Abstract
Clear cell renal cell carcinoma (RCC) is the most common and invasive adult kidney cancer. The genetic and biological mechanisms that drive metastatic spread of RCC remain largely unknown. We have investigated the molecular signatures and underlying genomic aberrations associated with RCC metastasis, using an approach that combines a human xenograft model, expression profiling of RNA, DNA and microRNA (miRNA), functional verification, and clinical validation. We show that increased metastatic activity is associated with acquisition of a myofibroblast-like signature in both tumor cell lines and in metastatic tumor biopsies. Our results also show that the mesenchymal trait did not provide an invasive advantage to the metastatic tumor cells. We further show that some of the constituents of the mesenchymal signature, including the expression of the well characterized myofibroblastic marker S100A4, are functionally relevant. Epigenetic silencing and miRNA-induced expression changes accounted for the change in expression of a significant number of genes, including S100A4, in the myofibroblastic signature; however, DNA copy number variation did not affect the same set of genes. These findings provide evidence that widespread genetic and epigenetic alterations can lead directly to global deregulation of gene expression and contribute to the development or progression of RCC metastasis culminating in a highly malignant myofibroblast-like cell with a mesenchymal phenotype.
Introduction
Cancer involves multistep changes in the genome that lead to deregulation of gene profiles and disruption of molecular networks. A variety of genomic approaches have been used to identify the molecular profiles that contribute to and reflect cancer progression. Moreover, gene expression profiling has been useful in the prediction of clinical outcome, disease progression, and metastatic recurrence (1). During carcinogenesis, genetic and epigenetic alterations drive tumor evolution towards higher grades of malignancy; however, the extent to which these changes alter gene expression, and how these alterations influence each other, remains incompletely understood. Association between gene dosage and gene expression levels has been reported in a variety of tumors (2), and ~12% of gene expression variation can be explained by differences in DNA copy number (3). In addition, integrated analyses of genome copy number and gene expression have provided insights into the etiology and mechanisms by which DNA copy number changes contribute to tumor evolution (2). Genomic methylation patterns are clearly deranged in cancer cells (4); however, the impact of promoter methylation on genome-wide gene expression changes during the pathogenesis of individual cancers still is poorly defined. Recently, the impact of global cancer-related changes in DNA methylation, genomic imbalance, and gene expression has been analyzed using whole genomic profiling approaches (5–6). Alternative approaches using recent advances in computational biology have successfully deciphered the functional impact of transcription factor networks on tumor progression (7).
Kidney cancer is the 7th most common cancer in the US. Clear cell renal-cell carcinoma (RCC) arises from the renal epithelium and accounts for approximately 85% adult kidney cancers (8)(9). In approximately 30% of patients, metastases are detected at the time of primary diagnosis, an additional 30–50% of initially localized tumors progress to distant metastases (10). The genetic mechanisms of RCC progression, especially the ones that underlie metastatic conversion, are poorly understood.
The prevailing multi-step progression model of tumorigenesis assumes that random mutations followed by selection eventually give rise to the metastatic phenotype in a rare subpopulation of tumor cells (11–12). Early genomic studies suggested that the primary tumor may or may not carry a molecular signature indicative of high metastatic potential (13). More recently, Massague and colleagues characterized extensively molecular signatures that allow breast tumor cells to colonize specific secondary sites (14–16). It has also been proposed that metastasis could arise from early disseminated tumor cells based on studies that showed that disseminated tumor cells can be identified in the secondary site very early on, even before the primary tumor becomes clinically apparent (17–18). Following up on Paget’s “seed and soil” hypothesis, proposed more than a hundred years ago (19), it has been suggested that metastatic growth requires the existence of a “metastatic niche” (20).
We used an integrated genomic approach and performed a comprehensive analysis of in vitro and xenograft models of RCC metastasis. For this, we used the RCC-derived and non-metastatic SN12C cell line and a collection of derived cell variants capable of efficient spontaneous metastasis to the lung when injected orthotopically. Gene expression profiling of non-metastatic and metastatic tumor cell lines revealed that metastatic activity was associated with the acquisition of a mesenchymal signature. Many of the genes in this signature are specifically enriched in the prototypical fibrotic cell. Notably, some of these genes are also know to control metastatic activity. By associating gene expression profiles and genetic and epigenetic alterations in the metastatic cells we were able to gain new insights into the mechanisms by which the genome of RCC tumor cells is rearranged during metastasis, as well as identify functionally relevant genes in this model.
Materials and methods
Full methods are described in supplemental methods
Cell lines and the xenograft model
The SN12C cell line was established in culture from a primary RCC from a 43-year-old male patient and was described previously (21). Derivation of the lung-metastatic LM1 cells was also previously described (22). LM2 cells were isolated from lung metastasis of NOD/SCID mice injected with 106 LM1 cells in the kidney subcapsule and sacrified 2 months later. Lung nodules from each mouse were dispersed into separate culture wells to generate individual cell lines. The cells were cultured in MEM medium supplemented with 10% fetal bovine serum. All animal studies were performed in accordance with a protocol approved by the Institutional Animal Care and Use Committee. 6–8 week-old NOD/SCID mice (NCI) were used for all xenograft studies.
Histology of xenograft tumors
Mouse lungs were harvested at necropsy. For hematoxylin and eosin staining, tissues were fixed overnight in 10% neutral buffered formalin, washed with PBS, and dehydrated in 70% ethanol prior to paraffin embedding (Histoserv Inc., Gaithersburg, MD).
Human tumor specimens
Fresh-frozen paired primary and metastatic tumor biopsies were obtained from 16 patients (32 samples) with RCC evaluated at the Memorial Slona-Kettering Cancer Center following an an IRB-approved protocol.
RNA isolation, labelling and microarray hybridization
RNA was isolated from cell lines as well as tumor tissues using an RNeasy kit (Qiagen, Valencia, CA). For expression profiling, the HG-U133 Plus 2.0 array (Affymetrix, Santa Clara, CA) was hybridized with cRNA from the samples using standard protocols. For microRNA (miRNA) analysis, total RNA was Isolated using the Qiagen miRNeasy kit and hybridized to Agilent Human miRNA Microarray kit V2-G4470B (Agilent Technologies, Santa Clara, CA) following labeling and hybridization protocols recommended by the manufacturer.
Immunoblotting
Cells were lysed using sample buffer and proteins were separated by SDS-PAGE and blotted onto nitrocellulose membranes and hybridized with the anti-S100A4 antibdy ab27957 (Abcom, Cambridge, MA).
DNA extraction and array comparative genomic hybridization (a-CGH)
Genomic DNA was isolated using the Qiagen DNA extraction kit. DNA digestion, labeling, and hybridization were performed following Agilent’s protocol version 4.0 for Agilent Human Genome CGH 244A oligo microarrays.
5 aza 2’ deoxycytidine (5’-AZA) treatment of cells
Cells were seeded (1×106) in culture medium and maintained for 24h before treatment with 5 µmol/L 5’-AZA (Sigma-Aldrich, St. Louis, MO) for 3d (23) . Medium containing 5’-AZA was replaced every 24h during the treatment. Control cells were treated in the same way, but without the addition of 5’-AZA.
