Abstract
Mutation of the ubiquitous cytosolic enzyme Cu/Zn superoxide dismutase (SOD1) is hypothesized to cause familial amyotrophic lateral sclerosis (FALS) through structural destabilization leading to misfolding and aggregation. Considering the late onset of symptoms as well as the phenotypic variability among patients with identical SOD1 mutations, it is clear that nongenetic factor(s) impact ALS etiology and disease progression. Here we examine the effect of Cys-111 glutathionylation, a physiologically prevalent post-translational oxidative modification, on the stabilities of wild type SOD1 and two phenotypically diverse FALS mutants, A4V and I112T. Glutathionylation results in profound destabilization of SOD1WT dimers, increasing the equilibrium dissociation constant Kd to ~10−20 μM, comparable to that of the aggressive A4V mutant. SOD1A4V is further destabilized by glutathionylation, experiencing an ~30-fold increase in Kd. Dissociation kinetics of glutathionylated SOD1WT and SOD1A4V are unchanged, as measured by surface plasmon resonance, indicating that glutathionylation destabilizes these variants by decreasing association rate. In contrast, SOD1I112T has a modestly increased dissociation rate but no change in Kd when glutathionylated. Using computational structural modeling, we show that the distinct effects of glutathionylation on different SOD1 variants correspond to changes in composition of the dimer interface. Our experimental and computational results show that Cys-111 glutathionylation induces structural rearrangements that modulate stability of both wild type and FALS mutant SOD1. The distinct sensitivities of SOD1 variants to glutathionylation, a modification that acts in part as a coping mechanism for oxidative stress, suggest a novel mode by which redox regulation and aggregation propensity interact in ALS.
Amyotrophic lateral sclerosis (ALS) is a late-onset neurodegenerative disorder for which effective treatment is extremely limited. The majority of ALS cases have no known genetic cause, but substantial insights into the disease have been gained by the study of Cu/Zn superoxide dismutase (SOD1), mutations of which are linked to inherited, or familial, ALS (FALS). Mutant SOD1 appears to play a prominent role in FALS pathology through an acquired propensity to misfold and aggregate.1–5 Notably, misfolded and/or aggregated wild type SOD1 has also been documented in patients with sporadic ALS,6,7 suggesting that this enzyme can be induced to adopt toxic conformational states by nongenetic factors. Such a phenomenon was recently demonstrated for wild type SOD1: Bosco et al. found that oxidative modification of SOD1WT induced structural rearrangement and conformational similarity to the FALS-associated G93A mutant.6 Oxidative stress may thus represent a factor in the cellular environment that is capable of inducing SOD1 to adopt noxious misfolded conformations.
Oxidative stress is thought to be a factor in the pathogenesis of several neurodegenerative diseases, including Alzheimer's disease, Parkinson's disease, and ALS.8,9 Conditions of oxidative stress produce a shift in the cellular environment that is reflected in altered ratios of various redox couples, notably the tripeptide glutathione. Glutathione is a primary regulator of oxidizing species in the cell and protects against oxidative damage by acting as a reducing agent as well as by reversibly modifying proteins to prevent permanent oxidation.10,11 Mixed protein–glutathione disulfides may later be removed by glutaredoxins, making protein S-glutathionylation an important defense against irreversible oxidative damage to proteins.11 We recently found that SOD1 is heavily glutathionylated at cysteine-111 in human tissue, with the modified enzyme constituting nearly 50% of the pool of SOD1 in freshly drawn erythrocytes.12
SOD1 misfolding and aggregation is initiated by dissociation of the native homodimer, leaving monomers more prone to loss of the stabilizing zinc ion.13–15 As such, dimer dissociation is a critical first step in the pathway to SOD1 aggregate formation. In light of our finding that a major fraction of SOD1 in human cells is glutathionylated at cysteine-111, a residue proximal to the dimer interface,12 we considered it pertinent to evaluate the influence of this modification on SOD1 dimer stability. We find that cysteine-111 glutathionylation has a profound effect on the dimer stabilities of wild type SOD1 and the FALS mutant A4V and that this effect is attributable to decreased association rate (kon). Using discrete molecular dynamics (DMD) simulations, we show that glutathionylation affects specific interface contacts in the SOD1 dimer, offering structural insight into the experimental differences we observe in measurements of dimer stability.
MATERIALS AND METHODS
Expression and Purification of SOD1 Variants
Wild type and mutant variants of SOD1 were produced in S. cerevisiae and isolated as previously described.12 After the final remetalation step, samples were separated by anion-exchange chromatography using a MonoQ column (GE Healthcare), which resolves two populations of SOD1 isolated from S. cerevisiae. μ-ESI-FT-ICR-MS analysis shows that SOD1 eluted at lower ionic strength is primarily unmodified, while the protein eluted at higher ionic strength is enriched >8-fold in the glutathione modification.12 The degree of enrichment is estimated by comparing mass spectrum intensities of modified and unmodified SOD1 in the high- and low-charge populations. By comparing the ratio of intensities for unmodified and modified SOD1, we control for any differences in ionization of these two species. Assuming equal ionization of SOD1 with and without glutathione, ~75% of SOD1 monomers in the high-charge population are glutathionylated, and over 90% of SOD1 dimers are expected to be modified on one or both subunits. The glutathione-enriched population eluted at high ionic strength is referred to as glutathionylated SOD1 (GS-SOD1).
Size Exclusion Chromatography
Purified samples of SOD1 at 88 or 8.8 μM in a buffer containing 20 mM Tris, 150 mM NaCl, pH 7.8, were applied to a Superdex 200 PC 3.2/30 column (GE Healthcare) equilibrated in the sample buffer at 4 °C using a 20 μL sample loop. DTT treatment was administered by dialyzing samples overnight against sample buffer containing 1 mM DTT.
