Abstract
Introduction
Endometrial carcinoma is the one of the leading causes of terminal cancer death in women, and the best way to solve it is through an early diagnosis. Serum proteomic profiling is a promising approach to classify disease versus normal and matrix-assisted laser desorption/ionization time of flight mass spectrometry (MALDI-TOF). This analysis is a new potential tool for the diagnosis of human diseases. The objective of our study was to assess the feasibility of mass spectrometry, based on serum proteomic pattern for the discrimination of endometrial carcinoma patients from healthy control patients.
Material and methods
By using 30 pre-operative endometrial carcinoma serum samples and 30 healthy controls, we generated MALDI-TOF protein profiles, established the serum detection model, and analyzed the data. Furthermore, we validated the data and got the total sensitivity (97.6%) and specificity (100%), which could classify all the samples well and demonstrated the high, significant separation ability.
Results
Our results indicated that high sensitivity and specificity classify endometrial carcinoma by the serum detection model and showed the potential usefulness of serum protein profiles.
Conclusions
From these results, we could tell that MALDI-TOF MS is a potential tool for diagnosing diseases by the use of serum samples and will be widely used in the future for clinical work.
Keywords: serum proteomic analysis MALDI-TOF endometrial carcinoma serum detection model
Introduction
Endometrial carcinoma is the third most common cause of gynecologic cancer death, only behind ovarian and cervical cancer, and occurs only in females [1]. Endometrial carcinoma occurs in both premenopausal (about 25%) and postmenopausal women (about 75%).The most commonly affected age group is between 50 and 59 years [2]. The 5-year survival rate for endometrial cancer is usually as follows: 95% for stage 1, 50% for stage 2, 30% for stage 3, less than 5% for stage 4 [1]. There is a better probable outcome associated with this form of cancer as opposed to other types of gynecological cancers, such as cervical or ovarian cancer. In the United States, there are over 35,000 women diagnosed each year [3]. During the past two decades, the incidence of endometrial carcinoma has been improving steadily in China.
The common signs and symptoms of endometrial carcinoma are usually include the followings: abnormal uterine bleeding, abnormal menstrual menstrual periods, bleeding between normal periods in premenopausal women aged over 40, anemia caused by chronic loss of blood, lower abdominal pain or pelvic cramping, and thin white or clear vaginal discharge in postmenopausal women [4–6]. Additionally, there are several risk factors for endometrial carcinoma such as high levels of estrogen, endometrial hyperplasia, obesity, hypertension, polycystic ovary syndrome, nulliparity, infertility, early menarche, late menopause, tamoxifen, high intake of animal fat, family history, phenotype characteristics etc [7, 8].
Screening for endometrial carcinoma is always not as easy as screening for cervical cancer because the uterine cavity is well above the cervix. A pelvic examination is frequently normal, especially in the early stages of disease. Changes in the size, shape or consistency of the uterus and its surrounding and supporting structures may exist when the disease is more advanced.
The traditional diagnostic method is endometrial curettage. Both endometrial and endocervical material should be sampled. If endometrial curettage does not yield sufficient diagnostic material, we need a dilation and curettage for diagnosing the cancer.
Recently, the TruTest has been introduced as a new method of testing. It uses the small flexible Tao Brush to brush the entire lining of the uterus. This method is less painful than the usual pipelle biopsy and just like of procuring enough tissue for testing. As it is simpler and less invasive, the TruTest can be performed as often, and at the same time as a routine Pap smear, and now is allowed to use for early detection and treatment.
The tumor marker CA-125 is sometimes checked, since this can predict advanced stage disease [9].
