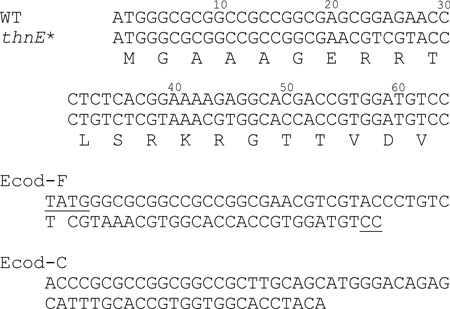Abstract
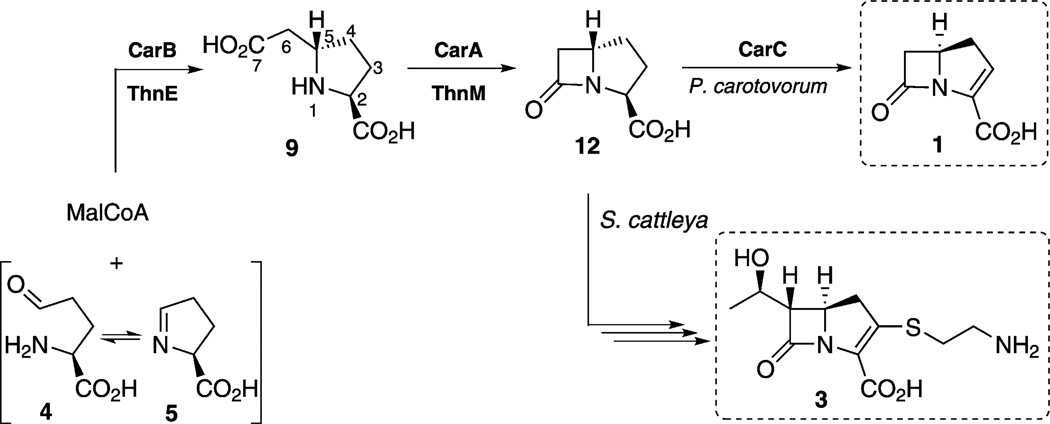
Approximately 50 naturally occurring carbapenem β-lactam antibiotics are known. All but one of these have been isolated from Streptomyces species and are disubstituted structural variants of a simple core that is synthesized by Pectobacterium carotovorum (Erwinia carotovora), a phylogenetically distant plant pathogen. While the biosynthesis of the simple carbapenem, (5R)-carbapen-2-em-3-carboxylic acid, is impressively efficient requiring only three enzymes, CarA, CarB and CarC, the formation of thienamycin, one of the former group of metabolites from Streptomyces, is markedly more complex. Despite their phylogenetic separation, bioinformatic analysis of the encoding gene clusters suggests that the two pathways could be related. Here we demonstrate with gene swapping, stereochemical and kinetics experiments that CarB and CarA and their S. cattleya orthologues, ThnE and ThnM, respectively, are functionally and stereochemically equivalent, although their catalytic efficiencies differ. The biosynthetic pathways, therefore, to thienamycin, and likely to the other disubstituted carbapenems, and to the simplest carbapenem, (5R)-carbapen-2-em-3-carboxylic acid, are initiated in the same manner, but share only two common steps before diverging.
Keywords: beta-lactam antibiotics, biosynthesis, carbapenems, enzyme catalysis, thienamycin
Introduction
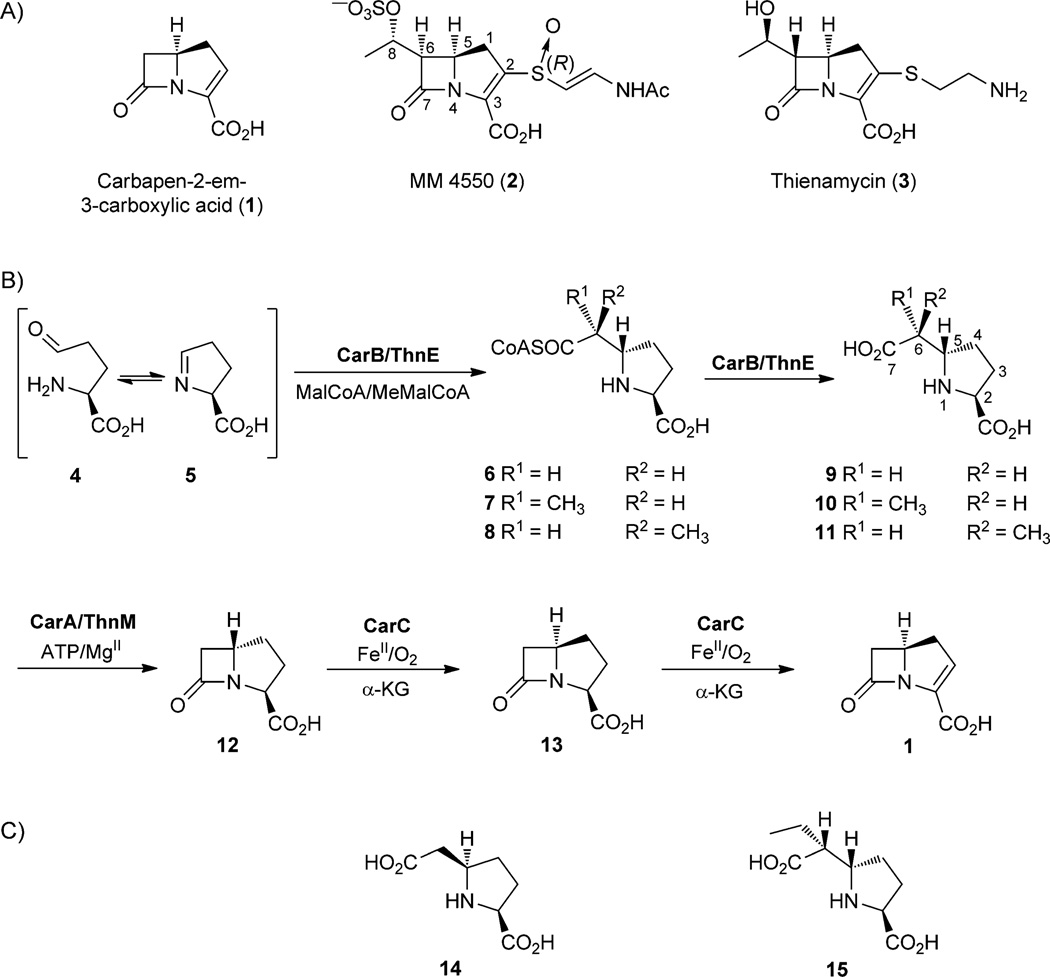
While semisynthetic penicillins and cephalosporins remain frequently prescribed drugs in human medicine, the carbapenem β-lactam antibiotics have assumed growing importance for their broad-spectrum activity, potency and effectiveness against resistant infections.[1] The family of naturally-occurring carbapenems consists of the simplest core metabolite, carbapen-2-em-3-carboxylic acid (1) on the one hand, and approximately 50 more highly elaborated structures that differ by the nature of the sulfur substituent at C2 and an alkyl group at C6, often present at an elevated oxidation state, for example, 2 (Scheme 1 A).[2] Prominent among the latter is thienamycin (3) the amidine derivative of which, imipenem, is marketed as Primaxin.[3] The two carbons of the hydroxyethyl side chain of 3 are known to be derived from methionine by presumably successive C1-transfers.[4–6] Recent work has established that the C2 side chain is derived by stepwise truncation of coenzyme A (CoA) rather than by direct incorporation of cysteamine or cysteine, as previously thought.[7] The timing, however, of C2 and C6 side-chain attachment among the overall biosynthetic events leading to thienamycin is unknown.