To estimate dissociation constants, A280 data from size exclusion chromatography (SEC) were deconvoluted to determine approximate concentrations of monomeric and dimeric SOD1. Diffusion and other band-broadening effects often create significant peak asymmetry in SEC chromatograms, and accurately modeling peak shapes is nontrivial.16 In all chromatograms from the Superdex 200 PC 3.2/30 column, even those of single-subunit standard proteins that do not self-associate, there is skewness toward the trailing edge of peaks that increases with the quantity of protein loaded.17 To deconvolute peaks corresponding to monomeric and dimeric SOD1 while taking this skewness into account, we assume the A280 curve for unmodified wild type SOD1 as the peak shape for completely dimeric SOD1. This assumption is justified since SEC experiments are performed at an SOD1 concentration well above the previously reported value of Kd, which is 10 nM.15 This “standard” dimeric SOD1 curve is subtracted from each data set to be deconvoluted to yield the signal attributable to monomeric SOD1. To control for the band-broadening effects of increased sample load, we subtract the curve for dimeric SOD1 collected at equivalent total protein concentration. The concentrations of monomeric and dimeric SOD1 in each sample are then calculated using
where [M] and [D] are the concentrations of SOD1 monomer and dimer, respectively, in the equilibrated sample; AM and AD are the areas under the curves corresponding to monomeric and dimeric SOD1, respectively; and [D]total is the starting concentration of dimeric SOD1 (88 or 8.8 μM). Kd is then calculated using Kd = [M]2/[D]. A Kd of 10 nM for unmodified SOD115 corresponds to monomer concentrations of 0.93 and 0.29 μM at equilibrium when the initial dimer concentrations are 88 and 8.8 μM, respectively, as calculated using
Hence, the maximum contribution of unmodified SOD1 dimer dissociation to the estimated monomer population for GS-SOD1 is less than 5% (see Figure 1b).
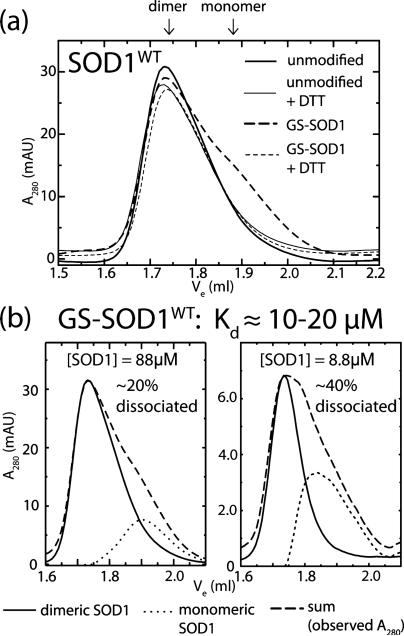
Figure 1.
Wild type SOD1 dimers are destabilized by Cys-111 glutathionylation. (a) Size exclusion chromatography at physiological [SOD1] (88 μM) shows marked destabilization of GS-SOD1WT that is reversed by treatment with DTT to remove the glutathione moeity. Solid lines indicate unmodified SOD1 while dashed lines represent GS-SOD1. Broad and fine lines show these species before and after treatment with DTT, respectively. The elution volumes corresponding to dimeric (~1.74 mL) and monomeric (~1.89 mL) SOD1 are indicated above the panel. Experiments were performed at least in duplicate. (b) Comparison of dimeric and monomeric populations of SOD1 at low and high concentration. Dashed lines show SEC data for GS-SOD1 at 88 μM (left) and 8.8 μM (right), which was deconvoluted as described in Materials and Methods. Solid and dotted lines show curves corresponding to dimeric and monomeric SOD1, respectively.
Determination of Dimer Dissociation Rate Constants Using Surface Plasmon Resonance
Wild type and mutant SOD1 dimers were biotinylated on a single subunit as previously described;13 briefly, SOD1 was incubated with a primary amine-reactive biotinylating agent (EZ-Link NHS-LC-LC Biotin, Pierce) for 30 min at 25 °C and brought to 20 mM Tris, 150 mM NaCl, pH 7.8, using a 1 mL Sephadex G-25 medium spin column. Dimer dissociation was monitored by surface plasmon resonance (SPR) using a Biacore 2000 instrument with biotinylated SOD1 dimers immobilized on a streptavidin-coated flow cell (sensor chip SA or Biotin CAPture Kit, GE Healthcare). Biotinylated SOD1 at ~40 μM was loaded onto the surface at 5 μL/min until achieving a signal gain of 1500–2500 response units (RU), at which point the dissociation reaction was initiated by flowing SOD1-free buffer over the surface. Biotinylation and SPR measurements were conducted at 25 °C, and biotinylated SOD1 was stored at 4 °C until use.
Measuring kinetics of very slow reactions using SPR is complicated by noncovalent interactions of buffer components with the chip surface, resulting in signal drift over time.18 We corrected for this drift by subtracting the signal from a reference streptavidin surface from each data set as well as by calculating rate constants using the Guggenheim method,19 which removes the need for an accurate infinite time value. For SOD1WT and SOD1I112T, 5000 s of Guggenheim data (dRU/dt) were fit using the equation for double-exponential decay, excluding the first 1000 s that were typically noisy. Since the A4V mutant dissociates rapidly, data fitting was performed for the first 1000 s only. While SOD1 dimer dissociation is a first-order process, an additional, fast decay was present in all reactions, evidenced by the comparatively poor fit of a single exponential. We therefore fit SPR data to a double-exponential decay, and the rate constant for dimer dissociation was taken to be that of the process that accounted for the majority of signal loss (>70%). The half-time of the minor exponential function was invariably between 1 and 15 min and accounted for approximately 5–30% of the signal loss during the reaction (Table S1). Because of the consistent presence of this process across all reactions, we conclude it to be an instrumental artifact or perhaps the dissociation of transient noncovalent interactions between nonimmobilized and immobilized SOD1 dimers following the transition from sample loading to buffer flow. Such observations have been made previously concerning SPR measurements.20
Comparison of SOD1 Monomer Stability Using Thermal Denaturation Monitored by Circular Dichroism (CD) Spectroscopy
SOD1 variants with and without Cys-111 glutathionylation were analyzed using a Jasco J-815 CD spectrometer (Jasco Inc., Easton, MD). Yeast-expressed SOD1 mutants were dialyzed overnight against 10 mM phosphate buffer and diluted to 0.2 mg/mL for analysis. Sample spectra were taken at 20 and 96 °C, and the major loss of signal occurred at 230 nm. Upon cooling to 20 °C, the decrease in ellipticity at 230 nm was reversible for all samples to within 65–85% of the initial value (Figure 4a). All subsequent unfolding experiments were temperature ramps from 20 to 96 °C monitored at 230 nm by 1 °C increments with a 5 s dwell time. Dialysis buffer was used as a blank. To obtain apparent melting temperature Tm (Tm*) values, blank-corrected thermal melting data were fit to a modified form of the van't Hoff equation, as previously described in ref 21. This equation includes parameters for the melting transition as well as the baselines corresponding to the native and denatured states:22
where θ(T) is the observed ellipticity at a given temperature T; an (ad) and bn (bd) are the slopes and intercepts, respectively, of the baselines corresponding to the native (and denatured) states; and K is the equilibrium constant for unfolding:
where ΔGu is the difference in Gibbs free energy between the native and denatured states at a given temperature T, and R is the universal gas constant. ΔGu was calculated according to the Gibbs–Helmholtz equation:23
where Tm is the temperature at which ΔGu = 0, and ΔHu and ΔCu are the changes in enthalpy and heat capacity, respectively, associated with thermal denaturation. Data were fit with the parameters an, ad, bn, bd, ΔHu, ΔCu, and Tm using nonlinear least-squares regression, and Tm values were reported as apparent Tm (Tm*) due to the incomplete reversibility of the unfolding transition.
