Abstract
Activation of the glucocorticoid receptor (GR) by endogenous and synthetic glucocorticoids regulates hundreds of genes to control regulatory networks in development, metabolism, cognition and inflammation. Elucidation of the mechanisms that regulate glucocorticoid action has highlighted the dynamic nature of hormone signalling and provides novel insights into genomic glucocorticoid actions. The major factors that regulate GR function include chromatin structure, epigenetics, genetic variation and the pattern of glucocorticoid hormone secretion. We review our current understanding of the mechanisms that contribute to GR signalling and how these contribute to glucocorticoid sensitivity, resistance and side effects.
Keywords: rhythms, pulsatile secretion, epigenetics, chromatin, glucocorticoid, genetic variation, glucocorticoid sensitivity
Introduction
Endogenous glucocorticoid steroid hormones—cortisol in humans and corticosterone in rodents—act to regulate transcriptional pathways in diverse cellular contexts to regulate development, homeostasis, metabolism, cognition and inflammation. Since glucocorticoids have anti-inflammatory and immunosuppressive properties, they are frequently used to treat many inflammatory conditions, from inflammatory arthritis and ulcerative colitis to asthma and skin diseases, while pro-apoptotic properties make them a major component of the treatment of many oncological disorders. Unfortunately, however, long-term and/or high-dose glucocorticoid administration is commonly associated with side effects, from hyperglycaemia, weight gain and hypertension to osteoporosis, depression and decreased immunological function. Furthermore, patients on glucocorticoids can develop reduced glucocorticoid sensitivity and even resistance. Although the prevalence of glucocorticoid resistance is unclear, in part due to poor definitions, some reports suggest up to one-third of patients with asthma [1], RA [2], ulcerative colitis [3] and SLE [4] show altered glucocorticoid sensitivity [5]. Furthermore, 10–30% of patients with acute lymphocytic leukaemia are glucocorticoid resistant, with a higher rate of resistance developing in patients who have had a relapse of leukaemia [6].
The actions of glucocorticoids are predominantly mediated through its receptor, the glucocorticoid receptor (GR). Upon exposure to glucocorticoids, GR undergoes a structural conformational change that drives translocation from the cell cytoplasm into the nucleus. Once in the nucleus, ligand-bound GR is available to interact with regulatory elements in the genome [7–12], where it is known to induce and repress transcription of hundreds of target genes [12, 13]. The expression of GR, a DNA-binding transcription factor of the nuclear receptor superfamily, is nearly ubiquitously expressed [14]. Its biological activities result predominately from interactions with chromatin. Chromatin is a complex of DNA with DNA packaging proteins called nucleosomes. Nucleosomes are made up of histone proteins, which act to compact the length of DNA into the nucleus. The availability of binding sites for GR in the DNA necessitates engagement with chromatin to access the underlying DNA concealed by nucleosomes. Through mechanisms that alter chromatin structure via histone modifications, and structural remodelling, as well as DNA methylation, the accessibility of the DNA sequence can be modified [15]. These factors do not alter the DNA sequence, but can provide epigenetic memory of transcriptional states [16], that is, non-genetic mechanisms that are heritable across cell division and generations. In this review we discuss the complexity of glucocorticoid signalling, in the context of GR interactions with chromatin, and the factors that contribute to steroid sensitivity, resistance and unwanted side effects.
Physiological and pathological patterns of glucocorticoid secretion
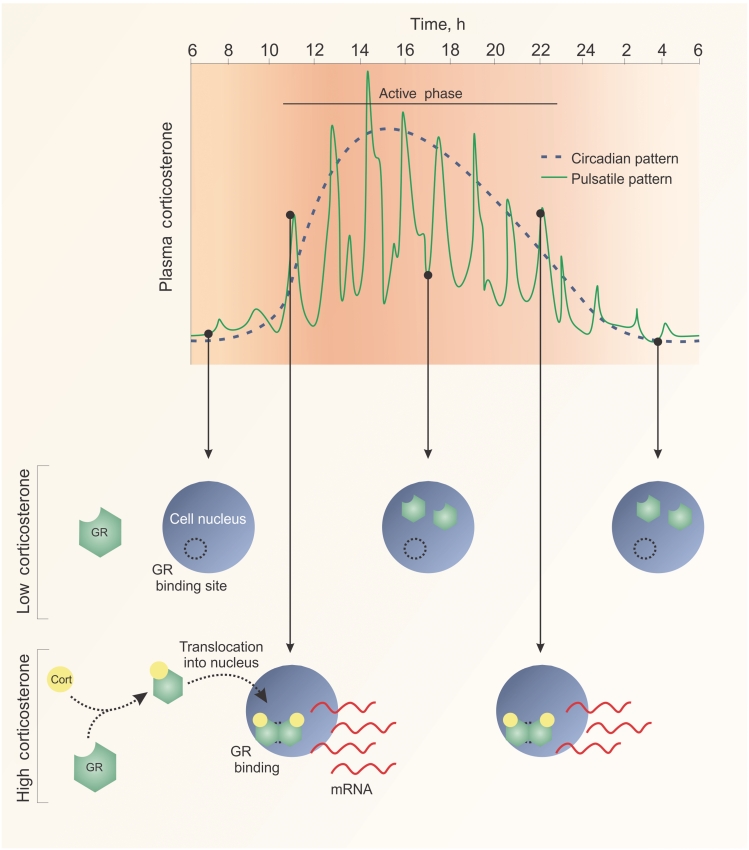
The secretion of glucocorticoid hormones from the adrenal gland is under the control of adrenocorticotropic hormone (ACTH) from the anterior pituitary and corticotropin-releasing hormone (CRH) from the hypothalamus. Additionally, the circulating level of cortisol is regulated through its negative feedback activity on both ACTH and CRH release [17]. These regulatory loops act to maintain normal physiological levels of hormone [17]. The pattern of cortisol secretion is classically circadian, with peak secretion early in the morning in anticipation of waking-related activities (Fig. 1). This circadian pattern, however, is actually made up from an underlying ultradian pattern of hormone secretion, with the largest pulses of secretion occurring early in the morning (Fig. 1). This ultradian pattern is evolutionarily conserved across mammals, implicating pulsatility as an important feature of glucocorticoid signalling [18, 19].
Fig. 1.
Glucocorticoid pulsatility drives transient activation of GR-responsive genes. Murine serum corticosteroid (cort) levels rise in anticipation of the active phase. Hormone levels follow a circadian pattern, although the underlying pattern of hormone secretion is ultradian, where glucocorticoids are released approximately every hour. During a pulse, exposure to hormone drives GR translocation into the nucleus, where it binds to genomic elements to drive transcription. Hormone troughs result in GR dissociation from chromatin, releasing the receptor into the nucleoplasm ready to initiate transcription during further rises in hormone levels. The dynamics of the receptor and hormone secretion patterns allow rapid response to rapidly changing cellular and physiological conditions.
