 17 | −0.08 | 22 | ||||||
| B6r | −0.16 | 27 | ||||||||||||
| G4r | −0.05 | 26 | −0.07 | 20 | ||||||||||
| F8r | −0.13 | 13 | −0.15 | 16 | −0.17 | 16 | ||||||||
| G2r | −0.13 | 16 | −0.12 | 25 | ||||||||||
2.4 Whole Cell Voltage Clamp
The voltage sensitivity was tested 1 or 2 days after the transfection. The cells were perfused with a bathing solution (NaCl 150mM, KCl 4mM, CaCl2 2mM, MgCl2 1mM, D-glucose 5mM and HEPES 5mM, pH 7.4) at 27–30 °C. The temperature was maintained by an in-line heater and a stage bath heater SH-27B (Warner Instruments, Hamden, CT). Whole cell voltage clamping was performed using a PC-505B amplifier (Warner Instruments). The pipette solution contained K-aspartate 120mM, NaCl 4mM, MgCl2 4mM, CaCl2 1mM, EGTA 10mM, Na2ATP 3mM and HEPES 5mM, pH7.2. Cells were held at −70 mV at rest, and the membrane potential was changed to test the probes’ voltage sensitivity.
2.5 Imaging
2.5.1 Wide field imaging
The voltage clamped cells were imaged with a water immersion objective, Nikon Fluor 60×/1.00W, on a Nikon Eclipse E6000FN microscope (Nikon, Melville, NY) with a 150W Xenon arc lamp (OptiQuip, Highland Mills, NY). The filter cube contained an excitation filter HQ480/30X (Chroma, Bellows Falls, VT), a dichroic mirror 505DCXR (Chroma) and an emission filter HQ520LP (Chroma). The image was demagnified by an Optem® zoom system A45699 (Qioptiq LINOS, Inc, Fairport, NY) and projected onto the 80×80 pixel chip of a NeuroCCD-SM camera controlled by NeuroPlex software (RedShirtImaging, Decatur, GA). The images were recorded at a frame rate of 1 kfps. The measured fluorescence is the average intensity of all the pixels receiving light from the patched cell. Averaging 10 to 64 trials was used to improve the signal to noise ratio. We applied low pass temporal filters using NeuroPlex for the results presented in Figures 4 and 5. A frame subtraction image was used to represent the signal polarity during the depolarization (Fig 5b). It was calculated by subtracting the average pixel values of 20 frames recorded at the resting potential from those of 50 frames during the depolarization.
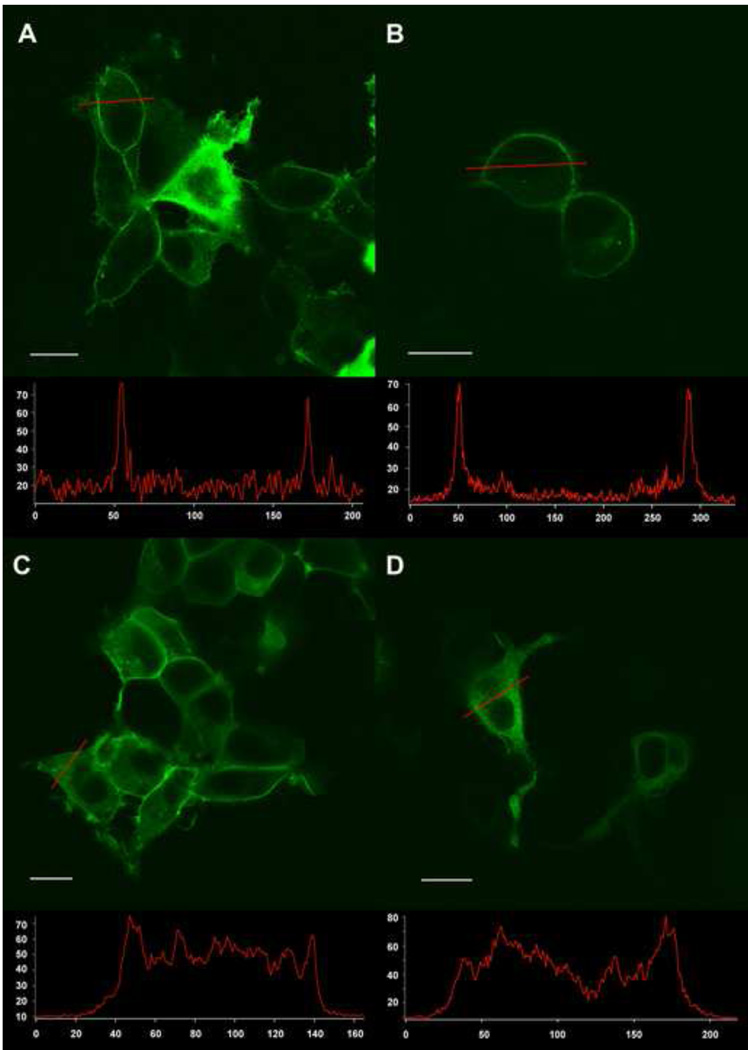
Figure 4. Examples of the location of fluorescence expression in images taken with a confocal microscope.
