Abstract
The product of the SALL2 protein p150Sal2 is a multi-zinc finger transcription factor with growth-arrest and pro-apoptotic functions that overlap those of p53. Its DNA binding properties are unknown. We have used a modified SELEX procedure with purified p150Sal2 and a pool of oligonucleotides of random sequence to identify those that are bound preferentially by p150Sal2. The consensus sequence for optimal binding in vitro is GGG(T/C)GGG, placing p150Sal2 among a large group of GC box-binding proteins including the Sp1 family of transcription factors. A triple zinc finger motif in p150Sal2 similar to that in Sp1 is required for DNA binding. p150Sal2 and Sp1 show evidence of co-operative binding in vitro and of interaction in vivo. p150Sal2, a known activator of the CDK inhibitor p21Cip1/Waf1 (p21), binds to regions of the human p21 promoter that contain variations of the consensus sequence in multiple copies. p150Sal2 is also shown to bind to the BAX promoter with similar elements and to activate its expression following an apoptotic stimulus. These results demonstrate binding of p150Sal2 to two natural promoters with GC elements related to the optimal binding sequence defined in vitro and whose regulation is important for suppression of tumor growth.
Keywords: SALL2 transcription factor, DNA binding, growth arrest, apoptosis, polyoma virus
1 INTRODUCTION
p150Sal2 is the product of the Sall2 gene, a member of the SALL (‘spalt-like’) gene family which encodes multi-zinc finger transcription factors. Orthologues of the homeotic gene Spalt in Drosophila [1], SALL genes are conserved from flies to man. SALL genes function in embryonic development in vertebrate as well as invertebrate species [2]. Sall4 is an important transcriptional regulator in mouse embryonic stem cells [3-5] and is essential for maintaining a pluripotent state ([6]. Sall4 also regulates growth and survival in human leukemic cells [7]. Sall2 is the only member of the family suggested to act as a tumor suppressor [8, 9]. Sall2 plays a role in neuronal development, affecting neurite outgrowth [10] and, along with Sall1 and Sall4, neural tube closure in mice [11]. Naturally occurring mutations in these genes give rise to developmental abnormalities in man [12-15]. The roles of SALL genes in development are not well understood at a molecular level.
p150Sal2 was first identified in the mouse as a binding target of the polyoma virus large T (tumor) antigen. This was done using a procedure designed to identify tumor suppressors or other cellular factors with which the virus must interact in order to replicate efficiently and which may undergo spontaneous loss or alteration in certain cancer cells [8]. p150Sal2 acts in some manner to inhibit DNA replication by the virus and the binding of p150Sal2 by large T overcomes this inhibition. A virus mutant unable to bind p150Sal2 is unable to replicate or induce tumors broadly in the mouse [8].
Though strongly expressed in the normal ovary, p150Sal2 is not expressed in some human ovarian carcinoma-derived cell lines. Restoration of expression in such tumor cells results in suppression of growth in SCID mice. Tumor suppression is accompanied by both a rise in apoptotic index and a fall in mitotic index. p150Sal2 binds within the extended promoter of p21Cip1/Waf1 (p21) and transactivates p21 in the absence of p53 [16]. Independent studies have shown that SALL2 expression is required for human fibroblasts to exit the cell cycle and to maintain a quiescent state under conditions of serum deprivation. Downregulation of SALL2 prevents G1 arrest in serum-deprived cells [17]. These observations establish p150Sal2 as a negative regulator of cell growth with functions in postnatal as well as embryonic tissues. They also suggest that it may act as a tumor suppressor based on its ability to regulate cell growth and survival.
Knowledge of the DNA-binding properties of p150Sal2 is presently lacking but essential to understanding its regulatory functions. A triple zinc finger motif in p150Sal2 is required for binding and transactivation of the p21 promoter [16] but the sequence specificity of binding is not known. The current investigation was undertaken to determine the DNA binding specificity of p150Sal2 by identifying the optimal DNA sequence for binding in vitro and to demonstrate that related sequence elements are present in natural promoters regulated by the endogenous protein. A modified SELEX procedure has been used to establish p150Sal2 as a GC-box-binding protein. Evidence is presented that p150Sal2 binds to GC-rich elements in the human p21 and BAX promoters consistent with its growth arrest and pro-apoptotic functions and ability to transactivate these genes.
2 MATERIAL AND METHODS
2. 1 Cell lines and plasmids
293 (human embryonic kidney cells) and HOSE (established human ovarian surface epithelial cells) [16] were cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum at 37 °C with 5% CO2. p150Sal2 and deletion mutants Z3D (deletion of amino acids of 631-711) and Z4D (deletion of amino acids of 911-956) have been reported previously [8]. Z1D (amino acids 35-55) and Z2D (amino acids 373-421) were made using QuikChange site directed mutagenesis kit (Stratagene) and cloning into the pEBG GST expression vector [18]. The G6TI luciferase vector containing 6 GC-boxes [19, 20] was a gift from Dr. G. Gill (Tufts University). The renilla luciferase vector (pTL-Renilla) was constructed by replacing the firefly luciferase gene in pTL (Panomics) with renilla luciferase from phRluc/SV40 (Promega) using the Hind III and Xba I sites.
2. 2 GST fusion protein purification
GST, GST-tagged p150Sal2 or zinc finger deletion protein expression vector was transfected into 293 cells using Lipofectamine 2000 (Invitrogen) and GST-fusion protein purification was performed using GST beads (Amersham). After elution, proteins for EMSA were dialyzed in HEPES (pH 7.9) buffer [21] and concentrated using Ultracel YM-10 centricon (Millipore). Proteins were quantified using the Bio-Rad Protein Assay and by SDS-PAGE electrophoresis and Coomassie Blue staining.
2. 3 Random sequence DNA library synthesis
A random pool of DNA [5’-ATGTGGATCCACTGACGG(N)20GCTACGCCTCGAGATTG- 3’] and two PCR primer sequences corresponding to the first 18 nucleotides (SELEX primer 1, 5’-ATGTGGATCCACTGACGG-3’) and complementary to the last 17 bases (SELEX primer 2, 5’-CAATCTCGAGGCGTAGC-3’) were purchased from Integrated DNA Technologies (IDT). The random DNA library was generated using 10 units Klenow fragment (New England Biolabs) in a 40 μl reaction containing 100 pmol DNA pool and 1 nM SELEX primer 2, as per the manufacturer’s protocol. Following a 30 min incubation at 25 °C, the reaction was stopped by heating at 75 °C for 20 min.
