Abstract
Serum retinol-binding protein 4 (RBP4) levels are increased in insulin-resistant humans and correlate with severity of insulin resistance in metabolic syndrome. Quantitative Western blotting (qWestern) has been the most accurate method for serum RBP4 measurements, but qWestern is technically complex and labor intensive. The lack of a reliable, high-throughput method for RBP4 measurements has resulted in variability in findings in insulin-resistant humans. Many commonly used ELISAs have limited dynamic range. Neither the current ELISAs nor qWestern distinguish among full-length and carboxyl terminus proteolyzed forms of circulating RBP4 that are altered in different medical conditions. Here, we report the development of a novel quantitative mass spectrometry immunoaffinity assay (qMSIA) to measure full-length and proteolyzed forms of RBP4. qMSIA and qWestern of RBP4 were performed in identical serum aliquots from insulin-sensitive/normoglycemic or insulin-resistant humans with impaired glucose tolerance or type 2 diabetes. Total RBP4 qMSIA measurements were highly similar to qWestern and correlated equally well with clinical severity of insulin resistance (assessed by clamp glucose disposal rate, r = −0.74), hemoglobin A1c (r = 0.63), triglyceride/high-density lipoprotein (r = 0.55), waist/hip (r = 0.61), and systolic blood pressure (r = 0.53, all P < 0.001). Proteolyzed forms of RBP4 accounted for up to 50% of total RBP4 in insulin-resistant subjects, and des(Leu)-RBP4 (cleavage of last leucine) correlated highly with insulin resistance (assessed by glucose disposal rate, r = −0.69). In multiple regression analysis, insulin resistance but not glomerular filtration rate was the strongest, independent predictor of serum RBP4 levels. Thus, qMSIA provides a novel tool for accurately measuring serum RBP4 levels as a biomarker for severity of insulin resistance and risk for type 2 diabetes and metabolic syndrome.
Retinol-binding protein 4 (RBP4) is an adipokine that contributes to insulin resistance in obesity and type 2 diabetes (T2D) in mice (1). A growing body of literature reveals that elevated circulating RBP4 levels in humans are associated with insulin resistance, body mass index (BMI), waist to hip ratio (WHR), dyslipidemia, and hypertension (2–8). RBP4 appears to be an early predictor of insulin resistance in nonobese, nondiabetic subjects with a strong family history of T2D (4, 5) and a biomarker for prediabetes [impaired fasting glucose or impaired glucose tolerance (IGT)] in the general population (9). In addition, serum RBP4 levels respond to treatments that enhance insulin sensitivity in insulin-resistant subjects, including exercise training and gastric bypass for obesity (5, 10, 11). Elevated serum RBP4 may have a causative role in the development of insulin resistance. Increased RBP4 impairs insulin-stimulated insulin receptor substrate-1 phosphorylation and phosphatidylinositol 3 kinase activity in muscle and increases hepatic glucose production (1). Incubation of human adipocytes with RBP4 impairs insulin-stimulated insulin receptor substrate-1 and ERK/MAPK phosphorylation (12). Elevated RBP4 may diminish insulin signaling through activation of Janus kinases 2 (Jak2)/signal transducer and activator of transcription 5 (Stat5)/suppressor of cytokine signaling 3 (Socs3) (13). Recently, we (Hosooka T., J. Norseed, and B.B. Kakhn, unpublished data) and others (14) found that RBP4 may induce insulin resistance by increasing proinflammatory cytokine secretion from macrophages. Human data are consistent with a causal role for elevated RBP4 in insulin resistance. In overweight women in a breast cancer prevention trial, treatment with fenretinide, a retinoid that decreases serum RBP4 levels by disrupting RBP4-transthyretin (TTR) binding, resulted in a 7-fold greater possibility of improving insulin sensitivity after 2 yr than tamoxifen treatment (15). Some of the most convincing data for RBP4 playing a causative role come from genetic studies showing that RBP4 variants are associated with elevated RBP4 levels and insulin resistance (7, 16–18). For instance, a gain of function single-nucleotide polymorphism in the RBP4 promoter, which increases RBP4 expression in adipose tissue, is associated with a 2-fold increased risk for T2D (17, 19). This indicates that RBP4 polymorphisms can contribute to increased diabetes risk in some populations.
However, not all studies show an association between elevated serum RBP4 levels and insulin resistance (20–22). The reasons for the discrepancies may relate to multiple factors, such as different study populations, cohort/sample size, and assay methods (18). A major factor for the discrepancies is lack of a standard method for measuring RBP4 levels. Quantitative Western blotting (qWestern) is the most accurate technique for RBP4 measurements in insulin-resistant subjects judged by its correlation with insulin sensitivity measured by euglycemic clamp (23). However, qWestern is technically complex and labor intensive. Developing a reliable and high-throughput method for measuring serum RBP4 levels is critical for large-scale human studies and potential clinical application. Currently, there are multiple commercially available (23) and also individually generated ELISAs (8), which produce variable results. For instance, some ELISAs are easily saturated and mask the elevated serum RBP4 levels in insulin-resistant states (23). Others overestimate the RBP4 levels in normal subjects (23). In addition, RBP4 forms a dimeric complex with TTR, which itself circulates as a homotetramer (24). This interaction with TTR may prevent certain antibodies from interacting with RBP4 in the ELISAs. Finally, several C-terminally proteolyzed forms of RBP4 are present in circulation (25, 26). The most prominent proteolyzed variants lack the last leucine [des(Leu)-RBP4 (RBP4-L)] or both C-terminal leucines [di-des(Leu)-RBP4 (RBP4-LL)] (26). ELISAs may have different affinities for these proteolyzed forms. The discrepancies in methodology for RBP4 measurements may at least partially explain why some studies fail to show correlations between circulating RBP4 levels and insulin resistance.
