Interleukin 2, STAT5, and Blimp-1 work together to suppress differentiation of follicular helper T cells in mice.
Abstract
Follicular helper T cells (TFH cells) constitute the CD4+ T cell subset that is specialized to provide help to germinal center (GC) B cells and, consequently, mediate the development of long-lived humoral immunity. TFH cell differentiation is driven by the transcription factor Bcl6, and recent studies have identified cytokine and cell–cell signals that drive Bcl6 expression. However, although TFH dysregulation is associated with several major autoimmune diseases, the mechanisms underlying the negative regulation of TFH cell differentiation are poorly understood. In this study, we show that STAT5 inhibits TFH cell differentiation and function. Constitutive STAT5 signaling in activated CD4+ T cells selectively blocked TFH cell differentiation and GCs, and IL-2 signaling was a primary inducer of this pathway. Conversely, STAT5-deficient CD4+ T cells (mature STAT5fl/fl CD4+ T cells transduced with a Cre-expressing vector) rapidly up-regulated Bcl6 expression and preferentially differentiated into TFH cells during T cell priming in vivo. STAT5 signaling failed to inhibit TFH cell differentiation in the absence of the transcription factor Blimp-1, a direct repressor of Bcl6 expression and TFH cell differentiation. These results demonstrate that IL-2, STAT5, and Blimp-1 collaborate to negatively regulate TFH cell differentiation.
The germinal center (GC) reaction is an essential step in the development of humoral immunity, in which B cells undergo affinity maturation and differentiation into memory cells and long-lived plasma cells (Gatto and Brink, 2010). Follicular helper T cells (TFH cells) are CD4+ T cells that migrate into B cell follicles and provide specialized help to GC B cells (Crotty, 2011). Impaired TFH cell differentiation results in a loss of GCs and T-dependent antibody responses (Johnston et al., 2009; Nurieva et al., 2009; Yu et al., 2009). Conversely, excessive TFH cell differentiation can drive the production of autoantibodies and is associated with several autoimmune diseases (Hu et al., 2009; Linterman et al., 2009).
Recent studies have investigated the signals that regulate TFH cell differentiation. TFH cells possess a distinctive gene program (Crotty, 2011), and the transcription factor Bcl6 is necessary for TFH cell differentiation in vivo (Johnston et al., 2009; Nurieva et al., 2009; Yu et al., 2009). Multiple rounds of interaction with antigen-presenting cells are necessary for TFH cell differentiation (Johnston et al., 2009; Deenick et al., 2010; Choi et al., 2011; Goenka et al., 2011), and multiple signals are involved. In particular, ICOS–ICOSL interaction (Akiba et al., 2005; Gigoux et al., 2009; Hu et al., 2009; Choi et al., 2011) and contributions from IL-6 (Nurieva et al., 2008, 2009; Eto et al., 2011; Harker et al., 2011) are important for Bcl6 expression and TFH cell differentiation in most murine in vivo conditions. However, the signals that negatively regulate TFH cell differentiation are not well understood. One repressor of Bcl6 is the transcription factor Blimp-1, which is expressed by non-TFH effector CD4+ T cells such as TH1, TH2, and induced regulatory T cells (Treg cells; Fazilleau et al., 2009; Johnston et al., 2009; Ma et al., 2009; Yusuf et al., 2010; Choi et al., 2011; Cretney et al., 2011). Bcl6 and Blimp-1 are mutually antagonistic; together, they constitute a regulatory axis that determines commitment to TFH or non-TFH effector CD4+ T cell differentiation (Johnston et al., 2009; Crotty et al., 2010). Consequently, negative regulators of TFH cell differentiation may act by directly targeting Bcl6 or by inducing Blimp-1 or other factors.
STAT-mediated cytokine signaling pathways are important regulators of effector lymphocyte differentiation (Zhu et al., 2010). In B cells, STAT5 and STAT3 regulate Bcl6 and Blimp-1, but in both cases, the type of regulation is controversial. STAT5 has been reported to induce Bcl6 expression (Scheeren et al., 2005) and, in other studies, to repress Bcl6 expression (Walker et al., 2007; Duy et al., 2011). Similarly, STAT3 signaling in B cells has been reported to drive Blimp-1 expression (Reljic et al., 2000; Diehl et al., 2008; Kwon et al., 2009) and to drive Bcl6 expression (Arguni et al., 2006). STAT5 has also recently been shown to play an important role in effector CD8+ T cell persistence (Tripathi et al., 2010). In CD4+ T cells, STAT3 signaling is required for TH17 cell differentiation (Hirahara et al., 2010) and may contribute to TFH cell differentiation (Nurieva et al., 2008, 2009; Poholek et al., 2010; Eto et al., 2011). STAT5 signaling represses TH17 cell differentiation (Yang et al., 2011) but enhances the differentiation of multiple effector CD4+ T cell subsets, including Treg cells (Wei et al., 2008), TH2 cells (Zhu et al., 2010), and TH1 cells (Liao et al., 2011).
In this study, we found that STAT5 signaling blocked TFH cell differentiation and that this inhibition was induced by IL-2 and dependent on Blimp-1. These results identify a key negative regulatory pathway of TFH cell differentiation.
RESULTS AND DISCUSSION
STAT5 signaling selectively inhibits TFH cell differentiation and function
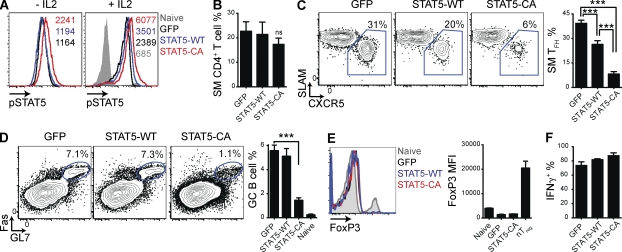
It remains unclear how diverse signals combine to specify commitment to TFH or effector (TH1, TH2, TH17, etc.) CD4+ T cell differentiation. Given the importance of STATs in regulating effector CD4+ T cell gene programs (Zhu et al., 2010) and conflicting reports of STAT5 regulating Bcl6 or Blimp-1 in B cells (Scheeren et al., 2005; Walker et al., 2007; Duy et al., 2011), we examined the role of STAT5 in TFH cell differentiation. Because the primary limiting factor for STAT activation and signaling is the availability of activating cytokines, we used retroviral expression vectors (RVs) expressing only GFP (“GFP”), GFP and WT STAT5b (STAT5-WT), or GFP and a constitutively active mutant of STAT5b (STAT5-CA; Onishi et al., 1998). SMARTA TCR transgenic CD4+ T cells (SM cells), specific for the lymphocytic choriomeningitis virus (LCMV) epitope GP66–77 bound by MHC class II I-Ab, were transduced with these RVs. STAT5-CA+ cells exhibited increased phospho-STAT5 protein in the absence of IL-2 (93% increase relative to GFP+ cells; Fig. 1 A; Onishi et al., 1998). Both STAT5-CA+ and STAT5-WT+ cells had augmented levels of phospho-STAT5 protein after stimulation with IL-2 (150% and 47% increase relative to GFP+ cells, respectively; Fig. 1 A). Sorted transduced cells were adoptively transferred into WT C57BL/6J host mice. Shortly thereafter, host mice were infected with Armstrong strain LCMV.