Generation of retrovirus and knockdown cells
For overexpression, S100A4 was subcloned into the pQCXIN retroviral-based vector (Clontech, Palo Alto, CA). For knockdown, the pLKO.1 plasmid (clone TRCN0000053608) encoding short hairpin RNA (shRNAs) targeting S100A4 was obtained from Open Biosystems (Open Biosystems, Huntsville, AL). Viral supernatants were generated by transfecting 293-FT cells with the shRNA constructs in combination with the packaging vectors pVSVG and pDR2.
Results
A xenograft mouse model for RCC metastasis
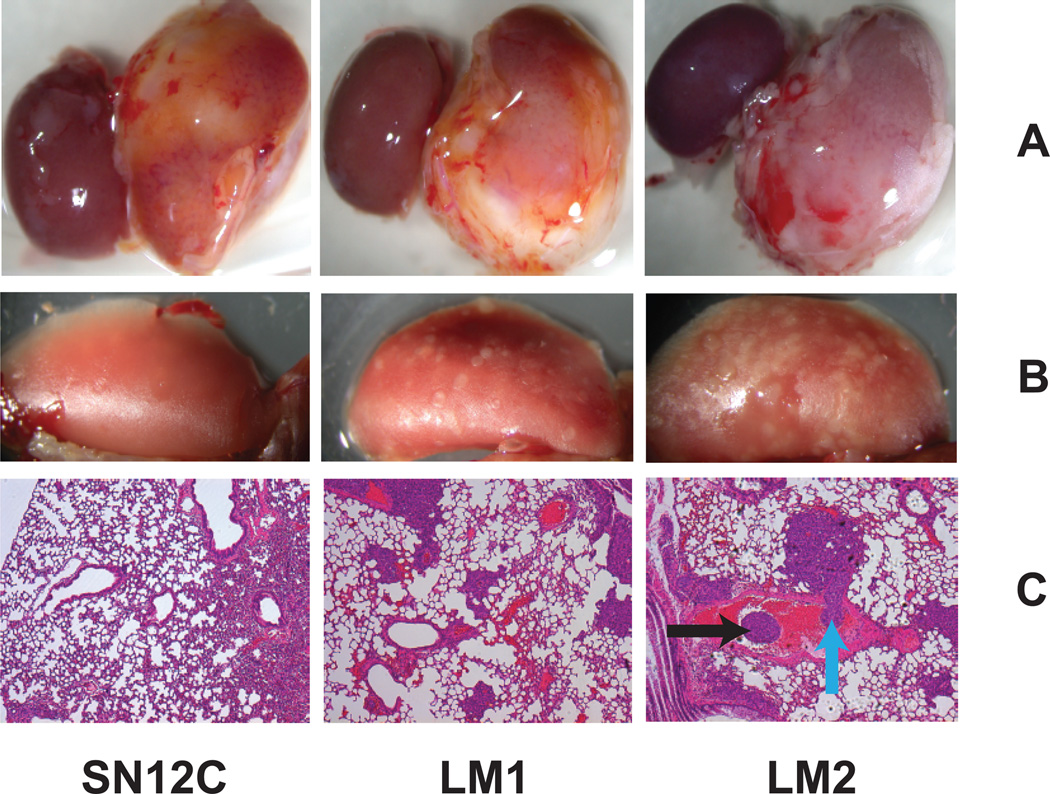
SN12C cells, when implanted in the renal sub-capsule of NOD/SCID mice, showed limited lung metastatic activity (3–10 nodules). Lung-derived nodules, when expanded in culture and reinoculated into the kidney capsule showed higher lung metastatic activity (80 nodules), designated LM1 (22). Another round of selection in vivo yielded a second generation of highly metastatic cells (400 nodules), designated LM2 (Fig. 1A). Interestingly, the LM cells also showed an increased metastatic spread to other organs ((22), our results). Of note, the growth rate of the highly metastatic cells at the primary site was similar to that of the parental SN12C cells (Fig. 1B). Histologic review of lung metastasis showed that the tumor cells, in addition to colonizing the lung parenchyma, associated in thrombus-like structures inside lymphatic as well as blood vessels (Fig. 1C).
Figure 1. A xenograft mouse model for RCC metastasis.
NOD/SCID mice were injected with 106 tumor cells as indicated in the kidney subcapsule. (A Primary tumor (B) Lung metastasis (C) H&E staining. Black arrow indicates thrombi inside the blood vessels. Blue arrow indicates vascular thrombus invading the vessel wall.
A mesenchymal expression signature correlates with RCC metastatic phenotype
To evaluate genome wide changes in gene expression during metastatic conversion we subjected the three cell types, SN12C, LM1, and LM2, to gene expression profiling using the Affymetrix U133 plus 2.0 arrays. Class comparison between SN12 and LM2 cells identified 349 unique genes that showed differential expression; 191 were upregulated and 158 were downregulated in the most aggressive metastatic cells. Analysis of the gene expression patterns revealed a mesenchymal signature as the dominant feature of the gene list that characterized the metastatic cells (Table 1) (Fig. S1). Epithelial-mesenchymal transition (EMT) has been proposed to be one of the initial steps during metastatic conversion that endows tumor cells with the ability to invade surrounding tissues (24). We, therefore, investigated if the observed mesenchymal phenotype of LM cells correlated with an increase in invasiveness. Surprisingly, the ability of LM cells to invade through both an artificial basement membrane (Boyden chamber assay) and an artifical endothelium (HUVECs) was not significantly altered when compared with parental SN12C cells (Fig. S2). This result suggested that the SN12C cells may already have undergone the initial stages of EMT. Consistent with this assumption, we found that epithelial markers such as E-cadherin were downregulated while mesenchymal markers such as N-cadherin and vimentin were already upregulated in the SN12C poorly metastatic cells (Fig. S2C). Emerging evidence suggests that epithelial cells are also an important source of myofibroblasts in fibrosis and cancer (25). Therefore, we reasoned that a significant function of tumor EMT would be to provide tumor cells with stromal myofibroblastic properties, which would bypass the requirement of an activated stroma during metastatic colonization. Detailed analysis of mesenchymal genes upregulated in the LM cells revealed a high enrichment for expression of genes related to fibrosis and desmoplastic response (S100A4, COL11A1, COL1A1, COL3A1, CALD1, and TNNC1) (26–27). Conversely, the expression of some of the genes characteristic of the epithelial lineage (KRT34, DSC3 and KRT33A) was lost in the metastatic cells (Fig.S3).
Table 1. A mesenchymal gene signature is upregulated during metastatic conversion of RCC cell lines.
