Abstract
The spatial heterogeneity in magnetic resonance (MR) metabolic and diffusion parameters and their relationship were studied for patients with treatment-naive grade 3 gliomas. MR data were evaluated from 51 patients with newly diagnosed grade 3 gliomas. Anatomic, diffusion, and metabolic imaging data were considered. Variations in metabolite levels, apparent diffusion coefficient (ADC), and fractional anisotropy (FA) were evaluated in regions of gadolinium enhancement and T2 hyperintensity as well as regions with abnormal metabolic signatures. Contrast enhancement was present in only 21 of the 51 patients. When present, the enhancing component of the lesion had higher choline-to-N-acetylaspartate index (CNI), higher choline, lower N-acetylaspartate, similar creatine, similar ADC and FA, and higher lactate/lipid than the nonenhancing lesion. Regions with CNI ≥ 4 had higher choline, lower N-acetylaspartate, higher lactate/lipid, higher ADC, and lower FA than normal-appearing white matter and regions with intermediate CNI values. For lesions that exhibited gadolinium enhancement, the metabolite levels and diffusion parameters in the region of enhancement were consistent with it corresponding to the most abnormal portion of the tumor. For nonenhancing lesions, areas with CNI ≥ 4 were the most abnormal in metabolic and diffusion parameters. This suggests that the region with the highest CNI might provide a good target for biopsies for nonenhancing lesions to obtain a representative histologic diagnosis of its degree of malignancy. Metabolic and diffusion parameter levels may be of interest not only for directing tissue sampling but also for defining the targets for focal therapy and assessing response to therapy.
Introduction
The incidence rate of brain tumors is increasing, with an estimated 22,020 new cases of primary brain tumors being detected each year in the United States [1]. For adults older than 45 years, 90% of such tumors are diagnosed as gliomas. These lesions are infiltrative in nature, which makes it difficult to define tumor margins and to specify which regions should be targeted for focal therapy. Grade 2 gliomas have the best prognosis with a median survival of 7.9 years [2], grade 3 gliomas have intermediate survival with a median of 33.5 months [3], and grade 4 gliomas are the most malignant with a median survival of 40.9 weeks [4]. Whereas the initial therapeutic strategy is maximal safe resection for all types of glioma, next-line therapies differ for patients with grade 2 versus grade 3 or 4 lesions, with radiation and, in some cases, chemotherapy being delayed until the time of recurrence. Making an accurate histologic diagnosis is especially critical for grade 3 lesions, which often look relatively uniform on T2-weighted magnetic resonance (MR) images and have little or no enhancement on post-gadolinium T1-weighted images. In practice, the underlying biological properties of different regions of the tumor may be quite variable, and there may be regions with characteristics of grade 2 and grade 3 histologic diagnosis within the same tumor. The development of noninvasive imaging methods that define tumor heterogeneity and can be used to direct tissue sampling to the most malignant region of the tumor is a priority for optimizing patient care.
One of the methods proposed for characterizing the heterogeneity of both enhancing and nonenhancing regions of glioma is metabolic imaging using MR spectroscopic imaging (MRSI). When used in combination with standard MR imaging (MRI), this has been has been shown to contribute to brain tumor diagnosis and localization [5–11], staging [12–15], assessment of progression [14,16–18], treatment planning [19–23], and response to therapy [24–29]. Long echo time MRSI, which is the most commonly applied method, provides arrays of spectra that are characterized by three major metabolite peaks in healthy brain tissue: choline (Cho), total creatine (Cr), and N-acetylaspartate (NAA). An increase in Cho provides a marker for increased proliferation or membrane turnover. The Cr peak reflects bioenergetic processes, and NAA is an indicator of neuronal viability that is observed to be significantly lower in tumors than regions of normal brain. Singlet lipid and doublet lactate peaks occur at similar spectral frequencies and may also be observed in the disease state. Lactate is a byproduct of anaerobic glycolysis and is present in regions of tumor as a result of poor oxygenation. Lipid is present in the bilayer of the cell membrane, and its presence indicates membrane breakdown and a more aggressive tumor phenotype.
Diffusion tensor imaging (DTI) is another MR technique that has been proposed as adding information on heterogeneity in the structural properties of different types of tissue. DTI acquires images with diffusion gradients applied in multiple directions, and uses these images to generate pixel by pixel estimates of the coefficients of the corresponding tensor matrix. The eigenvalues and eigenvectors derived from the tensor are used for computing maps of the apparent diffusion coefficient (ADC) and fractional anisotropy (FA). ADC values have been shown to increase in regions of infiltrative tumor and surrounding edema [30–34]. The values of FA provide an assessment of the directionality of water diffusion and have been shown to decrease in brain tumors due to disruptions in tissue architecture [30,31,34–36].
The current study evaluates data acquired from patients with treatment-naive grade 3 gliomas with respect to regional differences in MRSI and DTI parameters in enhancing and nonenhancing lesions. Of particular interest was to determine which MR parameters highlighted regions of the tumor that were likely to be malignant and might therefore be considered as targets for obtaining tissue during surgical resection to make an accurate histologic diagnosis.
Materials and Methods
MRI data of 51 patients with newly diagnosed supratentorial grade 3 glioma according to WHO stage II classification and who had been scanned 1 day before surgery were selected for this study (25 men and 26 women; median age = 40 years [range = 21–78 years]; 31 anaplastic astrocytomas [AAs], 7 anaplastic oligodendrogliomas [OD3s], 13 anaplastic oligoastrocytomas [OA3s]). The patients were scanned on a 1.5-T GE Signa Echospeed scanner (GE Medical Systems, Milwaukee, WI). All patients provided informed consent for participating in the study, based on a protocol approved by the Committee on Human Research at our institution. A biopsy was performed during the surgery. The biopsy specimens were taken from the contrast-enhanced portion of the contrast-enhancing lesions. For nonenhancing lesions, multiple specimens were taken from different areas within the T2 hyperintense area. The clinical diagnosis was made by a neuropathologist, and the diagnosis was independently approved by a second neuropathologist.
The MRI protocol included an axial T2-weighted fast spin echo (FSE) sequence (repetition time [TR] = 3000 milliseconds, echo time [TE] = 105 milliseconds, matrix size = 192 x 256 x 120, field of view = 195 x 260 x 180 mm, slice thickness = 1.5 mm), and post-Gd-diethylenetriaminepentacetate (DTPA) T1-weighted spoiled gradient recalled (SPGR) sequence (TR = 34 milliseconds, TE = 3 milliseconds, flip angle = 35 degrees, matrix size = 192 x 256 x 124, field of view = 195 x 260 x 186 mm, slice thickness = 1.5 mm).