During the development of proteomic study, the proteomic profiles generated by mass spectrometry have been thought of as potential tools for the early diagnosis of cancer and other diseases. Different protein profiles may be associated with varying responses to therapeutics. It has been postulated that on the basis of the presence (or absence) of multiple low-molecular-weight serum proteins using time of flight mass spectrometry technologies, biomarkers can be identified [10–13]. Although the data from these studies are encouraging, critical notes have been made on both study design and experimental procedures for proteomic profiling [14–16]. They are the substances that can be found in abnormal amounts in the blood, urine, or tissues of some patients with cancer. Additionally, they can be used to help diagnose cancer, predict a patient's response to particular therapies, check a patient's response to treatment, or determine if cancer has returned. Researchers continue to study tumor markers and to develop more accurate methods to detect, diagnose, and monitor cancer. For this reason, we hope to use proteomic techniques to find the best way for the early diagnosis of endometrial carcinoma. And at the very beginning of our research, we optimized of the conditions with MALDI-TOF MS, choosing the best kind of magnetic beads for the pre-analysis of the disease and normal serum samples [17].
The objective of this study was to assess the feasibility of applying mass spectrometry for the profiling of serum proteomic profiles in endometrial carcinoma patients from healthy control individuals, and to find some new early potential biomarkers and aspire to do something to help carry out the high sensitivity and specificity of early detection of endometrial carcinoma.
Material and methods
Patients
The study protocol was approved by the Research Ethics Committee of Chinese PLA General Hospital and with each patient's agreement. Serum samples were obtained from a total of 30 endo-metrial carcinoma patients the day before surgery. Surgical specimens and pathological validation historically confirmed endometrial carcinoma. The median age of the female patient group was 50 years old, with patients admitted from November 2006 until May 2008 in our hospital. The healthy control volunteers group consisted of 30 individuals. The median age of the healthy female control group was 47 years old, and all were recruited from our hospital healthy check center.
Chemicals and calibrators
Gradient-grade alcohol and acetone were obtained from J.T. Baker; p.a.trifluoroacetic acid and ammonium acetate were purchased from Sigma-Aldrich. The peptide calibrator containing angiotensin II, the protein calibrator, and the matrix α-cyano-4-hydroxycinnamic acid (HCCA) were purchased from Bruker Daltonics GmbH. For magnetic bead preparations, we used 0.2 ml of polypropylene tubes (8-tube strips) from Axygen Company. Multifly needle sets and polypropylene vacuette without anticoagulants (EDTA, heparin and citrate) were obtained from BD Company.
Blood samples
The blood samples were processed according to a standardized protocol. After sample collection, the vacuette serum (collected in a red-top glass tube containing no preservatives or anticoagulant) were allowed to clot or to sediment at room temperature (about 25°C) for at least half an hour and then centrifuged at 1500 g for 15 min. Samples were divided in 100 µl aliquots and immediately frozen at –80°C, refrigerated until use; for proteomic fractionation, samples were thawed at room temperature (about 25°C) for 15 min and processed immediately.
Proteomic fractionation (isolation of peptides)
By using functionalized magnetic bead-based kits, serum samples were fractionated (ClinProt Kits, Bruker Daltonics Inc. Germany). Magnetic beads were designed to capture proteins and peptides from serum and to obtain specific profiles based on cationic exchange chromatography. They were used according to the manufacturer's protocols. As recommended, we added 5 µl WCX magnetic beads with 10 µl of a binding solution in a 0.2 ml polypropylene PCR tube. Then, we added 5 µl of the sample and mixed thoroughly by pipetting up and down several times, incubated the tube for 5 min, and separated the unbound solution using a magnetic bead separator. After magnetic bead separation and three washings, the bound proteins or peptides in the serum samples were eluted from the magnetic beads according to the manufacturer's instructions (Profiling Kit WCX and Profiling, Bruker Daltonics GmbH, Germany). A portion of the sample was diluted 1 : 10 in a solution containing HCCA (0.6 g/l in 2 : 1 ethanol: acetone). Then, 1 µl of the resulting mixture was spotted onto the AnchorChip target (Bruker Daltonics Inc., CA) and allowed to air dry for approximately 5 min at room temperature. Usually, each sample for each conditioned experiment was conducted three times.