2. 4 SELEX procedure
The SELEX protocol was modified from [22]. Briefly, 4 pmol random DNA library and 1 pmol glutathione bead-bound GST-p150Sal2 were incubated for 30 min at room temperature in 1x binding buffer (20mM HEPES, pH. 7.5, 50mM KCl, 1mM dithiothreitol, 3 mM MgCl, 1mM EDTA, 5% glycerol and 0.5% NP-40). The beads were pelleted (5 min, 2000g) and washed four times with 1 ml of 1x binding buffer. The final pellet was suspended in 45 μl of Platinum PCR SuperMix (Invitrogen) and subjected to 20 cycles of PCR amplification (as per manufacturer’s protocol) using 125 ng each of SELEX primer 1 and 2. The resulting PCR products were purified using MicroSpin G-50 columns (Amersham biosciences) and subjected to the next cycle of SELEX. Seven additional cycles of SELEX were then performed and the final DNA fragments cloned into pGEM-T Easy vector (Promega) following the manufacturer’s protocol. Twenty-four individual clones were sequenced.
The random DNA library and PCR product from the last SELEX cycle were end-labeled with [α-32P]-dATP using T4 Polynucleotide Kinase (Invitrogen) as per the manufacturer’s protocol. Single strand oligonucleotides containing the Consensus Sequence (CS) 5’-GGATCACTGGGTGGGAATCACGCT-3’ Sp1 binding site [23] (5’-ATTCGATCGGGGCGGGGCGAGC-3’), Oct1 binding site [24] (5’-TGTCGAATGCAAATCACTAGAA-3’, appropriate mutants, and their complimentary sequences, were purchased from IDT. Double stranded DNA was generated by annealing complimentary oligonucleotides (buffer: 10 mM Tris pH. 8.0, 1mM EDTA and 50 mM NaCl). The DNA was radiolabeled either by T4 PNK as described above, or obtained from IDT with two additional Gs at their 5’ termini to form an overhang, thus allowing Klenow Fragment (Invitrogen) fill-in reaction with [α-32P] dCTP. All the probes were purified using MicroSpin G-50 columns (Amersham) following the manufacturer’s instructions
2. 5 EMSA procedure
Electrophoretic mobility shift assays were carried out using purified GT-p150Sal2 as described [21] with modification. Briefly, 100 ng purified GST or GST-p150sal2 and 200,000 cpm radiolabeled probe were incubated in 20 μl reaction (10 mM HEPES pH 7.5, 25 nM KCl, 2.5 mM MgCl, 5 μl ZnCl, 3% glycerol, 2 μg BSA and 200 ng poly dA-dT). For competition studies, 50 or 100 molar excess unlabeled oligonucleotide was added to the mixture. Recombinant human Sp1(rhSp1) protein was purchased from Promega. Supershift was performed by adding 300ng polyclonal antibody to Sp1 (PEP2, Santa Cruz). After 20 min incubation at room temperature, the mixtures were loaded onto 4% acrylamide (60:1 acrylamide: bisacrylamide gel). The oligonucleotides-protein complex was separated by running gels in 0.5X TBE buffer at 180 volts for 2.5 hours. Dried gels were exposed to a PhosphorImager screen (Molecular Dynamics) and individual bands were quantitated with ImageQuant. p21 promoter binding reactions were performed using nuclear extracts [25].
2. 6 Luciferase assays
Hela cells at 90% confluence were cotransfacted with Flag-Sp1 [20] and/or GST-p150Sal2 expression vectors as indicated, and 0.3 μg G6Tl-Luc construct and 20 ng pTL-Renilla control vector. For dosage response experiments empty expression vector was included to normalize the quantity of DNA transfected. Cells were harvested 36 hours after transfection, and 10 μl of lysate was measured for luciferase activity using Promega luciferase assay kit.
2. 7 Immunoprecipitation and pulldown assays
HOSE cells grown to 85 % confluence were harvested by scraping, washed with ice cold PBS, and lysed using 1 ml of NP-40 lysing buffer [26]. 50 μl of 50% Protein A-Sepharose slurry was used to precipitate proteins bound by 3 μg Sp1 polyclonal antibody (Santa Cruz Biotechnology). Protein complexes were washed five times using the lysis buffer. Immnuoprecipitates were collected and analyzed by western blot with antibody against p150Sal2. Hose cells were also transfected with constructs expressing GST-p150Sal2 or GST alone using Lipofectamine 2000. Cells were harvested 36 hours later and protein complexes collected on glutathione beads. Complexes were analyzed by western blot with antibody against Sp1. A flag-tagged Sp1 clone was also used in GST pulldown assays. HOSE cells were transfected with Flag-Sp1 and pEBG or GST-p150Sal2 vectors. GST pull-down and western blot were performed both in the presence and absence of Ethidium Bromide (20μg/ml) or Benzonase nuclease (25 U/ml; Novagen).
2. 8 ChIP assays
ChIP assay was perfomed as described [27] with slight modification. For interaction of p150sal2 with the BAX promoter, HOSE cells were first transfected with pcDNA or pcDNA-p150Sal2 for 48 hours. Interaction of the endogenous protein was also examined in HOSE cells following treatment with 25 ug/ml etoposide for 22 hours to induce apoptosis. Cross-linking was done with 1.4% formaldehyde. Cells were lysed with IP buffer [0.5% Triton X-100, 20 mM Tris, pH 7.5, 2 mM MgCl2, 1 mM dithiothreitol (DTT), 1 mM EGTA, 50 mM β-glycerophosphate, 25 mM NaF, 1 mM Na vanadate, 100 μg/ml PMSF, and protease inhibitor cocktail (Roche; Indianapolis, IN, USA)]. Chromatin was sheared by sonication and incubated with rabbit polyclonal antibody to an N-terminal fragment of p150Sal or normal rabbit IgG. Sheared chromatin was incubated with protein A/G beads (Santa Cruz; CA, USA) for 2 hours, washed five times with IP buffer. Chelex 100 slurry was added to the washed beads. Beads were boiled and incubated with Proteinase K (Invitrogen; Carlsbad, CA, USA) at 55°C for 30 min. Samples were boiled again, cleared by centrifugation and the supernatants taken for real-time PCR. The bound chromatin fraction was amplified with human BAX promoter-specific primers flanking GC boxes at −117 for 40 cycles. Forward primer - 5’-GGCGCCACTGCTGGCACTTA-3’; reverse primer - 5’-CTCCCCGGACCCGTCCATCA-3’. As a control, human Aldolase A gene was amplified with 5’-CGCAGAAGGGGTCCTGGTGA-3’ and 5’-CAGCTCCTTCTTCTGCTCCGGGGT-3’. Real time PCR was carried out on a Roche LightCycler 480 using SYBR Green Master Mix and quantitated as described [27]. The data was analyzed by the comparative CT (ΔΔCT) method and quantitated relative to the aldolase A gene and normalized to the control.