MRSI data were referenced to the SPGR images and were acquired using three-dimensional point-resolved spectroscopic (PRESS) [37] for volume localization with spectral spatial pulses [38] and very selective suppression [39] outer volume suppression bands (TR = 1000 milliseconds, TE = 144 milliseconds, matrix size = 12 x 12 x 8, field of view = 120 x 120 x 80 mm, 1-ml nominal spatial resolution). Spectral data from 30 patients were acquired with J-difference lactate editing that allowed separation of lactate and lipid resonance [40].
MRSI data were quantified offline to estimate the levels of Cho, Cr, NAA, combined lactate and lipid (LL) for regular spectra, and separate lactate (Lac) and lipid levels for lactate-edited spectra using in-house software described in detail in a previous report [41]. Spectral values were normalized relative to the noise levels of the right hand end of the spectra. The Cho-to-NAA index (CNI) [42], which defines the distance between the Cho and NAA values for a given voxel and a regression line that was fit to the normal voxels in a given subject, was also computed. CNI values are expected to increase in the tumor because of the reduction in NAA and increase in Cho.
DTI sequences with six gradient directions were acquired in the axial planes with a b value of 1000 sec/mm2 (TR = 5000 milliseconds, TE = 105 milliseconds, matrix size = 256 x 256 x 40, field of view = 440 x 440 x 84 mm, slice thickness = 2.1 mm). DTI images were quantified to calculate the ADC and FA maps using the standard formulas [43]. Diffusion maps were resampled to the resolution of spectral data and normalized relative to the mean of the voxels in normal-appearing white matter (NAWM) to form maps of nADC and nFA.
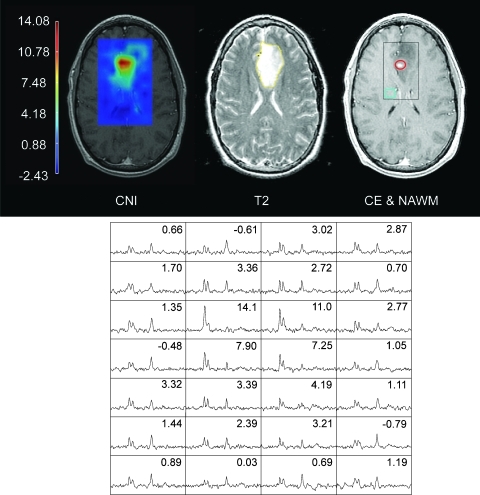
Regions of contrast enhancement (the CE lesion) and T2 hyperintensity were defined manually by tracing the abnormality borders on the SPGR and FSE slices using interactive in-house software by experienced research assistants and were verified by an experienced radiation oncologist. The NE lesion was defined as the regions of T2 hyperintensity that excluded the CE lesion. Metabolically abnormal regions were defined as voxels with CNI values of 2 or greater based on previous results [11]. CNI maps were then used to automatically generate three regions with distinct levels of metabolic abnormality, 2 ≤ CNI < 3 (CNI2–3), 3 ≤ CNI < 4 (CNI3–4), and CNI ≥ 4 (CNI > 4). Figure 1 shows an example of the regions analyzed for this study.
Figure 1.
A 40-year-old man diagnosed with a left frontal AA. Images from left to right display the distribution of CNI values, the FSE image with the NE LESION region (yellow), and T1-weighted post-Gd SPGR image with the CE LESION (red), NAWM (blue), and an axial representation of the MRSI-excited region (black box). Spectra from the black box are shown at the bottom with the corresponding CNI values displayed in each voxel.
A total of 11,861 voxels of data were considered from the PRESS-localized volumes, including 2286 from the NE lesion, 778 from the CNI2–3 region, 432 from the CNI3–4 region, 687 from the CNI > 4 region, and 68 from the CE lesion. A single median value was calculated for each spectral or diffusion parameter within a given region of interest. A Wilcoxon signed rank test was applied to detect paired significant differences of medians for spectral or diffusion values between the tumor regions and NAWM for all the patients. For comparison of values between patients, the metabolite levels were normalized to the levels in NAWM and Spearman rank correlation coefficients were calculated to detect correlations of diffusion and spectral parameters within the regions of interest. The Holm test was used for multiple comparisons correction. A sequentially increasing P value was used for pairwise region comparisons for each parameter, and a Pj value of less than .05 / (k - j + 1) was considered significant where j = 1, …, k and k was the total number of comparisons.
Results
Tumor Spatial Extent and Coverage
All the grade 3 patients included in this study exhibited a region of T2 hyperintensity. The median percentage of the NE lesion within the PRESS box was 54.2% (min = 27.9%, max = 83.3%). Of 51 patients, 21 exhibited CE on the SPGR images after the injection of Gd-DTPA. Five of these patients showed minimal CE, and in one patient, the PRESS-selected volume did not fully cover the CE lesion. This left 16 of 51 patients with CE lesions that could be evaluated. All of the patients had CNI2–3 regions, 48 patients had CNI3–4 regions, and 44 patients had regions with CNI > 4.
Overlap of Anatomic Tumor Regions and Metabolically Abnormal Areas
The CE lesion covered by the PRESS box was found to reside within the CNI > 4 region for five patients, the CNI3–4 region for two patients, and the CNI2–3 region for three patients. There were six patients who had CE voxels outside the CNI defined regions. For four of these patients, more than 80% of the CE lesion was within the CNI > 2 region, and the other two patients had 50% and 12.5% of their CE lesions within the CNI > 2 regions. The CNI regions were within the region of T2 hyperintensity for all patients. The median percentage of the NE lesion that was outside the CNI regions was 39.1% (min = 4.1%, max = 97.6%).