Scheme 1.
A) Representative carbapenems. B) The biosynthesis of carbapen-2-em-3-carboxylic acid (1). C) Carboxymethylprolines relevant to thienamycin (3) biosynthesis.
The unsubstituted carbapenem 1 is created in three highly efficient steps (Scheme 1B). Carboxymethylproline synthase (CarB), a member of the crotonase superfamily, mediates the decarboxylation of malonyl-CoA (MalCoA) and the stereospecific addition of the resulting enzyme-bound enolate to l-glutamate γ-semialdehyde (4) or l-pyrrolidine-5-carboxylic acid (l-P5C, 5). The resulting (2S,5S)-carboxymethylproline coenzyme A thioester (CMP-CoA, 6) partitions between hydrolysis and diffusion from the active site to give the acid 9. CarB is also capable of catalyzing the same series of reactions with methylmalonyl-CoA (MeMalCoA), albeit less efficiently, to give the 6-methyl (2S,5S)-CMP disasteriomers 10 and 11 in a 1.2:1 ratio.[8] Adenylation of 9 by carbapenam synthetase (CarA) is coupled to β-lactam formation by the coordinated participation of an active site Tyr–Glu dyad and a lysine residue to deftly catalyze formation of this more highly strained bicyclic intermediate.[9] In contrast to the strict stereospecificity of CarB, CarA will accept and process epimers of (2S,5S)-CMP (9).[8a, 9b] Finally, carbapenem synthase (CarC), a member of the nonheme iron α-ketoglutarate (α-KG)-dependent oxygenase superfamily, carries out the bridgehead epimerization of 12 to 13 and ring desaturation to give 1.[10]
Identification of the thienamycin biosynthetic gene cluster in Streptomyces cattleya opened the way to investigation of the more structurally complex members of the carbapenem antibiotic family.[6] It was immediately apparent that many more proteins were involved in the biosynthesis of 3 than of 1 and, while orthologues of CarA and CarB were encoded by the cluster, ThnE and ThnM bore only 37 and 25% amino acid sequence identity, respectively. Second, there was no orthologue of CarC, suggesting that different biochemical solutions had evolved to accomplish C2/3 desaturation and, by analogy to the formation of 1, to carry out bridgehead inversion—both essential to antibiotic activity. Support for at least one step in a common pathway to both 3 and the simple carbapenem 1 was recently reported when a truncated ThnE (Δ2–46) indeed catalyzed the formation of (2S,5S)-CMP (9, Scheme 1 B) from MalCoA, like CarB. A 4:1 diastereomeric mixture of 6-methyl-(2S,5S)-CMPs 10 and 11 was also observed from MeMalCoA.[11] We have gathered compelling gene swapping, stereochemical and kinetic evidence that 3 and 1 share two biosynthetic steps, despite substantial phylogenetic separation of the producing organisms, before diverging, and that introduction of the C6 side chain in thienamycin biosynthesis occurs after carbapenam 12 formation.