2. 9 Quantitative RT-PCR
Total RNA was isolated using RNeasy® kit (Qiagen), reversed transcribed using QuantiTect Reverse Transcription Kit (Qiagen) and quantitated by RT-PCR using specific primers: BAX - F: 5’-GGAGATGAACTGGATAGCAA-3’ and R: 5’- AGCCACAAAGATGGTCACT-3’, p21 – F: 5’-TCCCGTGGACAGTGAGCAGTTG-3’ and R: 5’- GACACACAGAGTGAGGGCTAAG-3’, and aldolase A the same as above.
3 RESULTS
3. 1 p150Sal2 binds selectively to DNA oligomers containing -GGG(T/C)GGG-
A modified SELEX procedure was used to identify DNA oligonucleotides with highest affinity binding to p150Sal2 (Figure 1A). A GST-p150Sal2 fusion protein containing full length (1005 aa) human p150Sal2 protein with alternative exon E1A [8, 16] was expressed and purified from 293 cells. Purified GST-p150Sal2 (Figure 1B; denoted hereafter in the text as p150Sal2) was incubated with double-stranded deoxyribonucleotides carrying inserts of random sequence twenty nucleotides long. A sufficient quantity of DNA from the library was used initially to allow theoretical representation of all possible 20-mers (4 pmoles, ~2.2X coverage). Bound oligonucleotides were separated from unbound DNA using glutathione beads. Bound DNA was amplified by PCR and subjected to seven additional rounds of selection. Results were analyzed by electrophoretic mobility gel shift assay (EMSA). Selective enrichment of bound sequences after eight rounds is evident from the level of binding (compare Figure 1C, lanes 1 and 3) and from the effects of unlabelled oligonucleotides used as competitor DNAs (Figure 1C, lanes 4, 5 and 6).
Figure 1. p150Sal2 binds selectively to DNA oligomers containing GGG(T/C)GGG.
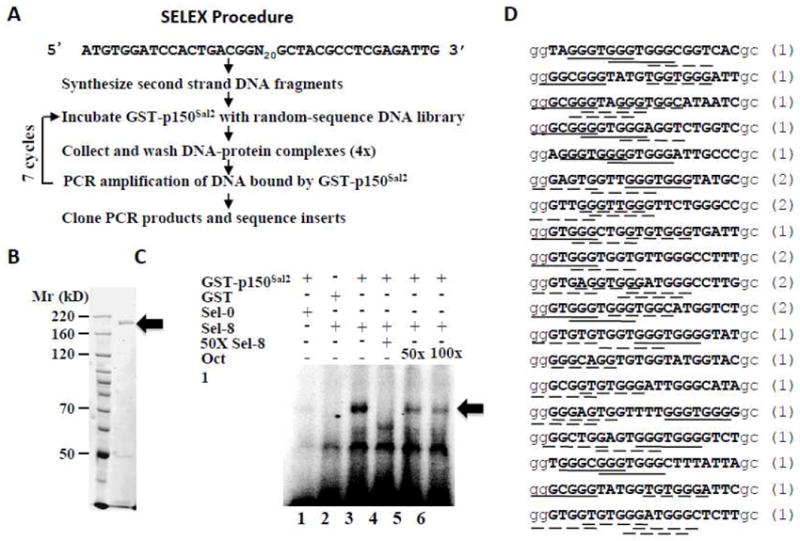
A - Outline of the SELEX protocol. B - Purified GST-p150Sal2 (◂) shown by SDS-PAGE stained with Coomassie Blue. C – EMSA (electromobility shift assay) was performed using 100ng purified GST-p150Sal2 or GST alone. Labeled oligonucleotide libraries before selection (Sel-0, lane 1) and after 8 cycles of selection (Sel-8, lane 2-6) were incubated with either GST (lane 2) or GST-p150Sal2 (lane 1, 3-6) for 20 min. Specific competition was conducted by adding 50 molar excess of unlabeled oligonucleotides from 8 cycles of selection (lane 4). Non-specific competition was performed with 50 and 100 -fold molar excess of unlabeled oligonucleotides containing Oct-1 binding site (lanes 5, 6). GST-p150Sal2 -DNA complexes are indicated (◂). D – Aligned sequences of 24 independent clones after 8 cycles of selection of GST-p150Sal2 binding. Insert sequences are in bold capital letters and vector sequences in small unbolded letters. Numbers in parentheses are the number of occurrences of each sequence. The consensus sequence (-GGGNGGG-) and sequences differing at a single position are indicated by solid and dotted lines, respectively.
Following the final round of selection, twenty-four clones were chosen at random and sequenced. Nineteen unique sequences were represented; five clones were represented twice (Figure 1D). Inspection of these oligonucleotides revealed a heptanucleotide -GGG(T/C)GGG- as a consensus sequence for binding to p150Sal2. The selected oligonucleotides contained at least one and more often multiple heptamer sequences that conformed either to the consensus itself (-GGGNGGG-) or to sequences differing at a single position (solid and dotted underlines, respectively, Figure 1D). In the majority of clones, the GG dinucleotide in the vector immediately 5’ to the inserts contributed to possible binding sequences.
DNA binding specificity was analyzed further using a set of twenty-one oligonucleotides with defined sequences. These contained internal heptanucleotide sequences representing three substitutions at each position of the consensus binding site. Flanking sequences in this set of oligonucleotides lacked Gs immediately adjacent to the heptanucleotides. Oligonucleotides were radiolabeled and evaluated for binding by EMSA and densitometry (Figure 2A). Binding strengths were assessed relative to the consensus sequence –GGGTGGG- (Figure 2B). Although T at position 4 was found most frequently in the set defined by SELEX (Figure 1D), oligonucleotides containing C at this position appeared to bind somewhat more strongly in the gel shift assay. Substitutions for G at the 5’ positions generally showed somewhat greater inhibitory effects than substitutions at the 3’ positions.
Figure 2. Determination of optimal binding sequence for p150Sal2.