Spectral and Diffusion Parameters within Anatomic Tumor Regions
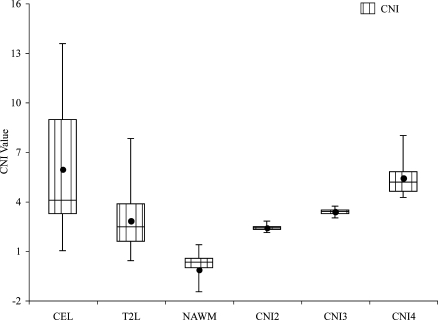
Table 1 shows the median values of the metabolite and diffusion values in the anatomic tumor regions and NAWM. Several differences in the median metabolite levels, CNI, and diffusion values were observed. Cho levels were significantly different in the NE lesion compared with NAWM (P < .001), and there were substantial increases in Cho within the CE lesion compared with both the NE lesion and NAWM (nCho = 1.83 in CE vs 1.2 in NE). Cr values were highly variable. There was a trend toward lower Cr values in the NE lesion and CE lesion relative to NAWM, but the difference was only significant for the NE lesion and this may have been driven by the higher number of sample points. Levels of NAA were significantly lower in the anatomic lesion regions compared with NAWM and in the CE lesion compared with the NE lesion (nNAA = 0.3 in CE vs 0.41 in NE). The highest Cho, lowest NAA, and, accordingly, highest CNI levels were observed in the CE lesion (Figure 2). Combined levels of LL were significantly higher in the CE lesion and the NE lesion than in NAWM and were also significantly higher in the CE lesion than in the NE lesion (LL/nNAA = 0.21 in CE vs 0.1 in NE and 0.06 in NAWM).
Table 1.
Differences of Spectral and Diffusion Parameters between NAWM, CE Lesion, and NE Lesion.
| Parameter | Region | Median | n | CE Lesion, P | NE Lesion, P |
| nCho | NAWM | 1 | 51 | .001* | <.001* |
| CE Lesion | 1.83 | 16 | .044* | ||
| NE Lesion | 1.2 | 51 | |||
| nCr | NAWM | 1 | 51 | NS | .006* |
| CE Lesion | 0.75 | 16 | NS | ||
| NE Lesion | 0.91 | 51 | |||
| nNAA | NAWM | 1 | 51 | <.001* | <.001* |
| CE Lesion | 0.3 | 16 | .005* | ||
| NE Lesion | 0.41 | 51 | |||
| CNI | NAWM | -0.02 | 51 | <.001* | <.001* |
| CE Lesion | 4.09 | 16 | .01* | ||
| NE Lesion | 2.47 | 51 | |||
| LL/nNAA | NAWM | 0.06 | 51 | .008* | <.001* |
| CE Lesion | 0.21 | 16 | .039* | ||
| NE Lesion | 0.1 | 51 | |||
| nADC | NAWM | 1 | 51 | <.001* | <.001* |
| CE Lesion | 1.59 | 16 | NS | ||
| NE Lesion | 1.67 | 51 | |||
| nFA | NAWM | 1 | 51 | .001* | <.001* |
| CE Lesion | 0.25 | 16 | NS | ||
| NE Lesion | 0.42 | 51 |
P values were calculated with the Wilcoxon signed rank test.
NS indicates not significant.
P < .05/(k-j+1).
Figure 2.
Box plots of median CNI values for all the patients. The borders of the box plots are 25th and 75th percentiles of the median values. The line inside the box depicts the median, and the black dot depicts the mean of the median values. Error bars extending outside the boxes indicate the minimum and maximum median values for this patient population.
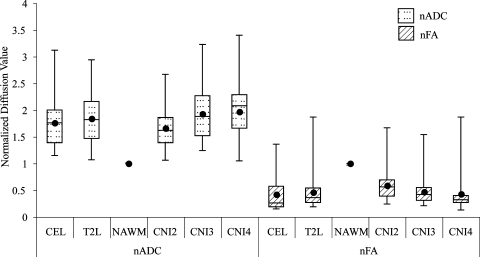
Normalized ADC values were significantly higher in the anatomic regions compared with NAWM but were not significantly different between the NE lesion and CE lesion (nADC = 1.59 in CE vs 1.67 in NE). Levels of FA were lower in the anatomic lesion regions than in NAWM (nFA = 0.25 in CE vs 0.42 in NE), and although the value in the CE lesion was lower than in the NE lesion, the difference was not significant. Box plots of the diffusion values are seen in Figure 3.
Figure 3.
Box plots of median normalized ADC and FA values for all the patients. The borders of the box plots are 25th and 75th percentiles of the median values. The line inside the box depicts the median, and the black dot depicts the mean of the median values. Error bars extending outside the boxes indicate the minimum and maximum median values for this patient population.
Spectral and Diffusion Parameters within Metabolically Abnormal Areas
There were clear differences in spectral and diffusion parameters between the metabolically abnormal areas and NAWM. Cho levels were significantly different between the NAWM and CNI2–3; there was an increase in Cho within the CNI3–4 (nCho = 1.37) and the CNI > 4 region (nCho = 1.83). NAA levels were significantly lower in all of the metabolically abnormal regions compared with NAWM and were significantly lower in regions with increasing CNI (nNAA = 0.55, 0.47, and 0.29). The CNI2–3 regions had significantly lower Cr levels (nCr = 0.97) than the CNI > 4 (nCr = 1.06). All abnormal metabolic regions had higher LL than NAWM with the level in the CNI > 4 region being the highest (LL/nNAA = 0.16 vs 0.06 in NAWM).
The ADC values were significantly higher in all of the metabolically abnormal regions compared with NAWM. The CNI2–3 had intermediate nADC levels (1.47), whereas the nADC in CNI3–4 and CNI > 4 (1.74 and 1.88) were increasing and were significantly higher than both NAWM and CNI2–3. The levels of FA were significantly lower in all of the metabolically abnormal regions than in NAWM, with values that significantly decreased within regions having higher CNI (nFA = 0.54, 0.41, and 0.34).