Results and Discussion
In vivo analysis of ThnE and ThnM
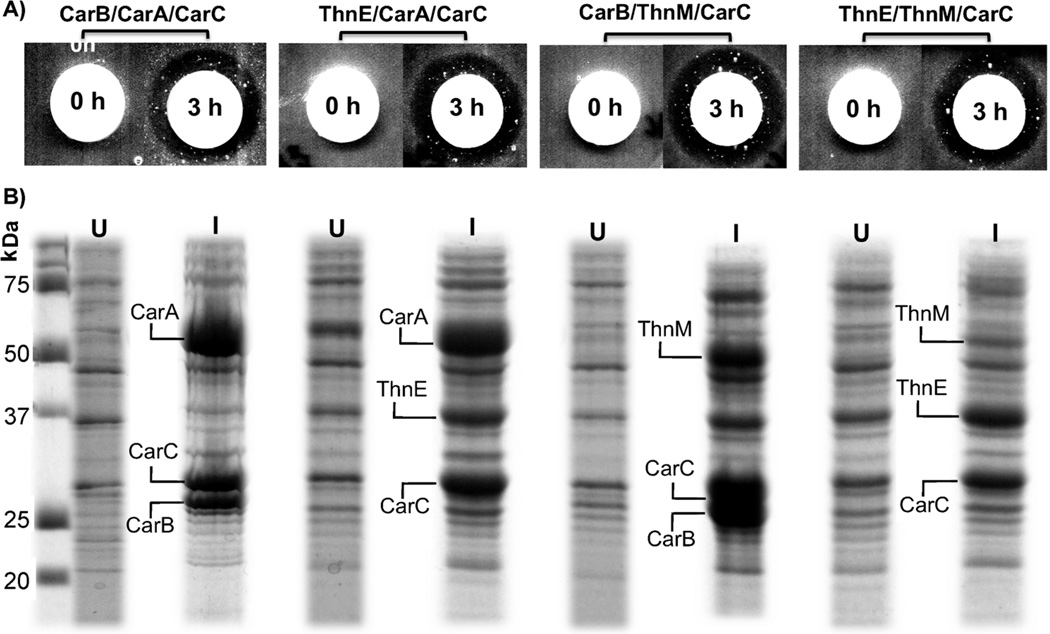
Despite the low overall identity between ThnE and CarB, or ThnM and CarA, many residues in their respective active sites appeared to be conserved, indicating that ThnE and ThnM could potentially carry out corresponding reactions in the simple carbapenem pathway. Their biosynthetic activities were first probed globally, therefore, by analyzing their abilities to substitute for their counterparts CarB and CarA and support the biosynthesis of (5R)-carbapen-2-em-3-carboxylic acid (1), in vivo (Figure 1). Genetic replacements by thnE and thnM were made individually and pairwise in the previously described plasmid, pET24a(+)/carABC.[12] In this construct, expression of carA, carB and carC were controlled by a T7 promoter and T7 terminator as a single operon. It had been shown that the coexpression of carB, carA and carC in E. coli BL21(DE3)(pLysS) reconstituted the synthesis of carbapenem 1 at a titer comparable to that of wild-type P. carotovorum, which could be readily detected by two sensitive bioassays by using a β-lactam super-sensitive E. coli and a β-lactamase induction assay.[9b, 13] To obtain higher expression and improved solubility of ThnE, the codons of the 21 N-terminal amino acids were optimized and the engineered ThnE (ThnE*) showed elevated expression profiles (data not shown). As illustrated in Figure 1, when substitutions were made in this construct by either or both thnE or thnM, approximately equivalent levels of antibiotic production were achieved in the optimized medium, suggesting overlap of their catalytic activities with both CarB and CarA, respectively.
Figure 1.
CarA and CarB orthologue replacement experiments. A) Inhibition of the growth of β-lactam super-sensitive E. coli SC12155. B) Coexpression of biosynthetic enzymes; U: uninduced cellular fraction, I: induced cellular fraction.
The ability, however, of CarB and truncated ThnE (Δ2–46) to carry out reactions with both MalCoA and MeMalCoA led us to address more rigorously the reactions catalyzed by ThnE.[8a] Similarly, the modest sequence identity between CarA and ThnM raised questions about the substrate preferences of both ThnE and ThnM, particularly with respect to C2 and C6 side-chain attachments and their timing in the biosynthetic pathway. The thnE gene was cloned from S. cattleya genomic DNA and the first 21 codons were optimized to favor expression in E. coli. The partially codon-optimized thnE* was inserted into pET28b encoding an N-terminal His6-tag. The thnM gene was similarly cloned and inserted into pET29b bearing a C-terminal His6-tag. The recombinant proteins were over-produced in E. coli Rosetta2(DE3) and purified by Ni-NTA affinity chromatography.
Preparation of substrates
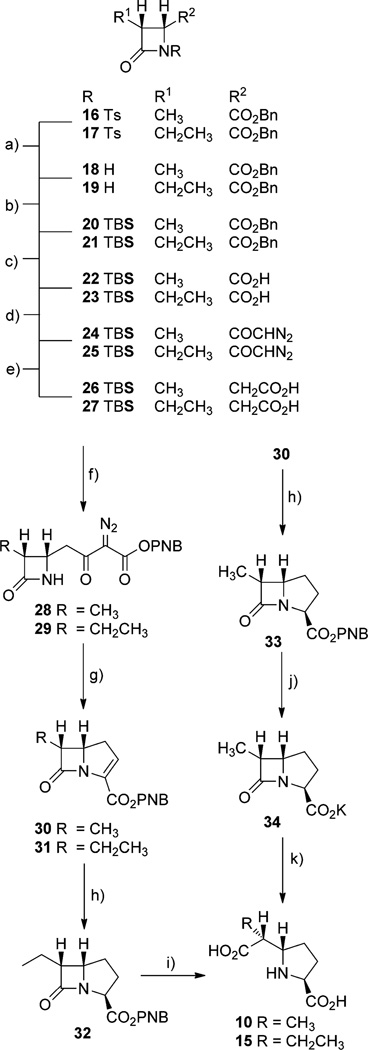
The substrates of CarB and CarA, l-P5C (5) and (2S,5S)-CMP (9), as well as (2S,5R)-CMP (14, Scheme 1 C) were synthesized by using established methods.[8a, 10b] In addition, (6R)-methyl and (6R)-ethyl CMPs (10 and 15) were synthesized by extension of a recently described route to carbapenems (Scheme 2).[14] 3-Methyl and 3-ethyl (3R,4R)-azetidinones 16 and 17 were prepared in catalytic, asymmetric reactions to set the absolute configuration at C4 that is produced by ThnE and the configuration at C3 that is observed in thienamycin (3) and all known carbapenem metabolites of S. cattleya. The cis relation of the C3/C4 substituents was preserved by Arndt–Eistert homologation of 24 and 25 in the synthesis of C2/3 unsubstituted carbapenems.[15,16] The α,β-unsaturated esters 30 and 31 thus obtained underwent reduction by selective conjugate hydride addition mediated by Stryker’s reagent to give the saturated carbapenams 32 and 33.[17] The newly created carbapenam C3 stereocenter was directed exclusively exo to the β-lactam ring, matching the orientation present in 9. The C3 acids could be deblocked by hydrogenolysis prior to hydrolysis of the β-lactam ring. Alternatively, the β-lactam ring and ester could be cleaved concurrently to give the C6-substituted carboxymethylprolines 10 and 15. Epimerization of the thermodynamically less stable cis-C5/C6 configuration to the more stable trans-C5/C6 configuration was not detected by 1H NMR spectroscopy.