MS analysis (protein profiling)
For the proteomic analysis, we used a linear MALDI-TOF mass spectrometer (Microflex; Bruker Daltonics) with the following settings: ion source 1, 20 kV; ion source 2, 18.40kV; lens, 7.50 kV; pulsed ion extraction, 120 ns; nitrogen pressure, 1700-2000 mbar. Ionization was achieved by irradiation with a nitrogen laser (λ = 337 nm) operating at 25 Hz.
The mass calibration was performed with the calibration mixture of peptides and proteins in a mass range of 1000-10000 Da. We measured 3 MALDI preparations from each magnetic bead fraction. For each MALDI spot, 400 spectra were acquired in analysis (50 laser shots at 8 different spot positions).
To increase the detection sensitivity, we usually use higher energy to shot the spot positions at first and then a lower energy to shot again and then save the spectra, which can remove the excess matrix with higher laser power before data acquisition.
Data processing
The spectra of all signals with a signal-to-noise (S/N) ratio > 5 in a mass range of 1000-10000 Da obtained from all of the samples, were analyzed by using ClinProTools software for spectrum processing and generation of proteomic fingerprints. Only the spectra obtained from WCX magnetic beads were used in the analysis of this report. We used the ClinProTools bioinformatics software (Ver. 2.1; Bruker Daltonics) for proteomic pattern recognition.
To increase the accuracy, we used the average of eight spots representing one serum sample. Specifically, all spectra were normalized to their own total ion count (summation of peak areas) and recalibrated using the most prominent peaks, followed by baseline subtraction, peak defining, and calculation of peak intensities and areas.
Then, we calculated the mean value of peak intensity, SD and CV (%) for each corresponding peak among the endometrial carcinoma and healthy control groups. The degree of variation on the basis of the whole spectrum was determined by calculating the CV values for some prominent peaks of the samples. We defined p < 0.01 as a significant difference.
Results
The whole correlation of mass spectrum
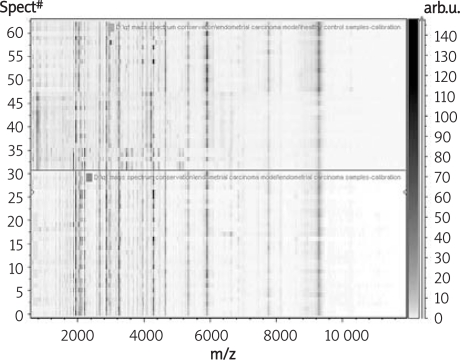
At first, before pre-processing and further analysis of each sample's mass spectra, all eight spots were measured for each sample. The simulated two-dimensional gel electrophoresis result was shown in Figure 1 to see the correlation of the whole correlation of the groups. From this, we determined that the coherence of each group performed very well, which laid the best foundation for our further research and analysis.
Figure 1.
The simulated two-dimensional gel electrophoresis result in endometrial carcinoma group and healthy control group (the upload – healthy control group, the download – the endometrial carcinoma group)
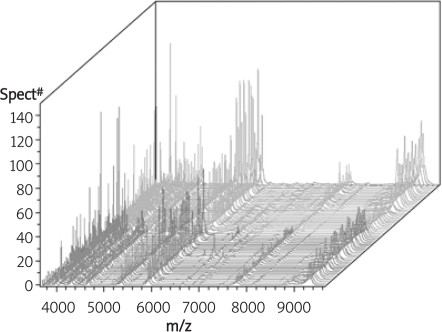
Then, we checked all the original mass spectra of the two groups, the endometrial carcinoma group and the healthy control group, to confirm the whole coherence of the raw mass data. From this result in Figure 2, we determined that the coherence of the raw mass data was promising enough to continue our further data analysis.
Figure 2.
