Abstract
A new species of the Stegana (Steganina) ornatipes species group (Diptera: Drosophilidae) is described from Hainan, China, S. (S.) xipengi sp. nov. Based on the mitochondrial ND2 and COI gene sequences, the relationships among eight species from mainland China of the ornatipes group, and their relationships to the undulata, nigrolimbata and shirozui species groups of the same subgenus, are investigated, using two species of the subgenus Stegana, S. emeiensis and S. quadrata, as outgroups. The result shows that S. (S.) mengla is debarred from the ornatipes group.
Keywords : Drosophilidae, molecular phylogeny, mitochondrial DNA, Stegana ornatipes group, Oriental region
Introduction
So far five species groups have been identified in the subgenus Steganina Wheeler (Diptera: Drosophilidae) of the genus Stegana Meigen: coleoptrata group (Laštovka and Máca 1982; Chen and Chen 2008), nigrolimbata group (Sidorenko 2002; Cao and Chen 2008), shirozui group (Chen et al. 2009), undulata group (Sidorenko 2002) and ornatipes group (Cheng et al. 2009), and they included 51 species; most of them were from the Oriental region except for some species of the coleoptrata group from the Palearctic region. The ornatipes group includes ten species from the Oriental region: S. (S.) vietnamensis Sidorenko, 1997 from Virtnam; S. (S.) albiventralis Cheng, Gao et Chen, 2009; S. (S.) angusigena Cheng, Gao et Chen, 2009; S. (S.) chitouensis Sidorenko, 1998; S. (S.) lingnanensis Cheng, Gao et Chen, 2009; S. (S.) mengla Cheng, Gao et Chen, 2009; S. (S.) nulliseta Cheng, Gao et Chen, 2009; S. (S.) ornatipes Wheeler et Takada, 1964; S. (S.) pilosella Cheng, Gao et Chen, 2009 and S. (S.) zhaofengi Cheng, Gao et Chen, 2009 from China. This group is supported by the following morphological characters as the diagnosis: surstylus large, with a strong prensiseta apically and several thin, long setae; 10th sternite mostly narrowed, nearly arcuate, with a pair of projections posterolaterally; gonopods with a pair of projections sublaterally. On the other hand, this group is similar to the nigrolimbata group in sharing the following morphological characters: palpus mostly black, sometimes yellow basally; gena yellow to brown, narrow (ch/o ≤ 0.10); aedeagus basally contiguous to aedeagal apodeme; which shows the both are more closely related to each other than other members of the subgenus Steganina.
Recently, some studies of molecular phylogeny were appeared to the subfamily Steganinae (Otranto et al. 2008; He et al. 2009a, b; Zhao et al. 2009; Li et al. 2010). Otranto et al. (2008) reconstructed the phylogenetic relationships among 13 species of 8 genera of Steganinae based on the DNA sequences of the cytochrome oxidase subunit I (COI) gene, however, in their phylogenetic analysis, only two Stegana species were sampled as the representative. Li et al. (2010) investigated the phylogenetic relationships among seven of the Chinese species of the subgenus Stegana (s.s.) based on the DNA sequences of the NADH dehydrogenase subunit 2 (ND2) gene, using two species of the subgenus Steganina (S. nigrilimbata Duda, 1924 and S. ctenaria Nishiharu, 1979) as outgroup taxa.
In the present study, we described a new species of the ornatipes group from Hainan, China. We also constructed the molecular phylogeny based on the mtDNA sequences of ND2 and COI genes. To investigate the relationships in ornatipes group and with the other species groups of subgenus Steganina, we employed the additional seven species from mainland China of this group and S. nigrolimbata Duda, S. xiaoleiae Cao and Chen, S. ctenaria Nishiharu, and S. undulata de Meijere which belong to nigrolimbata, shirozui and undulata species groups of the subgenus Steganina as ingroup taxa Two species from subgenus Stegana, S. emeiensis Sidorenko and S. quadrata Cao and Chen were chosen as outgroup taxa.