Differences in Levels of Lactate
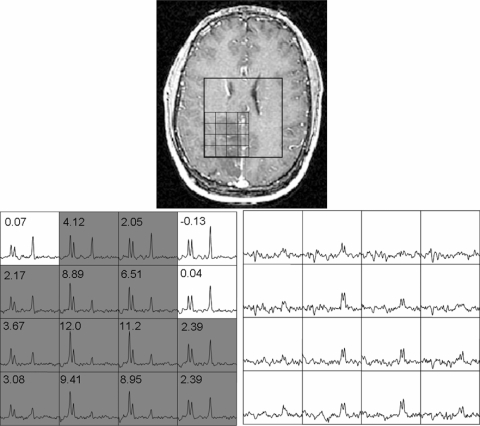
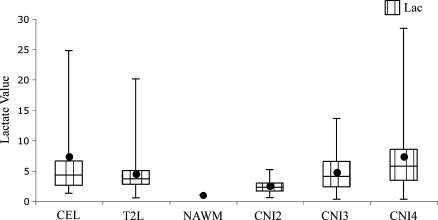
Lactate-edited spectroscopy data were acquired in 30 patients. In 20 of these patients, there were spectral voxels that had visible lactate peaks. Only nine patients in this lactate-edited cohort displayed CE and only six of these had visible lactate peaks. Figure 4 displays a grade 3 patient whose spectral data were acquired with lactate editing. The voxels with high Cho and low NAA intensities also displayed prominent lactate doublets at 1.3 ppm. Figure 5 shows the box plots for median lactate values estimated from the 20 patient data sets with visible lactate peaks. Lactate was significantly higher in the NE lesion (mean Lac = 5.37, P < .001) and in the CE lesion (mean Lac = 3.49, P = .03) compared with NAWM (mean Lac = 1.16). The CNI2–3 (mean Lac = 2.45, P < .001), CNI3–4 (mean Lac = 3.69, P = .005), and CNI > 4 (mean Lac = 5.65, P < .001) regions all had significantly higher lactate values than NAWM. The lactate levels were similar between CNI2–3 and CNI3–4 regions but significantly higher in the CNI > 4 region compared with CNI2–3 (P = .002) and CNI3–4 (P = .026) regions.
Figure 4.
Presence of lactate within the hypointense region in a 24-year-old man diagnosed with an OA3. The spectral data were acquired with J-difference lactate editing method. Top: Post-Gd-DTPA T1-weighted SPGR image with the spectral grid superimposed upon it. Bottom: Corresponding summed spectra is shown on the left with gray-shaded voxels that have CNI ≥ 2. The difference lactate spectra are shown on the right.
Figure 5.
Box plots of median lactate levels normalized by the standard deviation of noise estimated fromthe J-editing PRESS spectroscopy for the nine patients. The borders of the box plots are 25th and 75th percentiles of the median values. The line inside the box depicts the median, and the black dot depicts the mean of the median values. Error bars extending outside the boxes indicate the minimum and maximum median values for this patient population.
Tumor Subtype Differences
Differences in metabolic and diffusion parameters for the patients with AAs, OD3s, and OA3s were examined. Moreover, the 10th, 25th, 50th (median), 75th, and 90th percentile values for each parameter in each region of interest were considered. There were no statistically significant differences between the AAs and the OA3s for any of the imaging parameters in any given region. As is shown in Table 2, there were differences in some of the parameters that described levels of lipid between the OD3 compared with the AA and OA3 subtypes. This included significantly higher median, 75th, and 90th percentile lipid values in almost all regions of OD3 compared with AA lesions and in some of the regions of OD3 compared with OA3 lesions. The 25th percentile of NAA was also higher in AAs than OD3s in the CE lesion (P = .035, AA = 0.34, OD3 = 0.19). Although the median lactate levels were higher in the CNI2–3 region of OD3s than AAs (P = .03, AA = 2.18, OD3 = 3.68) and in the NE lesion of OA3s than OD3s (P = .036, median (OA) = 3.66, median (OD) = 3.50), there were only three patients with OD3 who had lactate-edited MRSI with visible lactate peaks, and so these differences need to be further verified in a larger population of patients.
Table 2.
Differences of Metabolic and Diffusion Parameters between AAs, OD3s, and OA3s.
| Parameter | Percentile | NE Lesion | CE Lesion | CNI2–3 | CNI3–4 | CNI > 4 |
| nNAA | 25th | P = .035 (AA = 0.34, OD3 = 0.19) | ||||
| LL/NAA | 50th (median) | P = .034 (AA = 0.04, OD3 = 0.08) | P = .05 (AA = 0.07, OD3 = 0.20) | P = .012 (AA = 0.08, OD3 = 0.16) | ||
| 75th | P = .013 (AA = 0.11, OD3 = 0.17) | P = .022 (AA = 0.14, OD3 = 0.29) | P = .034 (AA = 0.10, OD3 = 0.15) | P = .03 (AA = 0.11, OD3 = 0.20) | P = .018 (AA = 0.15, OD3 = 0.20) | |
| P = .048 (OD3 = 0.17, OA3 = 0.11) | P = .023 (OD3 = 0.15, OA3 = 0.08) | P = .007 (OD3 = 0.20, OA3 = 0.10) | ||||
| 90th | P = .023 (AA = 0.18, OD3 = 0.24) | P = .035 (AA = 0.27, OD3 = 0.33) | P = .034 (AA = 0.15, OD3 = 0.21) | P = .015 (AA = 0.16, OD3 = 0.27) | ||
| P = .018 (OD3 = 0.21, OA3 = 0.13) | P = .017 (OD3 = 0.27, OA3 = 0.14) | |||||
| Lac | 50th (median) | P = .036* (OA3 = 3.66, OD3 = 3.50) | P = .03* (AA = 2.18, OD3 = 3.68) |
P values were calculated with the Mann-Whitney rank sum test. Only the values that significantly differed between the grade 3 glioma subtypes are reported in this table.
There were only three patients with OD3 who had lactate-edited MRSI with visible lactate peaks, and so these differences were not considered significant.
Discussion
Whereas there have been several studies that focus on the applications of advanced MR techniques to the assessment of tumor grade and type, there have been relatively few studies that have provided a detailed analysis of the metabolic and physiological properties of grade 3 gliomas. Even when they are included, the number of grade 3 lesions is often small, and they are frequently grouped with a relatively large population of grade 4 lesions under the general designation of high-grade glioma. This means that the results presented are typically dominated by the findings for grade 4 glioma. Defining the metabolic and physiological characteristics that best represent grade 3 lesions is an important clinical problem that has so far not been adequately addressed. Determining where within the region of hyperintensity on T2-weighted MR images, it would be best to take tissue samples to have the highest probability of obtaining an accurate diagnosis is an important question that requires information about the functional properties of the lesion. It is for this reason that we have focused on the evaluation of spatial variations in parameters that were extracted from metabolic and physiological MR images of patients with grade 3 glioma.