Here, we report a novel adaptation of a quantitative mass spectrometry immunoaffinity assay (qMSIA) (27) that renders the assay highly accurate across a broad range of values and very useful for quantitation of RBP4 elevation in insulin-resistant subjects. RBP4 levels measured by the modified qMSIA are tightly correlated with levels measured by qWestern. RBP4 levels measured by both methods are highly correlated with insulin resistance and other metabolic parameters. Because some recent reports have suggested that renal function is the primary determinant of serum RBP4 levels in diabetic patients (20, 21), we also used multiple regression analysis to compare relative contributions of insulin resistance and renal function to elevated serum RBP4 levels. We found that insulin resistance measured by glucose disposal rate (GDR) during a euglycemic hyperinsulinemic clamp study is the strongest independent predictor for increased serum RBP4 levels in prediabetic and diabetic patients even after taking into account renal function.
Materials and Methods
Subjects
Subjects were described previously (5). Briefly, 59 white men and women with normal glucose tolerance (NGT) (n = 19), IGT (n = 20), or T2D (n = 20) determined by oral glucose tolerance test (OGTT) were randomly selected from the participants in a health survey (5, 28). None of the participants had been treated for diabetes at the time of the study. Subjects with NGT had no family history of diabetes. All subjects signed informed consents. All procedures were approved by the Human Research Committee of the University of Leipzig (28).
Measurements of serum RBP4 levels
qWestern method
Measurements of RBP4 levels with qWestern were described previously (23). RBP4 was detected with a polyclonal antibody to human RBP4 (DakoCytomation). RBP4 levels were calculated based on a standard curve of known amounts of purified full-length human RBP4 run on the same gel.
qMSIA method
Human serum samples were prepared with a previously described protocol (27), modified by addition of a 20-min sodium dodecyl sulfate (SDS) incubation to denature and liberate RBP4 from interacting molecules, such as TTR; 50 μl of a 1/100 dilution of serum were mixed in a 1:1 ratio with a 0.5% SDS solution (vol/vol) and incubated at room temperature for 20 min. Next, each sample was spiked with 50 μl of a 25 μg/μl solution of the β-lactoglobulin (β-Lac), which is used as the assay internal reference standard (IRS). Samples were diluted to 1 ml with HEPES-buffered saline (HBS), instead of the previously described method using a HBS dilution buffer containing EDTA and Polysorbate-20 as chelating agent and detergent (27). RBP4 and β-Lac IRS were coimmunoaffinity purified from duplicate samples using MSIA-Tips tailored to specifically retrieve both the IRS and RBP4 analytes from the samples. The MSIA-Tips were provided by Thermo Fisher Scientific and prepared as previously described (29, 30). The tips were outfitted with microfluidic solid supports (frits) manufactured with a molecular carboxyl scaffold. The carboxyl groups were activated with 1,1′-carbonyldiimidazole, which allows for the covalent coupling through the primary amines located on the antibodies. For high-throughput antibody coupling, the tips were loaded onto a 96-format robotic pipetting workstation (Multimek 96, Beckman Coulter, Fullerton, CA). RBP4 (Dako, Carpinteria, CA) and β-Lac (GeneTex, Inc., Irvine, CA) antibodies were coupled to the tips by repetitively flowing (aspirating and dispensing 400 times) 50-μl volumes of antibody solution [100 μl/well and 0.01 mg/ml in 10 mm sodium acetate (pH 4.8)] through each of the tips. The remaining active sites on the frits were blocked with ethanolamine [1 m (pH 8.5), 50 aspirations and dispensing, 100 μl each]. The tips were equilibrated in HBS buffer and stored at 4 C until ready for use. RBP4 measurements were performed using a standard 96-well plate format. Mass spectra were generated by matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) and analyzed with Zebra MS software (Intrinsic Bioprobes, Inc.) as previously described (27). In brief, RBP4 signal integrals corresponding to m/z of full-length RBP4, RBP-L, and RBP4-LL underwent single-point normalization to the integral of the IRS signal in each spectrum. Normalized RBP4 signal integrals from replicate samples were averaged and recorded. Total endogenous RBP4 was quantified by summing the averaged integrals of full-length and cleaved species in each sample. This summed or total integral was applied to the resultant line equation generated from different concentrations of a purified full-length RBP4 protein calibrant to calculate the concentration of total RBP4. Absolute concentrations of different RBP4 species were calculated by multiplying total RBP4 by the fraction of integral values for each form.
Other measurements
Insulin sensitivity was determined by GDR measured by euglycemic-hyperinsulinemic clamp with an insulin infusion rate of 20 mU/kg·min as previously described (28). Estimated glomerular filtration rate (eGFR) was calculated by the Modification of Diet in Renal Disease study equation (31).
Statistics
Statistical analyses were performed with SPSS (version 18) statistical package. Differences in outcome variables among the groups were determined by ANOVA with Tukey's post hoc testing or paired t test as indicated. Univariate correlation analyses were used to examine relationships between variables of interest. Multiple regression analyses were performed to determine independent associations between total RBP4 levels and insulin sensitivity. Statistical significance was set at the P < 0.05 level.
Results
Subject characteristics
The subject characteristics are described in Supplemental Table 1, published on The Endocrine Society's Journals Online web site at http://endo.endojournals.org. Subjects in IGT and T2D groups were older than subjects in the NGT group. IGT and T2D subjects had lower GDR, eGFR, and high-density lipoprotein cholesterol (HDL-C), and higher WHR, 2-h glucose levels during OGTT, hemoglobin A1c (HbA1c), triglyceride (Tg) to HDL-C ratio, and systolic blood pressure compared with NGT individuals. IGT and T2D subjects were similar in all of the subject characteristics except for HbA1c and 2-h OGTT, which were higher in the T2D group.