Comparison of gene expression of LM cells with parental SN12C cells. Genes upregulated 3.5 folds or higher in LM cells are shown. Genes in blue represent mesenchymal-associated genes.
| probe ID | gene name | FC | probe ID | gene name | FC |
|---|---|---|---|---|---|
| 37892_at | COL11A1 | 19.09992 | 213640_s_at | LOX | 4.70 |
| 231131_at | FAM133A | 16.84373 | 236548_at | GIPC2 | 4.54 |
| 205081_at | CRIP1 | 13.5026 | 203987_at | FZD6 | 4.50 |
| 212077_at | CALD1 | 12.05112 | 224646_x_at | H19 | 4.45 |
| 203186_s_at | S100A4 | 12.04302 | 229963_at | BEX5 | 4.38 |
| 209031_at | CADM1 | 12.02546 | 214217_at | GRM5 | 4.36 |
| 229523_at | TTMA | 11.19727 | 1552721_a_at | FGF1 | 4.35 |
| 227566_at | NTM | 10.8935 | 220205_at | TPTE | 4.34 |
| 220786_s_at | SLC38A4 | 9.601934 | 222686_s_at | CPPED1 | 4.17 |
| 205696_s_at | GFRA1 | 8.289183 | 49077_at | PPME1 | 4.11 |
| 211161_s_at | COL3A1 | 7.57812 | 201141_at | GPNMB | 4.09 |
| 235382_at | LVRN | 7.244645 | 211814_s_at | CCNE2 | 3.98 |
| 201417_at | SOX4 | 6.757759 | 212192_at | KCTD12 | 3.98 |
| 229427_at | SEMA5A | 6.694796 | 223629_at | PCDHB5 | 3.97 |
| 1554378_a_at | PDE1C | 6.610365 | 220062_s_at | MAGEC2 | 3.96 |
| 209656_s_at | TMEM47 | 6.538456 | 217841_s_at | NXPH2 | 3.92 |
| 229978_at | LOC729993 | 6.294449 | 230883_at | SLC18A3 | 3.90 |
| 222925_at | DCDC2 | 6.25821 | 207150_at | CYBA | 3.89 |
| 213816_s_at | MET | 6.173399 | 203028_s_at | CBX5 | 3.86 |
| 213849_s_at | PPP2R2B | 6.057762 | 226085_at | LOXL1 | 3.86 |
| 1556499_s_at | COL1A1 | 5.568215 | 203570_at | MKX | 3.84 |
| 201650_at | KRT19 | 5.493345 | 241902_at | HOXB8 | 3.83 |
| 204584_at | L1CAM | 5.118585 | 229667_s_at | C4BPB | 3.81 |
| 200800_s_at | HSPA1A | 4.843601 | 208209_s_at | TNNC1 | 3.77 |
| 204724_s_at | COL9A3 | 4.791276 | 209904_at | GALNT3 | 3.71 |
| 204749_at | NAP1L3 | 4.737973 | 203397_s_at | LOC404266 | 3.71 |
In order to validate the above observations from SN12C and its metastatic derivatives in the clinical disease, we subjected a cohort of paired primary and metastatic biopsies from 16 RCC patients to gene expression profiling using the Affymetrix U133 plus 2.0 arrays. The metastatic components in these pairs were also highly enriched for genes typically associated with the myofibroblast phenotype (SFRP2, COL11A1, OGN, COL14A1, COL1A2, DCN, ASPN, and CALD1) (26–27) (Table 2). Collectively, these results indicate that during RCC metastatic conversion, tumor cells acquire a myofibroblast-like phenotype.
Table 2. A mesenchymal gene signature is upregulated during metastatic conversion of RCC clinical samples.
Comparison of gene expression of primary tumors with matched metastasis from 16 individual RCC patients. Top ranked genes according to SAM analysis and fold change are shown. Genes in blue represent mesenchymal- associated genes.
| Gene Name | FC |
|---|---|
| SFRP2 | 2.57 |
| COL11A1 | 1.95 |
| HSPD1 | 1.93 |
| LOC339047 | 1.87 |
| OGN | 1.86 |
| ITGBL1 | 1.86 |
| S100A10 | 1.86 |
| PDZD2 | 1.78 |
| LOC653786 | 1.78 |
| PTN | 1.74 |
| COL14A1 | 1.72 |
| COL1A2 | 1.71 |
| MPPE1 | 1.70 |
| DCN | 1.68 |
| PLCE1 | 1.66 |
| PPFIBP1 | 1.66 |
| ASPN | 1.65 |
| KIAA1641 | 1.64 |
| SYNPO2 | 1.63 |
| PLAGL1 | 1.62 |
| DLC1 | 1.62 |
| FBXL21 | 1.61 |
| SFRP4 | 1.61 |
Components of the mesenchymal signature control metastatic activity
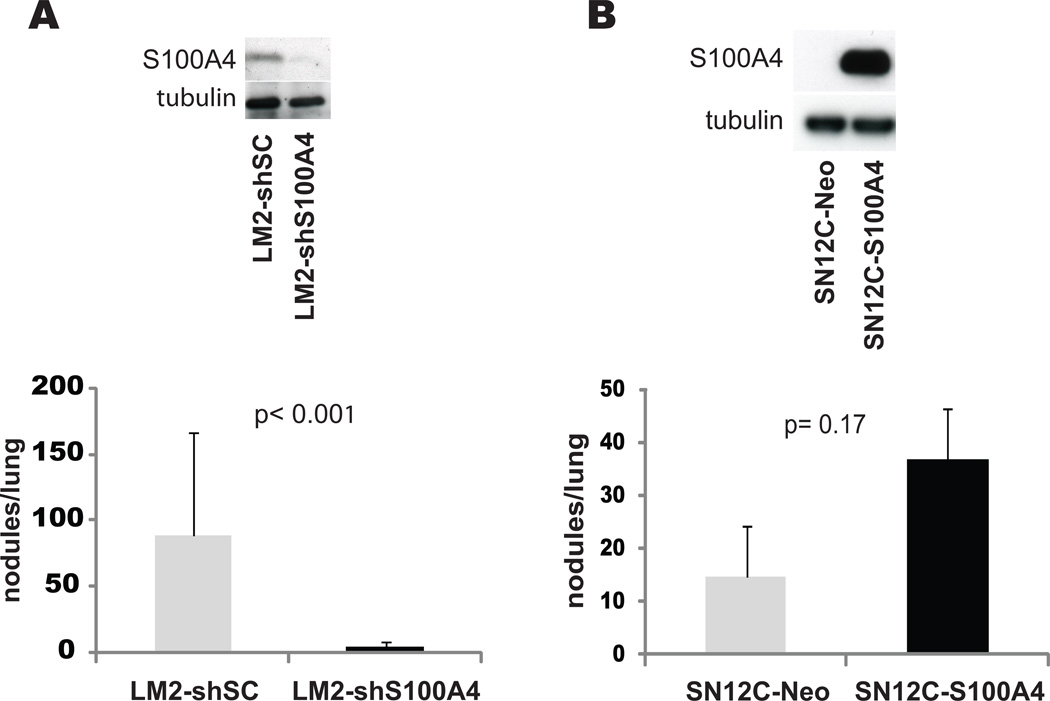
To formally demonstrate that the myofibroblast-like phenotype of the highly metastatic cells affects the metastatic behavior, we selected, from among a subset of genes biologically relevant to the mesenchymal phenotype, S100A4, a fibroblast marker for functional validation. We were particularly interested in S100A4 because this molecule could serve as a functional link between fibrosis and metastasis during kidney cancer progression, as strong evidence suggests S100A4 to be a crucial myofibroblast-expressed factor that regulates metastasis (28). To determine whether this gene plays a causal role in lung metastasis we stably decreased its expression in LM2 cells using shRNA and found that S100A4 levels had a profound affect on metastatic activity (Fig. 2A). To investigate if S100A4 was sufficient to induce metastatic spread, we engineered SN12C cells to overexpress S100A4 at levels comparable to those seen in LM2 cells. Injection of SN12C-S100A4 cells into the kidney sub-capsule showed a very modest lung colonization advantage in comparison with control SN12C cells (Fig. 2B) suggesting that S100A4 is required but not sufficient for LM2 cells to metastasize, and that cooperation of other genes may be required for the attainment of full metastatic competence. Globally, these results support a model where RCC cells co-opt a myofibroblastic gene expression program in order to colonize secondary sites during tumor progression.