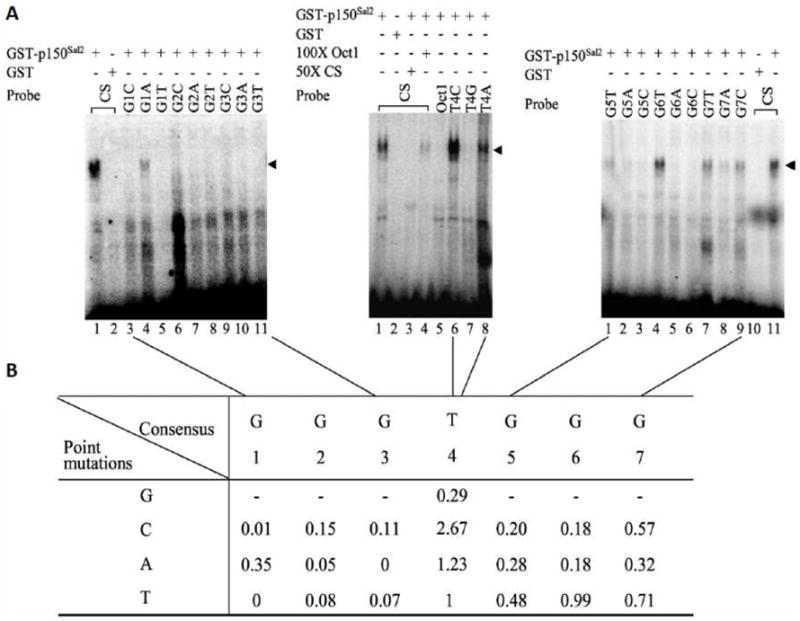
A - EMSA was performed to test the binding efficiency of GST-p150Sal2 towards [α-32P]dCTP-labeled consensus sequence (CS) from SELEX and single base substitutions at each position (left panel, lanes 3-11; middle panel, lanes 6-8; right panel, lanes 1-9). Equal amount (100ng) of GST-p150Sal2 or GST vector was used in the binding reaction. Competition was performed using 50 or 100 fold excess of unlabeled oligonucleotides as specific or non-specific competitor (middle panel, lanes 3 and 4, respectively). B - Summary of binding efficiencies of GST-p150Sal2 to oligonucleotides. Band densities of oligonuleotide-GST-p150Sal2 complexes were quantified using a PhosphorImager and normalized to the SELEX consensus sequence set as 1. GST-p150Sal2-DNA complexes are indicated (◂).
3. 2 p150Sal2 and Sp1 share DNA binding properties and show co-operative interactions
The consensus binding sequence for p150Sal2 is essentially the same as that for Sp1 [28]. p150Sal2 binds as expected to an oligonucleotide carrying a 12 bp GC-rich element known to serve as an Sp1 binding site (5’-ATTCGATCGGGGCGGGGCGAGC-3’) ([23] (Figure 3A, lane 1). Binding was inhibited by an excess of cold oligonucleotide carrying either the Sp1 site or the p150Sal2 consensus site but not by a control oligonucleotide (Oct1) (Figure 3A, lanes 2 thru 4). p150Sal2 carries a putative C2HC zinc finger motif near its N-terminus followed by clusters of two, three and two C2H2 fingers. Plasmids carrying deletions of each set of zinc fingers were constructed as GST fusions and the purified proteins tested for binding to the labeled Sp1 oligonucleotide (Figure 3B). Removal of the triple zinc finger motif greatly reduced binding while removal of the other regions had little or no effect (Figure 3C). This result extends earlier findings on the requirement for the triple zinc finger in p150Sal2 for binding to the p21 promoter ([16]. It also points to similarities between this motif in p150Sal2 and the triple zinc finger in Sp1 required for DNA binding [29, 30] with respect to spacing and linker sequences in the two proteins (Figure 3D). p150Sal2 and Sp1 share little homology outside of the triple zinc finger clusters.
Figure 3. The triple zinc finger cluster in p150Sal2 is essential for DNA binding.
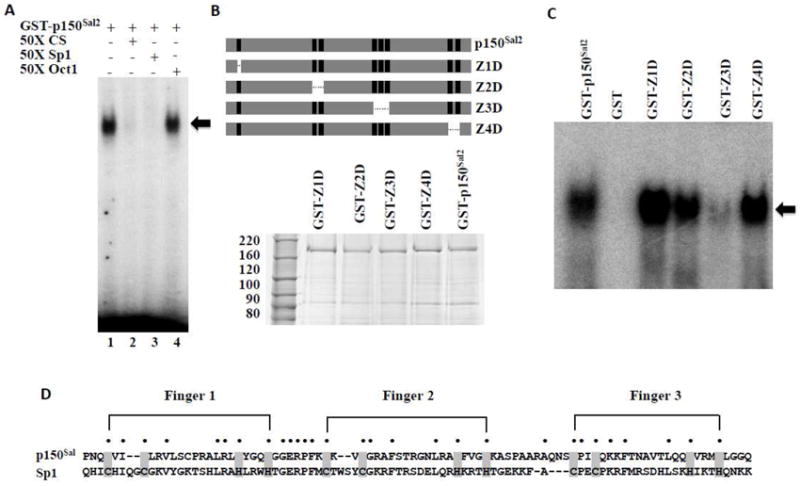
A - Binding of p150Sal2 to an oligonucleotide carrying a confirmed Sp1 binding site (5’-ATTCGATCGGGGCGGGGCGAGC-3’). Competition was conducted by adding 50 fold molar excess of unlabelled oligonucleotides containing the p150Sal2 consensus sequence (5’-GGATCACTGGGTGGGAATCACGCT-3’) (lane 2), Sp1 binding site (lane 3) or Oct 1 binding site (5’-TGTCGAATGCAAATCACTAGAA-3’) (lane 4) in the reaction. DNA-GST-p150Sal2 complexes are indicated (◂). B - Top: Schematic of p150Sal2 illustrating zinc finger deletion mutants. Bottom: Purified GST-p150Sal2 and zinc finger deletion mutants. C - Equal amounts of purified GST-p150Sal2 or mutant proteins were incubated with labelled Sp1 oligonucleotide and complexes analyzed by EMSA (◂). D - Alignment of the triple zinc finger motif in p150Sal2 and Sp1 illustrating similarities of spacing of the C2H2 fingers and linking sequences.
When incubated together with the labeled Sp1 oligonucleotide, p150Sal2 and Sp1 formed DNA-protein complexes efficiently, suggesting possible synergism in binding (Figure 4A). Complexes formed in the presence of both proteins were ‘supershifted’ with antibody to Sp1. Whether both factors make direct contact with a single dodecamer or bind in ‘piggyback’ fashion with only one of the factors making direct contact with the DNA is unclear. To investigate whether p150Sal2 and Sp1 interact functionally, HeLa cells were transfected with an Sp1-luciferase reporter containing 6 ‘GC box’ repeats (G6Tl) [20] to which both factors are expected to bind. Increasing amounts of a p150Sal2 expression vector resulted in a dose-dependent activation of this reporter (Figure 4B – left panel). Introduction of Sp1 and p150Sal2 together led to additive or possibly synergistic activation (Figure 4B - right panel).