Figure 1.
STAT5 signaling selectively inhibits TFH cell differentiation and function. CD45.1+ SMARTA TCR transgenic (SM) CD4+ T cells were transduced with RVs expressing GFP and WT STAT5b (STAT5-WT), GFP and a constitutively active form of STAT5b (STAT5-CA), or only GFP (GFP). (A) Representative histograms of phospho-STAT5 levels in transduced SM cells (GFP+), without stimulation (left) or after stimulation with IL-2 (right). Phospho-STAT5 MFIs are indicated. Data are representative of two independent experiments. (B–F) Transduced SM cells (those expressing GFP) were adoptively transferred into C57BL/6J mice that were subsequently infected with LCMV (see Materials and methods). Splenocytes were analyzed 8 d after infection. Data are a composite of four (B–D) or two (E and F) independent experiments and total n = 11–16/group (B–D) or 4/group (E and F). (B) Quantitation of SM cells as a percentage of all CD4+ T cells. (C) Representative FACS plots gated on SM cells (CD4+ CD45.1+), with TFH cells (SLAMlow CXCR5high) boxed. Quantitation of SM TFH cells as a percentage of all SM cells. GFP versus STAT5-WT: ***, P = 0.0002; GFP versus STAT5-CA: ***, P < 0.0001; STAT5-WT versus STAT5-CA: ***, P < 0.0001. (D) Representative FACS plots gated on B cells (B220+), with GC B cells (Fas+ GL7+) circled. Quantitation of GC B cells as a percentage of all B cells. Uninfected C57BL/6J mice (naive) are also shown. ***, P < 0.0001. (E) Representative histograms of FoxP3 expression in SM cells and in total CD4+ T cells from an uninfected C57BL/6J mouse (naive). Quantitation of FoxP3 MFI, with natural Treg cells (nTreg; CD4+ CD25+ FoxP3+) from a naive C57BL/6J mouse included as a control. (F) Quantitation of the percentage of SM cells that produced IFN-γ after PMA/ionomycin stimulation in vitro. Error bars depict the standard error of the mean.
GFP+, STAT5-WT+, and STAT5-CA+ SM cells all expanded normally in response to acute LCMV infection (Fig. 1 B). However, STAT5-CA+ SM cells largely failed to differentiate into TFH cells (78% fewer TFH cells; P < 0.0001; Fig. 1 C). Overexpression of WT STAT5 also reduced TFH cell differentiation (33% fewer TFH cells; P = 0.0002; Fig. 1 C). Mice that received STAT5-CA+ SM cells had fewer GC B cells (71% fewer GC B cells; P < 0.0001; Fig. 1 D), consistent with a substantial loss of TFH cell help.
One possible mechanism by which STAT5 signaling could impair effector CD4+ T cell differentiation was induction of Treg cell differentiation. In some settings, STAT5 signaling drives Treg cell differentiation via induction of FoxP3 (Wei et al., 2008). However, we found that SM cells transduced with the STAT5-WT RV or the STAT5-CA RV did not express FoxP3 and did not detectably suppress the endogenous immune response (Fig. 1 E and not depicted). Furthermore, STAT5-WT+ and STAT5-CA+ SM cells produced high levels of the TH1-associated cytokine IFN-γ (Fig. 1 F), suggesting that non-TFH effector cell differentiation was not impaired by enhanced STAT5 signaling. Collectively, these data indicated that STAT5 signaling selectively inhibited TFH cell differentiation during an acute viral infection.
Lack of STAT5 signaling enhances TFH cell differentiation
We hypothesized that STAT5 was a physiological inhibitor of TFH cell differentiation and consequently that a lack of STAT5 signaling during CD4+ T cell priming would enhance TFH cell differentiation. However, insufficient STAT5 signaling in the thymus results in a loss immunological self tolerance (Malek et al., 2002; Burchill et al., 2007). To avoid this complication, we conditionally deleted STAT5 in mature CD4+ T cells by transducing splenic STAT5fl/fl SM CD4+ T cells with a Cre-expressing RV (Cre). Phospho-STAT5 protein was absent in STAT5-deficient (STAT5fl/fl Cre+) cells (Fig. 2 A), in agreement with PCR data (not depicted). STAT5-deficient SM cells or untransduced but similarly treated (control) SM cells were adoptively transferred into C57BL/6J host mice that were subsequently infected with LCMV. STAT5-deficient SM cells expanded as well as control SM cells and expressed surface markers consistent with normal activation (Fig. 2, B and C). Although the STAT5 signaling cytokines IL-2 and IL-7 are drivers of T cell survival and proliferation (Rochman et al., 2009), these results show that deletion of STAT5 from mature CD4+ T cells is a viable approach to examine the role of STAT5 in effector CD4+ T cell differentiation, consistent with recent CD4 T cell studies using STAT5fl/fl (Tripathi et al., 2010) or IL-2−/− CD4 T cells (Liao et al., 2011).
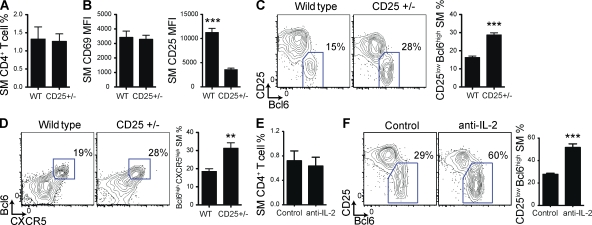
Figure 2.