The whole mass spectra of the endometrial carcinoma and healthy control group (red – endometrial carcinoma group, green – healthy control group)
Establishment of the endometrial carcinoma detection model
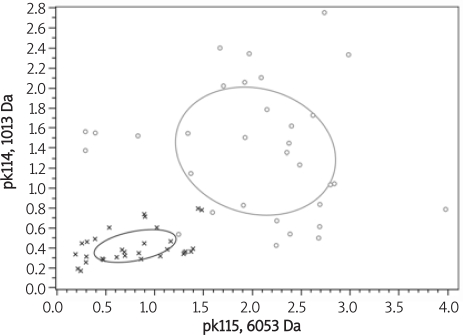
We chose 30 endometrial carcinoma patients and 30 patients with gender and age-matched healthy controls to establish the endometrial carcinoma serum detection model. With the help of ClinProTools software, we established the sample distribution coordinate by two differential proteins or peptides, which were the pk14, 1012.6 in the vertical axis, and the pk115, 6052.9 in the horizontal axis. The result of that was shown in Figure 3 as follows.
Figure 3.
The sample distribution map of endometrial carcinoma samples and healthy control individuals (red – endometrial carcinoma samples, green – healthy control individuals)
From the result shown in Figure 2, we can determine that by using these two proteins or peptides, there was no overlap region, which showed that we could differentiate the samples in the two groups well.
The prominent expression of proteins or peptides in the endometrial carcinoma serum detection model
There were a total of 147 peaks found in the endometrial carcinoma and healthy control group. After the statistical analysis in the established endometrial carcinoma serum detection model, we found that some proteins or peptides were higher expressed and some were lower expressed in the disease group. We can easily differentiate the endometrial carcinoma and healthy control samples by using either one of these proteins or peptides or using some of them together. If we use all these proteins or peptides together as a combined one, the sensitivity and specificity is much higher than the result that uses each individually.
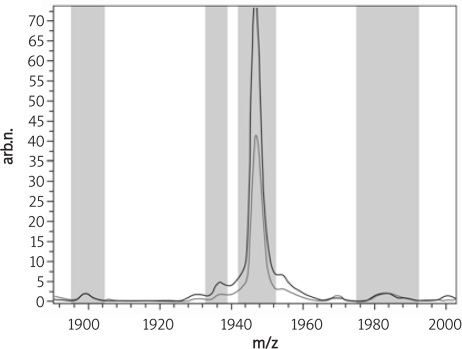
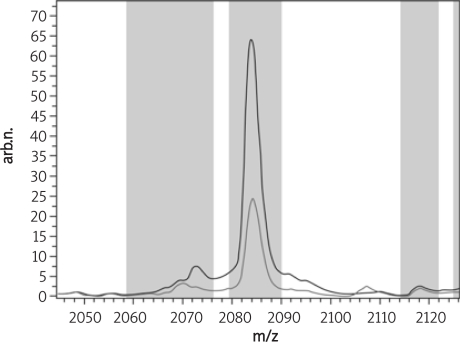
From all the peaks found in the disease and healthy control group, peak 40, 1946.71 m/z shown below in Figure 4 was higher expressed in the endometrial carcinoma group than expressed in the healthy control group, and peak 45, 2084.06 m/z just as peak 40 shown below in Figure 5 was also higher expressed in the endometrial carcinoma group than in the healthy control group.
Figure 4.
The average peak expression of peak 40, 1946.71 m/z in the endometrial carcinoma and the healthy control group (red – endometrial carcinoma group, green – healthy control group)
Figure 5.
The average peak expression of peak 45, 2084.06 m/z in the endometrial carcinoma and the healthy control group (red – endometrial carcinoma group, green – healthy control group)
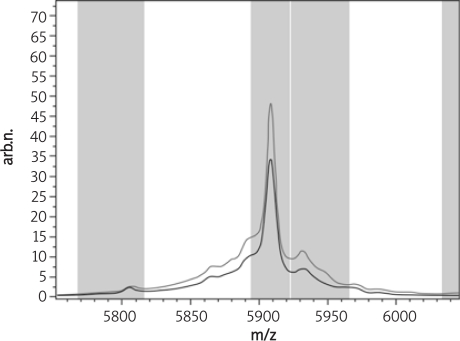
But peak 114, 5931.22 m/z (see in Figure 6) was lower expressed in the endometrial carcinoma group than in the healthy control group. And the p value in all these three proteins or peptides were < 0.01, with great significance.
Figure 6.