A) Type 1: A1fB5r in HEK294 cells, mainly membrane fluorescence; intracellular fluorescence in over expressing cells. B) Type 1: A1fF7r in NIE115 cells, mainly membrane expression . C) Type 2: A1fA1r in HEK293 cells, both membrane and intracellular fluorescence. D) Type 3: E7fF7r in HEK293 cells, mainly intracellular fluorescence. The white line in each panel is the scale bar of 10 µm. Bottom inserts: line scan data from individual cells demonstrate the relative intensity between the plasma membrane and the cytosol. The x-axis is pixel number. The y-axis is fluorescence intensity (arbitrary units).
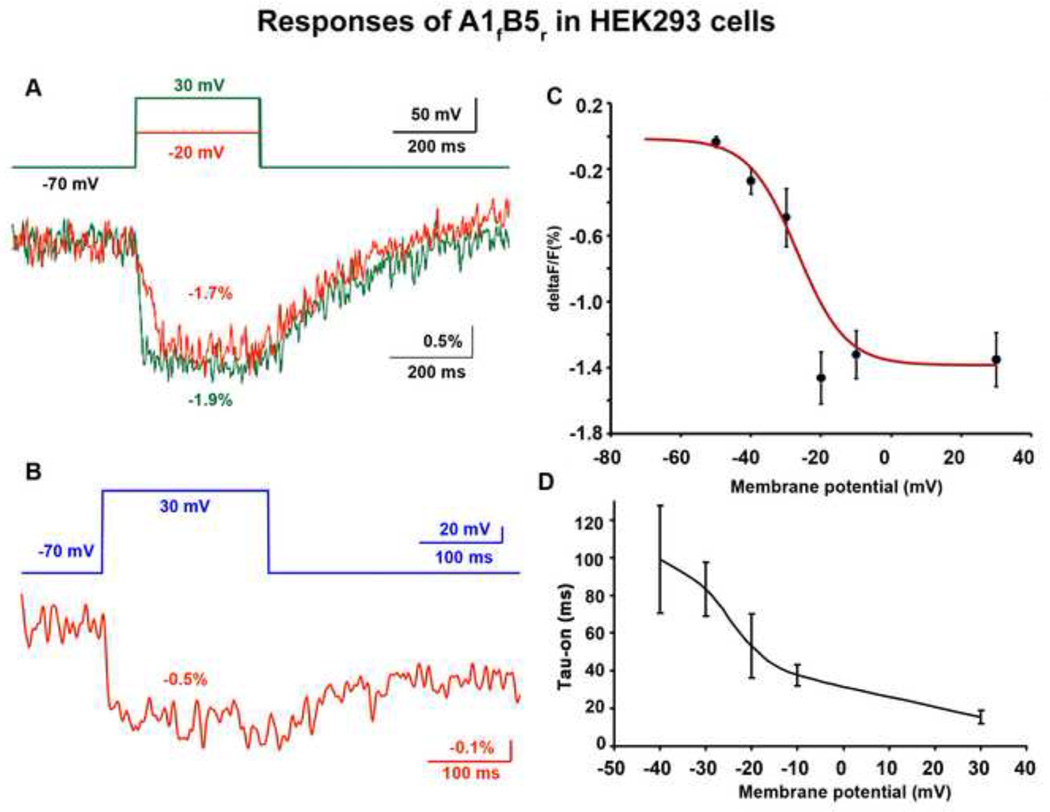
Figure 5. A1fB5r’s fluorescence signal in response to depolarization in HEK293 cells.
A) An example of A1fB5r’s fluorescence changes upon depolarizations of 50 mV and 100 mV, and their recovery after the repolarization. The optical traces were smoothed with five passes of a binomial low pass filter. B) An example of incomplete fluorescent recovery after the repolarization. The optical trace was smoothed with a 60 Hz Gaussian low pass filter. Ten trials were averaged for both A and B. C) ΔF/F of A1fB5r versus membrane potential. The numbers of cells used for each data point: −50 mV: 10, −40 mV: 7, −30 mV: 4, −20 mV: 5, −10 mV: 16, 30 mV: 12. D) On time constant of A1fB5r versus holding potentials from −40 mV to 30 mV. The numbers of cells used for each data point: −40 mV: 5, −30 mV: 3, −20 mV: 5, −10 mV: 16, 30 mV: 12.
2.5.2 Confocal imaging
Confocal images were obtained using an Olympus LUMFL 60X/1.10W objective and an Olympus Fluoview FV1000 LSM confocal microscope. The light source was a 515 nm argon ion laser. A DM405-440/515 dichroic mirror and a BA535-565 emission filter (Olympus, Center Valley, PA) were used to isolate the fluorescence emission.
3 Results
Eight fusion proteins with the front half split-can and fifteen fusion proteins with the rear half split-can were created. The constructs and their insertion sites are listed in table 1. The split-can half is inserted after the amino acid listed under the construct’s name. The insertion positions in the Shaker channel are illustrated in figure 3. HEK293 cells were co-transfected with all of the combinations shown. A portion of these combinations were co-transfected into NIE115 cells.
Table 1.