Scheme 2.
Synthesis of carboxymethylprolines. a) SmI2, iPrOH, Sm(0), THF, 80%; b) CH2Cl2, TBSOTf, Et3N, 92%; c) THF, H2, Pd/C, 99%; d) 1) CH2Cl2, cat. DMF, oxalyl chloride, 2) Et2O, diazomethane, 85%; e) THF, 10% H2O, hv, 96%; f) 1) CH3CN, carbonyl diimidazole, 2) Mg(mono-PNB malonate)2, 3) MeOH, 10% 1 m HCl, 4) CH3CN, mesyl azide, Et3N, 50%; g) 1) benzene, Rh2(OAc)4, 80°C, 2) THF/MeOH, NaBH4, −78°C, 3) CH2Cl2, mesyl chloride, Et3N, 65 %; h) benzene, MeOH, [(PPh3)CuH]6, polymethylhydrosiloxane, 50%; i) 2:1 THF/H2O, 2 equiv KOH, 99%; j) 2:1 THF/H2O, 1 equiv KHCO3, H2, Pd/C, 90%; k) H2O, 1 equiv KOH, 99%.
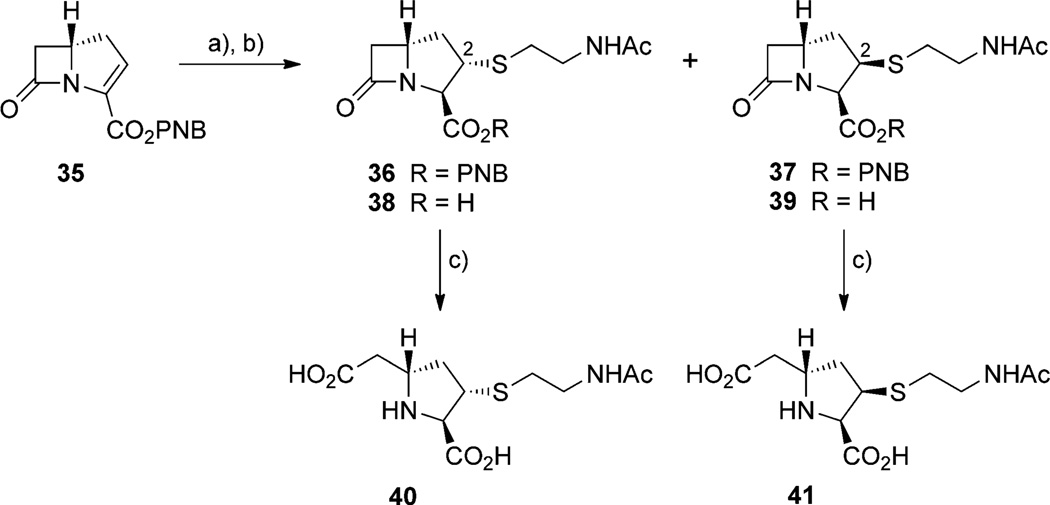
The known (5S)-carbapenem 35 was treated with N-acetylcysteamine to give a disastereotopic mixture of 36 and 37 in a 4:1 ratio, which was separated by chromatography on silica gel and deprotected by hydrogenolysis.[16] The carboxylic acids 38 and 39 were purified by HPLC. The low concentration of acid present during this step catalyzed β-lactam hydrolysis. The individual stereoisomers upon lyophilization yielded the corresponding cleaved (2S,3R,5S)- and (2R,3R,5S)-2-cysteaminyl-5-carboxy-ethylprolines 40 and 41 (Scheme 3 and the Supporting Information).
Scheme 3.
Synthesis of 2-N-acetylcysteaminyl-CMP diastereomers. a) N-acetylcysteamine, DBU, CH3CN; b) H2, 10% Pd/C, KHCO3, 2:1 THF/H2O; c) HPLC separation/lyophilization.
Alternate substrate studies and kinetics
A more detailed analysis of ThnE function began with a comparison of its substrate specificity for MalCoA, the native substrate of CarB, or MeMalCoA, a potential precursor of the thienamycin (3) hydroxyethyl group. The derivation of (R)- or (S)-MeMalCoA or ethylmalonyl-CoA by C1-transfers to MalCoA by S-adenosylmethionine is unprecedented, but is chemically feasible.[18] However remote, this possibility was examined by incubation of l-P5C (5) with MalCoA or MeMalCoA and monitoring the course of reaction by HPLC. The ThnE-catalyzed reaction between MalCoA and 5 was confirmed by Dowex purification and characterization of (2S,5S)-CMP (9) from the enzymatic reaction. ThnE also condensed MeMalCoA with 5 to give a 3:2 diastereomeric mixture of 6-methyl CMPs 10 and 11, respectively, an unselective product ratio similar to that seen previously with CarB.[8a] No reaction was observed with ethylmalonyl-CoA as previously with a truncated ThnE.[11]
With MeMalCoA as a substrate, ThnE also accumulated two other products observable by HPLC. HPLC purification and ESI mass spectrometric analysis (m/z 935.14 [M–H+]) identified them as CMP-CoA esters 7 and 8. The appearance of these CMP-CoA ester intermediates was also observed in the CarB-catalyzed reaction and could indicate a high degree of catalytic similarity between the enzymes. To further probe the extent of functional similarity, the kinetic constants for the ThnE-catalyzed reaction between MalCoA or MeMalCoA and 5 were determined (Table 1). ThnE is selective for MalCoA with a 66-fold higher specificity constant (kcat/KM) for the reaction with 5 and MalCoA than with MeMalCoA.
Table 1.