The average peak expression of peak 114, 5931.22 m/z in the endometrial carcinoma and the healthy control group (red – endometrial carcinoma group, green – healthy control group)
We thought all the proteins or peptides used to establish the endometrial carcinoma serum detection model were the prominent ones, this may warrant further investigation in the future to know to determine more specific roles.
Based on these results, we list some of the prominent proteins or peptides that were higher or lower expressed in the endometrial carcinoma model and with the satisfaction of p value < 0.01 and DAve > 80 as great significance. Then, we list these differential proteins or peptides found in the peak list in Table I.
Table I.
Higher or lower expressed proteins or peptides in the endometrial carcinoma serum detection model
| Peak number | Peak index | Mass | DAve | P value |
|---|---|---|---|---|
| 1 | 67 | 3243.51 | +87.08 | < 0.01 |
| 2 | 55 | 2662.68 | +119.58 | < 0.01 |
| 3 | 45 | 2084.06 | +160.63 | < 0.01 |
| 4 | 59 | 2864.98 | +89.21 | < 0.01 |
| 5 | 40 | 1946.77 | +134.65 | < 0.01 |
| 6 | 92 | 4285.06 | +185.73 | < 0.01 |
| 7 | 114 | 5931.22 | –111.97 | < 0.01 |
+ expressed higher in endometrial carcinoma group, – expressed lower in endometrial carcinoma group
And from the results in Table II, we could know that most of the differential proteins or peptides found in the detection model were higher expressed in the endometrial carcinoma group than those in the healthy control group.
Table II.
The sensitivity and specificity in the three algorithm methods
| Algorithm method | Sensitivity [%] | Specificity [%] |
|---|---|---|
| GA method | 97.60 | 100 |
| SNN method | 92 | 100 |
| QC method | 92.90 | 93.80 |
The MS data analysis by algorithm
With the help of ClinProTools software and the genetic algorithm (GA) method, we used four characteristic peaks that were pk61, 2902.49 m/z, pk100, 5068.89 m/z, pk115, 6052.9, and pk128, 7010.58 m/z as combined factors to establish the endometrial carcinoma serum detection model, and determined that the sensitivity and the specificity was 97.62 and 100%, respectively.
Then, we used the other two algorithm methods: supervised neural network (SNN) and quick classifier (QC) to analyze the MS data. The result showed that there were ten differential proteins or peptides used in the SNN method, and the sensitivity was 92.02% and the specificity was 100%. In the QC method, there were only two proteins or peptides used to classify the two group samples, they were pk14, 1012.6 m/z and pk16, 6093.12 m/z and got the sensitivity was 92.86% and the specificity was 93.75%, respectively.
From these results, we created a table to compare the proteins or peptides being used in these three algorithm methods, the results were shown in Table III as follows.
Table III.
Proteins or peptides used for classification in the three algorithm methods
| GA method | SNN method | QC method | ||||
|---|---|---|---|---|---|---|
| Peak number | Mass | Peak number | Mass | Peak number | Mass | |
| 128 | 7010.58 | 53 | 2585.78 | 14 | 1012.6 | |
| 115 | 6052.9 | 80 | 3938.26 | 116 | 6093.12 | |
| 100 | 5068.89 | 67 | 3243.51 | |||
| 61 | 2902.49 | 26 | 1501.02 | |||
| 116 | 6093.12 | |||||
| 105 | 5224.33 | |||||
| 25 | 1451.97 | |||||
| 131 | 7606.66 | |||||
| 115 | 6052.9 | |||||
| 18 | 1083.74 | |||||
From the results in Table III, we could propose that in the GA method, four proteins or peptides were used to classify all the samples in the endometrial carcinoma group and healthy group; in the SNN method, about ten differential proteins or peptides were used to do the classification; but in the QC method, just two proteins or peptides were used to do the classification. The same protein pk115, 6052.9 was used in both the GA method and SNN method and pk116, 6093.12 was also used to do the classification in the SNN method and QC method. But no same proteins or peptides were used in the GA and QC method and no same proteins or peptides were used to do the classification in all three-algorithm methods.