Figure 4. DNA binding and activation by p150Sal2 and Sp1.
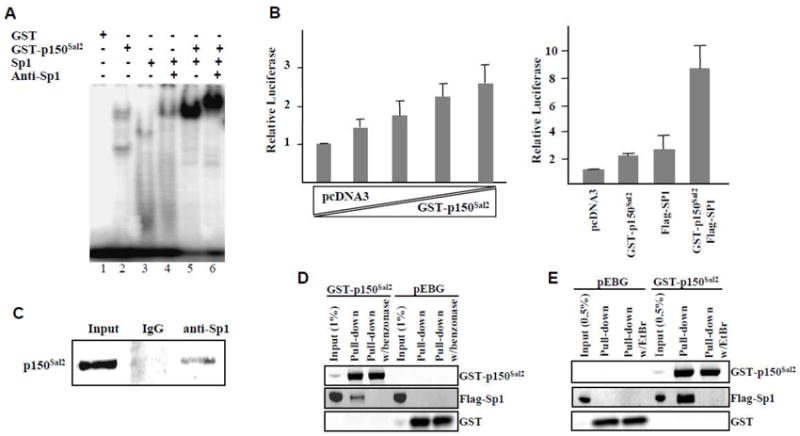
A – Binding of p150Sal2 and Sp1 to end-labeled Sp1 oligonucleotide by EMSA and supershift with anti-Sp1. B – Left: Relative activity of Sp1-luciferase reporter in HeLa cells transfected with increasing amounts of GST- p150Sal2 and decreasing amounts of empty vector. Right: Relative luciferase activity with empty vector, GST-p150Sal2, Flag-Sp1 or both. Vector DNA was added as needed to equalize total amounts of DNA. pTL-Renilla was used to normalize for transfection efficiencies. Results are mean ± .S.D. of three experiments measured 36 hours after transfection. C – Co-immunoprecipitation of endogenous p150Sal2 with anti-Sp1 in HOSE cells and blotting with anti-p150Sal2. D and E – HOSE cells were transfected with Flag-Sp1 and empty GST vector or GST-p150Sal2. Extracts prepared 36 hours post-transfection were analyzed by GST pull-downs either directly (C), after benzonase treatment to digest DNA (D), or after incubation with ethidium bromide (EtBr) to unwind DNA (E).
The apparent synergy between p150Sal2 and Sp1 acting on an artificial promoter raises the possibility of protein-protein interaction and co-operative action in vivo. This possibility was explored using established human ovarian surface epithelial (HOSE) cells which express both factors endogenously. Cell extracts were immunoprecipitated with anti-Sp1 and blotted with anti-p150Sal2 antibody. Results showed evidence consistent with direct interaction between the endogenous proteins (Figure 4C). This result could also have arisen by virtue of both proteins binding to DNA fragments carrying two or more copies of shared or overlapping binding sequences. To test this possibility, HOSE cells were transfected with expression vectors for GST-p150Sal2 and Flag-tagged Sp1 or empty vector controls. Extracts prepared 36 hours post-transfection were treated with benzonase nuclease which has preference for GC-rich sequences [31] or with ethidium bromide to partially unwind DNA [32]. Treated extracts were then analyzed by GST pulldown followed by western blot. No evidence for interaction was seen following these treatments that either digest DNA (Figure 4D) or disrupt protein-protein interaction on DNA (Figure 4E). We conclude that p150Sal2 and Sp1 associate in vivo by binding to DNA with multiple copies of their common or overlapping binding sites. Protein-protein interaction may occur once assembled on DNA. Weak or transient interaction may also occur in the absence of DNA.
3. 3 p150Sal2 binds to regions of the p21 promoter containing GC boxes
p150Sal2 was previously shown to bind to two regions of the long p21 promoter and to transactivate p21 in the absence of p53 [16]. To identify more precisely the locations of p150Sal2 binding sites within the ~2.7 kb p21 promoter and to verify the presence of GC boxes at those sites, the larger fragments previously shown to bind were digested to produce a series of subfragments approximately 140-220 base pairs long. End-labeled subfragments were incubated with nuclear extracts from 293 cells expressing endogenous p150Sal2 and the products analyzed by EMSA (Figure 5). A variable fraction of the subfragments showed evidence of protein binding. The shifts generally gave rise to diffuse bands suggesting binding by multiple factors in the extract. When antibody to p150Sal2 was added, mobility shifts resulted in more focused bands with three of the six subfragments, viz., F1.2, F1.3 and F2.1 (compare lanes 2 with lanes 4 and 5 in Figure 5B and C). The optimal consensus sequence defined in vitro is not found in the p21 promoter. However, each of the subfragments that underwent shift with anti-p150Sal2 contains a GC-box related to the consensus sequence. Specifically, F1.2 contains GGAGGG, F1.3 site GGTCGGG, and F2.1 site GGAGGGG. These results confirm the presence of p150Sal2 binding sequences in a naturally regulated promoter. Similar sequences present elsewhere in the promoter fragments showed no evidence of binding in this assay, presumably due to occupancy by other factors in the nuclear extract.
Figure 5. p150Sal2 binds in vitro to fragments of the p21Cip1/Waf1 promoter containing GC boxes.
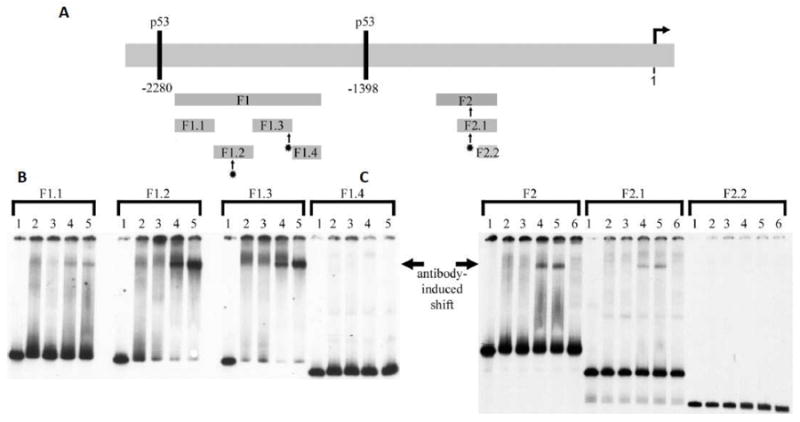
A – Schematic of the ~ 2.6 kb promoter showing fragments F1 and F2 and subfragments generated by Xho and HindIII analyzed by EMSA. B and C – EMSA results showing binding of p150Sal2 to subfragments F1.2, F1.3 and F2.1 as indicated (-). Radioactively labeled subfragments were incubated with nuclear extracts of 293 cells and the products analyzed by EMSA. Antibody to p150Sal2 was used to induce supershifts in subfragments F1.2, F1.3 and F2.1. Lanes 1: DNA probe alone. Lanes 2: probe + nuclear extract. Lanes 3: probe + nuclear extract + antibody to Sp1 (control). Lanes 4: probe + nuclear extract + antibody to N-terminus of p150Sal2. Lanes 5: probe + nuclear extract + antibody to C-terminus of p150Sal2. Lanes 6: probe + nuclear extract + antibody buffer control. Antibody-induced shifts in mobility and focusing of bands are indicated (¢¡). Subfragments F1.2, F1.3 and F2.1 contain GC boxes. [See text].