Lack of STAT5 signaling enhances TFH cell differentiation. STAT5fl/fl SM cells were transduced with an RV expressing Cre recombinase (Cre) or were not transduced but treated similarly (control). (A) Representative histograms of phospho-STAT5 levels in SM cells, with and without IL-2 stimulation. The percentage of cells that was phospho-STAT5+ is indicated. (B–J) Cre+ and control SM cells were adoptively transferred into C57BL/6J mice that were subsequently infected with LCMV. Splenocytes were analyzed 8 (B–F) or 4 (G–J) d after infection. Data are a composite of two independent experiments, and n = 8/group. (B) Quantitation of SM cells as a percentage of all CD4+ T cells. (C) Representative histograms gated on SM cells or on CD4+ T cells from an uninfected C57BL/6J mouse (naive). (D) Representative FACS plots gated on SM cells, with TFH cells (SLAMlow CXCR5high) boxed. Quantitation of SM TFH cells as a percentage of total SM cells. ***, P < 0.0001. (E) Representative FACS plots gated on SM cells, with GC TFH cells (CXCR5high GL7high) boxed. Quantitation of SM GC TFH cells as a percentage of total SM cells. **, P = 0.01. (F) IL-21 production by SM cells after PMA/ionomycin stimulation in vitro. Quantitation of IL-21+ SM cells as a percentage of total SM cells. (G) Quantitation of SM cells as a percentage of total CD4+ T cells. (H) Representative histograms of Bcl6 expression, gated on SM cells or on CD4+ T cells from an uninfected C57BL/6J mouse (naive). Quantitation of SM cell Bcl6 MFI. **, P = 0.0012. (I) Representative FACS plots gated on SM cells, with TFH (Bcl6high CXCR5high) boxed. Quantitation of SM TFH cells as a percentage of all SM cells. ***, P < 0.0001. (J) Representative FACS plots gated on SM cells, with non-TFH cells (CD25high CXCR5low) boxed. Quantitation of non-TFH SM cells as a percentage of total SM cells. ***, P < 0.0001. Error bars depict the standard error of the mean.
Deletion of STAT5 markedly enhanced SM TFH cell differentiation (80% more TFH cells; P < 0.0001; Fig. 2 D). TFH cell differentiation is a multistep, multistage process (Crotty, 2011), and TFH cells that have progressed into GCs, GC TFH cells, can be identified by PD-1 or GL7 staining (Haynes et al., 2007; Yusuf et al., 2010; Kitano et al., 2011; Lee et al., 2011; Goenka et al., 2011). GC TFH cell abundance was also increased in the absence of STAT5 (55% more GC TFH cells; P = 0.01; Fig. 2 E). STAT5-deficient TFH cells expressed normal levels of IL-21, a key TFH-produced cytokine that sustains the GC reaction (Fig. 2 F; Crotty, 2011). Production of IL-2 in both TFH and non-TFH effector cells was also maintained in the absence of STAT5 signaling (not depicted). These data showed that TFH proliferation, survival, and function were not dependent on STAT5 signaling and, more importantly, that STAT5 was a physiological inhibitor of TFH cell differentiation.
Recently, we and others found that activated CD4+ T cells rapidly bifurcate into TFH or non-TFH effector cell differentiation programs during priming (Choi et al., 2011; Kitano et al., 2011). Consequently, we examined the effect of STAT5 deficiency on commitment to TFH cell differentiation. 4 d after LCMV infection, STAT5-deficient (Cre+) SM cells had again expanded as well as control SM cells (Fig. 2 G), yet the absence of STAT5 signaling strongly enhanced Bcl6 expression and TFH cell differentiation (123% increase in Bcl6 mean fluorescence intensity [MFI; P = 0.0012] and 128% more TFH cells [P < 0.0001]; Fig. 2, H and I). These results indicate that STAT5 acts early in T cell priming during an acute viral infection to block Bcl6 expression and thereby prevent commitment to TFH cell differentiation.
A key function of STAT5 in T cells is to mediate signaling by IL-2. Intriguingly, activated CD4+ T cells that have recently begun TFH cell differentiation express lower levels of the high affinity subunit of the IL-2 receptor, IL-2Rα (CD25; Choi et al., 2011). We noted that STAT5 deficiency resulted in a reduction in IL-2Rα expression (82% reduction in CD25high CXCR5low non-TFH SM cells; P < 0.001; Fig. 2 J), consistent with previous studies (Nakajima et al., 1997; Malek et al., 2002). Overall, these results suggested that STAT5 inhibited commitment to TFH cell differentiation during T cell priming and that IL-2 may induce this pathway.
IL-2 signaling inhibits Bcl6 expression during T cell priming
To directly test the role of IL-2 on commitment to TFH cell differentiation, we first used SM cells that were heterozygous for deletion of IL-2Rα (CD25+/−). When transferred into C57BL/6J mice that were subsequently infected with LCMV, activation and expansion of CD25+/− SM cell was equivalent to that of WT SM cells (Fig. 3, A and B). Strikingly, the twofold reduction in IL-2Rα expression resulted in preferential TFH cell differentiation by CD25+/− cells as early as 48 h after LCMV infection (110% more CD25low Bcl6high cells; P < 0.001; Fig. 3 C). Similar results were obtained 72 h after LCMV infection (P < 0.01; Fig. 3 D), by which time SM cells had bifurcated into TFH and TH1 effector cells.
Figure 3.
IL-2 signaling inhibits TFH cell differentiation during T cell priming. (A–D) WT and CD25+/− SM CD4+ T cells were adoptively transferred into C57BL/6J mice that were infected with LCMV 1 d later. Splenocytes were analyzed 2 (A–C) or 3 (D) d after infection. Data are representative of two independent experiments, and n = 5/group. (A) Quantitation of SM cells as a percentage of total CD4+ T cells. (B) Quantitation of SM cell CD69 and CD25 MFI. ***, P < 0.0001. (C) Representative FACS plots gated on SM cells, with TFH-committed cells (CD25low Bcl6high) boxed. Quantitation of TFH-committed SM cells as a percentage of total SM cells. ***, P < 0.0001. (D) Representative FACS plots gated on SM cells, with TFH-committed cells (Bcl6high CXCR5high) boxed. Quantitation of TFH-committed SM cells as a percentage of total SM cells. **, P = 0.01. (E and F) WT SM CD4+ T cells were adoptively transferred into C57BL/6J mice. Host mice were treated with anti–IL-2 neutralizing antibodies or isotype-matched control antibodies and infected with LCMV 1 d later. Splenocytes were analyzed 2 d after infection. Data are representative of two independent experiments, and n = 5/group. (E) Quantitation of SM cells as a percentage of total CD4+ T cells. (F) Representative FACS plots gated on SM cells, with TFH-committed cells (CD25low Bcl6high) boxed. Quantitation of TFH-committed SM cells as a percentage of total SM cells. ***, P = 0.001. Error bars depict the standard error of the mean.