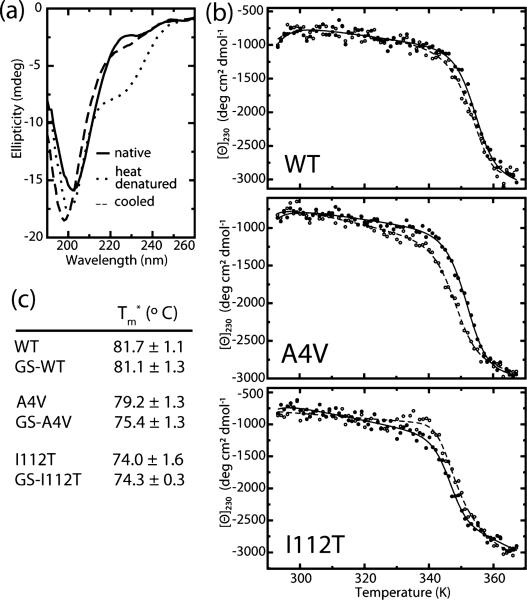
Figure 4.
Effect of glutathionylation on monomer thermal stability. (a) Representative CD spectra of SOD1 before and after cooling shows reversible decrease in ellipticity at 230 nm. (b) Representative curves for thermally induced unfolding of unmodified (closed symbols) and glutathionylated (open symbols) wild type and mutant SOD1 monitored by circular dichroism at 230 nm. (c) Apparent Tm (Tm*) values obtained by fitting blank-corrected thermal melting data as described in Materials and Methods. Experiments were performed at least in duplicate; Tm* values are reported as the mean of all experiments ± SD.
All-Atom DMD Simulations of Glutathionylated SOD1 Mutants
To obtain the structures of post-translationally modified mutant and wild type SOD1, we use the known X-ray crystallographic structure of wild type SOD1 (PDBID: 1SPD) as a reference structure and constrain glutathione molecules to their respective SOD1 residues. Mutations are made to these structures using the Eris suite,24 avoiding changes to residues participating in the metal-binding, glutathionylation, or disulfide bond interactions. The overall structure energy was minimized using an all-atom protein model with discrete molecular dynamics (DMD) simulations.25–27
We perform equilibration and production simulations using DMD. DMD is a molecular dynamics engine that uses discrete potentials in place of continuous potentials, which transforms the simulation into simple calculations of ballistic equations, increasing the speed and efficiency of the simulation and extending sampling of conformational space. Each system is equilibrated for 500 ps at 226 K with a heat exchange occurring every 5 fs. We conduct 50 ns equilibrium simulations of dimeric SOD1 277 K. We perform simulations for each case of mutant or wild type, both the glutathionylated and unmodified structures, resulting in 6 cases total (2 (glutathionylated or unmodified) × 3 (two mutants and wild type)).
Dimer Interface Contact Maps
In our DMD simulations, we define two residues as being in contact in the dimer interface if two Cα atoms of opposing chains are within 10 Å of each other. At each simulation snapshot (5 ps of simulation time), we evaluate the contacts present between the two monomers. We then normalize the count between every pair of residues over the entire simulation.
Calculation of Dimer Interface Area
We sample even intervals of single-temperature simulations for structure snapshots of unmodified and glutathionylated SOD1WT, SOD1A4V, and SOD1I112T and for each snapshot calculate the solvent accessible surface area (SASA) of each individual monomer and of the dimer. The SASA of the dimer is subtracted from the sum of the SASAs of the monomers, resulting in the total buried area of both monomers in the respective dimer structure. We divide this resulting total area by two (since two monomers form the interface) to obtain the dimer interface area. All SASAs are calculated using the Gaia suite.28
RESULTS
SOD1 Wild Type and Mutant Dimers Are Destabilized by Glutathionylation under Physiological Conditions
Size exclusion chromatography (SEC) analysis of GS-SOD1WT reveals the substantial destabilization of dimers by this physiologically prevalent modification (Figure 1). GS-SOD1 used in these assays was isolated from the endogenous pool of enzyme expressed in S. cerevisiae using ion-exchange chromatography, yielding a population that is heavily (~8-fold) enriched in glutathionylated protein.12 While some unmodified enzyme remains in this sample, we do not perform additional in vitro glutathionylation of SOD1 in order to avoid nonphysiological modification of cysteine-6.12 Hence, we report a lower limit for the destabilizing effect of cysteine-111 glutathionylation.
We examined the effect of cysteine-111 glutathionylation on the SOD1 monomer–dimer equilibrium by assaying the oligomeric state of unmodified and glutathionylated SOD1 at physiological pH and concentration (estimated as 50–100 μM in neurons29,30). Unmodified wild type SOD1 is completely dimeric under these conditions (thick solid curve, Figure 1a), in agreement with the previously reported Kd of 10 nM for wild type SOD1 expressed in S. cerevisiae.15 Glutathionylation of SOD1WT results in the appearance of a significant monomeric population (thick dashed curve, Figure 1a). To estimate Kd, we must deconvolute the overlapping peaks for dimeric and monomeric SOD1. We estimate the monomer contribution while accounting for peak skewness (see discussion in Materials and Methods) by assuming the peak shape for unmodified SOD1 (solid curves, Figure 1b) to be characteristic of dimeric SOD1. By removing the contribution of this curve from the observed A280 (dashed curves, Figures 1b and 2b), we obtain the signal attributable to monomeric SOD1 (dotted curves, Figures 1b and 2b). We estimate the Kd of the GS-SOD1WT homodimer to be approximately 10–20 μM, which represents an increase of ~1000-fold over that of the extremely stable unmodified enzyme (previously reported as 10 nM15). SOD1A4V is also destabilized by glutathionylation, experiencing an ~30-fold increase in Kd (Figure 2). In contrast, SOD1I112T stability is unaffected by this modification, remaining dimeric when glutathionylated (Figure 2a). The mean elution volume for monomeric SOD1A4V is slightly lower (~1.84 mL) compared to that of the wild type (~1.89 mL). This mutation is reported to increase the radius of gyration of monomeric SOD1,31 accounting for the decrease in mobility in SEC. The differences in oligomeric state between unmodified and glutathionylated SOD1A4V and SOD1WT are observed in replicate experiments and are abrogated by treatment with DTT to remove the glutathione moiety (thin curves, Figures 1a and 2a), implying that the observed destabilization is due to the presence of the glutathione modification at cysteine-111. All SOD1 species remained dimeric after DTT treatment (Figures 1a and 2a), demonstrating that only the mixed SOD1–glutathione disulfide was reduced, leaving the native intramolecular disulfide intact. This fact is unsurprising since SOD1 retains this disulfide (between Cys-57 and Cys-146) in the reducing environment of the cytosol.