Materials and Methods
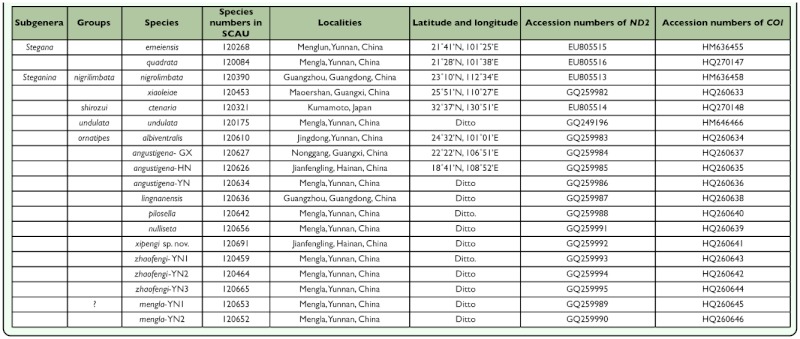
All materials were collected on tussock and tree trunks along streams in forest, preserved in 75% ethanol immediately and identified (Table 1). A small piece of tissue was removed from the fly abdomen and used for the DNA extraction; then, the body and terminalia parts were dried and deposited in the Department of Entomology, South China Agricultural University, Guangzhou, China (SCAU). McAlpine (1981) was followed for morphological terminology and Zhang and Toda (1992), and Chen and Toda (2001) for the definitions of measurements, indices and abbreviations.
Table 1.
Collecttion data of samples for DNA sequencing, and accession numbers of the ND2 and CO1 sequences.
DNA extraction and sequencing
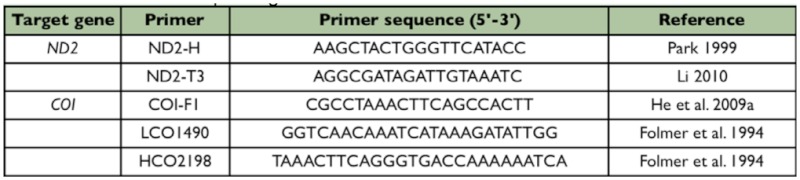
The total DNA was extracted using the DNA extraction Kit (TIANGEN®) according to the manufacture's protocol. The ND2 gene and the 5' end of COI gene were amplified. Primers used were given in table 2. The PCR cycle program comprised an initial 3 min of predenaturation at 94 °C, 35 cycles of amplification ( 50 s of denaturation at 94 °C; 1 min of annealing at 53 °C for ND2, 49 °C for COI; 1 min of extension at 72 °C), and a final elongation for 5 min at 72 °C. When possible, purified amplified products were directly run on an ABI 3730 sequencer for sequenceing, otherwise they were cloned into the pMD18-T plasimid vector (TAKARA®), and then sequenced. The related ND2 sequences of S. emeiensis, S. quadrata, S. ctenaria and S. nigrolimabata were retrieved from the National Center for Biotechnology Information (NCBI); the related COI sequences of emeiensis, S. nigrolimbata and S. undulata were also retrieved from the NCBI.
Table 2.
Primers used for PCR and sequencing.
Phylogenetic analyses
The sequences were aligned by the Clustal W (Thompson et al. 1994) method implemented in program MEGA 4.0 (Tamura et al. 2007) with default options. A partition homogeneity test (PHT) between the ND2 and COI sequences was performed with PAUP 4.0b10* (Swofford 2002). The program DAMBE 5.0.80 (Xia and Xie 2001) was used to measure the nucleotide substitution saturation using the method of Xia et al. (2003) as the substitution saturation masked the phylogenetic signal (Lopez et al. 1999; Philippe and Froterre 1999). Base compositions were investigated by means of the software PAUP 4.0b10* (Swofford 2002), and a χ2 test was also used to test the nucleotide composition homogeneity. Uncorrected pairwise divergence was estimated by program MEGA 4.0 (Tamura et al. 2007).