Next, we transferred WT SM cells into C57BL/6J mice treated with anti–IL-2 neutralizing antibodies and then infected with LCMV. IL-2 neutralization did not impair SM cell expansion (Fig. 3 E) but did significantly enhance commitment to TFH cell differentiation (86% increase; P = 0.0004; Fig. 3 F). Together, these data demonstrated that IL-2 is a key mediator of STAT5 signaling and inhibition of TFH cell differentiation during T cell priming and that reduced IL-2 signaling is sufficient to bias T cells away from TH1 effector cell differentiation and toward TFH cell differentiation.
STAT5-mediated inhibition of TFH cell differentiation is dependent on Blimp-1
Because IL-2 and STAT5 regulate myriad genes in lymphocytes, it was important to identify the STAT5 targets responsible for inhibiting TFH cell differentiation. Because IL-2 can induce Blimp-1 expression in CD8+ T cells (Martins and Calame, 2008; Kalia et al., 2010; Pipkin et al., 2010), we hypothesized that STAT5 signaling in CD4+ T cells inhibited TFH cell differentiation by also inducing expression of Blimp-1.
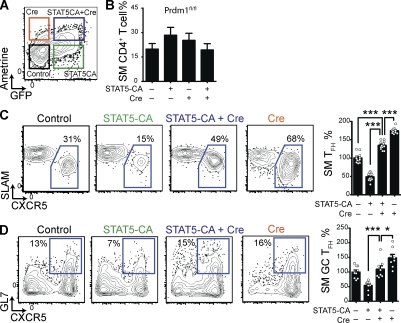
We tested the ability of STAT5 signaling to inhibit TFH cell differentiation in the absence of Blimp-1 (encoded by the gene Prdm1) by cotransducing Prdm1fl/fl SM cells with both STAT5-CA RV (expressing GFP) and Cre RV (expressing the fluorescent protein Ametrine). STAT5-CA+ Cre+, STAT5-CA+ Cre−, STAT5-CA− Cre+, and STAT5-CA− Cre− (control) SM cells were purified and adoptively transferred into C57BL/6J mice that were subsequently infected with LCMV (Fig. 4 A). All populations of SM cells expanded equivalently (Fig. 4 B). Deletion of Blimp-1 alone (Cre+) enhanced TFH cell differentiation (Fig. 4 C), as previously demonstrated (Johnston et al., 2009). Consistent with the experiments shown in Fig. 2, expression of STAT5-CA in Cre− (Blimp-1 intact) Prdm1fl/fl SM cells inhibited TFH cell differentiation (Fig. 4 C). However, STAT5-CA and Cre cotransduced Prdm1fl/fl SM cells, which possessed constitutive STAT5 signaling but lacked Blimp-1, readily differentiated into TFH cells (176% more TFH cells than for Blimp-1–intact STAT5-CA+ SM cells; P < 0.0001; Fig. 4 C). GC TFH cell differentiation was also restored in STAT5-CA+ SM cells by the absence of Blimp-1 (104% increase; P < 0.0001; Fig. 4 D). These data indicated that STAT5 inhibition of TFH cell differentiation was mediated by Blimp-1, consistent with the recent finding that STAT5 can directly bind the Prdm1 promoter in CD4+ T cells after IL-2 stimulation (Yang et al., 2011). Additional STAT5 targets may also contribute (Liao et al., 2011), as the contribution of Blimp-1 was not complete (Fig. 4, C and D).
Figure 4.
STAT5 inhibition of TFH cell differentiation is mediated by Blimp-1. Prdm1fl/fl SM cells were transduced with STAT5-CA RV and/or with a variant of Cre RV expressing the fluorescent protein mAmetrine or were not transduced but treated similarly (control). Data are a composite of three (A–C) or two (D) independent experiments, and n = 11–12/group (A–C) or 8/group (D). (A) Representative FACS plot gated on viable (7AADlow) cells, with untransduced, singly transduced, and cotransduced cells boxed. (B–D) Transduced or control SM cells were adoptively transferred into C57BL/6J mice that were infected with LCMV 3–5 d later. Splenocytes were analyzed 8 d after infection. (B) Quantitation of SM cells as a percentage of total CD4+ T cells. (C) Representative FACS plots gated on SM cells, with TFH cells (SLAMlow CXCR5high) boxed. Quantitation of SM TFH cells as a percentage of total SM cells. Data have been normalized so that the mean control SM TFH percentage for each experiment is 100%. ***, P < 0.0001. (D) Representative FACS plots gated on SM cells, with GC TFH cells (GL7high CXCR5high) boxed. Quantitation of SM GC TFH cells as a percentage of total SM cells. Data have been normalized so that the mean control SM GC TFH percentage for each experiment is 100%. *, P = 0.0330; ***, P < 0.0001. Error bars depict the standard error of the mean.
The signals that negatively regulate TFH cell differentiation have not been well characterized. In this study, we found that STAT5 is a key physiological inhibitor of Bcl6 expression and thereby an inhibitor of TFH cell differentiation. The absence of STAT5 resulted in increased Bcl6 expression and preferential TFH cell differentiation. This STAT5 function appears to be primarily induced by IL-2, as reduced IL-2 signaling substantially increased TFH cell differentiation. Because IL-2 and other STAT5 signaling cytokines are important mediators of T cell proliferation and survival, it was somewhat surprising that IL-2–deprived or STAT5-deficient TFH cells expanded normally. In agreement with our observations, the size of the antiviral CD4+ T cell response was unaffected by STAT5 deficiency in another study (Tripathi et al., 2010).
Our data suggest that STAT5 is not necessary for the CD4+ T cell effector response per se, but is required to properly balance TFH and TH1 effector CD4+ T cell differentiation. Importantly, our finding that a twofold reduction in IL-2Rα expression (CD25+/−) shifts CD4+ T cells toward TFH cell differentiation demonstrates that small changes in IL-2 availability can have a significant impact on T cell fate decisions in vivo. Bcl6 is also involved in the development of T cell memory (Ichii et al., 2004; Crotty et al., 2010; Pipkin et al., 2010; Pepper et al., 2011). Although in this study we focus on how STAT5 negatively regulates TFH cell differentiation and the development of GCs (Fig. 1 C), it is also intriguing to consider how these processes may impact CD4 T cell memory.