The pattern of glucocorticoid ultradian pulsatility is highly variable in frequency and amplitude. Studies in the rat show sex-specific differences, and both genetic and epigenetic modification of ultradian pattern and glucocorticoid response to stress [17, 18]. Altered patterns of glucocorticoid secretion also occur in response not only to acute stressors, but also to chronic stress states such as obstructive sleep apnoea in man [20], and of particular importance for this review, immune-mediated disease both in man and in experimental animal models [21, 22]. The significance of the hypothalamic-pituitary-adrenal (HPA) axis–immune interaction has been appreciated for many years [23]. As inflammation drives increased production of anti-inflammatory glucocorticoids, the HPA–immune axis acts in a classical negative feedback loop [24]. Evidence from animal experiments strongly implicates the HPA axis in regulating inflammation [25–27]. Accordingly, disruption of the HPA axis by adrenalectomy results in increased disease severity in an experimental model of adjuvant-induced arthritis [27–29]. Significantly, dysregulated rhythms of endogenous glucocorticoid secretion are a key characteristic of inflammation-associated chronic activation of the HPA axis, with loss of circadian rhythm [30] and altered ultradian rhythm [22].
There is also evidence that chronic exposure to altered glucocorticoid secretion following chronic stress can result in glucocorticoid resistance [31, 32]. Indeed, there is evidence that suggests this can be a feature of RA [33]. It is clear, therefore, that there are dynamic patterns of glucocorticoid secretion that respond to both physiological and pathological states, and suggest that during some of these pathological states the aberrant glucocorticoid secretory pattern could alter glucocorticoid signalling and resistance.
Another situation in which there is an altered pattern of plasma glucocorticoids is found during glucocorticoid therapy—either when it is given therapeutically as an anti-inflammatory or immune modulator, or when it is given as replacement therapy in Addison's disease, congenital adrenal hyperplasia or hypopituitarism. Unfortunately, current treatment protocols are unable to reproduce either the normal ultradian pattern of hormone secretion or the pre-awakening circadian rise of hormone secretion, although new delayed-release preparations are being developed that may improve the circadian pattern [34]. There is now considerable evidence that pulsatile changes in plasma glucocorticoid levels result in gene pulsing mediated by transient GR activation [35, 36]. Pulsatile patterns therefore exert homeostatic control through GR-dependent transcription regulation that rapidly responds to circulating hormone levels (Fig. 1) [36, 37]. Conversely, constant non-oscillatory hormone levels result in continuous transcription, aberrant mRNA accumulation and abnormal protein levels [36]. We suggest that a treatment regimen comprising pulsatile glucocorticoid patterns over a circadian rhythm might confer therapeutic benefits while minimizing undesired side effects.
Genomic actions and cell specificity of GR
The organization of DNA in eukaryotic cells as chromatin permits compaction of DNA into the nuclear space. DNA-binding factors, such as GR, must overcome the chromatin barrier to access the underlying DNA sequences in regulatory elements to drive transcription control. Accessibility of chromatin across the genome, however, is highly variable. Recent genome-wide data have shown that active regulatory elements, such as promoters and enhancers, are openly accessible chromatin domains [38, 39].
Upon ligand activation, GR acts as a sequence-specific transcription factor, binding to a consensus DNA motif, the glucocorticoid response element (GRE), to drive gene expression [40]. Despite widespread expression, GR regulation of transcriptional programmes is highly cell and tissue specific, binding to distinct genomic loci in different cellular contexts [10–12]. With hundreds of different cell types in complex eukaryotes containing identical genomic content, the selection of specific binding sites in different cells has remained an important question. Chromatin has emerged over the past few decades as a critical player in transcriptional regulation. Control of transcription is thought to occur through altering chromatin structure or epigenetics by mechanisms such as chromatin remodelling, DNA methylation and modifications of histones. Indeed, the accessibility of chromatin has been shown to direct the binding of GR to regulatory elements [10, 11]. Importantly, cell-specific GR-binding events are associated with cell-specific regulatory elements marked by accessible chromatin [10, 11]. The vast majority of interactions between GR and chromatin have been shown to be predetermined before hormone, that is, GR predominantly binds to the genome at chromatin where the DNA is accessible before hormone and thus independent of hormone action. These observations implicate open chromatin as a major player in GR binding and define the cell-specific landscape of GR action, suggesting the cellular context contributes to the pre-setting of cell-specific hormone action [11].
In addition to the direct actions of GR on DNA, the receptor interacts with a large cohort of other DNA-binding factors [41]. Known cooperative protein–protein interactions include nuclear factor 1 (NF1), CCAAT/enhancer-binding proteins (C/EBP) and octamer transcription factors [42–44]. Through interacting with these accessory factors, GR-dependent transcription is thought to be dynamic and variable, including cell-specific interactions with accessory factors (Fig. 2A) [11]. Therefore the regulation of cell-specific genes by glucocorticoids is dependent on the expression and activity of interacting partners. These interactions allow diversity in regulatory control over activated genes and also mean that alterations in accessory factor activity could modify the sensitivity and resistance to hormone action.
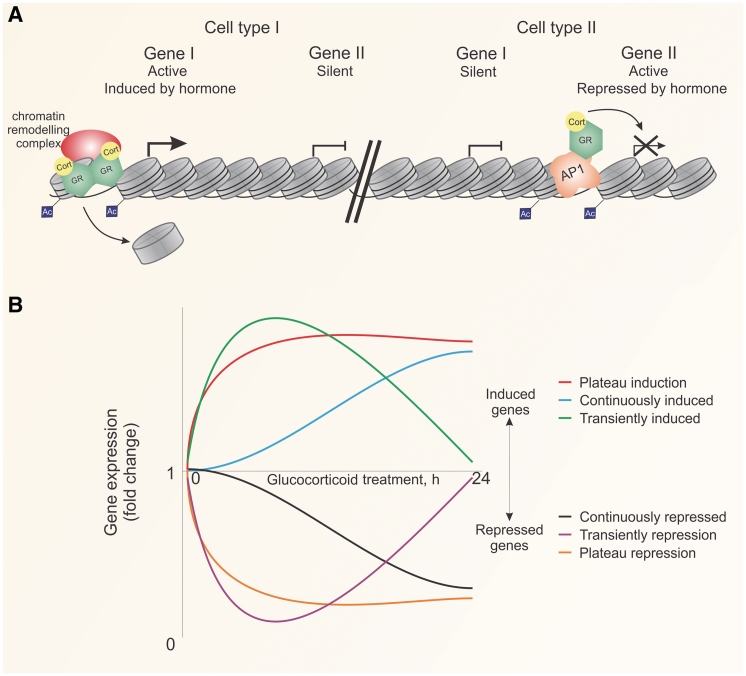
Fig. 2.