The current study indicates that the median nADC values were similar in enhancing and nonenhancing lesions from the patients with grade 3 lesions, with values of 1.59 and 1.67. These are considerably higher than the median values that we and others have reported in the literature for grade 4 lesions (typically 1.4–1.5) [44] and are in the same range that we have observed for grade 2 lesions (1.6 for oligodendrogliomas and of 2.0 for astrocytomas) [45]. The interpretation of these differences is complex because there have been a number of reports in the literature that there are inverse correlations between ADC and markers of tumor proliferation for brain tumors, including of Ki-67 labeling index [46], cell density [47], tumor cellularity [48], and malignancy [33]. When these studies are examined in the composition of the populations of patients being considered, it becomes clear that the results cannot be considered representative for grade 3 glioma because there were only 2 of 47 [46], 2 of 18 [47], 0 of 56 [48], and 6 of 59 [33] of the patients with this diagnosis. In a recent study that focused specifically on patients with grade 2 and 3 glioma, a positive rather than a negative correlation between ADC and total tumor cell number was reported for both types of lesions [49]. In our study of grade 4 glioma, the lower 10th percentile of ADC values and larger areas of nADC < 1.5 were both associated with a poor survival [44], which might indicate an increase in tumor burden. The median nADC levels in the contrast enhancing and T2 hyperintense areas of grade 3 patients were both found to be higher than 1.5 in our study, indicating smaller nADC < 1.5 volumes, that may contribute to the improved survival, which is typically observed in such patients. Our study is consistent with the diffusion properties in grade 3 glioma being more similar to those in lower-grade lesions and with the processes determining the observed levels of nADC and nFA being associated with demyelination, edema, and disruption of normal brain as opposed to being direct measures of tumor cell density.
There were 21 (41%) of the 51 patients in our study with areas of low to moderate contrast enhancement. These enhancing lesions were relatively small in volume and resided mainly within the metabolically abnormal regions. This is consistent with the findings of Pirzkall et al. [19], in which the enhancing lesion had the highest median nCho and the lowest median nNAA. This was not the case for grade 4 lesions, where the highest Cho levels were observed in the nonenhancing portion of the tumor [44]. This may be due to there being more necrosis in the contrast enhancement for grade 4 tumors, which would reduce overall tumor cellularity. The fact that the median nADC was significantly higher and the median nFA was significantly lower in the enhancement of grade 3 gliomas compared with NAWM may suggest that there was breakdown of the normal tissue architecture in these regions that allowed for more rapid diffusion. The high median LL/nNAA and low median nCr levels suggest that there was a tendency toward the enhancing regions being hypoxic, which may also be a factor in inducing the breakdown of the blood-brain barrier. Overall, these results suggest that, when present, the regions of enhancement are the most malignant portion of untreated grade 3 glioma and provide an appropriate target for directing biopsy or surgical sampling for diagnostic purposes.
The other anatomic region studied was the portion of the T2 hyperintensity that was nonenhancing. For 30 (59%) of 51 patients, this comprised the entire lesion. There were highly variable median levels of Cho in these regions, with 16 being lower and 35 being higher than the levels in NAWM. Although there were variations in median levels of NAA, these were all lower than the corresponding values in NAWM. Differences in the normalized metabolite levels reflect the heterogeneity in lesion characteristics between patients and underline the motivation for using the CNI index, which defines whether a spectrum is abnormal by combining the values of nCho and nNAA. The fact that the LL/nNAA was higher in both the nonenhancing and enhancing regions than the value in NAWM is consistent with the presence of regions with hypoxia. The relatively large increase in median nADC and decrease in median nFA in the nonenhancing regions may be attributed to a combination of demyelination, inflammation, edema, and tumor infiltration.
If variations in levels of CNI are used to define regions of abnormality, it is possible to obtain further insight into the relationships between metabolite and diffusion parameters. The three regions considered in this study represented areas that were mildly abnormal (CNI2–3), moderately abnormal (CNI3–4), and very abnormal (CNI > 4). In the mildly abnormal region, the Cho was higher than normal, the Cr was slightly reduced, the NAA was almost half of the normal value, and the LL/nNAA was significantly increased over normal. The reduction in nNAA was inversely correlated with nADC, which was increased to 1.47 and the nFA was decreased to 0.54. This suggests that the region included some tumor infiltration but that the changes in MR parameters were dominated by reduction in neuronal function and disruption of tissue architecture. In the moderately abnormal region, the Cho was significantly higher than normal but the Cr was similar to the values in the mildly abnormal region. The NAA was further reduced, the LL/nNAA further increased, the nADC further increased and the nFA further decreased. This is consistent with the presence of a larger number of tumor cells, together with a continued and more extensive disruption of normal tissue.
In the highly abnormal region, there were further significant increases in the median values of nCho, nCr, LL/nNAA, and nADC and a continued decrease in the median values of nFA and nNAA relative to the moderately abnormal region. This suggests that the highly abnormal region included a relatively large number of tumor cells with a higher likelihood of there being areas with reduced oxygenation. Taking into consideration the variations in metabolite and diffusion parameters, it seems that, for patients with grade 3 glioma, the area of the tumor likely to be the most malignant is at the location of the maximum CNI, irrespective of whether the lesion is enhancing or not. This is consistent with a recent study in non-enhancing grade 2 and grade 3 gliomas, which investigated correlations between spectroscopic parameters and histologic measures, and reported that both the ratio of Cho/NAA and the CNI correlated with MIB-1 proliferative index, cell density, and the ratio of proliferation to cell death [50].
In the 30 patients who received lactate-edited spectroscopy, the region with highly abnormal metabolism was observed to have the highest level of lactate. This suggests that these regions exhibit anaerobic glycolysis and are poorly oxygenated. The regions with mild and moderate metabolic abnormalities and the nonenhancing lesion also had lactate levels higher than NAWM, indicating poor oxygenation even in areas of relatively lower tumor invasion. This is consistent with recent studies from our group, which have noted voxels with lactate and lipid peaks in some patients with grade 3 glioma [31] and reported the presence of significant lactate and lipid peaks with volumes of these regions varying from 0.1 to 11.8 ml [15]. The findings from this study suggest that the MRSI data may be valuable in predicting sensitivity to radiation therapy by delineating such poorly oxygenated regions.
Some of the diffusion and metabolic imaging patterns were different for subtypes of grade 3 gliomas than had previously been reported for subtypes of treatment-naive grade 2 gliomas. In particular, it was found that the ADC values were similar for all 3 subtypes of grade 3 gliomas, whereas they were significantly higher for grade 2 astrocytomas and oligoastrocytomas compared with grade 2 oligodendrogliomas [45]. The higher lipid values in subregions of OD3 compared with the other two subtypes of grade 3 glioma is also a novel finding and may indicate a higher tendency to cellular breakdown and the formation of necrosis than for other histologic subtypes.