Given the association of dysregulated TFH activity with autoantibody production, manipulation of STAT5 activity to attenuate TFH cell differentiation or function may be a useful tool in the treatment of autoimmune disease. Conversely, manipulation of this pathway may also be a valuable tool to augment TFH activity and thus the potency of candidate vaccines for a variety of infectious diseases.
MATERIALS AND METHODS
Mice.
C57BL/6J mice, as well as Prdm1fl/fl mice (Shapiro-Shelef et al., 2003) and CD25-deficient mice (Il2ra−/−, B6.129S4-Il2ratm1Dw/J; Willerford et al., 1995) fully backcrossed to C57BL/6J, were purchased from the Jackson Laboratory. CD4-cre mice were purchased from Taconic. SM TCR transgenic CD45.1+ mice with a C57BL/6J background were bred at the La Jolla Institute for Allergy and Immunology (LIAI; McCausland et al., 2007). STAT5fl/fl mice were generated by L. Hennighausen and colleagues (National Institutes of Health, Bethesda, MD; Cui et al., 2004) and backcrossed to the C57BL/6J background by M. Farrar and colleagues at the University of Minnesota (Minneapolis, MN), resulting in >10 generations of backcrossing to B6. STAT5fl/fl mice were further backcrossed to SM mice on the C57BL/6J background. 862 of 884 (97.5%) descriptive single nucleotide polymorphisms in backcrossed STAT5fl/fl mice were consistent with the C57BL/6J background, as determined by whole genome microsatellite analysis performed through the University of California, Los Angeles Southern California Genotyping Consortium on mice from LIAI. STAT5fl/fl mice, Prdm1fl/fl mice, and CD25−/− mice were crossed with SM mice at LIAI.
STAT5 deletion efficiency was determined by quantitative PCR (forward primer, 5′-ATGGACTCACACCCACAAGGA-3′; and reverse primer, 5′-CACTGCTACAAGGCTACACAAAACC-3′). For CD25 SM experiments, CD25+/+ littermate control SM mice were used with CD25+/− SM mice. All animal experiments were conducted in accordance with animal protocols approved by the LIAI Institutional Animal Care and Use Committee.
RVs, transductions, and cell transfers.
Gene expression experiments were performed with GFP-expressing RV pMIG as well as a modification of pMIG expressing the fluorescent protein mAmetrine1.1 (Ai et al., 2008) instead of GFP (provided by D. Vignali, St. Jude Children’s Research Hospital, Memphis, TN). STAT5-expressing retroviruses were designed using previously described sequences for WT and constitutively active STAT5b (H299R + S711F; Onishi et al., 1998). Cre recombinase–expressing retrovirus has been previously described (Johnston et al., 2009). Transduction of STAT5fl/fl SM CD4+ T cells with Cre RV resulted in a deletion efficiency of ∼93%, as measured by qPCR analysis.
Virions were produced using the Plat-E cell line as previously described (McCausland et al., 2007). For retroviral transduction of CD4+ T cells, CD4+ T cells were purified from the splenocytes of naive mice by magnetic bead negative selection (Miltenyi Biotec) and suspended in D-10 (DMEM + 10% fetal calf serum, supplemented with 2 mM GlutaMAX [Invitrogen] and 100 U/ml penicillin/streptomycin [Invitrogen]) with 10 ng/ml recombinant human IL-2 and 50 µM β-mercaptoethanol. 2 × 106 cells per well were stimulated in 24-well tissue culture plates precoated with 8 µg/ml anti-CD3 (clone 17A2; Bio X Cell) and anti-CD28 (clone 37.51; Bio X Cell). After 24 h, cells were transduced as described previously (McCausland et al., 2007). Where necessary, cells were cotransduced by simultaneous transduction with two separately prepared retrovirus stocks. After a total of 72 h of stimulation, CD4+ T cells were split and transferred into new wells with fresh D-10, IL-2, and β-mercaptoethanol. After an additional 72 h, transduced CD4+ T cells were highly purified by sorting for GFP and/or mAmetrine expression on a FACSDiva or FACSAria (BD). Transduction efficiencies ranged from 10–40% before sorting. 2.0 × 105 or 2.5 × 104 transduced SM cells were adoptively transferred into each C57BL/6J host mouse via the retroorbital sinus for day 4 and day 8 experiments, respectively. In some experiments, cells that were not transduced but treated similarly were used as control cells in parallel adoptive transfer experiments. For day 2 and day 3 experiments, 106 and 5 × 105 freshly isolated SM CD4+ T cells, respectively, were adoptively transferred into each C57BL/6J host mouse via the retroorbital sinus.
Infections.
LCMV stocks were prepared and quantified as previously described (McCausland et al., 2007). Infection doses were 106, 5 × 105, 2 × 105, and 5 × 104 plaque-forming units of LCMV Armstrong per mouse for day 2, day 3, day 4, and day 8 experiments, respectively. Infections were performed by intraperitoneal injection.
IL-2 neutralization in vivo.
Mice received either control rat IgG2a or rat anti–IL-2 (clone S4B6). Each mouse was treated with 1.0 mg antibody via intraperitoneal and retroorbital injection 24 h before LCMV infection and then again 24 h after LCMV infection.
Flow cytometry.
Single-cell suspensions of spleen were prepared by standard gentle mechanical disruption. Surface staining for flow cytometry was performed with monoclonal antibodies against SLAM (CD150; BioLegend) and CD4, CD8, CD45.1, CD44, CD62L, CD25, PD-1, CD69, B220, Fas, and GL7 (eBioscience). Surface stains were performed for 30–60 min at 4°C in PBS supplemented with 0.5% bovine serum albumin and 0.1% sodium azide, unless specified otherwise.
CXCR5 staining was performed as described previously (Johnston et al., 2009) for day 8 experiments using purified anti-CXCR5 (BD) for 60 min, followed by biotinylated anti–rat IgG (Jackson ImmunoResearch Laboratories, Inc.), and then by allophycocyanin (APC)- or PE-labeled streptavidin (Invitrogen) at 4°C in PBS supplemented with 0.5% bovine serum albumin, 2% fetal calf serum, and 2% normal mouse serum. For day 2–4 experiments, CXCR5 staining was performed using biotinylated anti-CXCR5 (BD) for 30 min, followed by APC- or PE-labeled streptavidin at 4°C (Choi et al., 2011).