A) Front half insertion site (constructs names with subscript f). B) Rear half insertion site (constructs names with subscript r). The split-can half is inserted after the amino acid listed under the construct’s name.
| (A) | ||||||||
|---|---|---|---|---|---|---|---|---|
| Construct | A1f | E7f | G1f | A8f | H6f | B6f | E3f | G2f |
| Insertion | 61G | 195G | 391A | 516H | 568S | 574I | 642M | 649A |
| (B) | ||||||||
|---|---|---|---|---|---|---|---|---|
| Construct | C3r | A1r | H5r | F7r | E7r | B5r | A8r | D2r |
| Insertion | 16H | 70A | 78G | 85H | 195G | 492E | 516H | 518K |
| Construct | H1r | H6r | B6r | G4r | F8r | E3r | G2r | |
| Insertion | 533D | 568S | 574I | 580H | 584Q | 642M | 649A | |
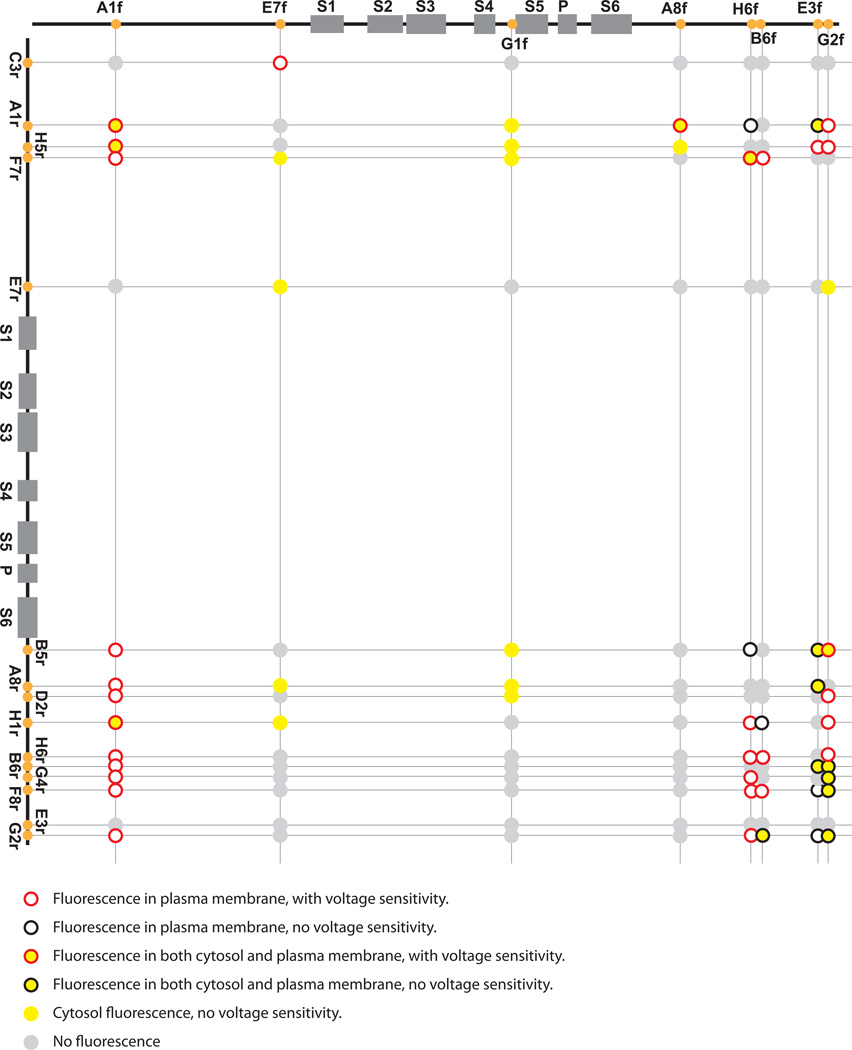
Figure 3. Insertion sites of the Venus split-cans in the Shaker channel, location of fluorescence and voltage sensitivity after co-transfection.
Top: Insertion sites of the split-can front half. Left: Insertion sites of the split-can rear half. The circles at the crossings represent the combinations of the two split-can halves. The colors of the circle surround and center indicate the fluorescence location and voltage-sensitivity. Red circle: fluorescence in plasma membrane, with voltage sensitivity; Black circle: fluorescence in plasma membrane, without voltage sensitivity; Red circle with yellow center: fluorescence in both cytosol and plasma membrane, with voltage sensitivity; Black circle with yellow center: fluorescence in both cytosol and plasma membrane, without voltage sensitivity; Solid yellow circle: cytosol fluorescence, without voltage sensitivity; Solid grey circle: No fluorescence.
We imaged cells transfected with split-can combinations to observe the location of fluorescence. According to our non-quantitative visual observation, we categorized the fluorescence of the split-can combinations into four types:
Type 1: Fluorescence mainly in the plasma membrane: The majority of the transfected cell population showed fluorescence mainly in the plasma membrane. Occasional cells with strong expression had intracellular expression. (Figure 4A and 4B). Twenty nine combinations were in this category (Figure3, red or black circles with empty centers; red, with voltage sensitive fluorescence; black, no voltage sensitivity).
Type 2: Fluorescence in both the plasma membrane and the cytosol: The majority of the transfected cell population had fluorescence in both the plasma membrane and the cytoplasm (Figure 4C). Fifteen combinations were in this category (Figure 3, red or black circles with yellow centers; red, with voltage sensitive fluorescence; black, no voltage sensitivity).