Finally, by using different proteins or peptides to do the classification we obtained different sensitivity and specificity in the three algorithm methods, and listed the results in Table II.
From the results in Table II we found that by using different proteins or peptides as the combination in these three algorithm method, we could get different sensitivity and specificity, and all the sensitivity and the specificity were of great results of more than 90%. In the QC method, we used only two proteins or peptides that were pk14, 1012.6 and pk116, 6093.12, and got the sensitivity and specificity were 92.90 and 93.80%, respectively which may tell us that we may can use just these two proteins or peptides to classify the samples in the endometrial carcinoma and healthy control group.
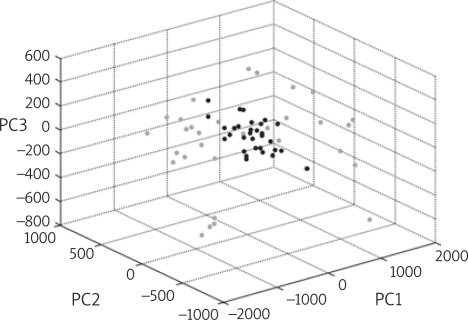
The Principle Component Analysis
In order to get more information about the variation within the serum endometrial carcinoma model generation classes and the homogeneity or heterogeneity of a spectra set, we can use a PCA. An external MATLAB software tool carried out the PCA. By the result of the PCA in Figure 7, we can identify the samples distribution in the model.
Figure 7.
Samples distribution in the Principle Component Analysis
From the results in Figure 7, we could potentially know the endometrial carcinoma and healthy control samples distribution with the Principle Component Analysis.
Discussion
Nowadays, endometrial carcinoma is becoming one of the common malignancy tumors and strongly threatening women's health as its incidence is growing at a rapid rate. The high risk factors of that include menopause delay, obesity, diabetes, family factors, and so on. For most cancers, endometrial carcinoma survival rates mainly depend on the early detection and early diagnosis of it. But up to now, there are no satisfactory biomarkers that could deal with this difficult task. In recent years, there have been great changes in the proteomic research field, new proteomic technologies have brought the hope to discovering differences between disease and healthy control in the proteomic pattern and the early cancer-specific biomarkers in the complex biologic samples.
The development of MALDI-TOF MS gives us an effective tool for protein profiling study and biomarker identification. The usual approaches for the discovery of disease-related proteins or peptides are two-dimensional polyacrylamide gel electrophoresis (2D-PAGE) and the mass spectrometry. However, there are some limitations in the classical methods such as extensive sample preparation, difficult pre-extraction or fractionation, the contaminants of samples etc. All these will hinder the use for high-throughput screening of disease from healthy ones. MALDI-TOF MS has overcome many of these limitations. By using this method, we can successfully find differential expression between disease and normal groups, and can discover potential serum diagnostic biomarker for future clinical work. MALDI-TOF MS has been widely used for breast cancer [18], lung cancer [13], bladder cancer [19], liver cancer [20], colon cancer [21], ovarian cancer [22] and gastric cancer [23]. Little has been reported about the proteomic study on endometrial carcinoma.
In our study, we chose 30 endometrial carcinoma patients and 30 with gender and age-matched healthy controls to establish the endometrial carcinoma serum detection model. With the help of ClinProTools software, we can just use two differential proteins or peptides pk14, 1012.6 and pk115, 6052.9 to classify all the samples, and tell us which was disease and which was healthy. Despite this, we still found some proteins or peptides differential expressed in the cancer and healthy control group. Some were highly expressed in the endometrial carcinoma group, and some were lower expressed in them, such as peak 40, 1946.71 m/z, peak 45, 2084.06 m/z, peak 114, 5931.22 m/z, peak 67, 3243.51 m/z, peak 59, 2864.98 m/z etc, and the p value were all < 0.01, with great significance. Then, by using three algorithm methods (GA, SNN and QC), we obtained the sensitivity and specificity analysis of our serum endometrial carcinoma model, and the results were very prosperous, the sensitivity and specificity were both more than 90%. Such results can lead the way to positively identify the candidate proteins or peptides markers found in the study to make a further understanding of the mechanisms of endometrial carcinoma genesis and development.