Cell- and gene-specific actions of GR. (A) Chromatin accessibility is a major determinant of receptor binding to chromatin. Active genes are shown to have accessible DNA and acetylated (Ac) histones. Genes induced by GR require homodimers that bind directly to DNA at GREs. Hormone-dependent gene repression is mediated by homomeric GR protein–protein interactions with other transcription factors such as AP1. (B) Transcription output in the presence of constant hormone is kinetically diverse. Induced and repressed genes can undergo phases of up- and down-regulation. Induced genes can be continuously (blue) or transiently (green) induced, or induced to a maintained plateau state (red). Similarly, repressed genes can be continuously (black) or transiently (purple) repressed, or repressed in a plateau state over time (orange). Lines represent classes of genes. The kinetics of transcription is therefore gene specific. Cort: corticosterone/cortisol, Ac: acetylation.
Temporal dynamics of gene regulation by the GR
GR is known to positively and negatively regulate transcription [12, 13, 45]. GR-dependent induction and repression of genes play critical roles in metabolic, circadian and inflammatory signalling networks. Inductive actions of homodimeric GR are mediated through direct binding to DNA at GREs [46, 47]. Upon DNA binding, GR promotes the recruitment of chromatin-modifying co-factors and the transcriptional machinery, including RNA polymerase II (Pol II) to drive transcription [48]. The recruitment of co-factors such as Brg1 and the histone acetyltransferase (HAT) CBP/p300 by GR alters chromatin structure to an accessible state poised for gene activation (Fig. 2) [49, 50]. For instance, the chromatin-remodelling activity of Brg1 is thought to alter the positioning and composition of nucleosomes, while HATs acetylate the histones H3 and H4 [51–53]. Both mechanisms are thought to alter the contacts between nucleosomes and DNA, exposing the DNA to regulatory factor binding [54, 55].
The binding of GR to chromatin and the hormone-dependent remodelling of chromatin is highly dynamic. Using fluorescently tagged receptor coupled with photobleaching experiments to monitor recovery times, GR was found to rapidly cycle chromatin on and off in living cells in seconds to minutes [56, 57]. In contrast to the classic model where binding of transcription factors to DNA is represented by stable complexes, the current model suggests transient transcription factor binding to chromatin [37]. Here, GR rapidly engages and disengages the chromatin template, during which Brg1-dependent chromatin remodelling concomitantly undergoes dynamic exchange [58, 59].
In addition to the rapid exchange of GR on chromatin, a slower cyclical activity is imposed on GR by the ultradian rhythm of ligand exposure, a phasic GR action regulated by the deterministic mechanism of the ligand pulse pattern [60]. The phasic action of GR is evident as cyclical shifts in the net equilibrium of GR at regulatory elements towards a bound state [61]. Precisely tracking the rhythm of pulsatile hormone and cyclical GR activity are cyclical changes in recruitment of the HAT complex CBP/P300. Phasic CBP/P300 recruitment results in rapid and reversible increases in histone acetylation at regulatory regions in glucocorticoid-target genes, again in phase with the ultradian hormone pattern [61, 62]. As histone acetylation is associated with recruitment of chromatin remodelling factors of the SWI/SNF family such as Brg1, allowing access of Pol II and auxiliary transcription machinery to the transcription start site (TSS), it is often closely correlated to transcriptional activity [52]. Consistent with this, Pol II recruitment also exhibits ultradian rhythm-directed cyclical activity at the TSS of glucocorticoid pulse regulated genes. These cyclical changes in acetylation status of glucocorticoid-target genes, and the concomitant cycling of GR, its regulatory co-factors and Pol II at the TSS, are therefore proposed as the mechanism for temporal regulation of the gene-pulsing phenomenon associated with ultradian glucocorticoid rhythm.
Therefore the two components—the stochastic receptor and the deterministic ligand secretory pattern—appear to have evolved together to establish and maintain an optimal system of transcriptional regulation. The dynamic action is not unique to GR, extending to other transcription factors, and represents a general mechanism that permits continuous sampling of the cellular milieu [37]. In the case of GR, the mechanism accommodates fluctuations in hormone levels and thus can rapidly respond to physiological and pathological glucocorticoid secretory patterns. The general requirement for accessible chromatin also suggests that chromatin structure, and factors that regulate chromatin accessibility, could influence hormone sensitivity and resistance.
Repressive actions of the GR
The interaction between GR and other transcription factors such as activator protein 1 (AP1) and nuclear factor kappa B (NF-κB) is known to repress gene activity through transrepression [45, 63–65]. GR-dependent perturbation of these pro-inflammatory factors is thought to occur through protein–protein interactions [45, 63, 64]. While transactivation requires GR homodimers, transrepression is mediated through GR monomers (Fig. 2A) [66, 67]. Knock-out of GR from mouse renders them unviable, although interestingly, GR dimerization mutants, where GR cannot form homodimers, are viable, suggesting that transrepression by monomeric GR is sufficient for survival [68]. In addition, GR and AP1 can interact at composite elements, where each factor binds directly to DNA, but their proximity results in hormone-dependent modulation of AP1 action [45]. The interactions between GR and pro-inflammatory factors involve altered recruitment of co-regulators that directly inhibit Pol II elongation [69, 70]. Additionally, GR is a target of histone deacetylase 2 (HDAC2), resulting in de-acetylation of the receptor and preferential interactions with NF-κB [71].
The activity of NF-κB and AP1 are critical to numerous inflammatory conditions, including arthritis and asthma [71, 72]. In addition to transrepression, gene induction contributes to anti-inflammation by inducing the expression of immune modulators in some cell types, including the negative regulator of NF-κB, IκB-α [73, 74]. The repressive action of glucocorticoids on NF-κB and AP1 activity is therefore context dependent, with the sensitivity of glucocorticoid action dependent on the cell type and the expression and activity of pro-inflammatory proteins and co-regulators. For example, over-expression of AP1 has been shown to impair GR repression, contributing to resistance [75]. Furthermore, GR-dependent induction of mitogen-activated protein kinase phosphatase 1 (MKP1) and glucocorticoid-induced leucine zipper (GILZ) have anti-inflammatory actions, where disruption of their regulation alters hormone sensitivity in RA [76–78]. In respiratory conditions such as chronic obstructive pulmonary disease (COPD), reduced expression and activity of HDAC2 is associated with increased inflammatory gene expression and confers glucocorticoid resistance [71].
The design of receptor ligands that dissociate GR-mediated induction and repression are of much interest, with the aim of reducing side-effect profiles [79]. However, the mechanisms regulating GR gene induction and repression are not always clear, as GR can also induce AP1 activity [45]. The effect of GR on AP1 activity, whether positive or negative, is thought to depend on the composition of the AP1 complex, a multi-subunit family that can form homo- and heterodimers, and the DNA sequence of regulatory elements [80]. Furthermore, some side-effect profiles are related to interactions between GR and pro-inflammatory factors. Indeed, the osteoporotic side effect of glucocorticoids is thought to involve GR and AP1 interactions that inhibit osteoblast differentiation [81]. It is therefore unclear what role selective GR modulators will have in the clinic, although the characterization of some compounds has suggested reduced side effects, including osteoporosis [82]. Their clinical efficacy and safety will need to be extensively evaluated.