Type 3: Intracellular fluorescence: There was no obvious plasma membrane fluorescence distinguishable from the intracellular expression. The intracellular expression either appeared uniform throughout the cytoplasm or as intracellular aggregates (Figure 4D). Twelve combinations were in this category; none had voltage sensitive fluorescence (Figure 3, solid yellow circles).
Type 4: No fluorescence. Sixty four combinations were in this category (Figure 3, solid grey circles).
To test the split-can combinations’ voltage sensitivity, transfected cells exhibiting fluorescence were simultaneously voltage clamped and imaged at a frame rate of 1 kfps. ΔF/F and Tau-on resulting from a 100 mV depolarization are summarized in table 2 (n≥3) for all the voltage sensitive combinations. The ΔF/F and Tau were calculated from the raw data without low pass filtering. Twenty nine combinations had an intensity decrease, and one combination had an intensity increase during depolarization in HEK293 cells. No optical signal was found during hyperpolarizing steps. For example, figure 6B shows that the combination A1fF7r changes its fluorescence in response to the 100 mV depolarization but not to the 70 mV hyperpolarization. None of the combinations with only intracellular fluorescence had detectable voltage sensitivity. Figure 3 summarizes the split-can insertion sites of all the combinations and their fluorescence location and voltage sensitivity. Voltage sensitive combinations have split-can halves inserted in N-terminal and/or C-terminal intracellular segments of the Shaker channel. None of the combinations containing G1f, whose insertion site is between S4 and S5, showed membrane fluorescence and voltage sensitivity.
Figure 6. A1fF7r’s fluorescence signals.
A) An example of A1fF7r ’s fluorescence change for a 100 mV depolarization in a HEK293 cell. Ten trials were averaged. The optical trace was smoothed with a Gaussian low pass filter of 16 Hz. B) A1fF7r’s fluorescence change in a NIE115 cell. Red trace: signals from the central region of the cell. Blue trace: signals from the edge of the cell. The left image is the resting light intensity. The right image is the frame subtraction image indicating the signal polarity resulting from the depolarization. The greyscale bar is for the subtraction image (arbitrary units). The optical trace was smoothed with a Gaussian low pass filter of 83 Hz. 64 trials were averaged.
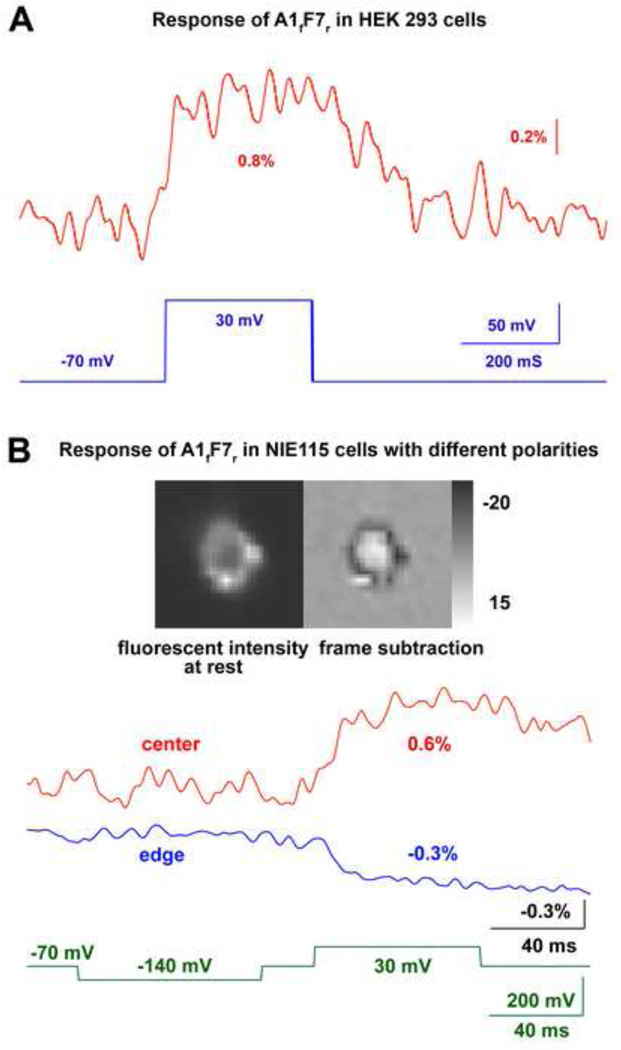
The combination A1fB5r gave the largest signal. Figure 5A shows examples of A1fB5r’s optical signals from a HEK293 cell. The signal size was −1.7% and −1.9% for the 50 mV and 100 mV depolarization steps respectively. Although the signal sizes are similar for the two depolarization steps, the on time constant of the 100 mV depolarization, 10 ms, is faster than 40 ms, the time constant for the 50 mV depolarization. The off time constants for the two depolarization steps are both about 200 ms. Averaged ΔF/F values and on time constants as a function of membrane potential in HEK293 cells are presented in Figure 5C and 5D. The ΔF/F vs Vm data was fit to a red curve by the Boltzman equation using Origin 8 (Figure 5C):
where constants define the maximum and minimum values: A1 = −0.015; A2 = −1.38; the midpoint of the curve: Vhalf = −26.69; and the slope factor: k = 6.90. This probe operates over the voltage range of −50 to 0 mV. Although the voltage sensitivity curve reaches its plateau around 0 mV, the on time constant continues to decrease up to +30 mV (Figure 5D). The average off-time constant of A1fB5r in HEK293 cells following a 100 mV depolarization step was 150 ± 10 ms (n = 12). Most other voltage sensitive combinations were only tested with 100 mV depolarizations; they have smaller fractional changes (Table 2) and signal-to-noise ratios (but similar on time constants).