Figure 2. S100A4 controls metastatic activity.
(A) LM2 cells were infected with lentiviruses encoding the indicated shRNAs. Equal amounts of total proteins were subjected to immunoblotting with antibodies to the indicated antigens. shSC, non-targeting shRNA; shS100A4, shRNAs targeting S100A4. (B) SN12C cells were infected with retroviruses bearing a vector encoding S100A4 or an empty vector. Total cellular lysis was subjected to immunoblotting using antibodies against the specified proteins. (A, B graphs) Groups of 5 mice were injected in the kidney subcapsule with the indicated cell populations and numbers of lung metastatic nodules were scored after 2 months.
DNA copy number variation-related gene expression changes
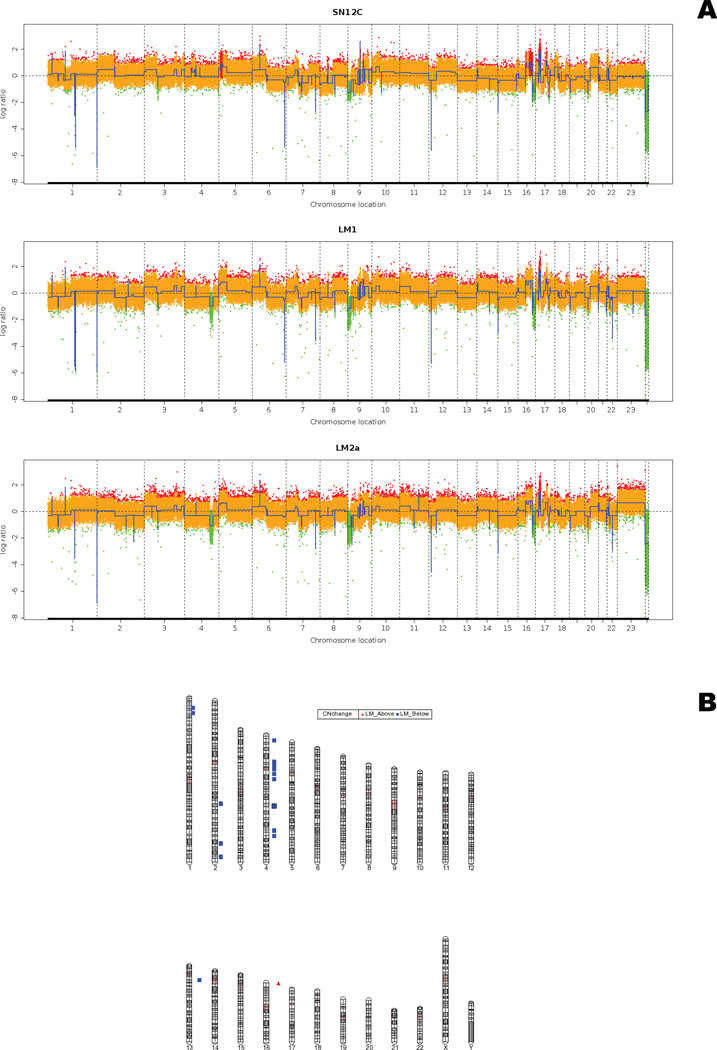
We performed high-resolution whole genome profiling of SN12C and derived metastatic LM1 and LM2 cells using the Agilent High-Density a-CGH Human 244A array to cross reference gene dosage and gene expression changes. The resultant data were analyzed with the Partek Genomic Suite. This analysis identified approximately 50 significant regions of chromosome copy number differences that distinguished the non-metastatic and metastatic cells, spanning from 2 Kbp to 80 Mbp. These regional changes in chromosome copy number were scattered over 14 chromosomes with a certain degree of enrichment on chromosomes 1, 2, and 4. Of the 50 aberrations, 10 were gains and 40 were losses (Fig. S4). Comparison of probe heat maps from the three different genomes showed complex differences (Fig. 3A). To identify genes belonging to regions of significant copy number change, the results of the segmentation algorithm of a-CGH profiles were annotated with the Agilent human genome annotation library and further aligned with the list of genes differentially expressed between the three groups using the Partek Genomic Suite. This analysis yielded a list of 57 genes that mapped to chromosomal regions of DNA copy number changes (Fig. 3B). Only 1 gene was mapped to a significant region of gain on chromosome 16, while 56 genes mapped to regions of significant loss on chromosomes 1, 2, 4, 10, 13, and 16 (Fig. S5). Interestingly, 50 of the 57 genes mapped to chromosome 4. None of the genes defining the myofibroblast-like phenotype mapped to regions of DNA copy number variation, arguing against a role for genomic rearrangements during the acquisition of the myofibroblastic phenotype.
Figure 3. DNA copy variation.
(A) Segmented copy numbers for each cell line were inferred with the GLAD (gain and loss analysis of DNA) algorithm and normalized to a median of two copies. Vertical dashed red lines represent the breakpoints detected with GLAD and the assigned statuses are indicated by a color code: green for loss, yellow for normal and red for gain. (B) Chromosomal mapping of regions of copy number alteration. Regions appearing increased in copy number are shown in red, and those decreased in copy number are shown in blue.