The distinction between GR inductive and repressive effects is a conventional, but limited, definition. Using genome-wide microarray approaches to evaluate gene expression over a long time course, the regulation of genes by GR displays complex kinetics [13]. In contrast to simple induction or repression, hundreds of genes undergo positive and negative regulatory phases in the presence of constant hormone (Fig. 2B) [13]. Over hours of constant glucocorticoid exposure, induced genes can be rapidly induced to a plateau phase, transiently induced or slowly and continuously induced. Repressed genes also exhibit similar kinetics patterns, showing rapid repression that plateaus transient repression or continuous repression (Fig. 2B). These classes of kinetic gene activity suggest gene-specific regulatory control by hormones. These classes are likely to be independent of hormone exposure pattern, constant or pulsatile, and independent of glucocorticoid type, endogenous or synthetic, although this has not been definitively shown. These kinetic implicate modifications to chromatin or regulatory factors, such as GR and recruited co-factors that act to modify the transcriptional output in a gene-specific manner. For instance, the acetylation of HDAC1 upon GR recruitment is thought to contribute to refractory phases in induced gene transcription [83]. The complexity and dynamics of glucocorticoid action are only beginning to be understood. The relevance of kinetic patterns of GR-dependent gene expression in vivo is not yet clear, particularly in underlying circadian and pulsatile glucocorticoid secretory modes. Nevertheless, we propose that the interplay between non-receptor transcription factors and co-regulators plays a critical role in GR action and provides a mechanism of plasticity in response the physiological and pathological signals.
Genetic and epigenetic factors in glucocorticoid sensitivity and resistance
Primary generalized glucocorticoid resistance is a rare condition associated with compensatory increases in plasma ACTH and cortisol [5]. Patients present with features of mineralocorticoid and adrenal androgen excess due to concomitant adrenal hyperplasia and elevated adrenal steroids [84]. The condition results from familial or sporadic mutations of the GR gene, resulting in impaired expression, ligand binding or nuclear translocation [84]. Mutations in co-regulator genes might, in principle, also confer glucocorticoid resistance. Indeed, defects in the steroid receptor co-activators (SRCs) were observed in a pan-steroid resistance syndrome [85]. Generally, genetic mutations are unlikely to contribute to resistance affecting specific cells in common inflammatory conditions such as arthritis. Sporadic genetic mutations are more likely in rapidly dividing cells, particularly cancers. GR mutations have been associated with leukaemic cell lines associated with resistance to chemotherapeutic apoptotic effects of glucocorticoid, although their contribution to resistance in patients is contentious [86]. Similarly, mutations in Brg1 have been found in lung cancers, and as an important co-factor for GR action, mutations in Brg1 could contribute to glucocorticoid resistance [87].
Interestingly, one-third of the normal population display in vitro resistance to glucocorticoids and will predictably fail to respond to clinical treatment [5]. Single nucleotide polymorphisms (SNPs) contribute extensively to human variation and possible glucocorticoid action. Polymorphisms of the CS-binding globulin (CBG), Toll-like receptors (TLRs) and macrophage migration inhibitory factor (MIF) genes are associated with altered serum cortisol levels and glucocorticoid sensitivity [5, 32]. At the level of DNA binding, SNPs have also been shown to cause aberrant transcription factor recruitment [88]. Whole-genome analyses of the DNA-binding factors NF-κB and CCCTC-binding factor (CTCF) have shown that genetic variation extensively contributes to differential factor occupancy [89, 90]. These observations suggest that SNPs could alter GR-binding patterns and alter sensitivity to glucocorticoids at specific target genes. Additionally, variation in the GRE sequence modifies GR structure by acting as an allosteric modulator [91]. The structural difference might promote specific interactions, with co-regulators affecting GR-dependent gene regulation (Fig. 3A). Therefore SNPs in GREs could modify GR action in a gene- and individual-specific fashion.
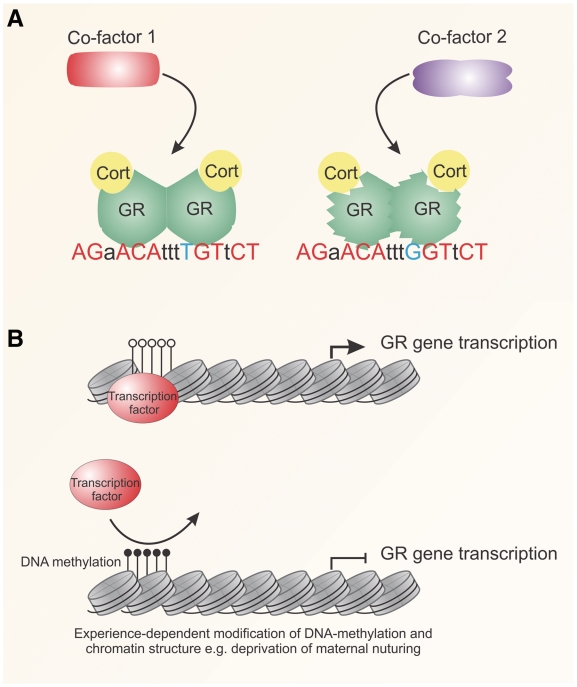
Fig. 3.
Genetic and epigenetic mechanisms in GR action. (A) DNA is an allosteric modulator, altering the structure of the receptor when bound to DNA. SNPs (blue) at GREs (red) could alter the interactions with co-factors by influencing GR structure. The effects might contribute to gene- and individual-specific gene regulation. (B) DNA methylation, an epigenetic mechanism, acts to silence gene transcription by altering chromatin structure. Individual-specific experiences, such as maternal nurturing, can alter DNA methylation patterns, producing expression patterns heritable across generations. The reduction in GR expression causes resistance to hormone action in specific tissues. Cort: cortisol.
DNA methylation, an epigenetic mechanism associated with chromatin condensation and gene silencing, has been shown to modify GR gene expression [92]. In an experience-dependent manner, DNA methylation of the GR promoter in specific brain regions is associated with reduced GR expression levels, and consequently resistance to glucocorticoids. In rodent models, deprivation of maternal nurturing increases DNA methylation of the GR promoter in the hippocampus, resulting in trans-generational inheritance of the methylation pattern (Fig. 3B) [92]. The observation suggests that DNA methylation is dynamic and programmable. Similarly, GR promoter methylation was observed in post-mortem hippocampi of suicide victims with a history of child abuse, suggesting interplay between methylation status and individual experience [93]. Experience-dependent epigenetic programming might therefore play a role in other physiological and pathological conditions. Genetic and epigenetic variation is therefore likely to contribute to glucocorticoid sensitivity, although the incidence of these variants in the general population is not fully understood.