Phylogenetic trees were constructed by using the maximum parsimony (MP) and maximum likelihood (ML) in PAUP 4.0b10* (Swofford 2002), the Bayesian inferring (BI) method performed in MrBayes 3.2.1 (Huelsenbeck and Ronquist 2001; Ronquist and Huelsenbeck 2003). The MP and ML trees were searched by the heuristic method, with initial trees obtained by randomly adding taxa, and the TBR algorithm was used in branching swapping. Branch support for each node in the MP and ML trees was assessed by 1000 bootstrap replicates. The nucleotide substitution models of ML and BI analyses were selected by MrModeltest 2.3 (Nylander 2004) using the hierarchical likelihood ratio test (hLRT) criterion (Posada and Crandall 1998). In the BI analyses, the site-specific models were assigned to dataset partitioned by locus (2 data partitions) and by codon positions (6 data partitions). Two independent runs with 2,000,000 generations were implemented in parallel, sampling frequency of every 100 generations was employed. When the average deviation of split frequencies fell well below 0.01, the two runs were stopped. For each running, the 5,000 early-phase samples were burn-in, the rest samples were used in summarizing and a majority rule tree showing all the compatible partitions was obtained.
Partition Bremer support (PBS) was used to show the contribution of each gene partition to the Bremer support of the simultaneous analysis (Baker and DeSalle 1997). Values can be positive, negative or zero and sum of all the partitioned Bremer support values at a node will equal the Bremer support value for that node. A positive PBS value suggests support for the node by that gene, whereas a negative PBS value indicates that the partition lends conflict to a given node, and zero indicate that the partition lends neither support nor conflict to a given node. The partitioned Bremer support values were calculated using the partitioned constraint file in TreeRot v3 (Sorenson 1999).
Results
Stegana (Steganina) xipengi sp. nov. (Figures 1,2)
-
Diagnosis
This species is related to S. (S.) albiventralis from Yunnan in having the entirety white katepisternum, but clearly distinguishable from it by the palpus yellow basally, black distally, the mesonotum brown, without stripe (in albiventralis: palpus entirely yellow; mesonotum brown, with yellow stripe medially).
-
Description
Male: Frons and face not rectangular in profile. Eyes red. Ocellar triangle black, with 1 pair of small setae above ocellar setae.
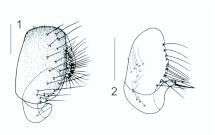
Postvertical setae slightly behind vertex ridge. Frons shiny, brown, with sporadic, minute setulae submedially, and a black, transverse band above ptilinal fissure. Proclinate orbital setae slightly nearer to ptilinal fissure than to inner vertical setae. Pedicel brown; first flagellomere yellow only basally, mostly black. Face black with yellow, transverse band medially, broadened ventrally; facial carina absent. Clypeus black medially, yellow laterally. Palpus yellow basally, black distally, with 1–2 longer setae distally and several shorter setae basally. Gena yellow, narrow. Vibrissa prominent; other orals small. Occiput glossy, yellow, but black around occipital foramen. Mesonotum brown. Mesopleuron with a black longitudinal stripe above (running from propleuron to base of halter). Postpronotal lobe brown on upper part, white on lower part, with 1 long and a few short setae. Acrostichal setulae approximately in 10 irregular rows. Prescutellar setae 1 pair. Katepisternum entirely white. Scutellum brown; basal setae divergent; apical setae crossing with each other. Wing dark brown anteriorly, pale posteriorly, curved downward on distal part. Basal medial-cubital crossvein present. C1 with 2 isometric setae. Costal vein with 9 minute spinules on ventral surface between veins R2+3 and R4+5. Vein R2+3 obviously curved to costa at tip; Veins R4+5 and M1 convergent distally. Halters white basally, greyish brown distally. Legs whitish yellow, brown on apical part of fore femur, and fore and hind tarsomeres, dark brown to black on medially on mid and hind femora, with 2 dark brown rings on fore and mid tibiae. Fore femur with 3–4 setae on distal part of ventral surface. Apical seta present on mid tibia. Preapical dorsal setae present on all tibiae. Mid tibia (misused to mid tarsus in Cao and Chen 2008; Cheng et al. 2009) with 5 strong setae on basal part of dorsal surface. Mid and hind tarsomeres with 2 and 1 row(s) of minute cuneiform setulae on ventral surface, respectively; fore and hind 1st tarsomeres slightly shorter than the rest combined; mid 1 st tarsomere longer than the rest combined. Abdominal all tergites dark brown. Sternites brown; 3rd to 5th broadened; 6th covered with 5th. Epandrium pubescent except for anteroventral margins, with approximately 21 setae near posterior margin on each side (Figure 1). Cercus separated from epandrium, setigerous, lacking pubescence (Figure 1). Surstylus separated from epandrium, with several thin, long setae on inner margin and surface (Figure 2), apically strongly curved and with 1 strong prensiseta (Figure 2). The hypandrium, gonopods, aedeagus and aedeagal apodeme were lost when clearing them in KOH solution.