Figure 2.
Effect of Cys-111 glutathionylation on Kd of selected FALS mutants. (a) Size exclusion chromatography at physiological [SOD1] (88 μM) shows destabilization of GS-SOD1A4V that is reversed by treatment with DTT to remove the glutathione moiety. The oligomeric state of SOD1I112T (bottom panel) is relatively unaffected by modification. Solid lines indicate unmodified SOD1 while dashed lines represent GS-SOD1. Broad and fine lines show these species before and after treatment with DTT, respectively. Experiments were performed at least in duplicate. (b) Comparison of dimeric and monomeric populations of SOD1A4V and GS-SOD1A4V. Dashed lines show A280 data for each species (reproduced from panel a), which was deconvoluted as described in Materials and Methods. Solid and dotted lines show curves corresponding to dimeric and monomeric SOD1, respectively.
Effects of Glutathionylation on Dimer Dissociation Kinetics
To measure the effect of glutathionylation on the rate of SOD1 dimer dissociation, we use surface plasmon resonance (SPR) to monitor the dissociation of biotinylated SOD1 dimers immobilized to a streptavidin-coated sensor chip.13 We observe a clear distinction between the effects of the A4V and I112T mutations on dissociation kinetics. SOD1112T dimers have an average dissociation half time of 1.10 h, compared to 1.29 h for the wild type, a difference that is within experimental error (Figure 3). SOD1A4V dimers, by contrast, dissociate significantly faster than the wild type, with an average half-time of 4.51 min (Figure 3). This observation is in stark agreement with the common classification of A4V as a mutation that particularly affects dimer stability,32,33 with the dimer dissociation rate constant koff for unmodified SOD1A4V nearly 20-fold greater than that of the unmodified wild type. Glutathionylation has a minimal effect on dissociation rate for all SOD1 variants studied: koff values for unmodified and glutathionylated dimers do not differ significantly for the wild type and the A4V mutant. SOD1I112T shows a significant, but small (20%), increase in dimer dissociation rate as a result of glutathionylation. The minimal effect of glutathionylation on the dissociation rate constants (koff) of SOD1WT and SOD1A4V dimers cannot account for the significant destabilization at equilibrium revealed by SEC (Figures 1 and 2); thus, the effects of this modification on Kd are attributable to decreases in the association rate constant (kon) of modified monomers.
Figure 3.
Dimer dissociation rate constants for unmodified and glutathionylated SOD1. Dissociation of immobilized dimers was monitored by surface plasmon resonance. Guggenheim plots (50 s intervals) of data from representative experiments are shown in gray; colored lines indicate double-exponential decay curves fitted to the data. All values are reported as the mean of triplicate experiments ± SD.
Glutathionylation Has Little Effect on SOD1 Monomer Stability
We assess the effect of mutations and glutathionylation on SOD1 monomer stability using thermal unfolding experiments monitored by circular dichroism (CD). Because CD primarily reflects protein secondary structure content, we expect that changes in signal upon thermal denaturation of SOD1 are mainly attributable to the loss of β-strand structure as monomers unfold, rather than dissociation of the homodimer. The effect of glutathionylation on monomer unfolding is minimal for the wild type protein, but results in a modest decrease in apparent Tm for SOD1A4V (Figure 4). These results, in agreement with a computational study by Proctor et al.,34 indicate that glutathionylation primarily exerts effects on dimer stability while leaving monomer stability largely unchanged.
Structural Effects of Glutathionylation on SOD1 Dimer Interface
Using DMD simulations, we show that dimer interface contacts are changed in the glutathionylated versus the unmodified structures. SOD1WT and SOD1A4V exhibit a general loss in overall interface Cα contacts, while the I112T mutant experiences a shift in Cα dimer interface contacts upon glutathionylation (Figure 5a,b and Figure S1). In SOD1I112T, residues that lose interface contacts are balanced by neighboring residues that gain contacts, resulting in an overall change in composition of the interface, without significantly changing the number of interface contacts. Interestingly, in wild type SOD1, we observe that, while the net number of Cα contacts decreases upon glutathionylation, the net number of Cβ contacts increases (Figure S2). This would indicate rearrangement of the side chains in the dimer interface in order to accommodate the glutathione moiety. We do not observe this effect in the A4V or I112T mutants, which have the same qualitative distribution of losses and gains in contact frequency upon glutathionylation in Cα and Cβ contacts.
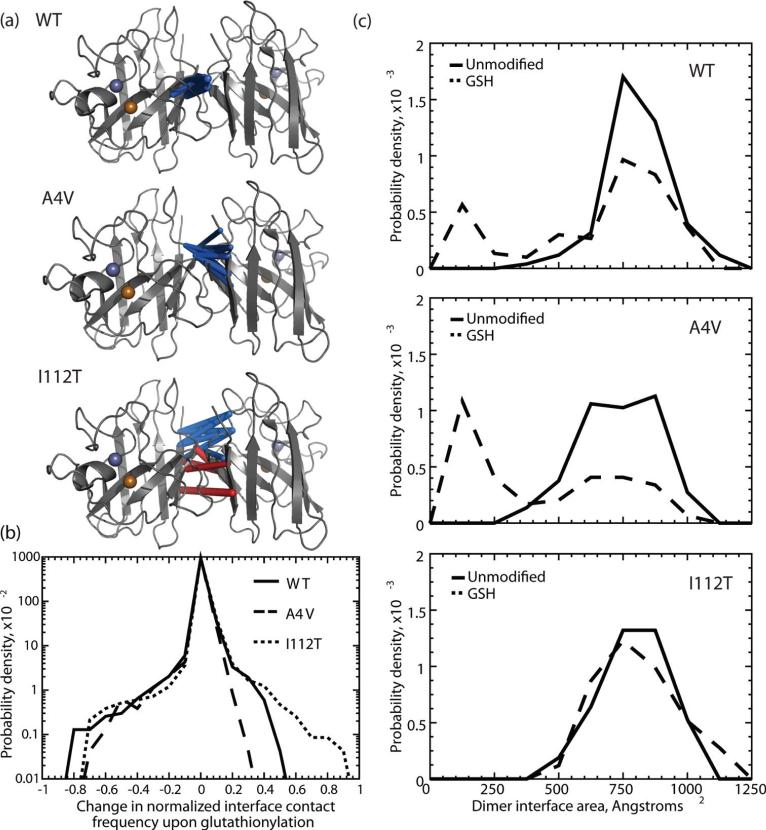
Figure 5.