Concluding remarks
The diverse and disparate transcription networks regulated by glucocorticoids result from ubiquitous receptor expression, but cell-specific actions [11]. In particular, the anti-inflammatory and immunosuppressive effects are extensively exploited clinically, making glucocorticoids one of the most commonly prescribed classes of therapeutics. The chronic nature of many of the inflammatory conditions, including RA, SLE, asthma and IBD, requires long-term glucocorticoid administration. The treatment paradigms frequently result in side effects that reduce their tolerance and, not uncommonly, induce the development of glucocorticoid resistance [5].
Factors that contribute to the dynamics and plasticity of glucocorticoid signalling, from the patterns and levels of hormone secretion to the binding of receptor at specific DNA sequences, represent potential mechanisms regulating sensitivity and resistance. Technological advances in molecular biology such as genome-wide techniques have advanced our understanding of the molecular mechanisms of GR [11]. It is vital that our current understanding is translated into physiological conditions and into the clinic.
Advances in drug design and therapeutics principles should extend beyond dissociating the desired anti-inflammatory effects and reducing side effects, targeting cell-specific functions of glucocorticoids. High-throughput technologies, including genomics, proteomics and metabolomics, as well as chemical and RNA interference screening, could be employed to discover important and novel targets in glucocorticoid action in multiple cell types under different physiological and pathological backgrounds. These studies will be crucial for understanding the networks regulating glucocorticoid action, sensitivity and resistance in health and disease.

Acknowledgements
Funding: We acknowledge the financial support provided by the Wellcome Trust, Wellcome Trust Programme grant reference 089647/Z/09/cZ (to S.L.L. and B.L.C.-C.).
Disclosure statement: The authors have declared no conflicts of interest.
References
- 1.Corrigan CJ, Brown PH, Barnes NC, et al. Glucocorticoid resistance in chronic asthma. Glucocorticoid pharmacokinetics, glucocorticoid receptor characteristics, and inhibition of peripheral blood T cell proliferation by glucocorticoids in vitro. Am Rev Respir Dis. 1991;144:1016–25. doi: 10.1164/ajrccm/144.5.1016. [DOI] [PubMed] [Google Scholar]
- 2.Kirkham BW, Corkill MM, Davison SC, Panayi GS. Response to glucocorticoid treatment in rheumatoid arthritis: in vitro cell mediated immune assay predicts in vivo responses. J Rheumatol. 1991;18:821–5. [PubMed] [Google Scholar]
- 3.Faubion WA, Jr, Loftus EV, Jr, Harmsen WS, Zinsmeister AR, Sandborn WJ. The natural history of corticosteroid therapy for inflammatory bowel disease: a population-based study. Gastroenterology. 2001;121:255–60. doi: 10.1053/gast.2001.26279. [DOI] [PubMed] [Google Scholar]
- 4.Greenstein B. Steroid resistance: implications for lupus. Lupus. 1994;3:143. doi: 10.1177/096120339400300302. [DOI] [PubMed] [Google Scholar]
- 5.Leung DY, Bloom JW. Update on glucocorticoid action and resistance. J Allergy Clin Immunol. 111:3–22. doi: 10.1067/mai.2003.97. quiz 23. [DOI] [PubMed] [Google Scholar]
- 6.Kaspers GJ, Pieters R, Klumper E, De Waal FC, Veerman AJ. Glucocorticoid resistance in childhood leukemia. Leuk Lymphoma. 1994;13:187–201. doi: 10.3109/10428199409056282. [DOI] [PubMed] [Google Scholar]
- 7.Rigaud G, Roux J, Pictet R, Grange T. In vivo footprinting of rat TAT gene: dynamic interplay between the glucocorticoid receptor and a liver-specific factor. Cell. 1991;67:977–86. doi: 10.1016/0092-8674(91)90370-e. [DOI] [PubMed] [Google Scholar]
- 8.Zaret KS, Yamamoto KR. Reversible and persistent changes in chromatin structure accompany activation of a glucocorticoid-dependent enhancer element. Cell. 1984;38:29–38. doi: 10.1016/0092-8674(84)90523-3. [DOI] [PubMed] [Google Scholar]
- 9.Richard-Foy H, Hager GL. Sequence-specific positioning of nucleosomes over the steroid-inducible MMTV promoter. EMBO J. 1987;6:2321–8. doi: 10.1002/j.1460-2075.1987.tb02507.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.John S, Sabo PJ, Johnson TA, et al. Interaction of the glucocorticoid receptor with the chromatin landscape. Mol Cell. 2008;29:611–24. doi: 10.1016/j.molcel.2008.02.010. [DOI] [PubMed] [Google Scholar]
- 11.John S, Sabo PJ, Thurman RE, et al. Chromatin accessibility pre-determines glucocorticoid receptor binding patterns. Nat Genet. 2011;43:264–8. doi: 10.1038/ng.759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.So AY, Chaivorapol C, Bolton EC, Li H, Yamamoto KR. Determinants of cell- and gene-specific transcriptional regulation by the glucocorticoid receptor. PLoS Genet. 2007;3:e94. doi: 10.1371/journal.pgen.0030094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.John S, Johnson TA, Sung MH, et al. Kinetic complexity of the global response to glucocorticoid receptor action. Endocrinology. 2009;150:1766–74. doi: 10.1210/en.2008-0863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bookout AL, Jeong Y, Downes M, Yu RT, Evans RM, Mangelsdorf DJ. Anatomical profiling of nuclear receptor expression reveals a hierarchical transcriptional network. Cell. 2006;126:789–99. doi: 10.1016/j.cell.2006.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Biddie SC. Chromatin architecture and the regulation of nuclear receptor inducible transcription. J Neuroendocrinol. 2011;23:94–106. doi: 10.1111/j.1365-2826.2010.02079.x. [DOI] [PubMed] [Google Scholar]
- 16.Bird A. Perceptions of epigenetics. Nature. 2007;447:396–8. doi: 10.1038/nature05913. [DOI] [PubMed] [Google Scholar]
- 17.Lightman SL, Conway-Campbell BL. The crucial role of pulsatile activity of the HPA axis for continuous dynamic equilibration. Nat Rev Neurosci. 2010;11:710–8. doi: 10.1038/nrn2914. [DOI] [PubMed] [Google Scholar]
- 18.Windle RJ, Wood SA, Lightman SL, Ingram CD. The pulsatile characteristics of hypothalamo-pituitary-adrenal activity in female Lewis and Fischer 344 rats and its relationship to differential stress responses. Endocrinology. 1998;139:4044–52. doi: 10.1210/endo.139.10.6238. [DOI] [PubMed] [Google Scholar]