Intracellular cytokine staining was performed as described previously (McCausland et al., 2007) after stimulation with 20 ng/ml PMA (Sigma-Aldrich) and 1 µM ionomycin (Sigma-Aldrich) in the presence of 2 µg/ml brefeldin-A (BD) for 4 h. Directly conjugated antibodies against IFN-γ and IL-2 (BD) were used. For IL-21, staining was performed using an IL-21R–FC chimeric protein (R&D Systems) followed by PE- or APC-labeled anti–human IgG (Jackson ImmunoResearch Laboratories, Inc.; Johnston et al., 2009). Intracellular staining for Bcl6 was performed as previously described (Choi et al., 2011) with an Alexa Fluor 647–conjugated monoclonal antibody to Bcl6 (clone K112-91; provided by E. O’Donnell and D. Ernst, BD) and the FoxP3 ICS kit buffers and protocol (eBioscience).
For phospho-STAT5 FACS, sorted GFP+ cells were fixed and permeabilized with Phosflow Lyse/Fix buffer and Phosflow Perm Buffer III (BD). Where indicated, cells were first stimulated with 20 ng/ml of recombinant human IL-2 for 30 min. Cells were stained with anti–phospho-STAT5 antibody (pY694; BD). Samples were acquired using a C6 Flow Cytometer (Accuri) or an LSRII (BD) and analyzed using FlowJo (Tree Star).
Statistical analysis.
Statistical tests were performed using Prism 5.0c (GraphPad Software). P-values were calculated by two-tailed unpaired Student’s t tests with a 95% confidence interval. Error bars depict the standard error of the mean. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Acknowledgments
We thank Mark Kroenke, Danelle Eto, Robin Kageyama, Lindsay Crickard, Cheryl Kim, Kurt Van Gunst, and Anthony Jose for technical advice and assistance. We thank Michael Farrar and Lothar Hennighausen for STAT5fl/fl mice. We thank Dario Vignali for providing mAmetrine1.1 DNA. We thank Erika O’Donnell, David Ernst, and BD for providing anti-Bcl6 antibodies.
This work was supported by a National Institutes of Health (NIH) training grant to R.J. Johnston and by NIH grants and LIAI institutional funds to S. Crotty.
R.J. Johnston is now a postdoctoral research fellow at Genentech. The authors have no additional conflicting financial interests.
Footnotes
Abbreviations used:
- APC
- allophycocyanin
- GC
- germinal center
- LCMV
- lymphocytic choriomeningitis virus
- MFI
- mean fluorescence intensity
- RV
- retroviral expression vector
References
- Ai H.W., Hazelwood K.L., Davidson M.W., Campbell R.E. 2008. Fluorescent protein FRET pairs for ratiometric imaging of dual biosensors. Nat. Methods. 5:401–403 10.1038/nmeth.1207 [DOI] [PubMed] [Google Scholar]
- Akiba H., Takeda K., Kojima Y., Usui Y., Harada N., Yamazaki T., Ma J., Tezuka K., Yagita H., Okumura K. 2005. The role of ICOS in the CXCR5+ follicular B helper T cell maintenance in vivo. J. Immunol. 175:2340–2348 [DOI] [PubMed] [Google Scholar]
- Arguni E., Arima M., Tsuruoka N., Sakamoto A., Hatano M., Tokuhisa T. 2006. JunD/AP-1 and STAT3 are the major enhancer molecules for high Bcl6 expression in germinal center B cells. Int. Immunol. 18:1079–1089 10.1093/intimm/dxl041 [DOI] [PubMed] [Google Scholar]
- Burchill M.A., Yang J., Vogtenhuber C., Blazar B.R., Farrar M.A. 2007. IL-2 receptor beta-dependent STAT5 activation is required for the development of Foxp3+ regulatory T cells. J. Immunol. 178:280–290 [DOI] [PubMed] [Google Scholar]
- Choi Y.S., Kageyama R., Eto D., Escobar T.C., Johnston R.J., Monticelli L., Lao C., Crotty S. 2011. ICOS receptor instructs T follicular helper cell versus effector cell differentiation via induction of the transcriptional repressor Bcl6. Immunity. 34:932–946 10.1016/j.immuni.2011.03.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cretney E., Xin A., Shi W., Minnich M., Masson F., Miasari M., Belz G.T., Smyth G.K., Busslinger M., Nutt S.L., Kallies A. 2011. The transcription factors Blimp-1 and IRF4 jointly control the differentiation and function of effector regulatory T cells. Nat. Immunol. 12:304–311 10.1038/ni.2006 [DOI] [PubMed] [Google Scholar]
- Crotty S. 2011. Follicular helper CD4 T cells (TFH). Annu. Rev. Immunol. 29:621–663 10.1146/annurev-immunol-031210-101400 [DOI] [PubMed] [Google Scholar]
- Crotty S., Johnston R.J., Schoenberger S.P. 2010. Effectors and memories: Bcl-6 and Blimp-1 in T and B lymphocyte differentiation. Nat. Immunol. 11:114–120 10.1038/ni.1837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui Y., Riedlinger G., Miyoshi K., Tang W., Li C., Deng C.X., Robinson G.W., Hennighausen L. 2004. Inactivation of Stat5 in mouse mammary epithelium during pregnancy reveals distinct functions in cell proliferation, survival, and differentiation. Mol. Cell. Biol. 24:8037–8047 10.1128/MCB.24.18.8037-8047.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deenick E.K., Chan A., Ma C.S., Gatto D., Schwartzberg P.L., Brink R., Tangye S.G. 2010. Follicular helper T cell differentiation requires continuous antigen presentation that is independent of unique B cell signaling. Immunity. 33:241–253 10.1016/j.immuni.2010.07.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diehl S.A., Schmidlin H., Nagasawa M., van Haren S.D., Kwakkenbos M.J., Yasuda E., Beaumont T., Scheeren F.A., Spits H. 2008. STAT3-mediated up-regulation of BLIMP1 Is coordinated with BCL6 down-regulation to control human plasma cell differentiation. J. Immunol. 180:4805–4815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duy C., Hurtz C., Shojaee S., Cerchietti L., Geng H., Swaminathan S., Klemm L., Kweon S.M., Nahar R., Braig M., et al. 2011. BCL6 enables Ph+ acute lymphoblastic leukaemia cells to survive BCR-ABL1 kinase inhibition. Nature. 473:384–388 10.1038/nature09883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eto D., Lao C., DiToro D., Barnett B., Escobar T.C., Kageyama R., Yusuf I., Crotty S. 2011. IL-21 and IL-6 are critical for different aspects of B cell immunity and redundantly induce optimal follicular helper CD4 T cell (Tfh) differentiation. PLoS ONE. 6:e17739 10.1371/journal.pone.0017739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fazilleau N., McHeyzer-Williams L.J., Rosen H., McHeyzer-Williams M.G. 2009. The function of follicular helper T cells is regulated by the strength of T cell antigen receptor binding. Nat. Immunol. 10:375–384 10.1038/ni.1704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatto D., Brink R. 2010. The germinal center reaction. J. Allergy Clin. Immunol. 126:898–907 10.1016/j.jaci.2010.09.007 [DOI] [PubMed] [Google Scholar]