Impact of promoter methylation on the gene expression profile of metastatic cells
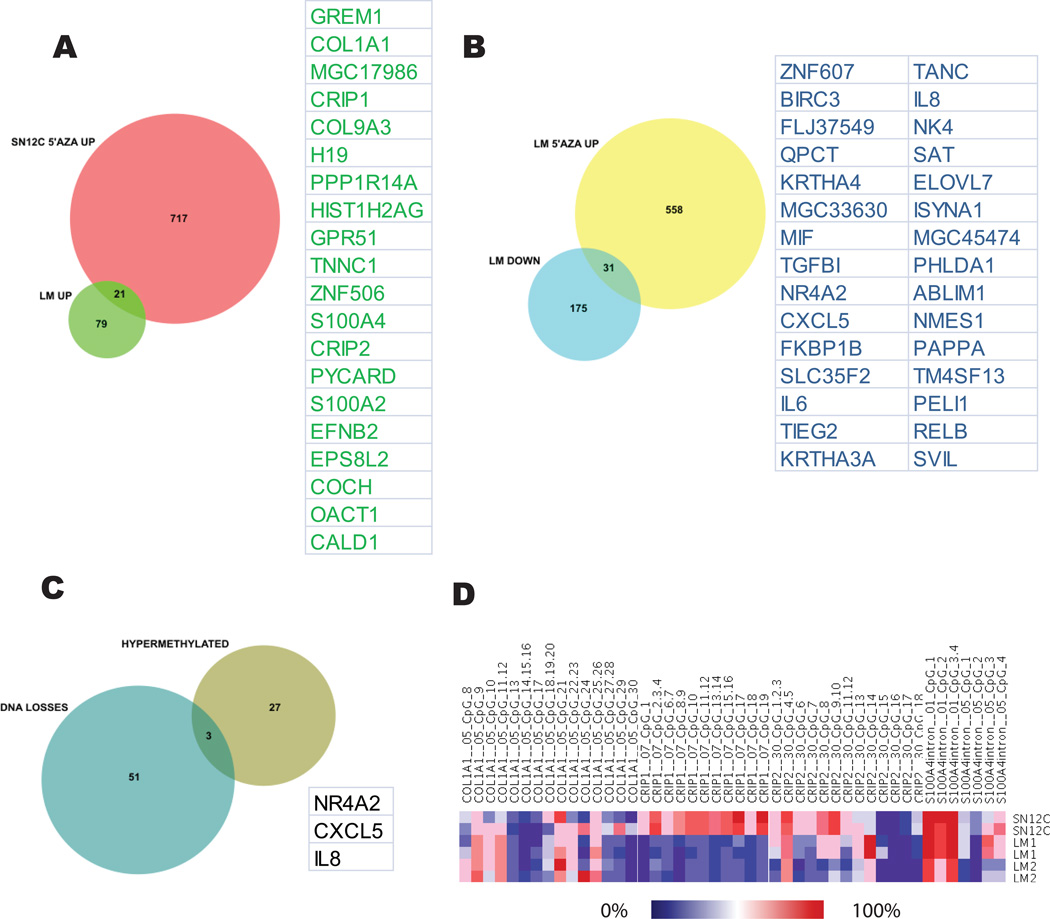
Since acquired changes in gene expression may be influenced by both genetic and epigenetic factors, we set out to study the contribution of promoter methylation in the establishment of the metastatic phenotype in our model of tumor progression. To identify epigenetically silenced genes we used an epigenetic reactivation strategy that combined treatment of cancer cells in vitro with the DNA methyltransferase inhibitor 5’-AZA, followed by global gene expression analysis using microarrays. Treatment of SN12C cells resulted in the upregulation of more than 600 unique genes. To uncover genes upregulated by promoter demethylation during the SN12C-LM transition, we performed a Venn analysis using a dataset of genes upregulated in SN12C cells after 5’-AZA treatment a n d a dataset of genes spontaneously upregulated in LM cells (Fig. 4A). By this method we identified 21 genes, including S100A4, CALD1, COL9A3, CRIP1, CRIP2, COL1A1, and TNNC1, as hypomethylated in the LM cells, supporting a metastasis promotion role for those genes. Conversely, treatment of LM cells with the same inhibitor followed by analysis of re-expressed genes and Venn analysis of overlapping genes with the list of genes downregulated during SN12C-LM transition uncovered 31 hypermethylated candidate genes (Fig. 4B). Remarkably, some of the hypermethylated and silenced genes in the LM cells (KRT34 and KRT33A) have been known to be associated with an epithelial phenotype (29). Overall, 52 out of 300 genes (~20%) deregulated during metastatic progression were upregulated after demethylation agent treatment and, therefore, had their expression likely regulated by changes in promoter methylation. A total of 48 genes harbored CpG islands in their promoters. Moreover, 70 and 10 genes were downregulated by drug treatment of SN12C and LM cells, respectively, probably reflecting the re-expression of a repressor (data not shown). To validate the results from the 5’-AZA re-expression experiment, we selected 4 genes predicted to be demethylated during metastatic conversion (S100A4, COL1A1, CRIP1, and CRIP2) and subjected them to quantitation of CpG methylation using the EpiTYPER (Sequenom) mass-spec analysis of bisulfite converted DNA (Figure 4D). 3 out of the 4 genes showed decreased CpG methylation in the LM cells, as predicted. S100A4 was also de-methylated in a cohort of clinical samples as the tumor progressed to a metastatic stage (Fig. S6).
Figure 4. Promoter methylation related gene expression changes.
(A) Venn diagram of genes predicted to be hypermethylated in SN12C and spontaneously demethylated in LM2 cells. (B) Venn analysis of genes predicted to be hypomethylated in SN12C and spontaneously hypermethylated in LM2 cells. (C) Genes predicted to be hypermethylated in LM2 cells and showing loss of copy number. (D) Quantitation of DNA methylation in CpG dinucleotides of 4 genes from panel A by EpiTYPER analysis. CpG specific methylation is showed in a heat map.
Taken together, these results suggest that a number of genes undergo methylation-associated silencing or re-expression in the course of RCC tumorigenesis and metastasis. Even more interestingly, many of the methylated genes are associated with a myofibroblastic phenotype, suggesting that promoter methylation may be one of the pivotal events controlling metastatic conversion.
Correlation between gene dosage and promoter methylation
Tumor-acquired promoter methylation often coincides with allele loss. To determine whether any of the 52 hypo- and hypermethylated candidate genes were also subject to DNA copy number changes, we cross-referenced this list of genes with the dataset of genes mapped to regions of significant DNA dosage changes, using the Venn diagram tool. We determined that only 3 out of the 52 genes whose expression was regulated by methylation belonged to regions of significant copy number losses (Fig. 4C). However, we did not identify any candidate methylated genes related to any of the regions of gene dosage gain. These results suggest a complex pattern in the genetic control of gene expression where a subset of genes is synergistically impacted by both promoter methylation and DNA copy variation. Parenthetically, the approach utilized here provides a new way to sort and prioritize functionally relevant genes.