The A1fB5r’s fluorescence recovered back to the baseline in some HEK293 cells (Figure 5A), but not completely in others (Figure 5B). This slow recovery is shared by many of the voltage sensitive combinations. In some cases, the fluorescence did not respond to the repolarization (Figure 6B).
Although data shown in Figure 5 was the average of 10 trials, averages of 64 trials were necessary for combinations with a small signal to noise ratio. More trials seemed to slow the recovery phase. In addition ΔF/Fs calculated from 10-trial recordings were usually somewhat larger than those from the 64-trial recordings. Furthermore, instances where the signals did not recover usually had a smaller ΔF/F.
In contrast to the negative signals of other combinations, A1fF7r had a net positive ΔF/F during depolarization in HEK293 cells (Figure 6A). Furthermore, In NIE115 cells, the voltage sensitive signal had a unique spatial pattern. The fluorescence from the cell outline decreased upon depolarization, but the fluorescence in the central pixels increased (Figure 6B). A similar signal distribution was also seen in six additional NIE115 cells. This kind of spatial distribution was not seen with 8 other split-can combinations tested in NIE115 cells.
4 Discussion
Our goal was to improve plasma membrane expression and voltage sensitivity of FlaSh-YFP in mammalian cell lines. This goal was achieved using the split-cans and the transposon reaction. With this strategy, thirty novel voltage sensitive probes were created, all of which had significant membrane expression.
However, the split-can method was not perfect in that 15 combinations had both membrane and intracellular fluorescence and another 12 combinations had only intracellular fluorescence. Possible explanations for the intracellular fluorescence are that these monomers are correctly folded and tetramerized, but they are retained intracellularly either as intracellular pools (Clay and Kuzirian, 2002), or that the cells lack sufficient trafficking partners. However, these hypotheses are unlikely because the 29 combinations with only plasma membrane fluorescence demonstrate that the HEK293 and NIE115 cell lines have the necessary trafficking partners and limited Shaker intracellular pools. Another explanation is an erroneous assumption of our split-can strategy. We assumed that the peptides are tetramerized after the completion of their translation and folding, and misfolded monomers will not tetramerize and the FP halves cannot combine. However, there is evidence that Shaker-type K channels are capable of tetramerizing via their N-terminal T1 domain while the nascent peptides are still being translated and translocated in the ER (Lu et al., 2001). Therefore, if the peptide is misfolded downstream of the N-terminal T1 domain, it is possible for misfolded monomers to be tetramerized and fluoresce.
We used the transposon reaction to insert the half-cans into the Shaker monomers at “random” locations. We hoped to find insertion sites that would bring the two half-cans close enough to reassemble and fluoresce in the tetramer. Among the 30 voltage sensitive combinations, the two with the largest fractional changes are A1fB5r and A1fF7r. The insertion sites of A1f and F7r are in the N-terminal intracellular segment, upstream to the S1 transmembrane domain of Shaker, and that of B5r is in the C-terminal intracellular segment, downstream of the S6 transmembrane domain. Because S4 is recognized as the voltage sensor, insertion sites close to the S4 domain are natural choices for FPs to sense S4’s translocation. Thus far the published FP voltage sensors have their FPs inserted downstream to either S4 or S6 of the voltage sensors (Ataka and Pieribone, 2002; Sakai et al., 2001; Siegel and Isacoff, 1997; Tsutsui et al., 2008). This is the first report showing that a fluorescent protein inserted in the N terminal intracellular segment of a voltage sensor exhibits optical signals upon voltage changes. So this study opens a new exploration space for possible insertion sites for future probe design.
The voltage dependence of A1fB5r’s fluorescence change was fitted with a Boltzmann equation. The curve had the midpoint at about −25 mV and a slope factor k of 6.9. The fluorescence is sensitive to the voltage change in the range of −50 to 0 mV. Perozo et al studied the wild-type Shaker channel’s characteristics in Xenopus oocytes. They compared the channel’s gating current and conductance’s voltage dependence after normalization (Perozo et al., 1992). The conductance curve’s midpoint is about −25 mV, which is more positive than that of the gating charge curve (− 35 mV), and the conductance curve has a steeper slope than the gating charge curve. They also reported that the mutation W434F did not change the voltage dependence of the gating charge movement (Perozo et al., 1993). The ΔF/F-voltage curve in Figure 5C, after normalizeation by the maximum ΔF/F value, overlaps with the conductance-voltage curve of the wild-type Shaker channel (Perozo et al., 1992). Thus we speculate that the fluorescence change is a result of the conformation changes inducing the channel opening, but not the gating charge movement. This speculation is different from that proposed for the FlaSh fluorescence signal (Siegel and Isacoff, 1997). Compared to A1fB5r, FlaSh operates over a narrower voltage range (−53 to −27 mV) which matches its own gating charge displacement . The different voltage dependence between FlaSh and A1fB5r may result from their different insertion sites. The GFP in FlaSh is inserted downstream of S6 transmembrane domain in a single subunit, while each venus in A1fB5r is composed of halves linked between two subunits at insertion sites, one upstream to S1 and the other downstream of S6. The split-can FP could sense either the conformation change of a single subunit, and/or changes in the relative angle and/or distance between two subunits in the tetramer.