RBP4 measurements with qMSIA
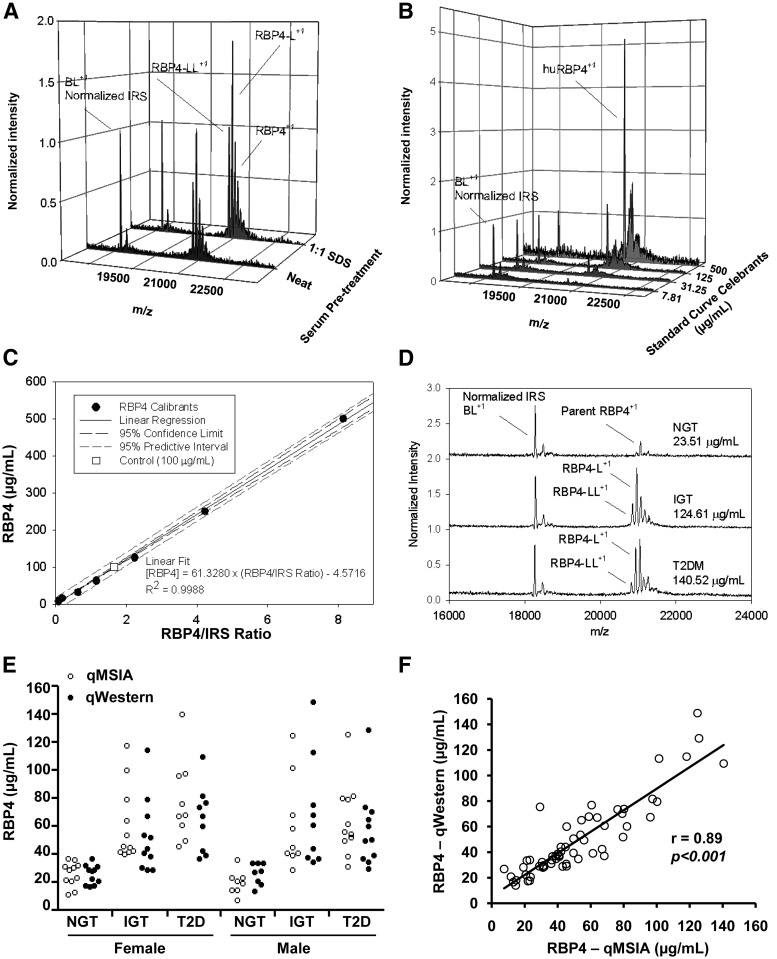
qMSIA with and without SDS treatment was performed on aliquots of the same sample to demonstrate the benefit gained from the incorporation of the SDS pretreatment. SDS treatment improved the recovery of all forms of RBP4 detected, including full-length RBP4 parent, m/z = 21,066.5; RBP4-L, m/z = 20,953.4; and RBP4-LL, m/z = 20,840.4 (Fig. 1A). The relative ratio between different RBP4 variants was preserved between both sample preparations (Fig. 1A), indicating that SDS treatment produces no bias among the different RBP4 forms present in the sample in terms of SDS effect to improve recovery.
Fig. 1.
RBP4 qMSIA and qWestern measurements. A, qMSIA RBP4 mass spectra in human serum samples prepared neat (without SDS) or with a SDS pretreatment. β-Lactoglobulin (BL) was added as an IRS spiked. +1 indicates the charge state for the m/z signal. B, Normalized mass spectra shows parity between the IRS signals and an observable increase in RBP4 abundance with increasing amounts of purified human RBP4 (huRBP4) calibrant protein. The MS signals next to huRBP4 peak are matrix adducts, which are normally seen in the MALDI process. These adducts do not interfere with the analysis. C, The observed response was linear and fit with a first order polynomial regression to produce a standard curve with an R2 = 0.9988 and an analytical error of 3.25%. D, Selected MS traces displaying endogenous full-length RBP4 (parent), RBP4-L, and RBP4-LL forms in serum samples from normal, IGT, and T2DM patients. E, RBP4 levels measured by qMSIA and qWestern in female and male subjects with NGT (n = 19), IGT (n = 20), and T2D (n = 20). F, Correlation between RBP4 levels measured by qMSIA and qWestern; r = 0.89, P < 0.001.
To make MSIA quantitative, purified human RBP4 protein was used as a calibrant to generate a standard curve (27). Different concentrations of purified RBP4 protein were pretreated with SDS and were subjected to MSIA analysis using β-Lac as IRS. The RBP4 spectra normalized to β-Lac increase in parallel with the increase in RBP4 concentrations (Fig. 1B). Plotting the RBP4/IRS ratio on the x-axis and RBP4 levels on the y-axis produces a highly linear relationship (R2 = 0.9988), even with RBP4 concentrations as high as 125–500 μg/ml (Fig. 1C). A control sample with RBP4 100 μg/ml (Fig. 1C, open square) displayed only a 3.25% error from the theoretical. Representative MS traces from NGT, IGT, and T2D patients show the relative abundance for each variant species detected within each sample (RBP4 parent, RBP4-L, and RBP4-LL) that are normalized to IRS (Fig. 1D). The RBP4 variants monitored in this study have previously been described in the literature (25). The masses of the variants were identified and confirmed using Protein Analysis Work Sheet software to compare the known primary structure with the observed mass. This has a mass accuracy within 150 parts per million. Analysis of many samples of human serum, plasma, and urine using both immunoaffinity purification followed by MS (25, 26, 32) and gel approaches (33, 34) have been in tight agreement regarding the identity of these RBP4 variants within human biological samples. The sum of these variants was used to calculate the total RBP4 using the equation in Fig. 1C, resulting in RBP4 levels ranging from 23.5 to 140.5 μg/ml. The sample spectra provides a typical example to demonstrate the capacity of qMSIA to distinguish different RBP4 forms and to quantify across a broad range of RBP4 levels. The intra- and interassay coefficients of variation for RBP4 qMSIA were 5.71 and 8.26%, respectively. Thus, qMSIA is a reliable and reproducible assay that is capable of accurately measuring high RBP4 levels in insulin-resistant patients, in contrast to some commercial assays, which have a limited dynamic range (23).