- 19.Young EA, Abelson J, Lightman SL. Cortisol pulsatility and its role in stress regulation and health. Front Neuroendocrinol. 2004;25:69–76. doi: 10.1016/j.yfrne.2004.07.001. [DOI] [PubMed] [Google Scholar]
- 20.Henley DE, Russell GM, Douthwaite JA, et al. Hypothalamic-pituitary-adrenal axis activation in obstructive sleep apnea: the effect of continuous positive airway pressure therapy. J Clin Endocrinol Metab. 2009;94:4234–42. doi: 10.1210/jc.2009-1174. [DOI] [PubMed] [Google Scholar]
- 21.Perry MG, Kirwan JR, Jessop DS, Hunt LP. Overnight variations in cortisol, interleukin 6, tumour necrosis factor alpha and other cytokines in people with rheumatoid arthritis. Ann Rheum Dis. 2009;68:63–8. doi: 10.1136/ard.2007.086561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Windle RJ, Wood SA, Kershaw YM, Lightman SL, Ingram CD, Harbuz MS. Increased corticosterone pulse frequency during adjuvant-induced arthritis and its relationship to alterations in stress responsiveness. J Neuroendocrinol. 2001;13:905–11. doi: 10.1046/j.1365-2826.2001.00715.x. [DOI] [PubMed] [Google Scholar]
- 23.Munck A, Guyre PM, Holbrook NJ. Physiological functions of glucocorticoids in stress and their relation to pharmacological actions. Endocr Rev. 1984;5:25–44. doi: 10.1210/edrv-5-1-25. [DOI] [PubMed] [Google Scholar]
- 24.Besedovsky H, del Rey A, Sorkin E, Dinarello CA. Immunoregulatory feedback between interleukin-1 and glucocorticoid hormones. Science. 1986;233:652–4. doi: 10.1126/science.3014662. [DOI] [PubMed] [Google Scholar]
- 25.Chowdrey HS, Larsen PJ, Harbuz MS, et al. Evidence for arginine vasopressin as the primary activator of the HPA axis during adjuvant-induced arthritis. Br J Pharmacol. 1995;116:2417–24. doi: 10.1111/j.1476-5381.1995.tb15089.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sarlis NJ, Stephanou A, Knight RA, Lightman SL, Chowdrey HS. Effects of glucocorticoids and chronic inflammatory stress upon anterior pituitary interleukin-6 mRNA expression in the rat. Br J Rheumatol. 1993;32:653–7. doi: 10.1093/rheumatology/32.8.653. [DOI] [PubMed] [Google Scholar]
- 27.Chover-Gonzalez AJ, Harbuz MS, Lightman SL. Effect of adrenalectomy and stress on interleukin-1 beta-mediated activation of hypothalamic corticotropin-releasing factor mRNA. J Neuroimmunol. 1993;42:155–60. doi: 10.1016/0165-5728(93)90005-j. [DOI] [PubMed] [Google Scholar]
- 28.Perretti M, Mugridge KG, Becherucci C, Parente L. Evidence that interleukin-1 and lipoxygenase metabolites mediate the lethal effect of complete Freund's adjuvant in adrenalectomized rats. Lymphokine Cytokine Res. 1991;10:239–43. [PubMed] [Google Scholar]
- 29.Yang YH, Hutchinson P, Leech M, Morand EF. Exacerbation of adjuvant arthritis by adrenalectomy is associated with reduced leukocyte lipocortin 1. J Rheumatol. 1997;24:1758–64. [PubMed] [Google Scholar]
- 30.Sarlis NJ, Chowdrey HS, Stephanou A, Lightman SL. Chronic activation of the hypothalamo-pituitary-adrenal axis and loss of circadian rhythm during adjuvant-induced arthritis in the rat. Endocrinology. 1992;130:1775–9. doi: 10.1210/endo.130.4.1312424. [DOI] [PubMed] [Google Scholar]
- 31.Silverman MN, Sternberg EM. Neuroendocrine-immune interactions in rheumatoid arthritis: mechanisms of glucocorticoid resistance. Neuroimmunomodulation. 2008;15:19–28. doi: 10.1159/000135620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Venkataraman S, Munoz R, Candido C, Witchel SF. The hypothalamic-pituitary-adrenal axis in critical illness. Rev Endocr Metab Disord. 2007;8:365–73. doi: 10.1007/s11154-007-9058-9. [DOI] [PubMed] [Google Scholar]
- 33.Harbuz MS, Jessop DS. Is there a defect in cortisol production in rheumatoid arthritis? Rheumatology. 1999;38:298–302. doi: 10.1093/rheumatology/38.4.298. [DOI] [PubMed] [Google Scholar]
- 34.Buttgereit F, Doering G, Schaeffler A, et al. Efficacy of modified-release versus standard prednisone to reduce duration of morning stiffness of the joints in rheumatoid arthritis [CAPRA-1]: a double-blind, randomised controlled trial. Lancet. 2008;371:205–14. doi: 10.1016/S0140-6736(08)60132-4. [DOI] [PubMed] [Google Scholar]
- 35.Conway-Campbell BL, Sarabdjitsingh RA, McKenna MA, et al. Glucocorticoid ultradian rhythmicity directs cyclical gene pulsing of the clock gene period 1 in rat hippocampus. J Neuroendocrinol. 2010;22:1093–100. doi: 10.1111/j.1365-2826.2010.02051.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stavreva DA, Wiench M, John S, et al. Ultradian hormone stimulation induces glucocorticoid receptor-mediated pulses of gene transcription. Nat Cell Biol. 2009;11:1093–102. doi: 10.1038/ncb1922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Biddie SC, Hager GL. Glucocorticoid receptor dynamics and gene regulation. Stress. 2009;12:193–205. doi: 10.1080/10253890802506409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Boyle AP, Davis S, Shulha HP, et al. High-resolution mapping and characterization of open chromatin across the genome. Cell. 2008;132:311–22. doi: 10.1016/j.cell.2007.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sabo PJ, Kuehn MS, Thurman R, et al. Genome-scale mapping of DNase I sensitivity in vivo using tiling DNA microarrays. Nat Methods. 2006;3:511–8. doi: 10.1038/nmeth890. [DOI] [PubMed] [Google Scholar]
- 40.Evans RM. The steroid and thyroid hormone receptor superfamily. Science. 1988;240:889–95. doi: 10.1126/science.3283939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schule R, Muller M, Kaltschmidt C, Renkawitz R. Many transcription factors interact synergistically with steroid receptors. Science. 1988;242:1418–20. doi: 10.1126/science.3201230. [DOI] [PubMed] [Google Scholar]