Overall, the results of the current study suggest that MRSI and diffusion parameters provide complementary information that may be valuable for characterizing nonenhancing regions of the anatomic lesion and possibly directing tissue sampling for accurate diagnosis of tumor grade. This is especially important for distinguishing patients with grade 3 glioma from those with lower-grade lesions. Regions of infiltrative tumor were differentiated by their increased water content (high nADC), altered normal tissue function (high Cho, low NAA), and structure (low nFA). Regions of macroscopic tumor could be identified by high cellularity (high CNI) and major disruptions in the tissue structure and function (low nFA and low NAA) and hypoxia (high Lac or LL/nNAA). These signatures may be of interest not only for directing tissue sampling but also for defining the targets for focal therapy and assessing response to therapy. Future studies that compare presurgery MRSI and diffusion parameters with the findings from image-guided tissue samples would be valuable for validating these biomarkers as clinical tools for managing patients with grade 3 glioma.
Footnotes
This study was partially supported by UC Discovery grants that were funded in conjunction with GE Healthcare and National Institutes of Health grants R01 CA059880 and P50 CA97257.
References
- 1.American Cancer Society, author. Cancer Facts and Figures. 2010. [August 18, 2010]. Available at: http://www.cancer.org/acs/groups/content/@epidemiologysurveilance/documents/document/acspc-026238.pdf.
- 2.Bauman G, Lote K, Larson D, Stalpers L, Leighton C, Fisher B, Wara W, MacDonald D, Stitt L, Cairncross JG. Pretreatment factors predict overall survival for patients with low-grade glioma: a recursive partitioning analysis. Int J Radiat Oncol Biol Phys. 1999;45:923–929. doi: 10.1016/s0360-3016(99)00284-9. [DOI] [PubMed] [Google Scholar]
- 3.Dehais C, Laigle-Donadey F, Marie Y, Kujas M, Lejeune J, Benouaich-Amiel A, Pedretti M, Polivka M, Xuan KH, Thillet J, et al. Prognostic stratification of patients with anaplastic gliomas according to genetic profile. Cancer. 2006;107:1891–1897. doi: 10.1002/cncr.22211. [DOI] [PubMed] [Google Scholar]
- 4.Laws ER, Parney IF, Huang W, Anderson F, Morris AM, Asher A, Lillehei KO, Bernstein M, Brem H, Sloan A, et al. Survival following surgery and prognostic factors for recently diagnosed malignant glioma: data from the Glioma Outcomes Project. J Neurosurg. 2003;99:467–473. doi: 10.3171/jns.2003.99.3.0467. [DOI] [PubMed] [Google Scholar]
- 5.Devos A, Simonetti AW, van der Graaf M, Lukas L, Suykens JA, Vanhamme L, Buydens LM, Heerschap A, Van Huffel S. The use of multivariate MR imaging intensities versus metabolic data from MR spectroscopic imaging for brain tumour classification. J Magn Reson. 2005;173:218–228. doi: 10.1016/j.jmr.2004.12.007. [DOI] [PubMed] [Google Scholar]
- 6.Stadlbauer A, Moser E, Gruber S, Buslei R, Nimsky C, Fahlbusch R, Ganslandt O. Improved delineation of brain tumors: an automated method for segmentation based on pathologic changes of 1H-MRSI metabolites in gliomas. Neuroimage. 2004;23:454–461. doi: 10.1016/j.neuroimage.2004.06.022. [DOI] [PubMed] [Google Scholar]
- 7.Sibtain NA, Howe FA, Saunders DE. The clinical value of proton magnetic resonance spectroscopy in adult brain tumours. Clin Radiol. 2007;62:109–119. doi: 10.1016/j.crad.2006.09.012. [DOI] [PubMed] [Google Scholar]
- 8.Moller-Hartmann W, Herminghaus S, Krings T, Marquardt G, Lanfermann H, Pilatus U, Zanella FE. Clinical application of proton magnetic resonance spectroscopy in the diagnosis of intracranial mass lesions. Neuroradiology. 2002;44:371–381. doi: 10.1007/s00234-001-0760-0. [DOI] [PubMed] [Google Scholar]
- 9.Pirzkall A, Li X, Oh J, Chang S, Berger MS, Larson DA, Verhey LJ, Dillon WP, Nelson SJ. 3D MRSI for resected high-grade gliomas before RT: tumor extent according to metabolic activity in relation to MRI. Int J Radiat Oncol Biol Phys. 2004;59:126–137. doi: 10.1016/j.ijrobp.2003.08.023. [DOI] [PubMed] [Google Scholar]
- 10.Nelson SJ. Multivoxel magnetic resonance spectroscopy of brain tumors. Mol Cancer Ther. 2003;2:497–507. [PubMed] [Google Scholar]
- 11.McKnight TR, von dem Bussche MH, Vigneron DB, Lu Y, Berger MS, McDermott MW, Dillon WP, Graves EE, Pirzkall A, Nelson SJ. Histopathological validation of a three-dimensional magnetic resonance spectroscopy index as a predictor of tumor presence. J Neurosurg. 2002;97:794–802. doi: 10.3171/jns.2002.97.4.0794. [DOI] [PubMed] [Google Scholar]
- 12.Toyooka M, Kimura H, Uematsu H, Kawamura Y, Takeuchi H, Itoh H. Tissue characterization of glioma by proton magnetic resonance spectroscopy and perfusion-weighted magnetic resonance imaging: glioma grading and histological correlation. Clin Imaging. 2008;32:251–258. doi: 10.1016/j.clinimag.2007.12.006. [DOI] [PubMed] [Google Scholar]
- 13.Howe FA, Barton SJ, Cudlip SA, Stubbs M, Saunders DE, Murphy M, Wilkins P, Opstad KS, Doyle VL, McLean MA, et al. Metabolic profiles of human brain tumors using quantitative in vivo 1H magnetic resonance spectroscopy. Magn Reson Med. 2003;49:223–232. doi: 10.1002/mrm.10367. [DOI] [PubMed] [Google Scholar]