Though, to some degree, the numbers of samples analyzed in our study limited the validity of the results. We will do further independent and blinded validation study with more serum samples to confirm the former results. In the future, we will perform a comparison study between early-stage cancer with later-stage cancer to discover the potential biomarkers to monitor the recurrence and metastasis of endometrial carcinoma. Then, we will go on to study the observation of serum proteomic profile expression following the development of the disease and still keep on working for the identification and validation of new biomarkers.
In conclusion, by the use of proteomic approach MALDI-TOF MS, with the combination of bio-informatics tools, we can find the differences in the endometrial carcinoma and the matched healthy control group and discover some new potential biomarkers. With the help of the panel of differential proteins or peptides found in our research, we could achieve the high sensitivity and specificity for the detection of endometrial carcinoma. In future work, we may attempt to validate and identify these potential biomarkers. From our research, we could make a conclusion that MALDI-TOF MS is a novel, high-throughput, sensitive, highly predictive and rapid method for the early detection, early diagnosis and prediction of endometrial carcinoma and will be widely used in future clinical screening work.
Acknowledgments
This work was supported by the Ministry of Science and Technology of China (2006FY230300).
References
- 1.Creasman WT. Endometrial cancer: incidence, prognostic factors, diagnosis, and treatment. Semin Oncol. 1997;24(1 Suppl 1) S1-140–50. [PubMed] [Google Scholar]
- 2.Feldman S, Cook EF, Harlow BL, Berkowitz RS. Predicting endometrial cancer among older women who present with abnormal vaginal bleeding. Gynecol Oncol. 1995;56:376–81. doi: 10.1006/gyno.1995.1066. [DOI] [PubMed] [Google Scholar]
- 3.Silver DF, Wheeless CR, Abbas FM. A vaginal and extraperitoneal approach to surgically stage patients with endometrial cancer. Gynecol Oncol. 2001;81:144–9. doi: 10.1006/gyno.2001.6123. [DOI] [PubMed] [Google Scholar]
- 4.Van Leeuwen FE, Benraadt J, Coebergh JW, et al. Risk of endometrial cancer after tamoxifen treatment of breast cancer. Lancet. 1994;343:448–52. doi: 10.1016/s0140-6736(94)92692-1. [DOI] [PubMed] [Google Scholar]
- 5.Fisher B, Costantino JP, Redmond CK, Fisher ER, Wickerham DL, Cronin WM. Endometrial cancer in tamoxifen-treated breast cancer patients: findings from the National Surgical Adjuvant Breast and Bowel Project (NSABP) B-14. J Natl Cancer Inst. 1994;86:527–37. doi: 10.1093/jnci/86.7.527. [DOI] [PubMed] [Google Scholar]
- 6.Wu Ming, Lang Jinghe. Treatment of endometrial cancer. Oncology Progress. 2006;1:13–8. [Google Scholar]
- 7.Ziel HK, Finkle WD. Increased risk of endometrial carcinoma among users of conjugated estrogens. N Engl J Med. 1975;293:1167–70. doi: 10.1056/NEJM197512042932303. [DOI] [PubMed] [Google Scholar]
- 8.Jick SS, Walker AM, Jick H. Estrogens, progesterone, and endometrial cancer. Epidemiology. 1993;4:20–4. doi: 10.1097/00001648-199301000-00005. [DOI] [PubMed] [Google Scholar]