- 42.Cordingley MG, Hager GL. Binding of multiple factors to the MMTV promoter in crude and fractionated nuclear extracts. Nucleic Acids Res. 1988;16:609–28. doi: 10.1093/nar/16.2.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Archer TK, Cordingley MG, Wolford RG, Hager GL. Transcription factor access is mediated by accurately positioned nucleosomes on the mouse mammary tumor virus promoter. Mol Cell Biol. 1991;11:688–98. doi: 10.1128/mcb.11.2.688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Belikov S, Astrand C, Wrange O. FoxA1 binding directs chromatin structure and the functional response of a glucocorticoid receptor-regulated promoter. Mol Cell Biol. 2009;29:5413–25. doi: 10.1128/MCB.00368-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Diamond MI, Miner JN, Yoshinaga SK, Yamamoto KR. Transcription factor interactions: selectors of positive or negative regulation from a single DNA element. Science. 1990;249:1266–72. doi: 10.1126/science.2119054. [DOI] [PubMed] [Google Scholar]
- 46.Tsai SY, Carlstedt-Duke J, Weigel NL, et al. Molecular interactions of steroid hormone receptor with its enhancer element: evidence for receptor dimer formation. Cell. 1988;55:361–9. doi: 10.1016/0092-8674(88)90059-1. [DOI] [PubMed] [Google Scholar]
- 47.Wrange O, Eriksson P, Perlmann T. The purified activated glucocorticoid receptor is a homodimer. J Biol Chem. 1989;264:5253–9. [PubMed] [Google Scholar]
- 48.Firzlaff JM, Diggelmann H. Dexamethasone increases the number of RNA polymerase II molecules transcribing integrated mouse mammary tumor virus DNA and flanking mouse sequences. Mol Cell Biol. 1984;4:1057–62. doi: 10.1128/mcb.4.6.1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fryer CJ, Archer TK. Chromatin remodelling by the glucocorticoid receptor requires the BRG1 complex. Nature. 1998;393:88–91. doi: 10.1038/30032. [DOI] [PubMed] [Google Scholar]
- 50.Chakravarti D, LaMorte VJ, Nelson MC, et al. Role of CBP/P300 in nuclear receptor signalling. Nature. 1996;383:99–103. doi: 10.1038/383099a0. [DOI] [PubMed] [Google Scholar]
- 51.Ito T, Ikehara T, Nakagawa T, Kraus WL, Muramatsu M. p300-mediated acetylation facilitates the transfer of histone H2A-H2B dimers from nucleosomes to a histone chaperone. Genes Dev. 2000;14:1899–907. [PMC free article] [PubMed] [Google Scholar]
- 52.Lee DY, Hayes JJ, Pruss D, Wolffe AP. A positive role for histone acetylation in transcription factor access to nucleosomal DNA. Cell. 1993;72:73–84. doi: 10.1016/0092-8674(93)90051-q. [DOI] [PubMed] [Google Scholar]
- 53.Dechassa ML, Sabri A, Pondugula S, et al. SWI/SNF has intrinsic nucleosome disassembly activity that is dependent on adjacent nucleosomes. Mol Cell. 2010;38:590–602. doi: 10.1016/j.molcel.2010.02.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bouazoune K, Miranda TB, Jones PA, Kingston RE. Analysis of individual remodeled nucleosomes reveals decreased histone-DNA contacts created by hSWI/SNF. Nucleic Acids Res. 2009;37:5279–94. doi: 10.1093/nar/gkp524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Reinke H, Horz W. Histones are first hyperacetylated and then lose contact with the activated PHO5 promoter. Mol Cell. 2003;11:1599–607. doi: 10.1016/s1097-2765(03)00186-2. [DOI] [PubMed] [Google Scholar]
- 56.Becker M, Baumann C, John S, et al. Dynamic behavior of transcription factors on a natural promoter in living cells. EMBO Rep. 2002;3:1188–94. doi: 10.1093/embo-reports/kvf244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.McNally JG, Muller WG, Walker D, Wolford R, Hager GL. The glucocorticoid receptor: rapid exchange with regulatory sites in living cells. Science. 2000;287:1262–5. doi: 10.1126/science.287.5456.1262. [DOI] [PubMed] [Google Scholar]
- 58.Nagaich AK, Walker DA, Wolford R, Hager GL. Rapid periodic binding and displacement of the glucocorticoid receptor during chromatin remodeling. Mol Cell. 2004;14:163–74. doi: 10.1016/s1097-2765(04)00178-9. [DOI] [PubMed] [Google Scholar]
- 59.Fletcher TM, Xiao N, Mautino G, et al. ATP-dependent mobilization of the glucocorticoid receptor during chromatin remodeling. Mol Cell Biol. 2002;22:3255–63. doi: 10.1128/MCB.22.10.3255-3263.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Conway-Campbell BL, Hager GL, Lightman SL. Molecular dynamics of ultradian glucocorticoid receptor action. Mol Cell Endocrinol. 2011 doi: 10.1016/j.mce.2011.08.014. in press. [DOI] [PubMed] [Google Scholar]
- 61.Conway-Campbell BL, George CL, Pooley JR, et al. The HSP90 molecular chaperone cycle regulates cyclical transcriptional dynamics of the glucocorticoid receptor and its co-regulatory molecules CBP/P300 during ultradian ligand treatment. Mol Endocrinol. 2011;25:944–54. doi: 10.1210/me.2010-0073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.George CL, Pooley JR, Knight DM, Lightman SL, Conway-Campbell BL. Ultradian glucocorticoid exposure results in rapid & distinct cycles of transcriptional co-activator protein recruitment and acetylation at the Period 1 promoter. The 93rd Annual Meeting of the Endocrine Society; Boston: 2011. [Google Scholar]
- 63.Yang-Yen HF, Chambard JC, Sun YL, et al. Transcriptional interference between c-Jun and the glucocorticoid receptor: mutual inhibition of DNA binding due to direct protein-protein interaction. Cell. 1990;62:1205–15. doi: 10.1016/0092-8674(90)90396-v. [DOI] [PubMed] [Google Scholar]
- 64.Schule R, Rangarajan P, Kliewer S, et al. Functional antagonism between oncoprotein c-Jun and the glucocorticoid receptor. Cell. 1990;62:1217–26. doi: 10.1016/0092-8674(90)90397-w. [DOI] [PubMed] [Google Scholar]
- 65.Caldenhoven E, Liden J, Wissink S, et al. Negative cross-talk between RelA and the glucocorticoid receptor: a possible mechanism for the antiinflammatory action of glucocorticoids. Mol Endocrinol. 1995;9:401–12. doi: 10.1210/mend.9.4.7659084. [DOI] [PubMed] [Google Scholar]
- 66.Adler S, Waterman ML, He X, Rosenfeld MG. Steroid receptor-mediated inhibition of rat prolactin gene expression does not require the receptor DNA-binding domain. Cell. 1988;52:685–95. doi: 10.1016/0092-8674(88)90406-0. [DOI] [PubMed] [Google Scholar]
- 67.Reichardt HM, Tuckermann JP, Gottlicher M, et al. Repression of inflammatory responses in the absence of DNA binding by the glucocorticoid receptor. EMBO J. 2001;20:7168–73. doi: 10.1093/emboj/20.24.7168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Reichardt HM, Kaestner KH, Tuckermann J, et al. DNA binding of the glucocorticoid receptor is not essential for survival. Cell. 1998;93:531–41. doi: 10.1016/s0092-8674(00)81183-6. [DOI] [PubMed] [Google Scholar]