-
Measurements
BL = 2.76 mm in holotype; ThL = 1.32 mm; WL = 2.58 mm; WW =1.12 mm. Indices: arb = 8/7, avd = 0.83, adf = 1.20, flw = 1.80, FW/HW = 0.36, ch/o = 0.08, prorb = 1.16, rcorb = 0.82, vb = 0.30, dcl = 0.40, presctl = 0.60, sctl = 1.80, sterno = 0.90, orbito = 2.20, dcp = 0.20, sctlp = 1.00, C = 1.86, 4c = 1.22, 4v = 1.74, 5x = 1.40, ac = 9.33, M = 0.61, C3F = 0.66.
-
Type
Holotype: ♂ (SCAU, No. 120589), CHINA: Jianfeng, Ledong, Hainan, 18°41′N, 108°52′E, alt. 750 m, 14.iv.2008, ex tussock, X.P. Chen.
-
Etymology
Patronym of the collector Xipeng Chen (SCAU).
-
Distribution
China (Hainan).
Figures 1–2.
Stegana (Steganina) xipengi sp. nov., ♂: 1. Epandrium, cercus and surstylus (lateral view); 2. surstylus (frontal view). Scale bars = 0.1 mm. High quality figures are available online.
Molecular analysis Data set analysis
The alignment was 1739 nucleotide positions (1029 for ND2 and 710 for COI, respectively) in length. There were end gaps in the ND2 sequence of S. undulata (sites 1–22) and in the COI sequence of S. xiaoleiae (sites 1–33). The base composition of ND2 and COI were generally AT rich with a mean of 83% and 69%, respectively. It contained high AT contents in the 3rd (94.2% and 94.2%, respectively) codon positions. Performance of the Chi-square test was showed in table 3. It yielded a homogeneous base composition in the ND2-alignments and COI-alignments or in the separate condon positions of the two mitochondrial genes.
Table 3.
Results of model selection, composition homogeneity test and test of substitution saturation.
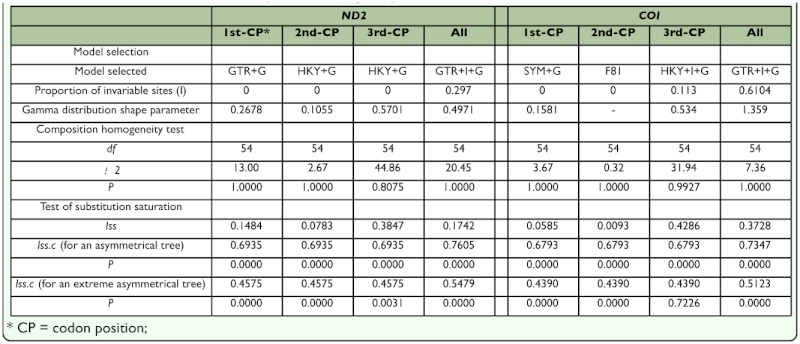
The test of substitution saturation showed that the observed index of substitution saturation (Iss) for ND2-alignments or for COIalignments was significantly lower than the corresponding critical index substitution saturation (Iss.c), indicating that there was little saturation in our sequences. However, when considering partitions separated by codon, we identified substitution saturation in the third codon position of the COI- alignments [Iss = 0.4286 < Iss.c = 0.4390 (for an extreme asymmetrical tree, p = 0.72)] (Table 3). Since none of the resulted trees of the present study are extremely asymmetric, there should be little substitution saturation in our sequence.