Steady state kinetic parameters for ThnE-catalyzed reactions.
| Substrate | KM [mm] | kcat [s−1] | kcat/KM [mm−1 s−1] |
|---|---|---|---|
| MalCoA | 0.017 ± 0.002 | 0.30 ± 0.01 | 17.4 ± 2.1 |
| MeMalCoA | 0.203 ± 0.008 | 0.05 ± 0.001 | 0.26 ± 0.01 |
A more likely point of side-chain methylation can be envisioned to occur at the stage of (2S,5S)-CMP (9) that would then be a substrate for the β-lactam-forming enzyme, ThnM. In a first experiment, potential substrates were screened in fixed time assays (3 h) at 10 mm, a high concentration 50-times greater than the KM of CarA with 9, its native substrate.[9b] Thus 9, 6-methyl (2S,5S,6R)-CMP (10) and 6-ethyl (2S,5S,6R)-CMP (15), the latter two bearing alkyl substituents matching the thienamycin (3) stereochemistry, were examined. As a further probe of pathway timing and C5 stereoisomerization, ThnM-catalyzed β-lactam formation with the epimeric (2S,5R)-CMP (14) as substrate was also investigated. Of 9, 10, 15 and 14, ThnM showed reaction only with 9 and 10. In an analogous manner the 2-cysteaminyl CMPs, 40 and 41 were assayed and also found to be inactive with ThnM. It is apparent that, while a methyl substituent is tolerated, more sterically demanding substitutions at C2 or C6 are not.
To investigate the reactions of ThnM with 9 and 10 more precisely, their kinetic constants were determined under optimized conditions from a pH/rate profile by using a continuous coupled-enzyme assay that linked production of the terminal byproduct AMP to a decrease in the concentration of NADH detectable at 340 nm (Table 2 and the Supporting Information).[19] Three notable observations emerged from this analysis. First, based on the selectivity constants (kcat/KM), the preferred substrate for ThnM, like CarA, is 9. Second, while the KM of 9 is nearly identical for ThnM and CarA, the kcat for the former is an order of magnitude lower, signaling a much less efficient enzyme.[9b] This lower kcat might result from selective pressure on the host to survive a more potent carbapenem product, or from downstream enzyme(s) that are overall rate-limiting to the flux of the biosynthesis, hence reducing evolutionary pressure on ThnM. Third, the kcat for 10 is approximately 50% greater than the unsubstituted substrate. We propose that it is the eclipsed relationship of the (6R)-methyl and the five-membered ring of the adenylated CMP imposed by the active site that engenders bond angle compression in the classic Thorpe–Ingold sense. This steric compression raises the ground state energy leading to β-lactam closure, hence accelerating the rate of this chemical step.[20] It is known from detailed kinetic analysis of CarA that both formation of the four-membered ring and a protein conformational change limit the overall rate of this enzyme.[9d] By analogy to ThnM, the intrinsic magnitude of the Thorpe–Ingold effect favoring β-lactam synthesis could be partially masked by a corresponding protein conformational change.
Table 2.
Steady state kinetic parameters for ThnM-catalyzed reactions.
| Substrate | KM [mm] | kcat [s−1] | kcat/KM [mm−1s−1] |
|---|---|---|---|
| (2S,5S)-CMP (9) | 0.115 ± 0.014 | 0.30 ± 0.01 | 0.260 ± 0.033 |
| (2S,5S,6R)-6-methyl CMP (10) | 5.78 ± 1.00 | 0.046 ± 0.004 | 0.008 ± 0.002 |
Conclusions
The combined results for ThnE and ThnM, particularly the kinetic discrimination of each enzyme for variously methyl- and ethyl-substituted potential substrates, reveal unambiguous preferences for the natural substrates of CarB and CarA involved in the synthesis of the structurally simpler (5R)-carbapen-2-em-3-carboxylic acid (1). The low sequence identity evident for each pair of homologous enzymes raised the question whether C1-alkylations or thioether side-chain introduction could take place at this early stage in the biosynthesis of thienamycin (3). One can envision that the active site(s) of CarB and ThnE or CarA and ThnM could have easily been altered by mutation to accommodate such charge–neutral steric changes in their substrates. The experiments strongly suggest, however, that this evolutionary alternative was not chosen despite the large phylogenetic separation of the respective hosts, and that thienamycin and simple carbapenem biosynthesis are functionally and stereochemically identical up to the formation of (3S,5S)-carbapenam (12). At this point the pathways to the simple 1 and 3 diverge (Scheme 4). In P. carotovorum the FeII/ α-ketoglutarate-dependent oxygenase CarC both inverts the bicyclic ring junction and desaturates C2/3 to give 1 (Scheme 1 B). By contrast a more complex series of events takes place to accomplish these steps and C2 and C6 side-chain modification in 3 formation.[2] Recently described insertional inactivation of thnL and thnP, two apparent radical SAM methyltransferases in the thienamycin biosynthetic gene cluster, gives rise to a detectable product the nominal mass of which is at least consistent with the accumulation of 12.[21] While this experiment does not strictly prove the identity or intermediacy of 12 in thienamycin biosynthesis, it is potentially supportive of the conclusions drawn here. C6 alkylation and C2 sulfur side-chain attachment each temper the reactivity of the simple carbapenem-3-carboxylic acid in ways desirable for antibiotic activity, a property that could account for the additional metabolic burden they entail. The timing and mechanism of these events, and of bicyclic ring inversion and C2/3 desaturation, remain to be determined.
Scheme 4.
Common biosynthetic steps to the carbapenem β-lactam antibiotics.
Experimental Section
Construction of co-overexpression vector pET24a(+)/carABC
RBS-carB generated as a XbaI–HindIII fragment from pET24a(+)/carB was blunted with T4 DNA polymerase and ligated into the blunt-ended NotI site located downstream of carA in pET24a(+)/carA. The resulting plasmid with the RBS-carB at the same orientation as the carA was named pET24a(+)/carA-carB. The RBS-carC was excised from pET24a(+)/carC as a XbaI–XhoI fragment. It was blunt-ended and inserted into the blunt-ended XhoI site downstream of the carB in pET24a(+)/carA-carB to form the final vector pET24a(+)/carABC. The co-overexpression of carA, carB and carC in pET24a(+)/carABC was controlled by the T7 promoter and T7 terminator as a single operon.