Effects of glutathionylation on the SOD1 dimer interface. (a) The top ten most frequently changed Cα interface contacts upon glutathionylation are highlighted with a three-dimensional rod representation for SOD1WT, SOD1A4V, and SOD1I112T. Rod thickness is proportional to the change in frequency of the interaction. Blue rods represent a loss in frequency of the interaction; red rods represent a gain in frequency of the interaction. (b) Distributions of changes in frequency of Cα interface contacts upon Cys-111 glutathionylation for SOD1WT, SOD1A4V, and SOD1I112T. Wild type SOD1 and the A4V mutant have distributions that are skewed toward loss of contacts, while the I112T mutant is balanced in loss and gain of contacts. (c) Distributions of dimer interface areas for unmodified (solid line) and glutathionylated (“GSH”, dashed line) SOD1WT, SOD1A4V, and SOD1I112T.
This phenomenon brings into question the size of the dimer interface in each SOD1 variant. We calculate the area of the dimer interface over the course of single-temperature simulations and find that all three unmodified SOD1 variants show a single population, featuring an approximately Gaussian distribution around a central value of 750–800 Å2 for the interface area (Figure 5c). However, upon glutathionylation, a second population is present in SOD1WT and SOD1A4V near 150 Å2, while the dimer interface area distribution of SOD1I112T is relatively unaffected. This finding agrees with that of decreased Cα (backbone) contacts found in SOD1WT and SOD1A4V upon glutathionylation.
DISCUSSION
SOD1 is abundantly glutathionylated at cysteine-111 in human tissue.12 Because of the proximity of this tripeptide moiety to the dimer interface, we hypothesized that it introduces steric clashes that favor dimer dissociation and/or hinder association of modified monomers. To distinguish these kinetic effects, we estimate the equilibrium dissociation constant Kd using size exclusion chromatography and measure the rate constant for dimer dissociation (koff) with surface plasmon resonance. Since the equilibrium dissociation constant Kd is equal to the ratio of the rate constants for dissociation and association, we can then deduce effects on dimer formation rate from these two parameters. Dimer dissociation precedes disulfide reduction and metal loss13 (Figure 6), so the contribution of the latter processes and irreversible aggregation is minimal compared to the dimer dissociation reaction in our SEC (Figures 1 and 2) and SPR (Figure 3) experiments.
Figure 6.
Summary of effects of Cys-111 glutathionylation on the stabilities of WT SOD1 and the FALS mutants I112T and A4V. Above, general schematic of SOD1 aggregation pathway. Below, effect of glutathionylation on dimer dissociation rate (koff: reaction 1), monomer association rate (kon: reaction 2), and monomer thermal stability for each SOD1 variant. For simplicity of representation, we condense metal loss, intramolecular disulfide reduction, and structural distortion of monomers into a single step (reaction 3). The effect of glutathionylation on kon is inferred from the measured effects on Kd (Figures 1 and 2) and koff (Figure 3) using the relationship Kd = koff/kon.
Glutathionylation has a dramatic effect on wild type dimer stability at equilibrium. The SOD1 homodimer is exceptionally stable, having low nanomolar binding affnity.12,15 In agreement with these findings, the unmodified wild type enzyme is dimeric under the conditions of our assay (Figure 1a). In contrast, the Kd of GS-SOD1WT is increased by several orders of magnitude, to approximately 10–20 μM, such that there is appreciable (~20%) dissociation at physiological con- centration (Figure 1b). Although GS-SOD1WT is destabilized at equilibrium relative to the unmodified enzyme, the rate of dimer dissociation does not differ significantly as a result of modification (Figure 3). We therefore conclude that glutathionylation destabilizes SOD1WT dimers by decreasing kon, the rate constant for monomer association. These results indicate a much greater destabilizing effect than we initially estimated for the glutathione modification.12 However, we previously estimated Kd using an activity assay to quantify SOD1 dissociation. This method of Kd estimation is predicated on the decreased activity of monomeric SOD1 due to loop disorder35 and is sensitive enough for use at low protein concentrations that are unobservable by A280.36 However, this method has the disadvantage of being an indirect measure of the oligomerization state and is susceptible to interference by factors unrelated to monomerization that influence the mobility of active site loops. It may be that altered loop mobilities caused by glutathionylation result in dismutase-active monomeric SOD1, which would cause an underestimation of Kd using this method. SEC analysis, by contrast, is a simple and direct method for assessing the extent of dimer dissociation and clearly demonstrates the striking destabilization of SOD1WT dimers by Cys-111 glutathionylation.
Glutathionylation also destabilizes SOD1A4V dimers. The Kd of this variant has previously been reported as 3 μM,32 which agrees with our calculated lower limit of 1 μM (Figure 2b). Modification by the glutathione moiety results in a significant shift toward monomeric SOD1 and an ~10-fold increase in Kd (Figure 2). As in the wild type, glutathionylation does not affect dissociation kinetics in SOD1A4V, indicating that destabilization of GS-SOD1A4V dimers occurs primarily through effects on kon. Glutathionylation may hinder monomer association in SOD1WT and SOD1A4V by sterically blocking the formation of certain interface contacts. Alternatively, glutathionylation at cysteine-111 may promote local structural rearrangements in these monomers that impede formation of interface contacts. The ~3.8 °C decrease in apparent melting temperature of GS-SOD1A4V monomers (Figure 4) may provide evidence for this; however, it is also possible that structural differences exist that do not significantly alter secondary structural elements.
To our knowledge, the stability of the I112T mutant of SOD1 has not previously been studied experimentally. Although SEC peaks for SOD1I112T show increased skewness compared to those of unmodified SOD1WT, all appear to be unimodal and centered at the elution volume of dimeric SOD1 (Figure 2a). This difference in peak shape may reflect increased conformational flexibility of SOD1I112T, resulting in a broader distribution of radii of gyration for the dimer. Alternatively, if this mutant has micromolar rather than nanomolar binding affinity, the peak could be skewed by the contribution of monomeric SOD1. Computational analysis of SOD1I112T thermodynamics showed that this mutation has increased dimer stability compared to the wild type,1 supporting the interpretation that this variant is solely dimeric under the conditions of our assay. Regardless of the effect of the I112T mutation itself, changes in dimer stability resulting from glutathionylation are clearly minimal (Figure 2a). In contrast to wild type SOD1 and the A4V mutant, glutathionylation of SOD1I112T dimers results in little to no effect on Kd despite a modest but statistically signifi-cant increase in the dissociation rate constant (Figure 3). Modification may exert opposing effects on this SOD1 mutant, destabilizing the dimer but facilitating the reassociation of modified monomers.