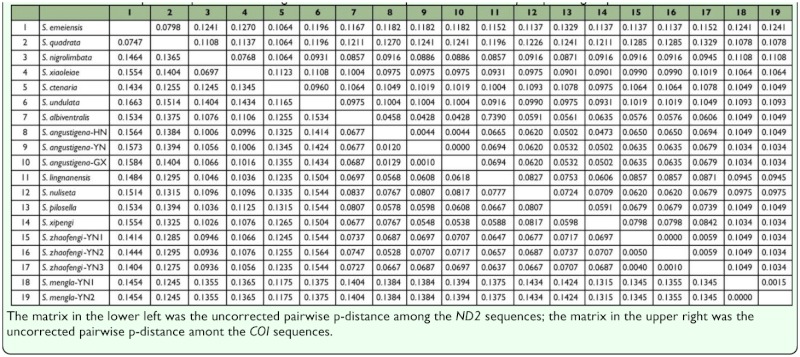
Table 4 shows the uncorrected pairwise p-distances for the ND2 and COI sequences. The genetic divergence of ND2 sequences of species within the ornatipes group ranged from 5.28% to 14.24%, and genetic divergence of COI sequences ranged from 4.28% to 10.49%, however, when we took no account of the S. mengla, the upper limits would declined to 8.37% and 8.42% for ND2 and COI, respectively. Within the ornatipes group, divergences between S. mengla and other species ranged from 9.36% to 15.64% for ND2 and from 8.57% to 10.93% for COI, whereas the genetic variance among groups ranged from 11.65% to 14.34% for ND2 and from 8.57% to 11.23% for COI.
Table 4.
Uncorrected pairwise p-distance among the ND2 and COI sequences of the ornatipes species group.
Phylogenetic analysis
The PHT resulted in a p value of 0.062, indicating that no significant incongruence was found between the ND2 and COI data sets. The best-fit models selected for the ML reconstruction and Bayesian inference were listed in table 3.
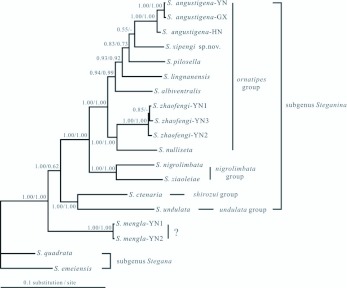
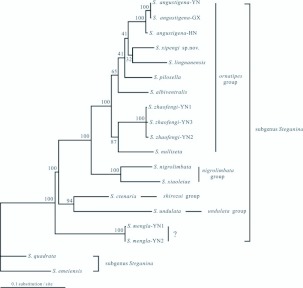
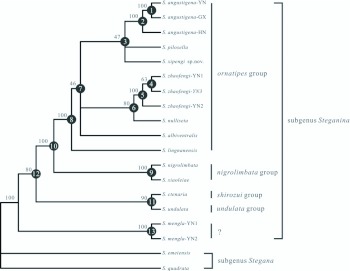
The relationships within the ornatipes group were not stable revealed by different treebuilding methods as the low supports for the basal nodes (Figures 3, 4 and 5), but it was surprising that S. mengla was debarred from the ornatipes group in all trees, and it was placed at the most basal clade of subgenus Steganina receiving great supports (MP BP, or bootstrap percentages of the MP analysis = 100; ML BP = 100; PP or posterior probability of the 2-/6-partition Bayesian inferring = 1.00/1.00). The remaining species of the ornatipes group were recovered as a monophyletic group with robust supports in all trees (MP BP = 100; ML BP = 100; PP = 1.00/1.00 in the 2-/6-partition Bayesian analyses, respectively). The nigrolimbata group appeared to be the closest relative to this monophyletic group with well supports (MP BP = 100; MLBP = 100; PP = 1.00/1.00) (Figures 3, 4 and 5). The Bayesian analysis yielded a general topology (Figure 3), which was mostly congruent with the result of the ML analysis (Figure 4). The monophyletic group diverged into two branches. One consist of S. zhaofengi triple and S. nulliseta, and the other further diverged into S. albiventralls, S. lingnanensis, S. pilosella, S. xipengi and S. angustigenai triple orderly in the Bayesian tree, whereas S. pilosella diverged prior to S. lingnanensis (ML BP = 41), leaving S. lingnanensis and S. xipngi as sister group in the ML reconstruction, but with a low support (ML BP = 32). The MP tree (Figure 5) differed from the ML and Bayesian tree at several points. It suggested a very basal position for S. lingnanensis in the ornatipes group and S. xipengi clustered with S. pilosella which was consistent with the Bayesian analysis. The Yunnan (-YN), Guangxi (-GX) and Hainan (-HN) samples of S. angustigena clustered together with well support (MP BP = 100; ML BP = 100; PP = 1.00/1.00), and so did in the YN1, YN2 and YN3 samples of S. zhaofengi (Figures 3, 4 and 5).