3. 4 p150Sal2 binds to and transactivates the BAX promoter
Restoration of p150Sal2 in an ovarian carcinoma-derived cell line was previously shown to result in elevated expression of the proapoptotic protein BAX as well as of p21. Suppression of tumor growth by p150Sal2 in these cells was accompanied by a roughly 3 fold increase in the apoptotic index along with a 3 fold reduction in mitoses [16]. The possibility that p150Sal2 binds and directly transactivates the BAX promoter was not examined. The human BAX promoter has two potential p150Sal2 binding sites located at positions −117 and -729 from the translational start site (Figure 6A). To investigate whether p150Sal2 binds to the BAX promoter, HOSE cells were first transfected with p150Sal2 and assayed by ChIP with anti-p150Sal2 antibody. Using primers flanking the proximal GC box at -117, binding of p150Sal2 to the BAX promoter increased 6-fold with no change in the level of binding to the aldolase promoter as control (Figure 6B). qRT-PCR was used to measure levels of BAX mRNA following transfection. Levels were elevated roughly 2.5 fold in transfected versus empty vector control (Figure 6C). Levels of BAX protein increased roughly 1.5 fold (Figure 6D).
Figure 6. p150Sal2 binds to the BAX promoter and upregulates its expression.
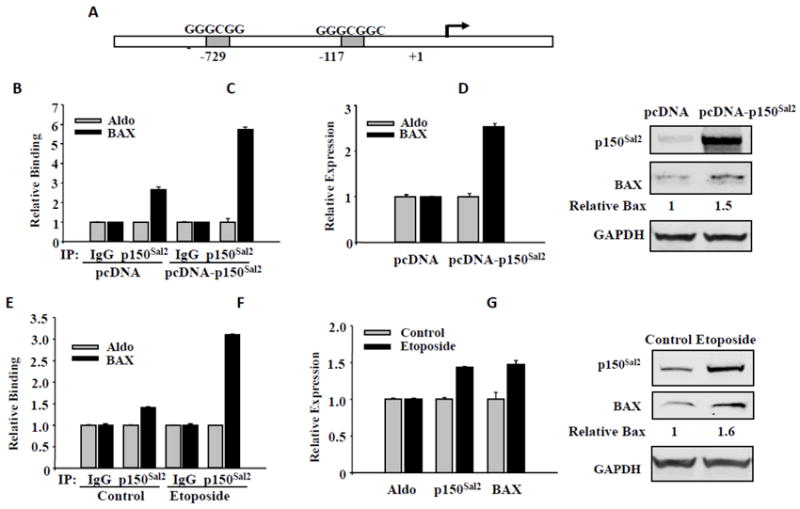
A - Schematic of the human BAX promoter showing GC boxes. B thru D - HOSE cells transfected with pcDNA-p150Sal2 or empty vector for 48 hours. B – ChIP assays for p150Sal2 binding to the BAX promoter. Assays were performed on chromatin fragments using antibody to p150Sal2 and normalized to preimmune rabbit IgG. Immunoprecipitated fractions were assayed by real time PCR for binding to the BAX and Aldolase promoters. Primers for BAX flanked the GC box at -117. C – Real time PCR on total cell RNA for BAX and Aldolase mRNAs. Results are normalized to Aldolase. D –Immunblots showing increased level of BAX protein in response to p150Sal2. E thru G - HOSE cells were treated with 25 ug/ml etoposide for 22 hours to induce apoptosis. E - ChIP assay showing increased binding of endogenous p150Sal2 to the BAX promoter in treated cells. F - Real time PCR on total cell RNA showing elevated levels of p150Sal2 and BAX mRNAs in apoptotic cells. G –Immunoblots showing elevated BAX protein in treated cells.
To investigate the effect of endogenous p150Sal2 on the BAX promoter, HOSE cells were first treated for 22 hours with etoposide to induce apoptosis. ChIP assay with anti-p150Sal2 antibody showed that binding of endogenous p150Sal2 to the BAX promoter increased 3-fold compared to the levels of binding to the aldolase promoter (Figure 6E). This treatment also resulted in increased levels of both p150Sal2 and BAX mRNAs (Figure 6F) and proteins (Figure 6G). These results demonstrate direct binding and regulation of BAX by endogenous p150Sal2 following an apoptotic stimulus. They also suggest that the effect of restoration of p150Sal2 on apoptosis in ovarian carcinoma cells is due to direct interaction [16].
4 DISCUSSION
We have investigated the sequence specificity of DNA binding by the human SALL2 transcription factor p150Sal2. The optimal sequence for binding in vitro is the heptanucleotide GGG(T/C)GGG. The binding specificity of p150Sal2 overlaps that of the Sp1 family of transcription factors. Though unrelated in overall amino acid sequence, p150Sal2 and Sp1 show similarity in their DNA binding domains, each marked by a cluster of three zinc fingers. Evidence for cooperative interaction between p150Sal2 and Sp1 comes from results of in vitro binding and synergistic action on an artificial promoter. Results of in vivo experiments suggest the likelihood of interaction at a distance by binding to separate GC elements found in natural promoters. Binding of p150Sal2 to two such promoters, BAX and p21, was analyzed further.
p150Sal2 was previously shown to bind and transactivate the p21 promoter [16]. Gel shift and ‘supershift’ experiments were carried out using short restriction fragments derived from the ~ 2.7 kb promoter to identify regions of binding and to confirm the expected presence of GC boxes. Three subfragments roughly 200 bp long were found to bind p150Sal2. Each contains a GC element related to the consensus sequence. Evidence implicating p150Sal2 as a possible direct regulator of BAX was inferred from elevated expression and increased apoptosis following restoration of p150Sal2 to ovarian carcinoma cells [16]. Chromatin immunoprecipitation, qRT-PCR and immunoblotting were used to demonstrate recruitment of endogenous p150Sal2 to the BAX promoter and activation following delivery of an apoptotic stimulus.