DMD simulations reveal a structural basis for the distinct effects of cysteine-111 glutathionylation on wild type and FALS mutant SOD1. SOD1WT and SOD1A4V experience a net loss of both dimer interface area and Cα interface contacts as a result of glutathionylation (Figure 5), and these dimers are both destabilized exclusively by decreased kon. Therefore, some losses in interface Cα contacts may be indicative of structural changes that hinder monomer association (kon) rather than directly impacting the rate constant for dissociation (koff). In particular, a net loss of Cα contacts specifically indicates backbone movements that separate the two monomers, rather than simple rearrangement of the residue side chains. The appearance of a significant smaller-interface population in the glutathionylated species of SOD1WT and SOD1A4V during simulations (Figure 5c) further indicates that this modification stabilizes a partially dissociated intermediate, as seen in ref 34. In the SOD1I112T dimer interface, glutathionylation results in a shift in interface composition rather than a net loss of Cα contacts (Figure 5a,b); likewise, no smaller-interface population is observed for GS-SOD1I112T (Figure 5c). For this variant, change in the dissociation constant Kd is minimal even though koff is increased. These trends raise the possibility that the identity, not quantity, of the residues participating in the dimer interface affects dissociation kinetics.
The late-onset nature of ALS suggests a connection to a natural process of aging that either allows the initiation of a previously suppressed pathology (e.g., SOD1 aggregation) or renders the organism less able to cope with an ongoing threat that was previously tightly regulated. While symptom onset occurs in midlife (>45 years) or later for the vast majority of ALS patients, disease duration is variable, even among patients with identical SOD1 mutations.2 Patients with the A4V mutation experience particularly aggressive motor function loss (<2 years average disease duration2) while I112T is apparently incompletely penetrant (not all individuals with this allele develop ALS37). The phenotypic heterogeneity of disease duration among those with identical SOD1 genotype implies that nongenetic environmental factors contribute significantly to mutant SOD1 pathogenicity.
Oxidative stress, manifested as a dysregulation of reactive oxygen species (ROS) or reactive nitrogen species (RNS), is one such process. The levels of ROS and RNS are normally tightly regulated by a variety of enzymes, such as SOD1, and small molecule or peptide redox couples. Glutathione, one such redox couple, is present at a high concentration in the cytosol (up to 12 mM38) and protects against oxidative damage by acting as a reducing agent, as well as by reversibly modifying proteins to prevent permanent oxidation.10,11 Protein S-glutathionylation occurs more frequently under conditions of oxidative stress as a result of two mechanisms. In the first, thiyl radicals generated by oxidizing species react with reduced glutathione (GSH). Under oxidizing conditions, there also exists a greater proportion of cellular glutathione in the disulfide-linked oxidized form (GSSG), which modifies free cysteine residues by disulfide exchange.
SOD1 is an enzyme that directly interacts with oxidizing species, converting superoxide to hydrogen peroxide, and glutathionylation is a common modification of SOD1 in human tissue, including that of ALS patients.12,39 SOD1 is glutathionylated at a steady state level that likely reflects the immediate degree of oxidative stress occurring in the individual organism, rather than accumulating over the entire lifespan (discussed in ref 12). Because a large fraction of SOD1 from a variety of healthy human donors is glutathionylated,12 SOD1 glutathionylation alone is unlikely to cause ALS. The substantial drop in glutathionylated wild type dimer stability to micromolar affinity (Figure 1b) has not previously been observed even though this modification is prevalent in SOD1 from both human tissue and recombinant sources.12,39,40 Since enrichment of the glutathionylated protein by ion exchange is necessary to observe this destabilizing effect, it may be that the decreased kon we report is only associated with formation of dimers of two modified subunits.
Given the central importance of dimer dissociation in the initiation of SOD1 aggregation,13,14 the high levels of glutathionylated SOD1 expected to be present in an oxidatively stressed motor neuron could trigger or exacerbate dysfunction by substantially increasing the monomer population. A prolonged shift in the monomer–dimer equilibrium, especially in harsh conditions, would result in increased populations of metal-free, misfolded, and aggregated SOD1 (Figure 6). It has been observed that Cys-111 mediates mutant SOD1 aggregation in a cell culture model of ALS and that overexpression of glutaredoxin-1 (which reduces both protein–protein and protein–glutathione disulfides) or mutation of Cys-111 attenuate this toxic process.41 While initially interpreted as further evidence of the involvement of intermolecular disulfide bonds in aggregate formation,30,42 these data also support the hypothesis that destabilization caused by Cys-111 glutathionylation promotes aggregation and cell death in ALS. The A4V and I112T mutant SODs are affected differently by glutathionylation, suggesting differing sensitivities of these SOD1 variants to an oxidizing intracellular environment. These differences could explain some of the variability in disease progression among the over 140 mutations implicated in the familial form of the disease. Furthermore, the significant destabilization of both wild type SOD1 and the FALS mutant A4V by glutathione suggests that this modification could promote formation of non-native SOD1 oligomers in both sporadic and familial ALS cases. The modulation of SOD1 dimer stability by cysteine-111 glutathionylation, a post-translational modification linked to redox status, suggests a novel mechanism by which oxidative stress and SOD1 aggregation are interconnected in ALS pathology.
Supplementary Material
ACKNOWLEDGMENTS
We thank Joan S. Valentine for graciously providing the EG118 yeast strain and yEP351:hwtSOD1 vector and Drs. Feng Ding, Ashutosh Tripathy, and Brian Kuhlman for helpful discussions.
Funding
This work was supported by the National Institutes of Health grant R01GM080742 and the ARRA supplement 3R01GM080742-03S1. R.L.R. was supported by the National Institutes of Health Predoctoral Fellowship F31NS073435-01 from the National Institute of Neurological Disorders and Stroke. E.A.P. was supported by the UNC Curriculum in Bioinformatics and Computational Biology and National Institutes of Health Predoctoral Fellowship F31AG039266-01 from the National Institute on Aging.