- 14.Dowling C, Bollen AW, Noworolski SM, McDermott MW, Barbaro NM, Day MR, Henry RG, Chang SM, Dillon WP, Nelson SJ, et al. Preoperative proton MR spectroscopic imaging of brain tumors: correlation with histopathologic analysis of resection specimens. AJNR Am J Neuroradiol. 2001;22:604–612. [PMC free article] [PubMed] [Google Scholar]
- 15.Li X, Lu Y, Pirzkall A, McKnight T, Nelson SJ. Analysis of the spatial characteristics of metabolic abnormalities in newly diagnosed glioma patients. J Magn Reson Imaging. 2002;16:229–237. doi: 10.1002/jmri.10147. [DOI] [PubMed] [Google Scholar]
- 16.Weber MA, Giesel FL, Stieltjes B. MRI for identification of progression in brain tumors: from morphology to function. Expert Rev Neurother. 2008;8:1507–1525. doi: 10.1586/14737175.8.10.1507. [DOI] [PubMed] [Google Scholar]
- 17.Graves EE, Nelson SJ, Vigneron DB, Verhey L, McDermott M, Larson D, Chang S, Prados MD, Dillon WP. Serial proton MR spectroscopic imaging of recurrent malignant gliomas after gamma knife radiosurgery. AJNR Am J Neuroradiol. 2001;22:613–624. [PMC free article] [PubMed] [Google Scholar]
- 18.Li X, Jin H, Lu Y, Oh J, Chang S, Nelson SJ. Identification of MRI and 1H MRSI parameters that may predict survival for patients with malignant gliomas. NMR Biomed. 2004;17:10–20. doi: 10.1002/nbm.858. [DOI] [PubMed] [Google Scholar]
- 19.Pirzkall A, McKnight TR, Graves EE, Carol MP, Sneed PK, Wara WW, Nelson SJ, Verhey LJ, Larson DA. MR-spectroscopy guided target delineation for high-grade gliomas. Int J Radiat Oncol Biol Phys. 2001;50:915–928. doi: 10.1016/s0360-3016(01)01548-6. [DOI] [PubMed] [Google Scholar]
- 20.Lin A, Bluml S, Mamelak AN. Efficacy of proton magnetic resonance spectroscopy in clinical decision making for patients with suspected malignant brain tumors. J Neurooncol. 1999;45:69–81. doi: 10.1023/a:1006387703127. [DOI] [PubMed] [Google Scholar]
- 21.Pirzkall A, Nelson SJ, McKnight TR, Takahashi MM, Li X, Graves EE, Verhey LJ, Wara WW, Larson DA, Sneed PK. Metabolic imaging of low-grade gliomas with three-dimensional magnetic resonance spectroscopy. Int J Radiat Oncol Biol Phys. 2002;53:1254–1264. doi: 10.1016/s0360-3016(02)02869-9. [DOI] [PubMed] [Google Scholar]
- 22.Nelson SJ, Graves E, Pirzkall A, Li X, Antiniw Chan A, Vigneron DB, McKnight TR. In vivo molecular imaging for planning radiation therapy of gliomas: an application of 1H MRSI. J Magn Reson Imaging. 2002;16:464–476. doi: 10.1002/jmri.10183. [DOI] [PubMed] [Google Scholar]
- 23.Chan AA, Lau A, Pirzkall A, Chang SM, Verhey LJ, Larson D, McDermott MW, Dillon WP, Nelson SJ. Proton magnetic resonance spectroscopy imaging in the evaluation of patients undergoing gamma knife surgery for grade IV glioma. J Neurosurg. 2004;101:467–475. doi: 10.3171/jns.2004.101.3.0467. [DOI] [PubMed] [Google Scholar]
- 24.Nelson SJ, Huhn S, Vigneron DB, Day MR, Wald LL, Prados M, Chang S, Gutin PH, Sneed PK, Verhey L, et al. Volume MRI and MRSI techniques for the quantitation of treatment response in brain tumors: presentation of a detailed case study. J Magn Reson Imaging. 1997;7:1146–1152. doi: 10.1002/jmri.1880070630. [DOI] [PubMed] [Google Scholar]
- 25.Alexander A, Murtha A, Abdulkarim B, Mehta V, Wheatley M, Murray B, Riauka T, Hanson J, Fulton D, McEwan A, et al. Prognostic significance of serial magnetic resonance spectroscopies over the course of radiation therapy for patients with malignant glioma. Clin Invest Med. 2006;29:301–311. [PubMed] [Google Scholar]
- 26.Howe FA, Opstad KS. 1H MR spectroscopy of brain tumours and masses. NMR Biomed. 2003;16:123–131. doi: 10.1002/nbm.822. [DOI] [PubMed] [Google Scholar]
- 27.Wald LL, Nelson SJ, Day MR, Noworolski SE, Henry RG, Huhn SL, Chang S, Prados MD, Sneed PK, Larson DA, et al. Serial proton magnetic resonance spectroscopy imaging of glioblastoma multiforme after brachytherapy. J Neurosurg. 1997;87:525–534. doi: 10.3171/jns.1997.87.4.0525. [DOI] [PubMed] [Google Scholar]
- 28.Nelson SJ. Imaging of brain tumors after therapy. Neuroimaging Clin N Am. 1999;9:801–819. [PubMed] [Google Scholar]
- 29.Nelson SJ, Vigneron DB, Dillon WP. Serial evaluation of patients with brain tumors using volume MRI and 3D 1H MRSI. NMR Biomed. 1999;12:123–138. doi: 10.1002/(sici)1099-1492(199905)12:3<123::aid-nbm541>3.0.co;2-y. [DOI] [PubMed] [Google Scholar]
- 30.Catalaa I, Henry R, Dillon WP, Graves EE, McKnight TR, Lu Y, Vigneron DB, Nelson SJ. Perfusion, diffusion and spectroscopy values in newly diagnosed cerebral gliomas. NMR Biomed. 2006;19:463–475. doi: 10.1002/nbm.1059. [DOI] [PubMed] [Google Scholar]
- 31.Nelson SJ, McKnight TR, Henry RG. Characterization of untreated gliomas by magnetic resonance spectroscopic imaging. Neuroimaging Clin N Am. 2002;12:599–613. doi: 10.1016/s1052-5149(02)00037-0. [DOI] [PubMed] [Google Scholar]
- 32.Oh J, Cha S, Aiken AH, Han ET, Crane JC, Stainsby JA, Wright GA, Dillon WP, Nelson SJ. Quantitative apparent diffusion coefficients and T2 relaxation times in characterizing contrast enhancing brain tumors and regions of peritumoral edema. J Magn Reson Imaging. 2005;21:701–708. doi: 10.1002/jmri.20335. [DOI] [PubMed] [Google Scholar]