RBP4 levels and parameters of metabolic syndrome
We performed parallel measurements of serum total RBP4 levels using qWestern and qMSIA in NGT, IGT, and T2D subjects. The range and mean of total RBP4 levels measured by qMSIA and qWestern are very similar (paired t test P > 0.05) (Fig. 1E). When comparing the measurements in individual subjects, there is a tight correlation between qMSIA and qWestern (r = 0.89, P < 0.001) (Fig. 1F).
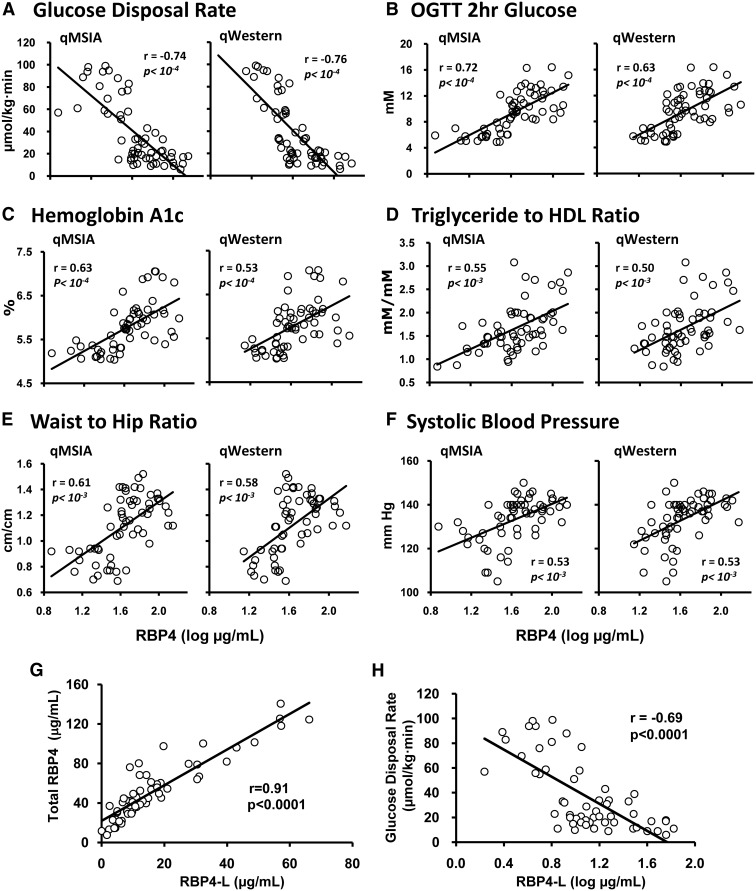
RBP4 levels measured by both qWestern and qMSIA are highly correlated with the severity of insulin resistance as assessed by GDR during clamp (Fig. 2A), 2-h glucose levels during OGTT (Fig. 2B), HbA1c (Fig. 2C), Tg to HDL-C ratio (Fig. 2D), WHR (Fig. 2E), and systolic blood pressure (Fig. 2F). The results indicate that qMSIA has a similar dynamic range and can be used instead of qWestern to measure serum RBP4 levels in insulin-resistant and NGT subjects.
Fig. 2.
A–F, Correlation between RBP4 levels measured by qMSIA or qWestern in serum from NGT, IGT, and T2D subjects and metabolic parameters as indicated in the panels; r and P values are indicated in panels. G, Correlation between serum total RBP4 levels and RBP4-L levels in NGT, IGT, and T2D subjects (r = 0.91, P < 0.0001). H, Negative correlation between serum RBP4-L levels and GDR (r = −0.69, P < 0.0001).
Different from qWestern, qMSIA has the capacity to separate full-length RBP4 and two proteolyzed forms of RBP4 (i.e. RBP4-L and RBP4-LL). RBP4-L is detected in subjects in all three groups, except one NGT subject. The mean percentage of RBP4-L of total RBP4 is 22% in NGT, 32% in IGT, and 31% in T2D individuals (Table 1). RBP4-L values are approximately 50% of total RBP4 in some IGT/T2D subjects. RBP4-L levels are elevated up to 4-fold in IGT and T2D groups compared with NGT group but are similar between the two insulin-resistant groups (Table 1). RBP4-L levels are highly correlated with total RBP4 levels (r = 0.91, P < 0.0001) (Fig. 2G) and inversely correlated with GDR (r = −0.69, P < 0.0001) (Fig. 2H). RBP4-LL is not detected in any of the NGT subjects but is found in 12 out of 20 subjects with IGT and 11 out of 20 subjects with T2D. In subjects with detectable RBP4-LL, it also correlates with total RBP4 (r = 0.77, P < 0.0001) and with RBP4-L (r = 0.89, P < 0.0001), but RBP4-LL does not correlate with GDR (data not shown), which may be due to the small number of subjects in which RBP4-LL was detected. These results indicate that the proteolyzed forms of RBP4 may contribute significantly to total RBP4 levels, especially in IGT and T2DM patients. In addition, because RBP4-LL is not detected in any of the NGT subjects, it may serve as a specific marker for IGT and T2D.
Table 1.