ABBREVIATIONS
- SOD1
Cu/Zn superoxide dismutase 1
- FALS
familial amyotrophic lateral sclerosis
- ALS
amyotrophic lateral sclerosis
- DMD
discrete molecular dynamics
- μ-ESI-FT-ICR-MS
microcapillary electrospray ionization Fourier transform ion cyclotron resonance mass spectrometry
- SPR
surface plasmon resonance
- ROS
reactive oxygen species
- RNS
reactive nitrogen species
- SEC
size exclusion chromatography
Footnotes
ASSOCIATED CONTENT
 Supporting Information
Supporting Information
Analysis of Cα–Cα (backbone) and Cβ–Cβ (side-chain) dimer interface contacts and additional information regarding SPR data fitting. This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.Khare S, Caplow M, Dokholyan N. FALS mutations in Cu, Zn superoxide dismutase destabilize the dimer and increase dimer dissociation propensity: a large-scale thermodynamic analysis. Amyloid. 2006;13:226–235. doi: 10.1080/13506120600960486. [DOI] [PubMed] [Google Scholar]
- 2.Wang Q, Johnson J, Agar N, Agar J. Protein aggregation and protein instability govern familial amyotrophic lateral sclerosis patient survival. PLoS Biol. 2008;6:e170. doi: 10.1371/journal.pbio.0060170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sandelin E, Nordlund A, Andersen PM, Marklund SSL, Oliveberg M. Amyotrophic Lateral Sclerosis-associated Copper/Zinc Superoxide Dismutase Mutations Preferentially Reduce the Repulsive Charge of the Proteins. J. Biol. Chem. 2007;282:21230–21236. doi: 10.1074/jbc.M700765200. [DOI] [PubMed] [Google Scholar]
- 4.Bystrom R, Andersen P, Grobner G, Oliveberg M. SOD1 mutations targeting surface hydrogen bonds promote amyotrophic lateral sclerosis without reducing apo-state stability. J. Biol. Chem. 2010;285:19544–19552. doi: 10.1074/jbc.M109.086074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lindberg MJ, Byström R, Boknäs N, Andersen PM, Oliveberg M. Systematically perturbed folding patterns of amyotrophic lateral sclerosis (ALS)-associated SOD1 mutants. Proc. Natl. Acad. Sci. U.S.A. 2005;102:9754–9759. doi: 10.1073/pnas.0501957102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bosco DA, Morfini G, Karabacak NM, Song Y, Gros-Louis F, Pasinelli P, Goolsby H, Fontaine BA, Lemay N, McKenna-Yasek D, Frosch MP, Agar JN, Julien J, Brady ST, Brown RH. Wild-type and mutant SOD1 share an aberrant conformation and a common pathogenic pathway in ALS. Nature Neurosci. 2010;13:1396–1403. doi: 10.1038/nn.2660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Forsberg K, Jonsson PA, Andersen PM, Bergemalm D, Graffmo KS, Hultdin M, Jacobsson J, Rosquist R, Marklund SL, Brännström T. Novel Antibodies Reveal Inclusions Containing Non-Native SOD1 in Sporadic ALS Patients. PLoS One. 2010;5:e11552. doi: 10.1371/journal.pone.0011552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Higgins G, Beart P, Shin Y, Chen M, Cheung N, Nagley P. Oxidative stress: emerging mitochondrial and cellular themes and variations in neuronal injury. J. Alzheimers Dis. 2010;20(Suppl 2):S453–S473. doi: 10.3233/JAD-2010-100321. [DOI] [PubMed] [Google Scholar]
- 9.Jellinger K. Basic mechanisms of neurodegeneration: a critical update. J. Cell. Mol. Med. 2010;14:457–487. doi: 10.1111/j.1582-4934.2010.01010.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.le-Donne I, Rossi R, Colombo G, Giustarini D, Milzani A. Protein S-glutathionylation: a regulatory device from bacteria to humans. Trends Biochem. Sci. 2009;34:85–96. doi: 10.1016/j.tibs.2008.11.002. [DOI] [PubMed] [Google Scholar]
- 11.Townsend D. S-glutathionylation: indicator of cell stress and regulator of the unfolded protein response. Mol Interv. 2007;7:313–324. doi: 10.1124/mi.7.6.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wilcox K, Zhou L, Jordon J, Huang Y, Yu Y, Redler R, Chen X, Caplow M, Dokholyan N. Modifications of superoxide dismutase (SOD1) in human erythrocytes: a possible role in amyotrophic lateral sclerosis. J. Biol. Chem. 2009;284:13940–13947. doi: 10.1074/jbc.M809687200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Khare S, Caplow M, Dokholyan N. The rate and equilibrium constants for a multistep reaction sequence for the aggregation of superoxide dismutase in amyotrophic lateral sclerosis. Proc. Natl. Acad. Sci. U.S.A. 2004;101:15094–15099. doi: 10.1073/pnas.0406650101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rakhit R, Crow J, Lepock J, Kondejewski L, Cashman N, Chakrabartty A. Monomeric Cu,Zn-superoxide dismutase is a common misfolding intermediate in the oxidation models of sporadic and familial amyotrophic lateral sclerosis. J. Biol. Chem. 2004;279:15499–15504. doi: 10.1074/jbc.M313295200. [DOI] [PubMed] [Google Scholar]
- 15.Doucette P, Whitson L, Cao X, Schirf V, Demeler B, Valentine J, Hansen J, Hart P. Dissociation of human copper-zinc superoxide dismutase dimers using chaotrope and reductant. Insights into the molecular basis for dimer stability. J. Biol. Chem. 2004;279:54558–54566. doi: 10.1074/jbc.M409744200. [DOI] [PubMed] [Google Scholar]
- 16.Felinger A, Pasti L, Dondi F, van HM, Schoenmakers P, Martin M. Stochastic theory of size exclusion chromatography: peak shape analysis on single columns. Anal. Chem. 2005;77:3138–3148. doi: 10.1021/ac050042b. [DOI] [PubMed] [Google Scholar]
- 17.Janson J, Rydén L. Protein Purification: Principles, High-Resolution Methods, and Applications. Wiley-VCH; New York: 1998. [Google Scholar]
- 18.Caplow M, Fee L. Dissociation of the tubulin dimer is extremely slow, thermodynamically very unfavorable, and reversible in the absence of an energy source. Mol. Biol. Cell. 2002;13:2120–2131. doi: 10.1091/mbc.E01-10-0089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Niebergall P, Sugita E. Utilization of the Guggenheim method in kinetics. J. Pharm. Sci. 1968;57:1805–1808. doi: 10.1002/jps.2600571045. [DOI] [PubMed] [Google Scholar]