GC elements related to the optimal binding sequence defined in vitro are present in the promoters of p21 and BAX regulated by p150Sal2. Sp1 also plays roles in the regulation of p21 [33, 34] and BAX [35-37], consistent with the possibility of co-regulation by Sp1 and p150Sal2 in a conditional and cell type-dependent manner. p21 and BAX are critical targets in the context of actions of transcription factors as tumor suppressors. They are also potentially important as targets of inhibition by DNA viruses whose replication is dependent on cell cycle progression and blocking of apoptosis. Knowledge of the DNA binding specificity together with genome-wide approaches to identify downstream targets and pathways should lead to a better understanding of the regulatory functions of p150Sal2 in development, as a potential tumor suppressor, and as an inhibitor of replication of an oncogenic virus.
Research Highlights.
DNA binding properties of p150Sal2 have been defined in vitro and in vivo.
p150Sal2 binds to GC-rich elements in the p21Cip1/Waf1 and BAX promoters.
p150Sal2 activates the BAX promoter following an apoptotic stimulus.
These DNA binding properties of p150Sal2 underlie its tumor suppressor-like functions.
Acknowledgments
The authors would like to thank Dr. Jean Dahl for helpful discussions throughout the course of this work. This work was supported by the National Cancer Institute of the National Institutes of Health [Grant RO1 CA-092520].
Abbreviations used
- ChIP
chromatin immunoprecipitation
- CS
optimal consensus sequence for DNA binding in vitro
- EMSA
electrophoretic mobility shift assay
- GST
glutathione S-transferase
- HOSE cells
established human ovarian surface epithelial cells
- p21
the cyclin-dependent kinase inhibitor p21Cip1/Waf1
- SALL2
the Spalt-like gene 2
- SELEX
systematic evolution of ligands by exponential enrichment
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Jurgens G. Head and tail development of the Drosophila embryo involves spalt, a novel homeotic gene. Embo J. 1988;7:189–196. doi: 10.1002/j.1460-2075.1988.tb02799.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sweetman D, Munsterberg A. The vertebrate spalt genes in development and disease. Dev Biol. 2006;293:285–293. doi: 10.1016/j.ydbio.2006.02.009. [DOI] [PubMed] [Google Scholar]
- 3.Zhou Q, Chipperfield H, Melton DA, Wong WH. A gene regulatory network in mouse embryonic stem cells. Proc Natl Acad Sci U S A. 2007;104:16438–16443. doi: 10.1073/pnas.0701014104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wu Q, Chen X, Zhang J, Loh YH, Low TY, Zhang W, Zhang W, Sze SK, Lim B, Ng HH. Sall4 interacts with Nanog and co-occupies Nanog genomic sites in embryonic stem cells. J Biol Chem. 2006;281:24090–24094. doi: 10.1074/jbc.C600122200. [DOI] [PubMed] [Google Scholar]
- 5.Yang J, Chai L, Fowles TC, Alipio Z, Xu D, Fink LM, Ward DC, Ma Y. Genome-wide analysis reveals Sall4 to be a major regulator of pluripotency in murine-embryonic stem cells. Proc Natl Acad Sci U S A. 2008;105:19756–19761. doi: 10.1073/pnas.0809321105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang J, Tam WL, Tong GQ, Wu Q, Chan HY, Soh BS, Lou Y, Yang J, Ma Y, Chai L, Ng HH, Lufkin T, Robson P, Lim B. Sall4 modulates embryonic stem cell pluripotency and early embryonic development by the transcriptional regulation of Pou5f1. Nat Cell Biol. 2006;8:1114–1123. doi: 10.1038/ncb1481. [DOI] [PubMed] [Google Scholar]
- 7.Yang J, Chai L, Gao C, Fowles TC, Alipio Z, Dang H, Xu D, Fink LM, Ward DC, Ma Y. SALL4 is a key regulator of survival and apoptosis in human leukemic cells. Blood. 2008;112:805–813. doi: 10.1182/blood-2007-11-126326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li D, Dower K, Ma Y, Tian Y, Benjamin TL. A tumor host range selection procedure identifies p150(sal2) as a target of polyoma virus large T antigen. Proc Natl Acad Sci U S A. 2001;98:14619–14624. doi: 10.1073/pnas.251447198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ma Y, Li D, Chai L, Luciani AM, Ford D, Morgan J, Maizel AL. Cloning and characterization of two promoters for the human HSAL2 gene and their transcriptional repression by the Wilms tumor suppressor gene product. J Biol Chem. 2001;276:48223–48230. doi: 10.1074/jbc.M106468200. [DOI] [PubMed] [Google Scholar]
- 10.Pincheira R, Baerwald M, Dunbar JD, Donner DB. Sall2 is a novel p75NTR-interacting protein that links NGF signalling to cell cycle progression and neurite outgrowth. Embo J. 2009 doi: 10.1038/emboj.2008.274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bohm J, Buck A, Borozdin W, Mannan AU, Matysiak-Scholze U, Adham I, Schulz-Schaeffer W, Floss T, Wurst W, Kohlhase J, Barrionuevo F. Sall1, sall2, and sall4 are required for neural tube closure in mice. Am J Pathol. 2008;173:1455–1463. doi: 10.2353/ajpath.2008.071039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Buck A, Kispert A, Kohlhase J. Embryonic expression of the murine homologue of SALL1, the gene mutated in Townes--Brocks syndrome. Mechanisms of development. 2001;104:143–146. doi: 10.1016/s0925-4773(01)00364-1. [DOI] [PubMed] [Google Scholar]
- 13.Kiefer SM, Ohlemiller KK, Yang J, McDill BW, Kohlhase J, Rauchman M. Expression of a truncated Sall1 transcriptional repressor is responsible for Townes-Brocks syndrome birth defects. Hum Mol Genet. 2003;12:2221–2227. doi: 10.1093/hmg/ddg233. [DOI] [PubMed] [Google Scholar]
- 14.Kohlhase J, Heinrich M, Liebers M, Frohlich Archangelo L, Reardon W, Kispert A. Cloning and expression analysis of SALL4, the murine homologue of the gene mutated in Okihiro syndrome. Cytogenet Genome Res. 2002;98:274–277. doi: 10.1159/000071048. [DOI] [PubMed] [Google Scholar]