Cloning and partial codon optimization of thnE
Wild-type thnE was amplified from genomic DNA (gDNA) of S. cattleya by using the forward primer: GCATATGGGC GCGGCCGCCG GCGAG, and reverse primer: CAAGCTTCAG CTCCGCCCGA TGACGCG. NdeI and HindIII restriction sites were introduced to facilitate downstream cloning experiments. The PCR reaction (100 mL) contained 10× cloned Pfu DNA polymerase buffer (10 mL), dNTPs (2.5 mL, 10 mm), each primer (2 mL, 20 nmol mL−1), DMSO (5 mL), gDNA (85 ngmL−1, 1 mL), and Pfu DNA polymerase (2.5 U mL−1, 1 mL). The reaction was preheated to 98°C for 2 min before the addition of Pfu DNA polymerase, then followed by 30 cycles of 45 s at 98°C, 45 s at 69°C, and 1 min at 72°C with the final extension time of 10 min at 72°C. The PCR product was purified with the GeneClean III kit (Q-BioGene, Solon, OH, USA) and ligated into pBluescript II SK+ (Stratagene, La Jolla, CA, USA) to obtain pBS/thnE. Once confirmed by DNA sequencing analysis, thnE was excised from pBS/thnE with NdeI/HindIII and ligated into pET24a(+) to generate pET24a(+)/thnE.
To optimize the codons of the first 21 amino acids of thnE, two oligonucleotides, Ecod-F and Ecod-R, harboring the optimized codons and NdeI and BsrI restriction ends were mixed in equal concentrations. After being heated at 100°C for 10 min, the mixture was left to cool slowly at room temperature to allow the oligonucleotides to anneal. Plasmid pET24a(+)/thnE was digested with HindIII/BsrI to generate a 821 bp fragment of thnE absent the 64 bp 5’ region. A three-way ligation reaction containing NdeI/HindIII digested pET24a(+), the 821 bp BsrI–HindIII thnE fragment and the 64 bp NdeI–BsrI annealed oligonucleotides was carried out to generate plasmid pET24a(+)/thnE*. Gene thnE* was excised with NdeI/HindIII from pET24a(+)/thnE* and inserted into pET28b(+) to give pET28b(+)/thnE* for expression of ThnE as N-His6 protein.
Construction of co-overexpression vector pET24a(+)/thnE*M-carC
RBS-carC was excised from pET24a(+)/carC with XbaI/HindIII, blunt-ended with Klenow DNA polymerase, and ligated into the blunt ended NotI site of pET24a(+)/thnM to give plasmid pET24a(+)/thnM-carC. The RBS-thnM-RBS-carC fragment was generated with XbaI/XhoI, blunt-ended and ligated downstream of thnE* at the blunt-ended XhoI site to obtain the final expression vector pET24a(+)/thnE*M-carC.
Construction of co-overexpression vector pET24a(+)/thnE*-carAC and pET28b(+)/thnE*-carAC
The RBS-carC fragment was generated by digesting pET24a(+)/carC with XbaI/XhoI. It was blunt-ended with T4 DNA polymerase and inserted into the blunt-ended NotI site of pET24a(+)/carA to give the co-overexpression vector pET24a(+)/carAC. RBS-carA-RBS-carC was excised as a XbaI–XhoI fragment from pET24a(+)/carAC and blunt-ended with Klenow DNA polymerase. Plasmids pET24a(+)/thnE* and pET28b(+)/thnE* were digested with NotI, blunt-ended with Klenow DNA polymerase and ligated with the RBS-carA-RBS-carC fragment to generate co-overexpression vectors pET24a(+)/thnE*-carAC and pET28b(+)/thnE*-carAC, respectively.
Construction of co-overexpression vector pET24a(+)/thnM-carBC
pET24a(+)/thnM-carC was digested with XbaI/XhoI to give a fragment containing RBS-thnM-RBS-carC. It was then blunt-ended with Klenow DNA polymerase and ligated into the blunt-ended XhoI site of pET24a(+)/carB to give the final plasmid pET24a(+)/thnM-carBC.
Small-scale co-overexpression of biosynthetic genes and heterologous production of carbapenem in E. coli
Seed medium (3 mL) containing kanamycin (50 mg mL−1) and chloramphenicol (25 mg mL−1) was inoculated with a single colony of freshly transformed Rosetta2(De3) or BL21 (DE3)(pLysS) harboring recombinant plasmid and grown, overnight, at 37°C. Seed culture (100 mL) was transferred into LB+ medium (50 mL; gL−1: Bacto-tryptone 10; Bacto-yeast extract 5; NaCl 10; glutamate 10; NaOAc 1; FeSO4·7H2O 0.25; CoCl2·6H2O 0.01, pH 7.5) or modified carbapenem production medium (g L−1: glutamate 5; NH4Cl 0.75; K2HPO4 2; NaCl 0.5; CaCO3 0.25; glucose 10, pH 7.6; 75 mg FeSO4·7H2O and 12.5 mL of 1 m MgSO4 were added after autoclaving). The secondary culture was grown at 37°C to OD600 = 0.3–0.4. Expression of proteins was induced with IPTG (1 mm) at 28°C for 5 h.
Detection of carbapenems
Samples were withdrawn from cultures every 60 min after co-overexpression was induced, and centrifuged at 16 000 g for 10 min. Supernatant (250 mL) was added to paper discs placed on a bioassay plate of Bacto-Nutrient agar seeded with β-lactam supersensitive E. coli SC12155.[13b] The bioassay plates were incubated at 37°C for 20 h. The production of carbapenems in the engineered E. coli was indicated by the inhibition of super-sensitive E. coli growth in zones around the paper discs. The production of carbapenem was also observed by Nitrocefin colorimetric assay.[13a] The recombinant E. coli supernatant (300 mL) was loaded on to paper discs sitting on BA2 agar (g L−1: BBL seed agar 30.5; NaCl 5) seeded with Bacillus lichniformis ATCC14580. The plates were incubated at 37 °C for 3 h and overlaid with Nitrocefin solution (1.5 mL of 300 mg mL−1). The production of carbapenems was indicated by the formation of red zones around the paper discs.