- 20.Schuck P. Reliable determination of binding affinity and kinetics using surface plasmon resonance biosensors. Curr. Opin. Biotechnol. 1997;8:498–502. doi: 10.1016/s0958-1669(97)80074-2. [DOI] [PubMed] [Google Scholar]
- 21.Wang Y, He H, Li S. Effect of Ficoll 70 on thermal stability and structure of creatine kinase. Biochemistry (Moscow) 2010;75:648–654. doi: 10.1134/s0006297910050160. [DOI] [PubMed] [Google Scholar]
- 22.Ramsay GD, Eftink MR. Part B: Numerical Computer Methods. Academic Press; New York: 1994. [27] Analysis of multidimensional spectroscopic data to monitor unfolding of proteins; pp. 615–645. [DOI] [PubMed] [Google Scholar]
- 23.Becktel WJ, Schellman JA. Protein stability curves. Biopolymers. 1987;26:1859–1877. doi: 10.1002/bip.360261104. [DOI] [PubMed] [Google Scholar]
- 24.Yin S, Ding F, Dokholyan N. Eris: an automated estimator of protein stability. Nature Methods. 2007;4:466–467. doi: 10.1038/nmeth0607-466. [DOI] [PubMed] [Google Scholar]
- 25.Ding F, Dokholyan N. Emergence of protein fold families through rational design. PLoS Comput. Biol. 2006;2:e85. doi: 10.1371/journal.pcbi.0020085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ding F, Tsao D, Nie H, Dokholyan N. Ab initio folding of proteins with all-atom discrete molecular dynamics. Structure. 2008;16:1010–1018. doi: 10.1016/j.str.2008.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dokholyan N, Buldyrev S, Stanley H, Shakhnovich E. Discrete molecular dynamics studies of the folding of a protein-like model. Fold. Des. 1998;3:577–587. doi: 10.1016/S1359-0278(98)00072-8. [DOI] [PubMed] [Google Scholar]
- 28.Kota P, Ding F, Ramachandran S, Dokholyan NV. Gaia: Automated Quality Assessment of Protein Structure Models. Bioinformatics. 2011 doi: 10.1093/bioinformatics/btr374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.DiDonato M, Craig L, Huff M, Thayer M, Cardoso R, Kassmann C, Lo T, Bruns C, Powers E, Kelly J, Getzoff E, Tainer J. ALS mutants of human superoxide dismutase form fibrous aggregates via framework destabilization. J. Mol. Biol. 2003;332:601–615. doi: 10.1016/s0022-2836(03)00889-1. [DOI] [PubMed] [Google Scholar]
- 30.Banci L, Bertini I, Durazo A, Girotto S, Gralla E, Martinelli M, Valentine J, Vieru M, Whitelegge J. Metal-free superoxide dismutase forms soluble oligomers under physiological conditions: a possible general mechanism for familial ALS. Proc. Natl. Acad. Sci. U.S.A. 2007;104:11263–11267. doi: 10.1073/pnas.0704307104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schmidlin T, Kennedy B, Daggett V. Structural changes to monomeric CuZn superoxide dismutase caused by the familial amyotrophic lateral sclerosis-associated mutation A4V. Biophys. J. 2009;97:1709–1718. doi: 10.1016/j.bpj.2009.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ray S, Nowak R, Strokovich K, Brown R, Walz T, Lansbury P. An intersubunit disulfide bond prevents in vitro aggregation of a superoxide dismutase-1 mutant linked to familial amytrophic lateral sclerosis. Biochemistry. 2004;43:4899–4905. doi: 10.1021/bi030246r. [DOI] [PubMed] [Google Scholar]
- 33.Hough M, Grossmann J, Antonyuk S, Strange R, Doucette P, Rodriguez J, Whitson L, Hart P, Hayward L, Valentine J, Hasnain S. Dimer destabilization in superoxide dismutase may result in disease-causing properties: structures of motor neuron disease mutants. Proc. Natl. Acad. Sci. U.S.A. 2004;101:5976–5981. doi: 10.1073/pnas.0305143101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Proctor EA, Ding F, Dokholyan NV. Structural and Thermodynamic Effects of Post-translational Modifications in Mutant and Wild Type Cu, Zn Superoxide Dismutase. J. Mol. Biol. 2011;408:555–567. doi: 10.1016/j.jmb.2011.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ferraroni M, Rypniewski W, Wilson K, Viezzoli M, Banci L, Bertini I, Mangani S. The crystal structure of the monomeric human SOD mutant F50E/G51E/E133Q at atomic resolution. The enzyme mechanism revisited. J. Mol. Biol. 1999;288:413–426. doi: 10.1006/jmbi.1999.2681. [DOI] [PubMed] [Google Scholar]
- 36.Heikkila RE, Cabbat F. A sensitive assay for superoxide dismutase based on the autoxidation of 6-hydroxydopamine. Anal. Biochem. 1976;75:356–362. doi: 10.1016/0003-2697(76)90089-0. [DOI] [PubMed] [Google Scholar]
- 37.Esteban J, Rosen D, Bowling A, Sapp P, Kenna-Yasek D, O'Regan J, Beal M, Horvitz H, Brown R. Identification of two novel mutations and a new polymorphism in the gene for Cu/Zn superoxide dismutase in patients with amyotrophic lateral sclerosis. Hum. Mol. Genet. 1994;3:997–998. doi: 10.1093/hmg/3.6.997. [DOI] [PubMed] [Google Scholar]
- 38.Dringen R. Metabolism and functions of glutathione in brain. Prog. Neurobiol. 2000;62:649–671. doi: 10.1016/s0301-0082(99)00060-x. [DOI] [PubMed] [Google Scholar]
- 39.Nakanishi T, Kishikawa M, Miyazaki A, Shimizu A, Ogawa Y, Sakoda S, Ohi T, Shoji H. Simple and defined method to detect the SOD-1 mutants from patients with familial amyotrophic lateral sclerosis by mass spectrometry. J. Neurosci. Methods. 1998;81:41–44. doi: 10.1016/s0165-0270(98)00012-0. [DOI] [PubMed] [Google Scholar]
- 40.Marklund S, Andersen P, Forsgren L, Nilsson P, Ohlsson P, Wikander G, Oberg A. Normal binding and reactivity of copper in mutant superoxide dismutase isolated from amyotrophic lateral sclerosis patients. J. Neurochem. 1997;69:675–681. doi: 10.1046/j.1471-4159.1997.69020675.x. [DOI] [PubMed] [Google Scholar]
- 41.Cozzolino M, Amori I, Pesaresi M, Ferri A, Nencini M, Carri M. Cysteine 111 affects aggregation and cytotoxicity of mutant Cu,Zn-superoxide dismutase associated with familial amyotrophic lateral sclerosis. J. Biol. Chem. 2008;283:866–874. doi: 10.1074/jbc.M705657200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Karch C, Prudencio M, Winkler D, Hart P, Borchelt D. Role of mutant SOD1 disulfide oxidation and aggregation in the pathogenesis of familial ALS. Proc. Natl. Acad. Sci. U.S.A. 2009;106:7774–7779. doi: 10.1073/pnas.0902505106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.