- 69.Luecke HF, Yamamoto KR. The glucocorticoid receptor blocks P-TEFb recruitment by NFkappaB to effect promoter-specific transcriptional repression. Genes Dev. 2005;19:1116–27. doi: 10.1101/gad.1297105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Nissen RM, Yamamoto KR. The glucocorticoid receptor inhibits NFkappaB by interfering with serine-2 phosphorylation of the RNA polymerase II carboxy-terminal domain. Genes Dev. 2000;14:2314–29. doi: 10.1101/gad.827900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Barnes PJ. Histone deacetylase-2 and airway disease. Ther Adv Respir Dis. 2009;3:235–43. doi: 10.1177/1753465809348648. [DOI] [PubMed] [Google Scholar]
- 72.Aikawa Y, Morimoto K, Yamamoto T, et al. Treatment of arthritis with a selective inhibitor of c-Fos/activator protein-1. Nat Biotechnol. 2008;26:817–23. doi: 10.1038/nbt1412. [DOI] [PubMed] [Google Scholar]
- 73.Auphan N, DiDonato JA, Rosette C, Helmberg A, Karin M. Immunosuppression by glucocorticoids: inhibition of NF-kappa B activity through induction of I kappa B synthesis. Science. 1995;270:286–90. doi: 10.1126/science.270.5234.286. [DOI] [PubMed] [Google Scholar]
- 74.Scheinman RI, Cogswell PC, Lofquist AK, Baldwin AS., Jr Role of transcriptional activation of I kappa B alpha in mediation of immunosuppression by glucocorticoids. Science. 1995;270:283–6. doi: 10.1126/science.270.5234.283. [DOI] [PubMed] [Google Scholar]
- 75.Jacques E, Semlali A, Boulet LP, Chakir J. AP-1 overexpression impairs corticosteroid inhibition of collagen production by fibroblasts isolated from asthmatic subjects. Am J Physiol Lung Cell Mol Physiol. 2010;299:L281–7. doi: 10.1152/ajplung.00360.2009. [DOI] [PubMed] [Google Scholar]
- 76.Ralph JA, Ahmed AU, Santos LL, et al. Identification of NURR1 as a mediator of MIF signaling during chronic arthritis: effects on glucocorticoid-induced MKP1. Am J Pathol. 2010;177:2366–78. doi: 10.2353/ajpath.2010.091204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Beaulieu E, Ngo D, Santos L, et al. Glucocorticoid-induced leucine zipper is an endogenous antiinflammatory mediator in arthritis. Arthritis Rheum. 2010;62:2651–61. doi: 10.1002/art.27566. [DOI] [PubMed] [Google Scholar]
- 78.Yang N, Zhang W, Shi XM. Glucocorticoid-induced leucine zipper [GILZ] mediates glucocorticoid action and inhibits inflammatory cytokine-induced COX-2 expression. J Cell Biochem. 2008;103:1760–71. doi: 10.1002/jcb.21562. [DOI] [PubMed] [Google Scholar]
- 79.Beck IM, Vanden Berghe W, Vermeulen L, Yamamoto KR, Haegeman G, De Bosscher K. Crosstalk in inflammation: the interplay of glucocorticoid receptor-based mechanisms and kinases and phosphatases. Endocr Rev. 2009;30:830–82. doi: 10.1210/er.2009-0013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Vinson C, Acharya A, Taparowsky EJ. Deciphering B-ZIP transcription factor interactions in vitro and in vivo. Biochim Biophys Acta. 2006;1759:4–12. doi: 10.1016/j.bbaexp.2005.12.005. [DOI] [PubMed] [Google Scholar]
- 81.Rauch A, Seitz S, Baschant U, et al. Glucocorticoids suppress bone formation by attenuating osteoblast differentiation via the monomeric glucocorticoid receptor. Cell Metab. 2010;11:517–31. doi: 10.1016/j.cmet.2010.05.005. [DOI] [PubMed] [Google Scholar]
- 82.Rauch A, Gossye V, Bracke D, et al. An anti-inflammatory selective glucocorticoid receptor modulator preserves osteoblast differentiation. FASEB J. 2011;25:1323–32. doi: 10.1096/fj.10-173393. [DOI] [PubMed] [Google Scholar]
- 83.Qiu Y, Zhao Y, Becker M, et al. HDAC1 acetylation is linked to progressive modulation of steroid receptor-induced gene transcription. Mol Cell. 2006;22:669–79. doi: 10.1016/j.molcel.2006.04.019. [DOI] [PubMed] [Google Scholar]
- 84.Lamberts SW, Huizenga AT, de Lange P, de Jong FH, Koper JW. Clinical aspects of glucocorticoid sensitivity. Steroids. 1996;61:157–60. doi: 10.1016/0039-128x(96)00005-0. [DOI] [PubMed] [Google Scholar]
- 85.New MI, Nimkarn S, Brandon DD, et al. Resistance to multiple steroids in two sisters. J Steroid Biochem Mol Biol. 2001;76:161–6. doi: 10.1016/s0960-0760(01)00045-0. [DOI] [PubMed] [Google Scholar]
- 86.Schmidt S, Rainer J, Ploner C, Presul E, Riml S, Kofler R. Glucocorticoid-induced apoptosis and glucocorticoid resistance: molecular mechanisms and clinical relevance. Cell Death Differ. 2004;11(Suppl. 1):S45–55. doi: 10.1038/sj.cdd.4401456. [DOI] [PubMed] [Google Scholar]
- 87.Rodriguez-Nieto S, Canada A, Pros E, et al. Massive parallel DNA pyrosequencing analysis of the tumor suppressor BRG1/SMARCA4 in lung primary tumors. Hum Mutat. 2011;32:E1999–2017. doi: 10.1002/humu.21415. [DOI] [PubMed] [Google Scholar]
- 88.Rahimov F, Marazita ML, Visel A, et al. Disruption of an AP-2alpha binding site in an IRF6 enhancer is associated with cleft lip. Nat Genet. 2008;40:1341–7. doi: 10.1038/ng.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.McDaniell R, Lee BK, Song L, et al. Heritable individual-specific and allele-specific chromatin signatures in humans. Science. 2010;328:235–9. doi: 10.1126/science.1184655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kasowski M, Grubert F, Heffelfinger C, et al. Variation in transcription factor binding among humans. Science. 2010;328:232–5. doi: 10.1126/science.1183621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Meijsing SH, Pufall MA, So AY, Bates DL, Chen L, Yamamoto KR. DNA binding site sequence directs glucocorticoid receptor structure and activity. Science. 2009;324:407–10. doi: 10.1126/science.1164265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Weaver IC, Cervoni N, Champagne FA, et al. Epigenetic programming by maternal behavior. Nat Neurosci. 2004;7:847–8. doi: 10.1038/nn1276. [DOI] [PubMed] [Google Scholar]
- 93.McGowan PO, Sasaki A, D'Alessio AC, et al. Epigenetic regulation of the glucocorticoid receptor in human brain associates with childhood abuse. Nat Neurosci. 2009;12:342–8. doi: 10.1038/nn.2270. [DOI] [PMC free article] [PubMed] [Google Scholar]