Figure 3.
Bayesian tree of the ornatipes group deduced from the ND2 and COI sequences. The numbers above the branches show the posterior probabilities of the corresponding node in the Bayesian inference. High quality figures are available online.
Figure 4.
ML tree of the ornatipes group deduced from the ND2 and COI sequences. The numbers above the branches show the bootstrap percentages (BPs) of the corresponding node in the ML analysis (-lnL = 7925.69531). High quality figures are available online.
Figure 5.
Strict consensus tree of two most parsimonious trees of the ornatipes group deduced form the ND2 and COI combined sequences. The numbers above the branches show the bootstrap percentages (BPs) of the corresponding node in the MP analysis [tree length = 1225, consistency index (Cl) = 0.6008, retention index (Rl) = 0.6362]. High quality figures are available online.
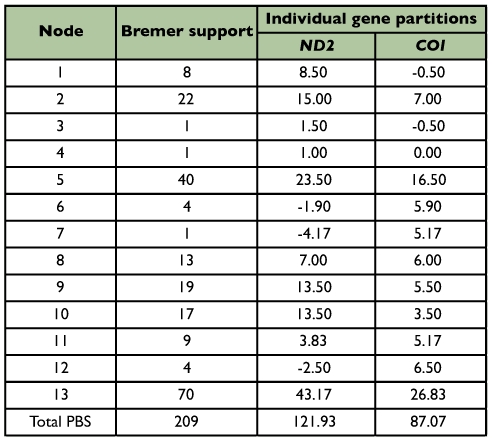
To determine the relative contributions of the two data partition to the combined analysis tree, partition Bremer supports were calculated and given in table 5. Support for combined analysis phylogeny came from ND2 was a little bit more than that from COI. Nodes 1, 3, 6, 7 and 12 showed a mixture of positive and negative PBS scores.
Table 5.
Partition Bremer support for all nodes in the MP tree (Fig. 5).
Discussion
The phylogenetic trees showed that the ornatipes group clearly appeared to be paraphyletic. To eliminate the effect of individual difference, another sample of S. mengla was included in the analysis, but the situation did not change. In morphological viewpoints, the S. mengla holds the same diagnostic characters of the onartipes group, which contradict with our molecular phylogeny as it formed a separate branch in the phylogenetic tree. The amount of genetic divergences between S. mengla and other species within the ornatipes group were high and overlapped to some extent with the divergence between species groups. Although speculative, the morphological convergence should be the reason for this situation. The convergent morphological evolution seems to be common in the subfamily Steganinae (Otranto et al. 2008), which is similar to the suggestion made in this research concerning convergent morphological evolution in S. mengla. Considering the closer relationship of S. mengla with the outgroup S. emiensis respect to the other species showed in the phylogenetic tree, it is possible that S. mengla is the interim species of the divergent between subgenus Steganina and subgenus Stegana. Of course, this hypothesis should be proved with analysis of suitable species of both the subgenus Steganina and subgenus Stegana. Except the S. mengla, the branch consist of the rest species of the ornatipes group showed the closer relationship with the nigrolimbata group than other species groups of subgenus Steganina was consistent with the morphological affinity in the two groups (Cao and Chen 2008).
In general, the NADH dehydrogenase subunit genes are rapidly evolving, but the cytochrome oxidase subunit is more slowly evolving (Simon 1994). It was supposed that the ND2 gene was better than the COI gene suited for species-level analysis, but the PBS analysis indicating that the contribution to the MP reconstruction in this research of the ND2 gene was nearly the same as the COI gene. Our PBS analysis had implications for the conflicts of the two genes at some nodes (e.g., nodes 1, 3, 6, 7 and 12), suggesting that the two partitions data (ND2 and COI) may be favoring an alternative tree topology. The relationships of these nodes should be viewed cautiously.