- 15.Sakaki-Yumoto M, Kobayashi C, Sato A, Fujimura S, Matsumoto Y, Takasato M, Kodama T, Aburatani H, Asashima M, Yoshida N, Nishinakamura R. The murine homolog of SALL4, a causative gene in Okihiro syndrome, is essential for embryonic stem cell proliferation, and cooperates with Sall1 in anorectal, heart, brain and kidney development. Development. 2006;133:3005–3013. doi: 10.1242/dev.02457. [DOI] [PubMed] [Google Scholar]
- 16.Li D, Tian Y, Ma Y, Benjamin T. p150(Sal2) is a p53-independent regulator of p21(WAF1/CIP) Mol Cell Biol. 2004;24:3885–3893. doi: 10.1128/MCB.24.9.3885-3893.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu H, Adler AS, Segal E, Chang HY. A transcriptional program mediating entry into cellular quiescence. PLoS Genet. 2007;3:e91. doi: 10.1371/journal.pgen.0030091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mayer B, Hirai H, Sakai R. Evidence that SH2 domains promote processive phosphorylation by protein-tyrosine kinases. Current Biology. 1995;5:296–305. doi: 10.1016/s0960-9822(95)00060-1. [DOI] [PubMed] [Google Scholar]
- 19.Pugh BF, Tjian R. Mechanism of transcriptional activation by Sp1: Evidence for coactivators. Cell. 1990;61:1187–1197. doi: 10.1016/0092-8674(90)90683-6. [DOI] [PubMed] [Google Scholar]
- 20.Ramos B, Gaudilliere B, Bonni A, Gill G. Transcription factor Sp4 regulates dendritic patterning during cerebellar maturation. Proceedings of the National Academy of Sciences. 2007;104:9882–9887. doi: 10.1073/pnas.0701946104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gebelein B, Urrutia R. Sequence-specific transcriptional repression by KS1, a multiple-zinc-finger-Kruppel-associated box protein. Mol Cell Biol. 2001;21:928–939. doi: 10.1128/MCB.21.3.928-939.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Blackwell TK, Weintraub H. Differences and similarities in DNA-binding preferences of MyoD and E2A protein complexes revealed by binding site selection. Science. 1990;250:1104–1110. doi: 10.1126/science.2174572. [DOI] [PubMed] [Google Scholar]
- 23.Hata Y, Duh E, Zhang K, Robinson GS, Aiello LP. Transcription factors Sp1 and Sp3 alter vascular endothelial growth factor receptor expression through a novel recognition sequence. J Biol Chem. 1998;273:19294–19303. doi: 10.1074/jbc.273.30.19294. [DOI] [PubMed] [Google Scholar]
- 24.Lei Z, Rao CV. cis-Acting elements and trans-acting proteins in the transcriptional inhibition of gonadotropin-releasing hormone gene by human chorionic gonadotropin in immortalized hypothalamic GT1-7 neurons. J Biol Chem. 1997;272:14365–14371. doi: 10.1074/jbc.272.22.14365. [DOI] [PubMed] [Google Scholar]
- 25.Dignam JD, Lebovitz RM, Roeder RG. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res. 1983;11:1475–1489. doi: 10.1093/nar/11.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dahl J, You J, Benjamin TL. Induction and utilization of an ATM signaling pathway by polyomavirus. J Virol. 2005;79:13007–13017. doi: 10.1128/JVI.79.20.13007-13017.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nelson JD, Denisenko O, Bomsztyk K. Protocol for the fast chromatin immunoprecipitation (ChIP) method. Nat Protoc. 2006;1:179–185. doi: 10.1038/nprot.2006.27. [DOI] [PubMed] [Google Scholar]
- 28.Letovsky J, Dynan WS. Measurement of the binding of transcription factor Sp1 to a single GC box recognition sequence. Nucleic Acids Res. 1989;17:2639–2653. doi: 10.1093/nar/17.7.2639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pabo CO, Sauer RT. Transcription factors: structural families and principles of DNA recognition. Annu Rev Biochem. 1992;61:1053–1095. doi: 10.1146/annurev.bi.61.070192.005201. [DOI] [PubMed] [Google Scholar]
- 30.Song J, Ugai H, Nakata-Tsutsui H, Kishikawa S, Suzuki E, Murata T, Yokoyama KK. Transcriptional regulation by zinc-finger proteins Sp1 and MAZ involves interactions with the same cis-elements. International journal of molecular medicine. 2003;11:547–553. [PubMed] [Google Scholar]
- 31.Meiss G, Friedhoff P, Hahn M, Gimadutdinow O, Pingoud A. Sequence preferences in cleavage of dsDNA and ssDNA by the extracellular Serratia marcescens endonuclease. Biochemistry. 1995;34:11979–11988. doi: 10.1021/bi00037a040. [DOI] [PubMed] [Google Scholar]
- 32.Lai JS, Herr W. Ethidium bromide provides a simple tool for identifying genuine DNA-independent protein associations. Proc Natl Acad Sci U S A. 1992;89:6958–6962. doi: 10.1073/pnas.89.15.6958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kardassis D, Papakosta P, Pardali K, Moustakas A. c-Jun transactivates the promoter of the human p21(WAF1/Cip1) gene by acting as a superactivator of the ubiquitous transcription factor Sp1. J Biol Chem. 1999;274:29572–29581. doi: 10.1074/jbc.274.41.29572. [DOI] [PubMed] [Google Scholar]
- 34.Koutsodontis G, Moustakas A, Kardassis D. The role of Sp1 family members, the proximal GC-rich motifs, and the upstream enhancer region in the regulation of the human cell cycle inhibitor p21WAF-1/Cip1 gene promoter. Biochemistry. 2002;41:12771–12784. doi: 10.1021/bi026141q. [DOI] [PubMed] [Google Scholar]
- 35.Thornborrow EC, Manfredi JJ. The tumor suppressor protein p53 requires a cofactor to activate transcriptionally the human BAX promoter. J Biol Chem. 2001;276:15598–15608. doi: 10.1074/jbc.M011643200. [DOI] [PubMed] [Google Scholar]
- 36.Igata E, Inoue T, Ohtani-Fujita N, Sowa Y, Tsujimoto Y, Sakai T. Molecular cloning and functional analysis of the murine bax gene promoter. Gene. 1999;238:407–415. doi: 10.1016/s0378-1119(99)00348-0. [DOI] [PubMed] [Google Scholar]
- 37.Schmidt T, Korner K, Karsunky H, Korsmeyer S, Muller R, Moroy T. The activity of the murine Bax promoter is regulated by Sp1/3 and E-box binding proteins but not by p53. Cell Death Differ. 1999;6:873–882. doi: 10.1038/sj.cdd.4400562. [DOI] [PubMed] [Google Scholar]