- 33.Bulakbasi N, Guvenc I, Onguru O, Erdogan E, Tayfun C, Ucoz T. The added value of the apparent diffusion coefficient calculation to magnetic resonance imaging in the differentiation and grading of malignant brain tumors. J Comput Assist Tomogr. 2004;28:735–746. doi: 10.1097/00004728-200411000-00003. [DOI] [PubMed] [Google Scholar]
- 34.Provenzale JM, McGraw P, Mhatre P, Guo AC, Delong D. Peritumoral brain regions in gliomas and meningiomas: investigation with isotropic diffusion-weighted MR imaging and diffusion-tensor MR imaging. Radiology. 2004;232:451–460. doi: 10.1148/radiol.2322030959. [DOI] [PubMed] [Google Scholar]
- 35.Tsuchiya K, Fujikawa A, Nakajima M, Honya K. Differentiation between solitary brain metastasis and high-grade glioma by diffusion tensor imaging. Br J Radiol. 2005;78:533–537. doi: 10.1259/bjr/68749637. [DOI] [PubMed] [Google Scholar]
- 36.Goebell E, Fiehler J, Ding XQ, Paustenbach S, Nietz S, Heese O, Kucinski T, Hagel C, Westphal M, Zeumer H. Disarrangement of fiber tracts and decline of neuronal density correlate in glioma patients—a combined diffusion tensor imaging and 1H-MR spectroscopy study. AJNR Am J Neuroradiol. 2006;27:1426–1431. [PMC free article] [PubMed] [Google Scholar]
- 37.Bottomley PA. Spatial localization in NMR spectroscopy in vivo. Ann N Y Acad Sci. 1987;508:333–348. doi: 10.1111/j.1749-6632.1987.tb32915.x. [DOI] [PubMed] [Google Scholar]
- 38.Star-Lack J, Vigneron DB, Pauly J, Kurhanewicz J, Nelson SJ. Improved solvent suppression and increased spatial excitation bandwidths for three-dimensional PRESS CSI using phase-compensating spectral/spatial spin-echo pulses. J Magn Reson Imaging. 1997;7:745–757. doi: 10.1002/jmri.1880070422. [DOI] [PubMed] [Google Scholar]
- 39.Tran TK, Vigneron DB, Sailasuta N, Tropp J, Le Roux P, Kurhanewicz J, Nelson S, Hurd R. Very selective suppression pulses for clinical MRSI studies of brain and prostate cancer. Magn Reson Med. 2000;43:23–33. doi: 10.1002/(sici)1522-2594(200001)43:1<23::aid-mrm4>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- 40.Star-Lack J, Spielman D, Adalsteinsson E, Kurhanewicz J, Terris DJ, Vigneron DB. In vivo lactate editing with simultaneous detection of choline, creatine, NAA, and lipid singlets at 1.5 T using PRESS excitation with applications to the study of brain and head and neck tumors. J Magn Reson. 1998;133:243–254. doi: 10.1006/jmre.1998.1458. [DOI] [PubMed] [Google Scholar]
- 41.Nelson SJ. Analysis of volume MRI and MR spectroscopic imaging data for the evaluation of patients with brain tumors. Magn Reson Med. 2001;46:228–239. doi: 10.1002/mrm.1183. [DOI] [PubMed] [Google Scholar]
- 42.McKnight TR, Noworolski SM, Vigneron DB, Nelson SJ. An automated technique for the quantitative assessment of 3D-MRSI data from patients with glioma. J Magn Reson Imaging. 2001;13:167–177. doi: 10.1002/1522-2586(200102)13:2<167::aid-jmri1026>3.0.co;2-k. [DOI] [PubMed] [Google Scholar]
- 43.Basser PJ, Pierpaoli C. A simplified method to measure the diffusion tensor from seven MR images. Magn Reson Med. 1998;39:928–934. doi: 10.1002/mrm.1910390610. [DOI] [PubMed] [Google Scholar]
- 44.Crawford FW, Khayal IS, McGue C, Saraswathy S, Pirzkall A, Cha S, Lamborn KR, Chang SM, Berger MS, Nelson SJ. Relationship of pre-surgery metabolic and physiological MR imaging parameters to survival for patients with untreated GBM. J Neurooncol. 2009;91:337–351. doi: 10.1007/s11060-008-9719-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bian W, Khayal IS, Lupo JM, McGue C, Vandenberg S, Lamborn KR, Chang SM, Cha S, Nelson SJ. Multiparametric characterization of grade 2 glioma subtypes using magnetic resonance spectroscopic, perfusion, and diffusion imaging. Transl Oncol. 2009;2:271–280. doi: 10.1593/tlo.09178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Calvar JA, Meli FJ, Romero C, Calcagno ML, Yanez P, Martinez AR, Lambre H, Taratuto AL, Sevlever G. Characterization of brain tumors by MRS, DWI and Ki-67 labeling index. J Neurooncol. 2005;72:273–280. doi: 10.1007/s11060-004-3342-2. [DOI] [PubMed] [Google Scholar]
- 47.Gupta RK, Cloughesy TF, Sinha U, Garakian J, Lazareff J, Rubino G, Rubino L, Becker DP, Vinters HV, Alger JR. Relationships between choline magnetic resonance spectroscopy, apparent diffusion coefficient and quantitative histopathology in human glioma. J Neurooncol. 2000;50:215–226. doi: 10.1023/a:1006431120031. [DOI] [PubMed] [Google Scholar]
- 48.Kono K, Inoue Y, Nakayama K, Shakudo M, Morino M, Ohata K, Wakasa K, Yamada R. The role of diffusion-weighted imaging in patients with brain tumors. AJNR Am J Neuroradiol. 2001;22:1081–1088. [PMC free article] [PubMed] [Google Scholar]
- 49.Stadlbauer A, Ganslandt O, Buslei R, Hammen T, Gruber S, Moser E, Buchfelder M, Salomonowitz E, Nimsky C. Gliomas: histopathologic evaluation of changes in directionality and magnitude of water diffusion at diffusion-tensor MR imaging. Radiology. 2006;240:803–810. doi: 10.1148/radiol.2403050937. [DOI] [PubMed] [Google Scholar]
- 50.McKnight T, Love T, Lamborn K, Berger M, Chang S, Dillon W, Bollen A, Nelson S. Correlation of MR spectroscopic and growth characteristics of grades II and III glioma. J Neurosurg. 2006;106(4):660–666. doi: 10.3171/jns.2007.106.4.660. [DOI] [PubMed] [Google Scholar]