- Gigoux M., Shang J., Pak Y., Xu M., Choe J., Mak T.W., Suh W.K. 2009. Inducible costimulator promotes helper T-cell differentiation through phosphoinositide 3-kinase. Proc. Natl. Acad. Sci. USA. 106:20371–20376 10.1073/pnas.0911573106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goenka R., Barnett L.G., Silver J.S., O’Neill P.J., Hunter C.A., Cancro M.P., Laufer T.M. 2011. Cutting edge: dendritic cell-restricted antigen presentation initiates the follicular helper T cell program but cannot complete ultimate effector differentiation. J. Immunol. 187:1091–1095 10.4049/jimmunol.1100853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harker J.A., Lewis G.M., Mack L., Zuniga E.I. 2011. Late interleukin-6 escalates T follicular helper cell responses and controls a chronic viral infection. Science. 334:825–829 10.1126/science.1208421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haynes N.M., Allen C.D., Lesley R., Ansel K.M., Killeen N., Cyster J.G. 2007. Role of CXCR5 and CCR7 in follicular Th cell positioning and appearance of a programmed cell death gene-1high germinal center-associated subpopulation. J. Immunol. 179:5099–5108 [DOI] [PubMed] [Google Scholar]
- Hirahara K., Ghoreschi K., Laurence A., Yang X.P., Kanno Y., O’Shea J.J. 2010. Signal transduction pathways and transcriptional regulation in Th17 cell differentiation. Cytokine Growth Factor Rev. 21:425–434 10.1016/j.cytogfr.2010.10.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y.L., Metz D.P., Chung J., Siu G., Zhang M. 2009. B7RP-1 blockade ameliorates autoimmunity through regulation of follicular helper T cells. J. Immunol. 182:1421–1428 http://www.jimmunol.org/content/182/3/1421.long [DOI] [PubMed] [Google Scholar]
- Ichii H., Sakamoto A., Kuroda Y., Tokuhisa T. 2004. Bcl6 acts as an amplifier for the generation and proliferative capacity of central memory CD8+ T cells. J. Immunol. 173:883–891 [DOI] [PubMed] [Google Scholar]
- Johnston R.J., Poholek A.C., DiToro D., Yusuf I., Eto D., Barnett B., Dent A.L., Craft J., Crotty S. 2009. Bcl6 and Blimp-1 are reciprocal and antagonistic regulators of T follicular helper cell differentiation. Science. 325:1006–1010 10.1126/science.1175870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalia V., Sarkar S., Subramaniam S., Haining W.N., Smith K.A., Ahmed R. 2010. Prolonged interleukin-2Ralpha expression on virus-specific CD8+ T cells favors terminal-effector differentiation in vivo. Immunity. 32:91–103 10.1016/j.immuni.2009.11.010 [DOI] [PubMed] [Google Scholar]
- Kitano M., Moriyama S., Ando Y., Hikida M., Mori Y., Kurosaki T., Okada T. 2011. Bcl6 protein expression shapes pre-germinal center B cell dynamics and follicular helper T cell heterogeneity. Immunity. 34:961–972 10.1016/j.immuni.2011.03.025 [DOI] [PubMed] [Google Scholar]
- Kwon H., Thierry-Mieg D., Thierry-Mieg J., Kim H.P., Oh J., Tunyaplin C., Carotta S., Donovan C.E., Goldman M.L., Tailor P., et al. 2009. Analysis of interleukin-21-induced Prdm1 gene regulation reveals functional cooperation of STAT3 and IRF4 transcription factors. Immunity. 31:941–952 10.1016/j.immuni.2009.10.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S.K., Rigby R.J., Zotos D., Tsai L.M., Kawamoto S., Marshall J.L., Ramiscal R.R., Chan T.D., Gatto D., Brink R., et al. 2011. B cell priming for extrafollicular antibody responses requires Bcl-6 expression by T cells. J. Exp. Med. 208:1377–1388 10.1084/jem.20102065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao W., Lin J.X., Wang L., Li P., Leonard W.J. 2011. Modulation of cytokine receptors by IL-2 broadly regulates differentiation into helper T cell lineages. Nat. Immunol. 12:551–559 10.1038/ni.2030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linterman M.A., Rigby R.J., Wong R.K., Yu D., Brink R., Cannons J.L., Schwartzberg P.L., Cook M.C., Walters G.D., Vinuesa C.G. 2009. Follicular helper T cells are required for systemic autoimmunity. J. Exp. Med. 206:561–576 10.1084/jem.20081886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma C.S., Suryani S., Avery D.T., Chan A., Nanan R., Santner-Nanan B., Deenick E.K., Tangye S.G. 2009. Early commitment of naïve human CD4(+) T cells to the T follicular helper (T(FH)) cell lineage is induced by IL-12. Immunol. Cell Biol. 87:590–600 10.1038/icb.2009.64 [DOI] [PubMed] [Google Scholar]
- Malek T.R., Yu A., Vincek V., Scibelli P., Kong L. 2002. CD4 regulatory T cells prevent lethal autoimmunity in IL-2Rbeta-deficient mice. Implications for the nonredundant function of IL-2. Immunity. 17:167–178 10.1016/S1074-7613(02)00367-9 [DOI] [PubMed] [Google Scholar]
- Martins G., Calame K. 2008. Regulation and functions of Blimp-1 in T and B lymphocytes. Annu. Rev. Immunol. 26:133–169 10.1146/annurev.immunol.26.021607.090241 [DOI] [PubMed] [Google Scholar]