- 9.Gornall RJ, Anthony FW, Coombes EJ, Hogston, Woolas RP. Investigation of women with endometrial carcinoma using serum vascular endothelial growth factor (VEGF) measurement. Int J Gynecol Cancer. 2001;11:164–6. doi: 10.1046/j.1525-1438.2001.011002164.x. [DOI] [PubMed] [Google Scholar]
- 10.Adam BL, Qu Y, Davis JW, et al. Serum protein fingerprinting coupled with a pattern-matching algorithm distinguishes prostate cancer from benign prostate hyperplasia and healthy men. Cancer Res. 2002;62:3609–14. [PubMed] [Google Scholar]
- 11.Petricoin EF, 3rd, Ornstein DK, Paweletz CP, et al. Serum proteomic patterns for detection of prostate cancer. J Natl Cancer Inst. 2002;94:1576–8. doi: 10.1093/jnci/94.20.1576. [DOI] [PubMed] [Google Scholar]
- 12.Rai AJ, Zhang Z, Rosenzweig J, et al. Proteomic approaches to tumor marker discovery. Arch Pathol Lab Med. 2002;126:1518–26. doi: 10.5858/2002-126-1518-PATTMD. [DOI] [PubMed] [Google Scholar]
- 13.Yanagisawa K, Shyr Y, Xu BJ, et al. Proteomic patterns of tumor subsets in non-small-cell lung cancer. Lancet. 2003;362:433–9. doi: 10.1016/S0140-6736(03)14068-8. [DOI] [PubMed] [Google Scholar]
- 14.Hu J, Coombes KR, Morris JS, Baggerly KA. The importanceof experimental design in proteomic mass spectrometry experiments: some cautionary tales. Brief Funct Genomic Proteomic. 2005;3:322–31. doi: 10.1093/bfgp/3.4.322. [DOI] [PubMed] [Google Scholar]
- 15.Coombes KR, Morris JS, Hu J, Edmonson SR, Baggerly KA. Serum proteomics profiling – a young technology begins to mature. Nat Biotech. 2005;23:291–2. doi: 10.1038/nbt0305-291. [DOI] [PubMed] [Google Scholar]
- 16.Ransohoff DF. Rules of evidence for cancer molecular-marker discovery and validation. Nat Rev Cancer. 2004;4:309–14. doi: 10.1038/nrc1322. [DOI] [PubMed] [Google Scholar]
- 17.Qiu F, Liu HY, Zhang XJ, Tian YP. Optimization of magnetic beads for MALDI-TOF MS analysis. Front Biosci. 2009;14:3712–23. doi: 10.2741/3482. [DOI] [PubMed] [Google Scholar]
- 18.Dekker LJ, Boogerd W, Stockhammer G, et al. MALDI-TOF Mass Spectrometry Analysis of Cerebrospinal Fluid Tryptic Peptide Profiles to Diagnose Leptomeningeal Metastases in Patients with Breast Cancer. Mol Cell Proteomics. 2005;4:1341–9. doi: 10.1074/mcp.M500081-MCP200. [DOI] [PubMed] [Google Scholar]
- 19.Mohammed SI, Rahman M. Proteomics and genomics of urinary bladder cancer. Proteomics Clin Appl. 2008;2:1194–207. doi: 10.1002/prca.200780089. [DOI] [PubMed] [Google Scholar]
- 20.Tong A, Zhang H, Li Z, et al. Proteomic analysis of liver cancer cells treated with suberonylanilide hydroxamic acid. Cancer Chemother Pharmacol. 2008;61:791–802. doi: 10.1007/s00280-007-0536-2. [DOI] [PubMed] [Google Scholar]
- 21.de Noo ME, Mertens BJ, Ozalp A, et al. Detection of colorectal cancer using MALDI-TOF serum protein profiling. Eur J Cancer. 2006;42:1068–76. doi: 10.1016/j.ejca.2005.12.023. [DOI] [PubMed] [Google Scholar]
- 22.Leung SM, Dikler S, Mok SC. A new approach for profiling ovarian cancer serum using functionalizied magnetic beads, anchor chip technology, MALDI-TOF MS, MALDI-TOF/TOF MS, and bioinformatics. Sixth International Symposium on Mass Spectrometry in the Health and Life Science, August 2003. [Google Scholar]
- 23.Ebert MP, Röcken C. Molecular screening of gastric cancer by proteome analysis. Eur J Gastroenterol Hepatol. 2006;18:847–53. doi: 10.1097/00042737-200608000-00007. [DOI] [PubMed] [Google Scholar]