MiRNA dosage-related gene expression changes
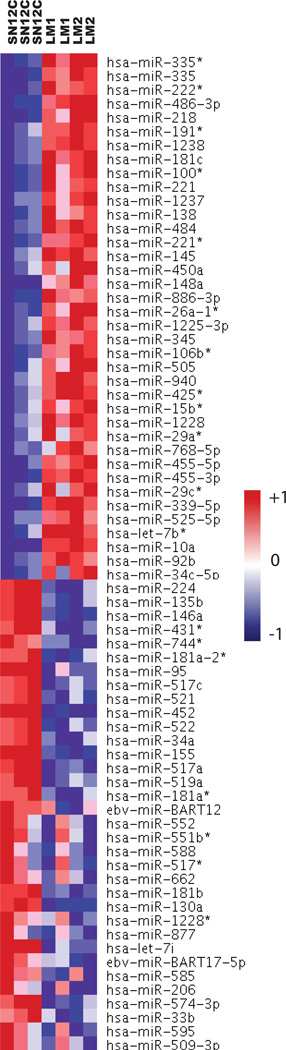
MiRNAs are attractive candidates as upstream regulators of metastatic progression because they can regulate entire sets of genes. Therefore, we reasoned that miRNA deregulation may account for at least some of the gene expression changes that distinguish the highly metastatic LM cells from the poorly metastatic SN12C cells. We performed array-based miRNA profiling of SN12C, LM1, and LM2 cells. Hierarchical clustering based on the expression of the differentially expressed miRNAs correctly classified the cell populations into 2 groups (Fig. 5). Of 700 human miRNAs assayed, 38 were upregulated and 34 were downregulated in the highly metastatic cells, using a 1.5-fold threshold. On the assumption that the expression profiles of miRNA genes and their targets are inversely correlated, we identified their putative targets using TargetScan 4.2 and integrated the target genes with expression data from the same experiment. Using this methodology, we identified a network of putative functional miRNA-target regulatory relations involving 100 genes (Fig. S7). We also validated the changes in expression of the 2 top miRNA, LM2 vs SN12C (Fig. S8). A large proportion of genes included in the miRNAdown-mRNAup-predicted network have reported mesenchymal functions (CALD1, COL1A1, COLA1A2, COL3A1, COL9A3, GREM1 and MYLK). Next, we sought to identify the underlying mechanisms that can explain the miRNA expression changes associated with the transition to a highly metastatic state in our model. MiRNAs are frequently located in cancer-associated regions of the human genome (30). Thus, in order to determine if genomic gains and losses might have impacted upon miRNA expression, we correlated gains and losses of genomic regions, as determined by a-CGH on each cell line, with relative miRNA expression values. Notably, we found that only 2 miRNA (hsa-miR-574-3p and hsa-miR-95) mapped to a region of gene-dosage change (Fig. S9). In addition to genetic alterations, the expression of specific miRNAs could be dysregulated by epigenetic aberrations. To identify miRNAs with putative DNA methylation-related inactivation involved in metastasis we treated SN12C and LM2 cell lines with 5’-AZA followed by hybridization to an expression microarray. By comparison of the list of genes re-expressed by 5’-AZA and the list of genes differentially expressed in SN12C and LM2 we found that hsa-miR-224 and hsa-miR-34c-5p miRNAs undergo specific hypermethylation-associated silencing (Fig. S10)
Figure 5.
Identification of miRNAs altered in RCC cell derivatives. Clustering of normalized miRNA expression levels in SN12C and LM cell lines.
Discussion
This study describes a comprehensive analysis of gene expression and genomic alterations in in vitro and xenograft models of RCC metastasis in order to elucidate the genetic mechanisms and associated biological functions that empower tumor cells with the ability to colonize distant sites. Molecular profiling of tumor cells in the xenograft model and RCC tumor biopsies uncovered a mesenchymal signature as the most significant feature linked to high metastatic activity. Unexpectedly, tumor cell invasion was not significantly increased in the highly metastatic populations. These results prompted us to search for alternative mechanisms by which a mesenchymal phenotype may be of advantage at the metastatic site. Detailed analysis of the gene functions associated with the mesenchymal signature revealed a substantial enrichment for pro-fibrotic genes suggesting that metastatic spread imposes a selective pressure for cells with a myofibroblastic makeup. We therefore suggest that an important function of tumor EMT is to generate cells with properties of stromal fibroblasts. It is logical to presume that a tumor cell endowed with such a phenotype would be highly effective at colonizing distant organs because it could bypass the requirement of an activated “compatible” stroma in the initial stages of metastatic establishment. Myofibroblasts produce and modify the extracellular matrix (ECM), secrete angiogenic and proinflammatory factors, and stimulate epithelial cell proliferation and invasion. Myofibroblasts were originally characterized by their role in wound healing (31) and its induction has been associated with diverse types of organ fibrosis (32). Myofibroblasts are also abundant in the reactive tumor stroma and are an established source of tumor promoting factors such as cell surface proteins, secreted growth factors, and extracellular matrix proteins (33–34). A direct pro-metastatic effect of tumor associated myofibroblasts was also recently implied (35). Consistent with our hypothesis, It has also been suggested that myofibroblasts are derived from malignant or normal epithelial cells undergoing EMT (36). Additional support for this hypothesis comes from the fact that increased levels of collagens and laminins have been associated with an increased likelihood of clinical metastasis of multiple human solid tumors (13). In order to substantiate the relevance of the mesenchymal signature we selected S100A4 for functional validation. Enforced repression of this factor profoundly affected metastatic activity in agreement with previous studies that demonstrated that S100A4 is a crucial myofibroblast-expressed factor regulating metastasis (37). Furthermore, the expression of S100A4 in renal tubular epithelium undergoing EMT during fibrosis (38), together with the fact that proximal tubules arise from the differentiation of mesenchymal cells, suggest a model wherein RCC epithelial cells reverse this developmental process back to the original mesenchymal state, in order to metastasize. The plasticity of the renal epithelial cell has been further supported by studies of the role of the developmental gene, GREM1, which is reactivated in adult renal fibrotic disease (39). Remarkably, GREM1 is also a component of the myofibroblastic signature identified in our in vitro model of RCC progression. Although deregulation of gene expression during tumor progression is relatively well characterized, the associated genetic and molecular mechanisms are largely unknown. In addition, while the sequence of genetic events that drive the establishment of primary tumors has begun to emerge, the subsequent events that lead to metastasis have remained generally obscure. As a consequence, few mutations, genomic alterations, or allelic imbalances are currently known to distinctively endow tumor cells with metastatic functions (40). We reasoned that by studying the mechanisms by which RCC genomes are deranged during metastatic conversion, we would be able to improve the ability to pinpoint critical genes involved in this process. By analyzing mRNA expression levels and DNA copy number changes in parallel, we found that changes in gene dosage have a modest effect on gene expression. These results suggest that during the evolution toward a metastatic phenotype, these cells acquire a set of chromosomal losses and gains that are perhaps associated with retention or otherwise of specific cell clones within the cell population. Although, none of the genes mapped to regions of genomic imbalance related to mesenchymal functions, we cannot exclude that these changes do not indirectly impact the mesenchymal signature.
In addition to changes in DNA sequence, gene expression can be modulated by aberrations in the patterns of DNA methylation. However, the pattern of DNA methylation abnormalities in cancer cells seems paradoxical. Compared with normal cells, cancer cells are concomitantly hypermethylated at specific CpG island sequences but hypomethylated at CpGs found in most other sites, resulting in a net loss of genomic 5meC content (41). Understanding the relative timing of DNA hypermethylation and hypomethylation alterations in cancer is crucial to understanding the importance of these changes during tumor progression. We found that a number of mesenchymal- associated genes were reactivated upon 5’-AZA treatment suggesting that they were hypermethylated in the poorly metastatic cells. S100A4 showed complete methylation of the intronic CpG sites analyzed in the poorly metastatic cells, consistent with its epigenetic transcriptional silencing. Some of those genes, including S100A4, COL1A1 and GREM1, have previously been shown to be methylated in other tumor models as well as in RCC (42–44). The proposed impact of promoter methylation on EMT is not unprecedented in the literature as altered methylation of some gene promoters have been reported to be one of the principal causes of EMT during tumor progression (45) or during epithelial to myofibroblast transition (46).