All of the genetically encoded voltage sensitive probes reported so far have slower kinetics compared with the fastest organic voltage sensitive dyes. Some organic voltage sensitive dyes respond with time constants faster than 1 µsec (Loew et al., 1985).These differences result from the different mechanisms of voltage sensitivity. We take di-8-ANEPPS as an example for voltage sensitive dyes. The electric field influences its chromophore’s electron distribution directly, and its spectra respond to the membrane voltage change very rapidly. On the other hand, the genetically encoded probes’ chromophores do not respond to the voltage change directly, but to the environment change caused by the conformation transition of the voltage sensor during gating, activation or inactivation. The probe inherits some properties of the voltage sensor, such as voltage sensitive dynamics and range. The slower dynamics reflects the speed of the conformation transition of the sensor and the indirect environment change of the chromophore.
The FP voltage sensors generated in this project all have relatively fast on rates and slower off rates. Compared with FlaSh-YFP, whose on-time constant is about 300 ms and off-time constant is larger than 1 s in Xenopus oocytes (Guerrero et al., 2002), the split-can constructs have improved dynamics, but their slow off-rate would still limit the application of these FP voltage sensors in certain physiological studies. One of the second generation voltage sensitive phosphatase based probes, VSFP2.1, has an on-time constant of 20 ms and off-time constant of 80 ms for depolarization steps of 90mV in PC12 cells at 35 °C (Dimitrov et al., 2007). A1fB5r is as fast as VSFP2.1 on depolarization, but has slower recovery after repolarization. In a few cases, the split-can’s fluorescence change had no appreciable recovery hundreds of ms after the repolarization. As a result, we have not reported the Tau-off values of all combinations.
The combination of A1fF7r is unique in that it exhibited a fluorescence increase during depolarization in HEK293 cells. Furthermore, in NIE115 cells, A1fF7r had a fluorescence increase in the central pixels but a fluorescence decrease in the pixels from the edge of the cell. Because the confocal image indicates that A1fF7r is mainly expressed in the plasma membrane (Figure 4B), we believe that the center pixels recorded fluorescence signals from the top and bottom plasma membrane. Compared to NIE115 cells, HEK293 cells are smaller in size and flatter in geometry, which will result in relatively reduced fluorescence from vertically oriented membrane. This, together with the limited spatial resolution of the NeuroCCD camera, may explain why we did not detect a decreasing edge signal in HEK293 cells.
Although A1fF7r’s unique spatially localized signals would be problematic for monitoring membrane potential, it may provide important clues for further biophysical studies on the response mechanism of FP voltage sensors. A hypothesis involving chromophore dipole rotation can explain the opposite signals in the center and at the edge of A1fF7r transfected NIE115 cells. We suggest that at resting potential the chromophore’s dipole is nearly perpendicular to the plasma membrane. The chromophore dipoles on the side membrane are nearly perpendicular to the light path, while those on the top and bottom membrane are nearly parallel to the light path. Chromophores with dipoles more perpendicular to the light path are better excited and emit stronger fluorescence. If depolarization causes the dipoles to be less perpendicular to the plasma membrane, the fluorescence from the top and bottom membrane will increase, and that from the edge will decrease.
In summary, we used split-can and transposon technologies to improve the localization and voltage sensitivity of FlaSh-YFP based FP voltage sensors in mammalian cells. It is clear from Figure 3 that we have not exhaustively explored this possibility in the Shaker potassium channel. Furthermore, other kinds of split-can FPs and other oligomeric voltage sensor proteins could be investigated in an effort to find larger and faster signals.
Highlight.
Split cans of FP (Venus) were inserted into many locations of K+ channel subunits.
These were expressed pairwise in HEK293 and NI115 cells.
This procedure often resulted in improved membrane localization of the fluorescence.
The largest fractional change was −1.4% in ΔF/F for a 100 mV depolarization.