HPLC analysis of ThnE-catalyzed reactions
A previously reported method was modified.[8a] HPLC conditions: Phenomenex Prodigy 5µ ODS-3 analytical column, λ = 260 nm, 1 mL min−1; buffer A = 100 mm NH4H2PO4, 75 mm ammonium acetate, pH 4.65 with acetic acid, buffer B = 70% solvent A and 30% methanol. Method 1, t = 0 min 40% B, t = 28 min 85% B, t = 29 min 100% B, t = 39 min 100% B, t = 42 min 40% B, t = 52 min 40% B. Method 2, t = 0 min 40% B, t = 20 min 80% B, t = 21 min 40% B, t = 30 min 40% B.
Relative rates of ThnE-catalyzed reactions with l-P5C (5) and MalCoA or MeMalCoA
Reactions containing l-P5C (5, 0.5 mm), 0.3 mm MalCoA or MeMalCoA and ThnE (2.4 mg mL−1) as well as the corresponding reactions without 5 and no-enzyme reactions were assembled in potassium phosphate (100 mm, pH 7.8).[8a] At 0, 15, 30, 60, 120 min and overnight, samples (200 µL) were quenched in HCl (200 µL, 0.2 m) then vortexed with CCl4 (400 µL), the top layer (300 µL) was removed from the CCl4 for HPLC analysis.
Purification and ESI-MS of CMP-CoA esters from ThnE reactions
New products observable in the HPLC trace were collected from multiple runs, lyophilized and desalted with a STRATA X column (33 µm, 30 mg mL−1).[8a] The column was prewashed with methanol (10 mL) and 0.1% TFA (10 mL). The lyophilized sample was taken up in water (1 mL) and 0.1% TFA (2 mL) was added. The sample was applied to the column and washed with 0.1% TFA (2 mL). It was then eluted with methanol/0.1% TFA 4:1 (5 mL). Fractions (1 mL) containing the desired CoA ester were identified by their characteristic absorbance at 260 nm and lyophilized. The resulting powder was taken up in 1:1 CH3CN/water with 0.1% NH4OH or 0.1% TFA and analyzed by ESI-MS.
HPLC analysis of ThnM-catalyzed reactions
Reactions for the determination of product formation (200 µL) were run in a buffer containing HEPES (100 mm) and piperazine (80 mm) at pH 8.2 with μ = 100 mm (KCl).[9b] Each reaction contained ATP (2 mm), DTT (1 mm), MgCl2 (12.5 mm), substrate (10 mm) and ThnM (15.0 µm). The reactions were run at 25°C for 2 h and immediately frozen in liquid N2 until analysis. Analysis was performed on an Agilent 1100 HPLC by using a Phenomenex Luna 5 μ phenyl–hexyl analytical column. Injections (50 µL) were run in an isocratic mobile phase consisting of a phosphate buffer (50 mm) at pH 6.5 with a flow rate of 2 mL min−1. Reactions were monitored at λ = 210 and 230 nm. Compounds produced in ThnM-catalyzed reactions were identified by comparison to authentic product standards.
Supplementary Material
Acknowledgements
We are grateful to Dr. Anthony Stapon for preparation of CMPs, and to Prof. Barbara Gerratana and Dr. Samantha O. Arnett for guidance and early insightful discussions. We thank Dr. Mary L. Raber for critical review of the kinetic data and the NIH (AI014937) for financial support.
Footnotes
Supporting information for this article is available on the WWW under http://dx.doi.org/10.1002/cbic.201100366.
References
- 1.a) Dalhoff A, Janjic N, Echols R. Biochem. Pharmacol. 2006;71:1085–1095. doi: 10.1016/j.bcp.2005.12.003. [DOI] [PubMed] [Google Scholar]; b) Kumagai ST, Tamai S, Abe T, Hikda M. Curr. Med. Chem. Anti-Infect. Agents. 2002;1:1–14. [Google Scholar]
- 2.Bodner MJ, Phelan RM, Freeman MF, Li R, Townsend CA. J. Am. Chem. Soc. 2010;132:12–13. doi: 10.1021/ja907320n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Birnbaum J, Kahan FM, Kropp H, MacDonald JS. Am. J. Med. 1985;78:3–21. doi: 10.1016/0002-9343(85)90097-x. [DOI] [PubMed] [Google Scholar]
- 4.Williamson JM, Inamine E, Wilson KE, Douglas AW, Liesch JM, Albers-Schönberg G. J. Biol. Chem. 1985;260:4637–4647. [PubMed] [Google Scholar]
- 5.a) Houck DR, Kobayashi K, Williamson JM, Floss HG. J. Am. Chem. Soc. 1986;108:5365–5366. [Google Scholar]; b) Zydowsky TM, Courtney LF, Frasca V, Kobayashi K, Shimizu H, Yuen LD, Matthews RG, Benkovic SJ, Floss HG. J. Am. Chem. Soc. 1986;108:3152–3153. [Google Scholar]; c) Arigoni D. Ciba Found. Symp. 1978:243–261. doi: 10.1002/9780470720424.ch14. [DOI] [PubMed] [Google Scholar]; d) Floss HG, Tsai MD. Adv. Enzymol. Relat. Areas Mol. Biol. 1979;50:243–302. doi: 10.1002/9780470122952.ch5. [DOI] [PubMed] [Google Scholar]