Serum levels of total and C-terminal proteolyzed RBP4 determined by qMSIA
| NGT (n = 19) | IGT (n = 20) | T2D (n = 20) | |
|---|---|---|---|
| Total RBP4, μg/ml (range) | 23.5 ± 2.0 (7.5–36.9) | 61.0 ± 6.4a (29.2–124.6) | 70.6 ± 6.2a (31.4–140.5) |
| Full-length RBP4, μg/ml (range) | 18.1 ± 1.6 (5.8–34.3) | 34.5 ± 2.4a (19.6–62.4) | 41.8 ± 3.5a (23.3–72.6) |
| RBP4-L, μg/ml (range) | 5.3 ± 0.7 (0.0–11.2) | 21.5 ± 3.9a (6.7–66.2) | 23.0 ± 3.4a (8.1–57.1) |
| RBP4-LL, μg/ml (range) | 0.0 ± 0.0 (0.0–0.0) | 4.9 ± 1.7b (0.0–24.6) | 5.8 ± 2.1a (0.0–41.2) |
| RBP4-L, % (range) | 22 ± 2 (0–33) | 32 ± 3a (16–53) | 31 ± 2a (12–49) |
| RBP4-LL, % (range) | 0.0 ± 0.0 (0–0) | 5.8 ± 1.5a (0–20) | 6.3 ± 1.8a (0–33) |
Data are mean ± se. %, Percentage of total RBP4.
P < 0.05 vs. NGT.
P = 0.086 vs. NGT.
RBP4 and kidney function
Recently, it was reported that elevated serum RBP4 in T2D is primarily caused by impaired renal function, not by diabetes (20, 21). Because the patients in the current study are newly diagnosed IGT and T2D, their renal function is relatively preserved. The average eGFR in NGT, IGT, and T2D is 107, 76, and 75 ml/min per 1.73 m2, respectively (Supplemental Table 1). Multiple regression analyses show that RBP4 levels measured by both qMSIA (β-coefficients −0.653, P < 0.001) and qWestern (β-coefficients −0.805, P < 0.001) remain highly associated with GDR after correcting for age, BMI, WHR, Tg/HDL-C, and systolic blood pressure (Table 2, model 1). Adding eGFR to the analysis (Table 2, model 2) has little impact on standardized β-coefficients, suggesting that insulin resistance is the strongest, independent predictor for increased RBP4 levels. eGFR is not a predictor for serum RBP4 levels measured by qMSIA or qWestern (Table 2). When creatinine is used in the multiple regression analysis instead of eGFR, GDR is still the strongest predictor for serum RBP4 levels measured by qMSIA (β-coefficients −0.491, P = 0.003) and by qWestern (β-coefficents −0.674, P < 0.001). In this case, creatinine levels predict, but to a lesser extent than GDR, serum RBP4 levels measured by qMSIA (β-coefficents 0.319, P = 0.008) and by qWestern (β-coefficents 0.257, P = 0.032). These data strongly demonstrate that insulin resistance (GDR) is the strongest determinant of elevated serum RBP4 levels in IGT and T2D.
Table 2.
Multiple regression analyses
| qMSIA |
qWestern |
|||
|---|---|---|---|---|
| Standardized β-coefficients | P values | Standardized β-coefficients | P values | |
| No eGFR | ||||
| Model 1 | ||||
| Age | −0.13 | 0.373 | 0.009 | 0.952 |
| BMI | −0.018 | 0.918 | −0.199 | 0.249 |
| WHR | −0.031 | 0.889 | −0.031 | 0.890 |
| Tg/HDL-C | 0.298 | 0.003 | 0.256 | 0.009 |
| LDL-C | 0.147 | 0.134 | 0.108 | 0.263 |
| SBP | 0.039 | 0.774 | −0.039 | 0.772 |
| GDR | −0.653 | <0.001 | −0.805 | <0.001 |
| Added eGFR | ||||
| Model 2 | ||||
| Age | −0.206 | 0.165 | −0.017 | 0.912 |
| BMI | −0.086 | 0.619 | −0.206 | 0.248 |
| WHR | 0.127 | 0.586 | −0.014 | 0.953 |
| Tg/HDL-C | 0.24 | 0.018 | 0.25 | 0.016 |
| LDL-C | 0.13 | 0.174 | 0.106 | 0.277 |
| SBP | −0.002 | 0.989 | −0.044 | 0.753 |
| GDR | −0.549 | 0.001 | −0.794 | <0.001 |
| eGFR | −0.226 | 0.062 | −0.024 | 0.846 |
Dependent variable, log MSIA or log qWestern. Independent variables, Age, BMI, WHR, Tg/HDL-C, low-density lipoprotein cholesterol (LDL-C), SBP, GDR (model 1), and eGFR (model 2). Multiple regression analyses were performed with SPSS (version 18) statistical package. SBP, Systolic blood pressure.
Discussion
Measuring serum RBP4 has been problematic, because many commercial assays were originally developed for detecting below-normal levels of RBP4 as a surrogate for vitamin A deficiency and were never optimized to measure the elevated levels characteristically found in insulin-resistant subjects (23, 35). We have developed a novel and highly accurate method for quantifying serum RBP4 levels designed for making measurements in insulin-resistant subjects. We adapted the MALDI-TOF-MS method (25) to make it quantitative. The most important modification was adding SDS to denature the RBP4 protein, similar to the condition used in qWestern. This eliminates epitope masking that can be caused by binding of RBP4 to TTR or other serum proteins. Thus, it increases RBP4 immunodetection and results in enhanced signal. The linear range of this assay is at least from approximately 7 to 500 μg/ml (Fig. 1C). qMSIA has many advantages over qWestern, such as high throughput (96 well), stability of assay conditions, and the capacity to distinguish different forms of RBP4. Because serum RBP4 levels measured by qMSIA highly correlate with the levels measured by qWestern, qMSIA provides a needed, high-throughput method that will enable use of RBP4 as an indicator of severity of insulin resistance among diabetic and prediabetic subjects in both research and clinical settings (8, 9).