Acknowledgement
This project is supported by NIH grants U24NS057631, R01DC005259, and NINDS grant R21NS054270.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Akemann W, Mutoh H, Perron A, Rossier J, Knöpfel T. Imaging brain electric signals with genetically targeted voltage-sensitive fluorescent proteins. Nat Methods. 2010;7:643–649. doi: 10.1038/nmeth.1479. [DOI] [PubMed] [Google Scholar]
- Ataka K, Pieribone V. A genetically targetable fluorescent probe of channel gating with rapid kinetics. Biophys J. 2002;82:509–516. doi: 10.1016/S0006-3495(02)75415-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker B, Lee H, Pieribone V, Cohen L, Isacoff E, Knopfel T, Kosmidis E. Three fluorescent protein voltage sensors exhibit low plasma membrane expression in mammalian cells. J Neurosci Methods. 2007;161:32–38. doi: 10.1016/j.jneumeth.2006.10.005. [DOI] [PubMed] [Google Scholar]
- Baker B, Mutoh H, Dimitrov D, Akemann W, Perron A, Iwamoto Y, Jin L, Cohen L, Isacoff E, Pieribone V, Hughes T, Knöpfel T. Genetically encoded fluorescent sensors of membrane potential. Brain Cell Biol. 2008;36:53–67. doi: 10.1007/s11068-008-9026-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown J, Cohen L, De Weer P, Pinto L, Ross W, Salzberg B. Rapid changes in intracellular free calcium concentration. Detection by metallochromic indicator dyes in squid giant axon. Biophys J. 1975;15:1155–1160. doi: 10.1016/S0006-3495(75)85891-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canepari M, Vogt K, Zecevic D. Combining voltage and calcium imaging from neuronal dendrites. Cell Mol Neurobiol. 2008;28:1079–1093. doi: 10.1007/s10571-008-9285-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter M, Shieh JC. Guide to Research Techniques in Neuroscience. New York: Academic Press; 2010. Visualizing Nervous System Function; pp. 169–189. [Google Scholar]
- Clay J, Kuzirian A. Trafficking of axonal K+ channels: potential role of Hsc70. J Neurosci Res. 2002;67:745–752. doi: 10.1002/jnr.10182. [DOI] [PubMed] [Google Scholar]
- Davila H, Salzberg B, Cohen L, Waggoner A. A large change in axon fluorescence that provides a promising method for measuring membrane potential. Nat New Biol. 1973;241:159–160. doi: 10.1038/newbio241159a0. [DOI] [PubMed] [Google Scholar]
- Dimitrov D, He Y, Mutoh H, Baker BJ, Cohen L, Akemann W, Knöpfel T. Engineering and characterization of an enhanced fluorescent protein voltage sensor. PLoS One. 2007;2:e440. doi: 10.1371/journal.pone.0000440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grinvald A, Lieke E, Frostig R, Gilbert C, Wiesel T. Functional architecture of cortex revealed by optical imaging of intrinsic signals. Nature. 1986;324:361–364. doi: 10.1038/324361a0. [DOI] [PubMed] [Google Scholar]
- Grinvald A, Manker A, Segal M. Visualization of the spread of electrical activity in rat hippocampal slices by voltage-sensitive optical probes. J Physiol. 1982;333:269–291. doi: 10.1113/jphysiol.1982.sp014453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grinvald A, Ross W, Farber I. Simultaneous optical measurements of electrical activity from multiple sites on processes of cultured neurons. Proc Natl Acad Sci U S A. 1981;78:3245–3249. doi: 10.1073/pnas.78.5.3245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerrero G, Siegel M, Roska B, Loots E, Isacoff E. Tuning FlaSh: redesign of the dynamics, voltage range, and color of the genetically encoded optical sensor of membrane potential. Biophys J. 2002;83:3607–3618. doi: 10.1016/S0006-3495(02)75361-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta R, Salzberg B, Grinvald A, Cohen L, Kamino K, Lesher S, Boyle M, Waggoner A, Wang C. Improvements in optical methods for measuring rapid changes in membrane potential. J Membr Biol. 1981;58:123–137. doi: 10.1007/BF01870975. [DOI] [PubMed] [Google Scholar]
- Humphrey W, Dalke A, Schulten K. VMD: visual molecular dynamics. J Mol Graph. 1996;14:33–38. 27–28. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- Jackrel M, Cortajarena A, Liu T, Regan L. Screening libraries to identify proteins with desired binding activities using a split-GFP reassembly assay. ACS Chem Biol. 2010;5:553–562. doi: 10.1021/cb900272j. [DOI] [PubMed] [Google Scholar]
- Kwong K, Belliveau J, Chesler D, Goldberg I, Weisskoff R, Poncelet B, Kennedy D, Hoppel B, Cohen M, Turner R. Dynamic magnetic resonance imaging of human brain activity during primary sensory stimulation. Proc Natl Acad Sci U S A. 1992;89:5675–5679. doi: 10.1073/pnas.89.12.5675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindman S, Johansson I, Thulin E, Linse S. Green fluorescence induced by EF-hand assembly in a split GFP system. Protein Sci. 2009;18:1221–1229. doi: 10.1002/pro.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loew L, Cohen L, Salzberg B, Obaid A, Bezanilla F. Charge-shift probes of membrane potential. Characterization of aminostyrylpyridinium dyes on the squid giant axon. Biophys J. 1985;47:71–77. doi: 10.1016/S0006-3495(85)83878-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J, Robinson J, Edwards D, Deutsch C. T1-T1 interactions occur in ER membranes while nascent Kv peptides are still attached to ribosomes. Biochemistry. 2001;40:10934–10946. doi: 10.1021/bi010763e. [DOI] [PubMed] [Google Scholar]