Several bioinformatic algorithms have been constructed to predict miRNA gene targets. These algorithms predict hundreds of potential gene targets, which cannot all be experimentally validated. Integration of putative targets with mRNA expression date provides a rational method to prioritize functionally relevant targets. Interestingly, the most significant correlation in this study was found between Let-7 and COL1A1, COLA1A2, COL3A1, COL9A3. In consonance with our results, Let-7 is widely viewed as a tumor suppressor miRNA (47–48).
Our findings revealing the lack of major genetic and epigenetic alterations accounting for the direct deregulation of miRNA expression suggest that miRNAs alterations are likely to be induced indirectly as a consequence of the dysregulation of specific transcription factors. Moreover, this observation argues that miRNA may be downstream targets of pathways that are commonly dysregulated in cancer and not initiating events during tumor progression.
In summary, in this study, we have proposed and provided proof of principle for a new mechanism of tumor progression in RCC based on acquisition of a myofibroblastic trait by tumor cells. We also showed that this approach that cross-references multiple whole-genome data sets can identify targets and genetic mechanisms important for tumor progression.
Supplementary Material
Acknowledgments
We are grateful to I. Fidler for providing the SN12C cell lines.
Grant Support
This study was supported by grants from the V-Foundation, a Syms kidney cancer award, Pfizer, Inc., and the NIH (CA-121327).
Footnotes
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- 1.van't Veer LJ, Bernards R. Enabling personalized cancer medicine through analysis of gene-expression patterns. Nature. 2008;452:564–570. doi: 10.1038/nature06915. [DOI] [PubMed] [Google Scholar]
- 2.Chin K, DeVries S, Fridlyand J, et al. Genomic and transcriptional aberrations linked to breast cancer pathophysiologies. Cancer Cell. 2006;10:529–541. doi: 10.1016/j.ccr.2006.10.009. [DOI] [PubMed] [Google Scholar]
- 3.Pollack JR, Sorlie T, Perou CM, et al. Microarray analysis reveals a major direct role of DNA copy number alteration in the transcriptional program of human breast tumors. Proc Natl Acad Sci U S A. 2002;99:12963–12968. doi: 10.1073/pnas.162471999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bestor TH. Unanswered questions about the role of promoter methylation in carcinogenesis. Ann N Y Acad Sci. 2003;983:22–27. doi: 10.1111/j.1749-6632.2003.tb05959.x. [DOI] [PubMed] [Google Scholar]
- 5.Sadikovic B, Yoshimoto M, Al-Romaih K, Maire G, Zielenska M, Squire JA. In vitro analysis of integrated global high-resolution DNA methylation profiling with genomic imbalance and gene expression in osteosarcoma. PLoS One. 2008;3:e2834. doi: 10.1371/journal.pone.0002834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Andrews J, Kennette W, Pilon J, et al. Multi-platform whole-genome microarray analyses refine the epigenetic signature of breast cancer metastasis with gene expression and copy number. PLoS One. 2010;5:e8665. doi: 10.1371/journal.pone.0008665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Taylor RC, Acquaah-Mensah G, Singhal M, Malhotra D, Biswal S. Network inference algorithms elucidate Nrf2 regulation of mouse lung oxidative stress. PLoS Comput Biol. 2008;4:e1000166. doi: 10.1371/journal.pcbi.1000166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Motzer RJ, Hutson TE, Tomczak P, et al. Sunitinib versus interferon alfa in metastatic renal-cell carcinoma. N Engl J Med. 2007;356:115–124. doi: 10.1056/NEJMoa065044. [DOI] [PubMed] [Google Scholar]
- 9.Banks RE, Tirukonda P, Taylor C, et al. Genetic and epigenetic analysis of von Hippel-Lindau (VHL) gene alterations and relationship with clinical variables in sporadic renal cancer. Cancer Res. 2006;66:2000–2011. doi: 10.1158/0008-5472.CAN-05-3074. [DOI] [PubMed] [Google Scholar]
- 10.Cohen HT, McGovern FJ. Renal-cell carcinoma. N Engl J Med. 2005;353:2477–2490. doi: 10.1056/NEJMra043172. [DOI] [PubMed] [Google Scholar]
- 11.Fidler IJ, Kripke ML. Metastasis results from preexisting variant cells within a malignant tumor. Science. 1977;197:893–895. doi: 10.1126/science.887927. [DOI] [PubMed] [Google Scholar]
- 12.Nguyen DX, Bos PD, Massague J. Metastasis: from dissemination to organ-specific colonization. Nat Rev Cancer. 2009;9:274–284. doi: 10.1038/nrc2622. [DOI] [PubMed] [Google Scholar]
- 13.Ramaswamy S, Ross KN, Lander ES, Golub TR. A molecular signature of metastasis in primary solid tumors. Nat Genet. 2003;33:49–54. doi: 10.1038/ng1060. [DOI] [PubMed] [Google Scholar]
- 14.Kang Y, Siegel PM, Shu W, et al. A multigenic program mediating breast cancer metastasis to bone. Cancer Cell. 2003;3:537–549. doi: 10.1016/s1535-6108(03)00132-6. [DOI] [PubMed] [Google Scholar]
- 15.Minn AJ, Gupta GP, Siegel PM, et al. Genes that mediate breast cancer metastasis to lung. Nature. 2005;436:518–524. doi: 10.1038/nature03799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bos PD, Zhang XH, Nadal C, et al. Genes that mediate breast cancer metastasis to the brain. Nature. 2009;459:1005–1009. doi: 10.1038/nature08021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Husemann Y, Geigl JB, Schubert F, et al. Systemic spread is an early step in breast cancer. Cancer Cell. 2008;13:58–68. doi: 10.1016/j.ccr.2007.12.003. [DOI] [PubMed] [Google Scholar]
- 18.Podsypanina K, Du YC, Jechlinger M, Beverly LJ, Hambardzumyan D, Varmus H. Seeding and propagation of untransformed mouse mammary cells in the lung. Science. 2008;321:1841–1844. doi: 10.1126/science.1161621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Paget S. The distribution of secondary growths in cancer of the breast. 1889. Cancer Metastasis Rev. 1989;8:98–101. [PubMed] [Google Scholar]
- 20.Psaila B, Lyden D. The metastatic niche: adapting the foreign soil. Nat Rev Cancer. 2009;9:285–293. doi: 10.1038/nrc2621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Naito S, von Eschenbach AC, Giavazzi R, Fidler IJ. Growth and metastasis of tumor cells isolated from a human renal cell carcinoma implanted into different organs of nude mice. Cancer Res. 1986;46:4109–4115. [PubMed] [Google Scholar]
- 22.Naito S, Walker SM, Fidler IJ. In vivo selection of human renal cell carcinoma cells with high metastatic potential in nude mice. Clin Exp Metastasis. 1989;7:381–389. doi: 10.1007/BF01753659. [DOI] [PubMed] [Google Scholar]