Furthermore, qMSIA separates and quantifies the two major proteolyzed forms of RBP4 in human serum (RBP4-L and RBP4-LL), which together can account for more than 50% of total RBP4 levels in some insulin-resistant subjects. RBP4-L correlates highly with insulin resistance (Fig. 2H), but RBP4-LL does not (data not shown). However, RBP4-LL is detected only in IGT and T2D subjects and, therefore, could provide a specific, although not highly sensitive, marker for insulin resistance. If an assay has decreased affinity for RBP4-L and RBP4-LL compared with full-length RBP4, it would significantly underestimate the RBP4 elevation in insulin resistance (23). On the other hand, some ELISAs exhibit greater reactivity for proteolyzed RBP4 compared with full-length RBP4 (21, 23). This may lead to the overestimation of RBP4 in some conditions, such as renal failure, in which renal excretion of proteolyzed forms is diminished (20, 21, 36). Thus unbiased measurements of the different forms of RBP4 are of critical importance for the accurate quantification of RBP4 in different medical conditions.
Chronic renal insufficiency is a common complication of T2D. Recently, some studies have shown that renal dysfunction, but not diabetes, is the primary cause for the elevated serum RBP4 levels in T2D (20, 21). However, we found that insulin resistance is the strongest predictor of serum RBP4 levels in IGT and T2D subjects (Table 2). The discrepancies among studies may be due to several factors. First, for many studies investigating RBP4 levels in renal failure, the subjects had advanced or end stage renal disease, which causes many medical problems, including insulin resistance secondary to uremia (37). In the current article, we studied newly diagnosed IGT and T2D patients who have normal or mild renal insufficiency, enabling us to dissect the individual contributions of insulin resistance and renal function to elevated RBP4 levels. Our multivariate analysis data show that insulin resistance is the strongest determinant of elevated serum RBP4 levels in IGT and T2D (Table 2). Renal function is a weaker contributor. A second factor is the parameter(s) used to assess insulin resistance. Many studies investigating RBP4 levels in renal failure used glucose levels or HbA1c (20), whereas others used homeostasis model assessment-estimated insulin resistance (HOMA-IR) or fasting insulin (21) as indicators of insulin resistance. We used the “gold-standard” clamp GDR to measure insulin resistance and show that insulin resistance is an independent predictor of serum RBP4 levels. Although glucose levels and HbA1c correlate with serum RBP4 levels (Fig. 2, B and C); multiple regression analysis shows that they are not independent predictors of serum RBP4 levels (data not shown). This is consistent with the fact that the correlation between RBP4 and insulin resistance or other metabolic parameters is equally strong in IGT and T2D subjects in spite of the difference in glucose and HbA1c levels. HOMA-IR or insulin levels may not reflect the extent of insulin resistance, and HOMA-IR is not reliable in diabetic subjects (38). Finally, RBP4 levels may be overestimated in the presence of renal failure (21, 23), because some RBP4 assays have higher affinity for cleaved forms of RBP4 that are retained in renal failure as discussed above. Thus, our data indicate that insulin resistance is the strongest predictor for serum RBP4 levels. Impaired renal function may also contribute to elevated serum RBP4 levels.
In summary, we report the development of a novel quantitative method for measuring serum RBP4 in insulin-resistant subjects. qMSIA has the capacity to accurately measure a large dynamic range of total and proteolyzed forms of RBP4 in normal, IGT, and T2D subjects. The accuracy of qMSIA in assessing the severity of insulin resistance is very similar to that of qWestern, but qMSIA is much less labor intensive and more high throughput. Application of qMSIA will greatly facilitate clinical research for further assessment of RBP4 as a biomarker for metabolic syndrome and diabetes risk.
Supplementary Material
Acknowledgments
We thank Frank Hu for expert advice on statistical analysis and Jincheng Yang for preparing figures.
Present address for T.E.G.: Molecular Medicine Program and Division of Endocrinology, Metabolism and Diabetes, Department of Medicine, University of Utah, Salt Lake City, Utah 84112.
This work was supported by National Institutes of Health Grants R37 DK43051 and R24 DK083938 (to B.B.K.), T32 DK07516 (to B.B.K. and I.E.), KO8 DK090149 (to Q.Y.), R42 DK071290 (to U.A.K.), and RO3 DK080195 (to T.E.G) and by a Doris Duke Clinical Scientist Development Award (T.E.G).
Disclosure Summary: U.A.K. and D.A.P. are employees of Intrinsic Bioprobes, Inc./Thermo Fisher Scientific, and U.A.K. has equity in that company. B.B.K. had a research grant from Takeda Pharmaceutical. Q.Y., T.E.G., and B.B.K. are inventors on a patent on RBP4 and insulin resistance (United States patent 7553631). I.E. and M.B. have nothing to disclose.
Footnotes
- BMI
- Body mass index
- eGFR
- estimated glomerular filtration rate
- GDR
- glucose disposal rate
- HbA1c
- hemoglobin A1c
- HBS
- HEPES-buffered saline
- HDL-C
- high-density lipoprotein cholesterol
- HOMA-IR
- homeostasis model assessment-estimated insulin resistance
- IGT
- impaired glucose tolerance
- IRS
- internal reference standard
- β-Lac
- β-lactoglobulin
- MALDI-TOF
- matrix-assisted laser desorption/ionization time-of-flight
- NGT
- normal glucose tolerance
- OGTT
- oral glucose tolerance test
- qMSIA
- quantitative mass spectrometry immunoaffinity assay
- qWestern
- quantitative Western blotting
- RBP4
- retinol-binding protein 4
- RBP4-L
- des(Leu)-RBP4
- RBP4-LL
- di-des(Leu)-RBP4
- SDS
- sodium dodecyl sulfate
- T2D
- type 2 diabetes
- Tg
- triglyceride
- TTR
- transthyretin
- WHR
- waist to hip ratio.