The genetic distances between Yunnan, Guangxi and Hainan samples of S. angustigena [p-distance of ND2 = 0.0129 (GX vs. -HN), 0.0120 (-HN vs. -YN), 0.001 (GX vs. -YN); p-distance of COI = 0.0044 (GX vs. -HN), 0.0044 (-HN vs. -YN), 0.0000 (-GX vs. -YN)] were among the mean intraspecific variability of Meier et al. 2008 (1.3 ± 1.6%) for Diptera. In addition, no essential morphological character was found to distinguish the specimens of these three samples, indicating that they should be taken as conspecific ones. It was the same as the case of the YN1, YN2 and YN3 samples of S. zhaofengi. The genetic data [p-distance of ND2 = 0.0050 (-YN1 vs. -YN2), 0.0040 (YN2 vs. -YN3), 0.0010 (-YN1 vs. -YN3); p-distance of COI = 0.0000 (-YNl vs. -YN2), 0.0059 (-YN2 vs. -YN3), 0.0059 (-YN1 vs. YN3)] also indicated the conspecific status of the three samples of S. zhaofengi.
Some relationships within the ornatipes group were not well resolved, especially the alternative placement of S. lingnanensis, S. xipengi and S. pilosella. Therefore, it may be worthy to increase either the genetic markers (such as nuclear markers) or the number of samples in the future phylogenetic analysis of the ornatipes group.
Acknowledgements
We thank Dr. JJ Gao (Yunnan University, China) for helping in fieldwork; Ms. Y Cheng for providing the figures 1–2. This work was supported by the National Natural Science Foundation of China (No. 30970396).
Editor's note: Paper copies of this article will be deposited in the following libraries. The date of publication is given in ‘About the Journal’ on the JIS website. Universitaetsbibliothek Johann Christian Senckenberg, Frankfurt Germany; National Museum of Natural History, Paris, France; Field Museum of Natural History, Chicago, Illinois USA; University of Wisconsin, Madison, USA; University of Arizona, Tucson, Arizona USA; Smithsonian Institution Libraries, Washington D.C. USA; The Linnean Society, London, England.
References
- Baker RH, DeSalle R. Multiple sources of character information and the phylogeny of Hawaiian drosophilids. Systematic Biology. 1997;46:654–673. doi: 10.1093/sysbio/46.4.654. [DOI] [PubMed] [Google Scholar]
- Cao HZ, Chen HW. Revision of the Stegana (Steganina) nigrolimbata species group from the Oriental region (Diptera, Drosophilidae). Zootaxa. 2008;1848:27–36. [Google Scholar]
- Chen XP, Chen HW. The Stegana coleoptrata species group (Diptera, Drosophilidae) from mainland China. Zootaxa. 2008;1891:55–65. [Google Scholar]
- Chen XP, Gao JJ, Chen HW. The Stegana shirozui species group (Diptera, Drosophilidae). Journal of Natural History. 2009;43:1909–1927. [Google Scholar]
- Chen HW, Toda MJ. A revision of the Asian and European species in the subgenus Amiota Loew (Diptera, Drosophilidae) and establishment of speciesgroups based on phylogenetic analysis. Journal of Natural History. 2001;35:1517–1563. [Google Scholar]
- Cheng Y, Gao JJ, Chen HW. The Stegana ornatipes species group from the Oriental Region (Diptera, Drosophilidae). Zootaxa. 2009;2216:37–48. [Google Scholar]
- Folmer O, Black M, Hoeh W, Lutz R, Vrijenhoek R. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Molecular Marine Biology and Biotechnology. 1994;3:294–299. [PubMed] [Google Scholar]
- He XF, Gao JJ, Cao HZ, Zhang XL, Chen HW. Taxonomy and molecular phylogeny of the Phortica hani species complex (Diptera: Drosophilidae). Zoological Journal of the Linnean Society. 2009a;157:359–372. [Google Scholar]
- He XF, Jiang JJ, Cao HZ, Chen HW. Taxonomy and molecular phylogeny of the Amiota nagatai species group (Diptera, Drosophilidae). Zootaxa. 2009b;2193:53–61. [Google Scholar]
- Huelsenbeck JP, Ronquist FR. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- Laštovka P, Máca J. European and North American species of the genus Stegana (Diptera, Drosophilidae). Annotations Zoologicae et Botanicae. 1982;149:1–38. [Google Scholar]