- 6.Nunez LE, Mendez C, Brana AF, Blanco G, Salas JA. Chem. Biol. 2003;10:301–311. doi: 10.1016/s1074-5521(03)00069-3. [DOI] [PubMed] [Google Scholar]
- 7.Freeman MF, Moshos KA, Bodner MJ, Li RF, Townsend CA. Proc. Natl. Acad. Sci. USA. 2008;105:11128–11133. doi: 10.1073/pnas.0804500105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.a) Gerratana B, Arnett SO, Stapon A, Townsend CA. Biochemistry. 2004;43:15936–15945. doi: 10.1021/bi0483662. [DOI] [PubMed] [Google Scholar]; b) Batchelar ET, Hamed RB, Ducho C, Claridge TDW, Edelmann MJ, Kessler B, Schofield CJ. Angew. Chem. 2008;120:9462–9465. doi: 10.1002/anie.200803906. Angew. Chem. Int. Ed.2008, 47, 9322–9325. [DOI] [PubMed] [Google Scholar]; c) Hamed RB, Batchelar ET, Clifton IJ, Schofield CJ. Cell. Mol. Life Sci. 2008;65:2507–2527. doi: 10.1007/s00018-008-8082-6. [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Sleeman MC, Sorensen JL, Batchelar ET, McDonough MA, Schofield CJ. J. Biol. Chem. 2005;280:34956–34965. doi: 10.1074/jbc.M507196200. [DOI] [PubMed] [Google Scholar]
- 9.a) Miller MT, Gerratana B, Stapon A, Townsend CA, Rosenzweig AC. J. Biol. Chem. 2003;278:40996–41002. doi: 10.1074/jbc.M307901200. [DOI] [PubMed] [Google Scholar]; b) Gerratana B, Stapon A, Townsend CA. Biochemistry. 2003;42:7836–7847. doi: 10.1021/bi034361d. [DOI] [PubMed] [Google Scholar]; c) Raber ML, Castillo A, Greer A, Townsend CA. ChemBioChem. 2009;10:2904–2912. doi: 10.1002/cbic.200900389. [DOI] [PMC free article] [PubMed] [Google Scholar]; d) Raber ML, Arnett SO, Townsend CA. Biochemistry. 2009;48:4959–4971. doi: 10.1021/bi900432n. [DOI] [PMC free article] [PubMed] [Google Scholar]; e) Raber ML, Freeman MF, Townsend CA. J. Biol. Chem. 2009;284:207–217. doi: 10.1074/jbc.M805390200. [DOI] [PMC free article] [PubMed] [Google Scholar]; f) Arnett SO, Gerratana B, Townsend CA. Biochemistry. 2007;46:9337–9345. doi: 10.1021/bi0618464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.a) Stapon A, Li RF, Townsend CA. J. Am. Chem. Soc. 2003;125:8486–8493. doi: 10.1021/ja034248a. [DOI] [PubMed] [Google Scholar]; b) Stapon A, Li RF, Townsend CA. J. Am. Chem. Soc. 2003;125:15746–15747. doi: 10.1021/ja037665w. [DOI] [PubMed] [Google Scholar]; c) Sleeman MC, Smith P, Kellam B, Chhabra SR, Bycroft BW, Schofield CJ. ChemBioChem. 2004;5:879–882. doi: 10.1002/cbic.200300908. [DOI] [PubMed] [Google Scholar]; d) Clifton IJ, Doan LX, Sleeman MC, Topf M, Suzuki H, Wilmouth RC, Schofield CJ. J. Biol. Chem. 2003;278:20843–20850. doi: 10.1074/jbc.M213054200. [DOI] [PubMed] [Google Scholar]
- 11.Hamed RB, Batchelar ET, Mecinovic J, Claridge TDW, Schofield CJ. ChemBioChem. 2009;10:246–250. doi: 10.1002/cbic.200800652. [DOI] [PubMed] [Google Scholar]
- 12.Li RF, Stapon A, Blanchfield JT, Townsend CA. J. Am. Chem. Soc. 2000;122:9296–9297. [Google Scholar]
- 13.a) Sykes RB, Wells JS. J. Antibiot. 1985;38:119–121. doi: 10.7164/antibiotics.38.119. [DOI] [PubMed] [Google Scholar]; b) Aoki H, Sakai H, Kohsaka M, Konomi T, Hosoda J, Kubochi Y, Iguchi E, Imanaka H. J. Antibiot. 1976;29:492–500. doi: 10.7164/antibiotics.29.492. [DOI] [PubMed] [Google Scholar]
- 14.Bodner MJ, Phelan RM, Townsend CA. Org. Lett. 2009;11:3606–3609. doi: 10.1021/ol901269d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fetter J, Lempert K, Gizur T, Nyitrai J, Kajtanperedy M, Simig G, Hornyak G, Doleschall G. J. Chem. Soc. Perkin Trans. 1986;1:221–227. [Google Scholar]
- 16.Ueda Y, Damas CE, Vinet V. Can. J. Chem. 1983;61:2257–2263. [Google Scholar]
- 17.a) Mahoney WS, Brestensky DM, Stryker JM. J. Am. Chem. Soc. 1988;110:291–293. [Google Scholar]; b) Lipshutz BH, Chrisman W, Noson K, Papa P, Sclafani JA, Vivian RW, Keith JM. Tetrahedron. 2000;56:2779–2788. [Google Scholar]
- 18.Erfle JD. Biochim. Biophys. Acta. 1973;316:143–155. doi: 10.1016/0005-2760(73)90004-0. [DOI] [PubMed] [Google Scholar]
- 19.Bachmann BO, Townsend CA. Biochemistry. 2000;39:11187–11193. doi: 10.1021/bi000709i. [DOI] [PubMed] [Google Scholar]
- 20.Jung ME, Piizzi G. Chem. Rev. 2005;105:1735–1766. doi: 10.1021/cr940337h. [DOI] [PubMed] [Google Scholar]
- 21.Rodriguez M, Nunez LE, Brana AF, Mendez C, Salas JA, Blanco G. Antimicrob. Agents Chemother. 2011;55:1638–1649. doi: 10.1128/AAC.01366-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.