References
- 1. Yang Q, Graham TE, Mody N, Preitner F, Peroni OD, Zabolotny JM, Kotani K, Quadro L, Kahn BB. 2005. Serum retinol binding protein 4 contributes to insulin resistance in obesity and type 2 diabetes. Nature 436:356–362 [DOI] [PubMed] [Google Scholar]
- 2. Balagopal P, Graham TE, Kahn BB, Altomare A, Funanage V, George D. 2007. Reduction of elevated serum retinol binding protein in obese children by lifestyle intervention: association with subclinical inflammation. J Clin Endocrinol Metab 92:1971–1974 [DOI] [PubMed] [Google Scholar]
- 3. Gavi S, Qurashi S, Melendez MM, Mynarcik DC, McNurlan MA, Gelato MC. 2007. Plasma retinol-binding protein-4 concentrations are elevated in human subjects with impaired glucose tolerance and type 2 diabetes: response to Cho et al. Diabetes Care 30:e7; author reply e8 [DOI] [PubMed] [Google Scholar]
- 4. Gavi S, Stuart LM, Kelly P, Melendez MM, Mynarcik DC, Gelato MC, McNurlan MA. 2007. Retinol-binding protein 4 is associated with insulin resistance and body fat distribution in nonobese subjects without type 2 diabetes. J Clin Endocrinol Metab 92:1886–1890 [DOI] [PubMed] [Google Scholar]
- 5. Graham TE, Yang Q, Blüher M, Hammarstedt A, Ciaraldi TP, Henry RR, Wason CJ, Oberbach A, Jansson PA, Smith U, Kahn BB. 2006. Retinol-binding protein 4 and insulin resistance in lean, obese, and diabetic subjects. N Engl J Med 354:2552–2563 [DOI] [PubMed] [Google Scholar]
- 6. Klöting N, Graham TE, Berndt J, Kralisch S, Kovacs P, Wason CJ, Fasshauer M, Schön MR, Stumvoll M, Blüher M, Kahn BB. 2007. Serum retinol-binding protein is more highly expressed in visceral than in subcutaneous adipose tissue and is a marker of intra-abdominal fat mass. Cell Metab 6:79–87 [DOI] [PubMed] [Google Scholar]
- 7. Kovacs P, Geyer M, Berndt J, Klöting N, Graham TE, Böttcher Y, Enigk B, Tönjes A, Schleinitz D, Schön MR, Kahn BB, Blüher M, Stumvoll M. 2007. Effects of genetic variation in the human retinol binding protein-4 gene (RBP4) on insulin resistance and fat depot-specific mRNA expression. Diabetes 56:3095–3100 [DOI] [PubMed] [Google Scholar]
- 8. Qi Q, Yu Z, Ye X, Zhao F, Huang P, Hu FB, Franco OH, Wang J, Li H, Liu Y, Lin X. 2007. Elevated retinol-binding protein 4 levels are associated with metabolic syndrome in Chinese people. J Clin Endocrinol Metab 92:4827–4834 [DOI] [PubMed] [Google Scholar]
- 9. Meisinger C, Rückert IM, Rathmann W, Döring A, Thorand B, Huth C, Kowall B, Koenig W. 2011. Retinol-binding protein 4 is associated with prediabetes in adults from the general population: the cooperative health research in the region of Augsburg (KORA) F4 study. Diabetes Care 34:1648–1650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kelly KR, Kashyap SR, O'Leary VB, Major J, Schauer PR, Kirwan JP. 2010. Retinol-binding protein 4 (RBP4) protein expression is increased in omental adipose tissue of severely obese patients. Obesity 18:663–666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Tschoner A, Sturm W, Engl J, Kaser S, Laimer M, Laimer E, Weiss H, Patsch JR, Ebenbichler CF. 2008. Retinol-binding protein 4, visceral fat, and the metabolic syndrome: effects of weight loss. Obesity 16:2439–2444 [DOI] [PubMed] [Google Scholar]
- 12. Ost A, Danielsson A, Lidén M, Eriksson U, Nystrom FH, Strålfors P. 2007. Retinol-binding protein-4 attenuates insulin-induced phosphorylation of IRS1 and ERK1/2 in primary human adipocytes. FASEB J 21:3696–3704 [DOI] [PubMed] [Google Scholar]
- 13. Berry DC, Jin H, Majumdar A, Noy N. 2011. Signaling by vitamin A and retinol-binding protein regulates gene expression to inhibit insulin responses. Proc Natl Acad Sci USA 108:4340–4345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Deng ZB, Poliakov A, Hardy RW, Clements R, Liu C, Liu Y, Wang J, Xiang X, Zhang S, Zhuang X, Shah SV, Sun D, Michalek S, Grizzle WE, Garvey T, Mobley J, Zhang HG. 2009. Adipose tissue exosome-like vesicles mediate activation of macrophage-induced insulin resistance. Diabetes 58:2498–2505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Johansson H, Gandini S, Guerrieri-Gonzaga A, Iodice S, Ruscica M, Bonanni B, Gulisano M, Magni P, Formelli F, Decensi A. 2008. Effect of fenretinide and low-dose tamoxifen on insulin sensitivity in premenopausal women at high risk for breast cancer. Cancer Res 68:9512–9518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Craig RL, Chu WS, Elbein SC. 2007. Retinol binding protein 4 as a candidate gene for type 2 diabetes and prediabetic intermediate traits. Mol Genet Metab 90:338–344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. van Hoek M, Dehghan A, Zillikens MC, Hofman A, Witteman JC, Sijbrands EJ. 2008. An RBP4 promoter polymorphism increases risk of type 2 diabetes. Diabetologia 51:1423–1428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wu Y, Li H, Loos RJ, Qi Q, Hu FB, Liu Y, Lin X. 2009. RBP4 variants are significantly associated with plasma RBP4 levels and hypertriglyceridemia risk in Chinese Hans. J Lipid Res 50:1479–1486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Munkhtulga L, Nagashima S, Nakayama K, Utsumi N, Yanagisawa Y, Gotoh T, Omi T, Kumada M, Zolzaya K, Lkhagvasuren T, Kagawa Y, Fujiwara H, Hosoya Y, Hyodo M, Horie H, Kojima M, Ishibashi S, Iwamoto S. 2010. Regulatory SNP in the RBP4 gene modified the expression in adipocytes and associated with BMI. Obesity 18:1006–1014 [DOI] [PubMed] [Google Scholar]