- Li T, Cao HZ, Gao JJ, Chen HW. A revision of the subgenus Stegana (s. str.) (Diptera, Drosophilidae) from mainland China. Zoological Journal of the Linnean Society. 2010;158:726–739. [Google Scholar]
- Lopez P, Forterre P, Philippe H. The root of the tree of life in the light of the covarion model. Journal Molecular Evolution. 1999;49:496–508. doi: 10.1007/pl00006572. [DOI] [PubMed] [Google Scholar]
- McAlpine JF. Morphology and terminology – adults. In: McAlpine J. F., editor. Manual of Nearctic Diptera, 1: 9–64. Research Branch Agriculture Canada Monograph, 27. Research Branch, Agriculture Canada; Ottawa: 1981. [Google Scholar]
- Meier R, Zhang G, Ali F. The use of mean instead of smallest interspecific distances exaggerates the size of the “Barcoding Gap” and leads to misidentification. Systematic Biology. 2008;57:809–813. doi: 10.1080/10635150802406343. [DOI] [PubMed] [Google Scholar]
- Nylander J A A. MrModeltest v2. Program distributed by the author. Evolutionary Biology Centre, Uppsala University; 2004. [Google Scholar]
- Otranto D, Stevens JR, Testini G, Cantacessi C, Máca J. Molecular characterization and phylogenesis of Steganinae (Diptera, Drosophilidae) inferred by the mitochondrial cytochrome c oxidase subunit 1. Medical and Veterinary Entomology. 2008;22:37–47. doi: 10.1111/j.1365-2915.2008.00714.x. [DOI] [PubMed] [Google Scholar]
- Park J. Molecular phylogenetic studies of the Drosophila (Drosophila) virilis section (Diptera, Drosophilidae). PhD. Thesis, Tokyo Metropolitan University; 1999. [Google Scholar]
- Philippe H, Forterre P. The rooting of the universal tree of life is not reliable. Journal Molecular Evolution. 1999;49:509–523. doi: 10.1007/pl00006573. [DOI] [PubMed] [Google Scholar]
- Posada D, Crandall KP. Modeltest: testing the model of DNA substitution. Bioinformatics. 1998;14:817–818. doi: 10.1093/bioinformatics/14.9.817. [DOI] [PubMed] [Google Scholar]
- Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- Sidorenko VS. Phylogeny of the tribe Steganini Hendel and some related taxa (Diptera, Drosophilidae). Far Eastern Entomologist. 2002;111:1–20. [Google Scholar]
- Simon C, Frati F, Beckenbach A, Crespi B, Liu H, Flook P. Evolution, weighting and phylogenetic utility of mitochondrial gene sequences and a compilation of conserved PCR primers. Annals of the Entomological society of America. 1994;87:651–710. [Google Scholar]
- Sorenson MD. TreeRot, vesion 2. Boston University; Boston, Massachusetts: 1999. [Google Scholar]
- Swofford DL. PAUP*: Phylogenetic analysis using parsimony (*and other methods), Version 4. Sinauer Associates; Sunderland, MA: 2002. [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software28 version 4.0. Molecular Biology and Evolution. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. Clustal W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position specific gap penalties and weight matrix choice. Nucleic Acids Research. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia XH, Xie ZH. DAMBE: Data analysis in molecular biology and evolution. Journal of Heredity. 2001;92:371–373. doi: 10.1093/jhered/92.4.371. [DOI] [PubMed] [Google Scholar]
- Xia XH, Xie ZH, Salemi M, Chen L, Wang Y. An index of substitution saturation and its application. Molecular Phylogenetics and Evolution. 2003;26:1–7. doi: 10.1016/s1055-7903(02)00326-3. [DOI] [PubMed] [Google Scholar]
- Zhang WX, Toda MJ. A new species-subgroup of the Drosophila immigrans species-group (Diptera, Drosophilidae), with description of two new species from China and Revision of Taxonomic Terminology. Japan Journal Entomology. 1992;60:839–850. [Google Scholar]
- Zhao F, Gao JJ, Chen HW. Taxonomy and molecular phylogeny of the Asian Paraleucophenga Hendel (Diptera, Drosophilidae). Zoological Journal of the Linnean Society. 2009;155:615–629. [Google Scholar]