- 23.Suzuki H, Gabrielson E, Chen W, et al. A genomic screen for genes upregulated by demethylation and histone deacetylase inhibition in human colorectal cancer. Nat Genet. 2002;31:141–149. doi: 10.1038/ng892. [DOI] [PubMed] [Google Scholar]
- 24.Yang J, Weinberg RA. Epithelial-mesenchymal transition: at the crossroads of development and tumor metastasis. Dev Cell. 2008;14:818–829. doi: 10.1016/j.devcel.2008.05.009. [DOI] [PubMed] [Google Scholar]
- 25.Selman M, Pardo A. Role of epithelial cells in idiopathic pulmonary fibrosis: from innocent targets to serial killers. Proc Am Thorac Soc. 2006;3:364–372. doi: 10.1513/pats.200601-003TK. [DOI] [PubMed] [Google Scholar]
- 26.Allinen M, Beroukhim R, Cai L, et al. Molecular characterization of the tumor microenvironment in breast cancer. Cancer Cell. 2004;6:17–32. doi: 10.1016/j.ccr.2004.06.010. [DOI] [PubMed] [Google Scholar]
- 27.Chambers RC, Leoni P, Kaminski N, Laurent GJ, Heller RA. Global expression profiling of fibroblast responses to transforming growth factor-beta1 reveals the induction of inhibitor of differentiation-1 and provides evidence of smooth muscle cell phenotypic switching. Am J Pathol. 2003;162:533–546. doi: 10.1016/s0002-9440(10)63847-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schneider M, Hansen JL, Sheikh SP. S100A4: a common mediator of epithelial-mesenchymal transition, fibrosis and regeneration in diseases? J Mol Med. 2008;86:507–522. doi: 10.1007/s00109-007-0301-3. [DOI] [PubMed] [Google Scholar]
- 29.Rogers MA, Winter H, Wolf C, Heck M, Schweizer J. Characterization of a 190-kilobase pair domain of human type I hair keratin genes. J Biol Chem. 1998;273:26683–26691. doi: 10.1074/jbc.273.41.26683. [DOI] [PubMed] [Google Scholar]
- 30.Calin GA, Sevignani C, Dumitru CD, et al. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc Natl Acad Sci U S A. 2004;101:2999–3004. doi: 10.1073/pnas.0307323101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gabbiani G, Ryan GB, Majne G. Presence of modified fibroblasts in granulation tissue and their possible role in wound contraction. Experientia. 1971;27:549–550. doi: 10.1007/BF02147594. [DOI] [PubMed] [Google Scholar]
- 32.Desmouliere A, Chaponnier C, Gabbiani G. Tissue repair, contraction, and the myofibroblast. Wound Repair Regen. 2005;13:7–12. doi: 10.1111/j.1067-1927.2005.130102.x. [DOI] [PubMed] [Google Scholar]
- 33.Orimo A, Gupta PB, Sgroi DC, et al. Stromal fibroblasts present in invasive human breast carcinomas promote tumor growth and angiogenesis through elevated SDF-1/CXCL12 secretion. Cell. 2005;121:335–348. doi: 10.1016/j.cell.2005.02.034. [DOI] [PubMed] [Google Scholar]
- 34.Hwang RF, Moore T, Arumugam T, et al. Cancer-associated stromal fibroblasts promote pancreatic tumor progression. Cancer Res. 2008;68:918–926. doi: 10.1158/0008-5472.CAN-07-5714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Karnoub AE, Dash AB, Vo AP, et al. Mesenchymal stem cells within tumour stroma promote breast cancer metastasis. Nature. 2007;449:557–563. doi: 10.1038/nature06188. [DOI] [PubMed] [Google Scholar]
- 36.Kalluri R, Zeisberg M. Fibroblasts in cancer. Nat Rev Cancer. 2006;6:392–401. doi: 10.1038/nrc1877. [DOI] [PubMed] [Google Scholar]
- 37.Grum-Schwensen B, Klingelhofer J, Berg CH, et al. Suppression of tumor development and metastasis formation in mice lacking the S100A4(mts1) gene. Cancer Res. 2005;65:3772–3780. doi: 10.1158/0008-5472.CAN-04-4510. [DOI] [PubMed] [Google Scholar]
- 38.Iwano M, Plieth D, Danoff TM, Xue C, Okada H, Neilson EG. Evidence that fibroblasts derive from epithelium during tissue fibrosis. J Clin Invest. 2002;110:341–350. doi: 10.1172/JCI15518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Roxburgh SA, Murphy M, Pollock CA, Brazil DP. Recapitulation of embryological programmes in renal fibrosis--the importance of epithelial cell plasticity and developmental genes. Nephron Physiol. 2006;103:p139–p148. doi: 10.1159/000092453. [DOI] [PubMed] [Google Scholar]
- 40.Nguyen DX, Massague J. Genetic determinants of cancer metastasis. Nat Rev Genet. 2007;8:341–352. doi: 10.1038/nrg2101. [DOI] [PubMed] [Google Scholar]
- 41.Feinberg AP, Vogelstein B. Hypomethylation distinguishes genes of some human cancers from their normal counterparts. Nature. 1983;301:89–92. doi: 10.1038/301089a0. [DOI] [PubMed] [Google Scholar]
- 42.Rosty C, Ueki T, Argani P, et al. Overexpression of S100A4 in pancreatic ductal adenocarcinomas is associated with poor differentiation and DNA hypomethylation. Am J Pathol. 2002;160:45–50. doi: 10.1016/S0002-9440(10)64347-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.van Vlodrop IJ, Baldewijns MM, Smits KM, et al. Prognostic significance of Gremlin1 (GREM1) promoter CpG island hypermethylation in clear cell renal cell carcinoma. Am J Pathol. 2010;176:575–584. doi: 10.2353/ajpath.2010.090442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Morris MR, Ricketts C, Gentle D, et al. Identification of candidate tumour suppressor genes frequently methylated in renal cell carcinoma. Oncogene. 2010;29:2104–2117. doi: 10.1038/onc.2009.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dumont N, Wilson MB, Crawford YG, Reynolds PA, Sigaroudinia M, Tlsty TD. Sustained induction of epithelial to mesenchymal transition activates DNA methylation of genes silenced in basal-like breast cancers. Proc Natl Acad Sci U S A. 2008;105:14867–14872. doi: 10.1073/pnas.0807146105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mann J, Oakley F, Akiboye F, Elsharkawy A, Thorne AW, Mann DA. Regulation of myofibroblast transdifferentiation by DNA methylation and MeCP2: implications for wound healing and fibrogenesis. Cell Death Differ. 2007;14:275–285. doi: 10.1038/sj.cdd.4401979. [DOI] [PubMed] [Google Scholar]
- 47.Takamizawa J, Konishi H, Yanagisawa K, et al. Reduced expression of the let-7 microRNAs in human lung cancers in association with shortened postoperative survival. Cancer Res. 2004;64:3753–376. doi: 10.1158/0008-5472.CAN-04-0637. [DOI] [PubMed] [Google Scholar]
- 48.O'Hara AJ, Wang L, Dezube BJ, Harrington WJ, Jr, Damania B, Dittmer DP. Tumor suppressor microRNAs are underrepresented in primary effusion lymphoma and Kaposi sarcoma. Blood. 2009;113:5938–5941. doi: 10.1182/blood-2008-09-179168. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.