- 20. Henze A, Frey SK, Raila J, Tepel M, Scholze A, Pfeiffer AF, Weickert MO, Spranger J, Schweigert FJ. 2008. Evidence that kidney function but not type 2 diabetes mellitus determines retinol-binding protein 4 (RBP4) serum levels. Diabetes 57:3323–3326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ziegelmeier M, Bachmann A, Seeger J, Lossner U, Kratzsch J, Blüher M, Stumvoll M, Fasshauer M. 2007. Serum levels of adipokine retinol-binding protein-4 in relation to renal function. Diabetes Care 30:2588–2592 [DOI] [PubMed] [Google Scholar]
- 22. Khovidhunkit W, Pruksakorn P, Plengpanich W, Tharavanij T. 2012. Retinol-binding protein 4 is not associated with insulin resistance in pregnancy. Metabolism 61:65–69 [DOI] [PubMed] [Google Scholar]
- 23. Graham TE, Wason CJ, Blüher M, Kahn BB. 2007. Shortcomings in methodology complicate measurements of serum retinol binding protein (RBP4) in insulin-resistant human subjects. Diabetologia 50:814–823 [DOI] [PubMed] [Google Scholar]
- 24. Mody N, Graham TE, Tsuji Y, Yang Q, Kahn BB. 2008. Decreased clearance of serum retinol-binding protein and elevated levels of transthyretin in insulin-resistant ob/ob mice. Am J Physiol Endocrinol Metab 294:E785–E793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kiernan UA, Tubbs KA, Nedelkov D, Niederkofler EE, Nelson RW. 2002. Comparative phenotypic analyses of human plasma and urinary retinol binding protein using mass spectrometric immunoassay. Biochem Biophys Res Commun 297:401–405 [DOI] [PubMed] [Google Scholar]
- 26. Nedelkov D, Kiernan UA, Niederkofler EE, Tubbs KA, Nelson RW. 2005. Investigating diversity in human plasma proteins. Proc Natl Acad Sci USA 102:10852–10857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kiernan UA, Phillips DA, Trenchevska O, Nedelkov D. 2011. Quantitative mass spectrometry evaluation of human retinol binding protein 4 and related variants. PLoS ONE 6:e17282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Blüher M, Fasshauer M, Tönjes A, Kratzsch J, Schön MR, Paschke R. 2005. Association of interleukin-6, C-reactive protein, interleukin-10 and adiponectin plasma concentrations with measures of obesity, insulin sensitivity and glucose metabolism. Exp Clin Endocrinol Diabetes 113:534–537 [DOI] [PubMed] [Google Scholar]
- 29. Niederkofler EE, Tubbs KA, Gruber K, Nedelkov D, Kiernan UA, Williams P, Nelson RW. 2001. Determination of β-2 microglobulin levels in plasma using a high-throughput mass spectrometric immunoassay system. Anal Chem 73:3294–3299 [DOI] [PubMed] [Google Scholar]
- 30. Tubbs KA, Nedelkov D, Nelson RW. 2001. Detection and quantification of β-2-microglobulin using mass spectrometric immunoassay. Anal Biochem 289:26–35 [DOI] [PubMed] [Google Scholar]
- 31. Levey AS, Coresh J, Greene T, Stevens LA, Zhang YL, Hendriksen S, Kusek JW, Van Lente F. 2006. Using standardized serum creatinine values in the modification of diet in renal disease study equation for estimating glomerular filtration rate. Ann Intern Med 145:247–254 [DOI] [PubMed] [Google Scholar]
- 32. Frey SK, Nagl B, Henze A, Raila J, Schlosser B, Berg T, Tepel M, Zidek W, Weickert MO, Pfeiffer AF, Schweigert FJ. 2008. Isoforms of retinol binding protein 4 (RBP4) are increased in chronic diseases of the kidney but not of the liver. Lipids Health Dis 7:29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Jaconi S, Rose K, Hughes GJ, Saurat JH, Siegenthaler G. 1995. Characterization of two post-translationally processed forms of human serum retinol-binding protein: altered ratios in chronic renal failure. J Lipid Res 36:1247–1253 [PubMed] [Google Scholar]
- 34. Jaconi S, Saurat JH, Siegenthaler G. 1996. Analysis of normal and truncated holo- and apo-retinol-binding protein (RBP) in human serum: altered ratios in chronic renal failure. Eur J Endocrinol 134:576–582 [DOI] [PubMed] [Google Scholar]
- 35. Gamble MV, Ramakrishnan R, Palafox NA, Briand K, Berglund L, Blaner WS. 2001. Retinol binding protein as a surrogate measure for serum retinol: studies in vitamin A-deficient children from the Republic of the Marshall Islands. Am J Clin Nutr 73:594–601 [DOI] [PubMed] [Google Scholar]
- 36. Henze A, Rohn S, Gericke B, Raila J, Schweigert FJ. 2008. Structural modifications of serum transthyretin in rats during protein-energy malnutrition. Rapid Commun Mass Spectrom 22:3270–3274 [DOI] [PubMed] [Google Scholar]
- 37. D'Apolito M, Du X, Zong H, Catucci A, Maiuri L, Trivisano T, Pettoello-Mantovani M, Campanozzi A, Raia V, Pessin JE, Brownlee M, Giardino I. 2010. Urea-induced ROS generation causes insulin resistance in mice with chronic renal failure. J Clin Invest 120:203–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wallace TM, Levy JC, Matthews DR. 2004. Use and abuse of HOMA modeling. Diabetes Care 27:1487–1495 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.