- Mealer R, Butler H, Hughes T. Functional fusion proteins by random transposon-based GFP insertion. Methods Cell Biol. 2008;85:23–44. doi: 10.1016/S0091-679X(08)85002-9. [DOI] [PubMed] [Google Scholar]
- Mealer R, Pralle A, Isacoff E, Hughes T. Scanning the Shaker channel subunit with fragments of GFP: Can complementing fragments on adjacent subunits produce a fluorophore?; Biophysics Annual Conference; 2006. Abstract r 2266. [Google Scholar]
- Nagai T, Ibata K, Park E, Kubota M, Mikoshiba K, Miyawaki A. A variant of yellow fluorescent protein with fast and efficient maturation for cell-biological applications. Nat Biotechnol. 2002;20:87–90. doi: 10.1038/nbt0102-87. [DOI] [PubMed] [Google Scholar]
- Nagaya N, Papazian D. Potassium channel alpha and beta subunits assemble in the endoplasmic reticulum. J Biol Chem. 1997;272:3022–3027. doi: 10.1074/jbc.272.5.3022. [DOI] [PubMed] [Google Scholar]
- Ozawa T. Designing split reporter proteins for analytical tools. Anal Chim Acta. 2006;556:58–68. doi: 10.1016/j.aca.2005.06.026. [DOI] [PubMed] [Google Scholar]
- Ozawa T. Protein reconstitution methods for visualizing biomolecular function in living cells. Yakugaku Zasshi. 2009;129:289–295. doi: 10.1248/yakushi.129.289. [DOI] [PubMed] [Google Scholar]
- Perozo E, MacKinnon R, Bezanilla F, Stefani E. Gating currents from a nonconducting mutant reveal open-closed conformations in Shaker K+ channels. Neuron. 1993;11:353–358. doi: 10.1016/0896-6273(93)90190-3. [DOI] [PubMed] [Google Scholar]
- Perozo E, Papazian D, Stefani E, Bezanilla F. Gating currents in Shaker K+ channels. Implications for activation and inactivation models. Biophys J. 1992;62:160–168. doi: 10.1016/S0006-3495(92)81802-7. discussion 9–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pralle A, Mealer R, Hughes T, Isacoff E. Optical voltage sensor from random insertion of GFP fragments into the Shaker channel; Biophysics Annual Conference; 2006. Abstract r 2271. [Google Scholar]
- Reddy P, Corley R. Assembly, sorting, and exit of oligomeric proteins from the endoplasmic reticulum. Bioessays. 1998;20:546–554. doi: 10.1002/(SICI)1521-1878(199807)20:7<546::AID-BIES5>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- Rekas A, Alattia J, Nagai T, Miyawaki A, Ikura M. Crystal structure of venus, a yellow fluorescent protein with improved maturation and reduced environmental sensitivity. J Biol Chem. 2002;277:50573–50578. doi: 10.1074/jbc.M209524200. [DOI] [PubMed] [Google Scholar]
- Robinson J, Deutsch C. Coupled tertiary folding and oligomerization of the T1 domain of Kv channels. Neuron. 2005;45:223–232. doi: 10.1016/j.neuron.2004.12.043. [DOI] [PubMed] [Google Scholar]
- Sakai R, Repunte-Canonigo V, Raj C, Knöpfel T. Design and characterization of a DNA-encoded, voltage-sensitive fluorescent protein. Eur J Neurosci. 2001;13:2314–2318. doi: 10.1046/j.0953-816x.2001.01617.x. [DOI] [PubMed] [Google Scholar]
- Salzberg B, Grinvald A, Cohen L, Davila H, Ross W. Optical recording of neuronal activity in an invertebrate central nervous system: simultaneous monitoring of several neurons. J Neurophysiol. 1977;40:1281–1291. doi: 10.1152/jn.1977.40.6.1281. [DOI] [PubMed] [Google Scholar]
- Siegel MS, Isacoff EY. A Genetically Encoded Optical Probe of Membrane Voltage. Neuron. 1997;19:735–741. doi: 10.1016/s0896-6273(00)80955-1. [DOI] [PubMed] [Google Scholar]
- Tolkovsky A, Richards C. Na+/H+ exchange is the major mechanism of pH regulation in cultured sympathetic neurons: measurements in single cell bodies and neurites using a fluorescent pH indicator. Neuroscience. 1987;22:1093–1102. doi: 10.1016/0306-4522(87)92984-8. [DOI] [PubMed] [Google Scholar]
- Tsutsui H, Karasawa S, Okamura Y, Miyawaki A. Improving membrane voltage measurements using FRET with new fluorescent proteins. Nat Methods. 2008;5:683–685. doi: 10.1038/nmeth.1235. [DOI] [PubMed] [Google Scholar]
- Wilson C, Magliery T, Regan L. Detecting protein-protein interactions with GFP-fragment reassembly. Nat Methods. 2004;1:255–262. doi: 10.1038/nmeth1204-255. [DOI] [PubMed] [Google Scholar]
- Zecević D, Wu J, Cohen L, London J, Höpp H, Falk C. Hundreds of neurons in the Aplysia abdominal ganglion are active during the gill-withdrawal reflex. J Neurosci. 1989;9:3681–3689. doi: 10.1523/JNEUROSCI.09-10-03681.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zochowski M, Cohen L. Oscillations in the olfactory bulb carry information about odorant history. J Neurophysiol. 2005;94:2667–2675. doi: 10.1152/jn.00328.2005. [DOI] [PubMed] [Google Scholar]