- McCausland M.M., Yusuf I., Tran H., Ono N., Yanagi Y., Crotty S. 2007. SAP regulation of follicular helper CD4 T cell development and humoral immunity is independent of SLAM and Fyn kinase. J. Immunol. 178:817–828 [DOI] [PubMed] [Google Scholar]
- Nakajima H., Liu X.W., Wynshaw-Boris A., Rosenthal L.A., Imada K., Finbloom D.S., Hennighausen L., Leonard W.J. 1997. An indirect effect of Stat5a in IL-2-induced proliferation: a critical role for Stat5a in IL-2-mediated IL-2 receptor alpha chain induction. Immunity. 7:691–701 10.1016/S1074-7613(00)80389-1 [DOI] [PubMed] [Google Scholar]
- Nurieva R.I., Chung Y., Hwang D., Yang X.O., Kang H.S., Ma L., Wang Y.H., Watowich S.S., Jetten A.M., Tian Q., Dong C. 2008. Generation of T follicular helper cells is mediated by interleukin-21 but independent of T helper 1, 2, or 17 cell lineages. Immunity. 29:138–149 10.1016/j.immuni.2008.05.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nurieva R.I., Chung Y., Martinez G.J., Yang X.O., Tanaka S., Matskevitch T.D., Wang Y.-H., Dong C. 2009. Bcl6 mediates the development of T follicular helper cells. Science. 325:1001–1005 10.1126/science.1176676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onishi M., Nosaka T., Misawa K., Mui A.L., Gorman D., McMahon M., Miyajima A., Kitamura T. 1998. Identification and characterization of a constitutively active STAT5 mutant that promotes cell proliferation. Mol. Cell. Biol. 18:3871–3879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pepper M., Pagán A.J., Igyártó B.Z., Taylor J.J., Jenkins M.K. 2011. Opposing signals from the Bcl6 transcription factor and the interleukin-2 receptor generate T helper 1 central and effector memory cells. Immunity. 35:583–595 10.1016/j.immuni.2011.09.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pipkin M.E., Sacks J.A., Cruz-Guilloty F., Lichtenheld M.G., Bevan M.J., Rao A. 2010. Interleukin-2 and inflammation induce distinct transcriptional programs that promote the differentiation of effector cytolytic T cells. Immunity. 32:79–90 10.1016/j.immuni.2009.11.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poholek A.C., Hansen K., Hernandez S.G., Eto D., Chandele A., Weinstein J.S., Dong X., Odegard J.M., Kaech S.M., Dent A.L., et al. 2010. In vivo regulation of Bcl6 and T follicular helper cell development. J. Immunol. 185:313–326 10.4049/jimmunol.0904023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reljic R., Wagner S.D., Peakman L.J., Fearon D.T. 2000. Suppression of signal transducer and activator of transcription 3–dependent B lymphocyte terminal differentiation by BCL-6. J. Exp. Med. 192:1841–1848 10.1084/jem.192.12.1841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rochman Y., Spolski R., Leonard W.J. 2009. New insights into the regulation of T cells by gamma(c) family cytokines. Nat. Rev. Immunol. 9:480–490 10.1038/nri2580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheeren F.A., Naspetti M., Diehl S., Schotte R., Nagasawa M., Wijnands E., Gimeno R., Vyth-Dreese F.A., Blom B., Spits H. 2005. STAT5 regulates the self-renewal capacity and differentiation of human memory B cells and controls Bcl-6 expression. Nat. Immunol. 6:303–313 10.1038/ni1172 [DOI] [PubMed] [Google Scholar]
- Shapiro-Shelef M., Lin K.I., McHeyzer-Williams L.J., Liao J., McHeyzer-Williams M.G., Calame K. 2003. Blimp-1 is required for the formation of immunoglobulin secreting plasma cells and pre-plasma memory B cells. Immunity. 19:607–620 10.1016/S1074-7613(03)00267-X [DOI] [PubMed] [Google Scholar]
- Tripathi P., Kurtulus S., Wojciechowski S., Sholl A., Hoebe K., Morris S.C., Finkelman F.D., Grimes H.L., Hildeman D.A. 2010. STAT5 is critical to maintain effector CD8+ T cell responses. J. Immunol. 185:2116–2124 10.4049/jimmunol.1000842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker S.R., Nelson E.A., Frank D.A. 2007. STAT5 represses BCL6 expression by binding to a regulatory region frequently mutated in lymphomas. Oncogene. 26:224–233 10.1038/sj.onc.1209775 [DOI] [PubMed] [Google Scholar]
- Wei L., Laurence A., O’Shea J.J. 2008. New insights into the roles of Stat5a/b and Stat3 in T cell development and differentiation. Semin. Cell Dev. Biol. 19:394–400 10.1016/j.semcdb.2008.07.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willerford D.M., Chen J., Ferry J.A., Davidson L., Ma A., Alt F.W. 1995. Interleukin-2 receptor alpha chain regulates the size and content of the peripheral lymphoid compartment. Immunity. 3:521–530 10.1016/1074-7613(95)90180-9 [DOI] [PubMed] [Google Scholar]
- Yang X.P., Ghoreschi K., Steward-Tharp S.M., Rodriguez-Canales J., Zhu J., Grainger J.R., Hirahara K., Sun H.W., Wei L., Vahedi G., et al. 2011. Opposing regulation of the locus encoding IL-17 through direct, reciprocal actions of STAT3 and STAT5. Nat. Immunol. 12:247–254 10.1038/ni.1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu D., Rao S., Tsai L.M., Lee S.K., He Y., Sutcliffe E.L., Srivastava M., Linterman M., Zheng L., Simpson N., et al. 2009. The transcriptional repressor Bcl-6 directs T follicular helper cell lineage commitment. Immunity. 31:457–468 10.1016/j.immuni.2009.07.002 [DOI] [PubMed] [Google Scholar]
- Yusuf I., Kageyama R., Monticelli L., Johnston R.J., Ditoro D., Hansen K., Barnett B., Crotty S. 2010. Germinal center T follicular helper cell IL-4 production is dependent on signaling lymphocytic activation molecule receptor (CD150). J. Immunol. 185:190–202 10.4049/jimmunol.0903505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J., Yamane H., Paul W.E. 2010. Differentiation of effector CD4 T cell populations. Annu. Rev. Immunol. 28:445–489 10.1146/annurev-immunol-030409-101212 [DOI] [PMC free article] [PubMed] [Google Scholar]