Abstract
Melanocortin signaling plays a central role in the regulation of phenotypes related to body weight and energy homeostasis. To specifically target and study the function of proopiomelanocortin (POMC) neurons, Pomc promoter elements have been utilized to generate reporter and Cre recombinase transgenic reagents. Across gestation, we find that Pomc is dynamically expressed in many sites in the developing mouse forebrain, midbrain, hindbrain, spinal cord, and retina. Although Pomc expression in most embryonic brain regions is transient, it is sufficient to direct Cre-mediated recombination of floxed alleles. We visualize the populations affected by this transgene by crossing Pomc-Cre mice to ROSA reporter strains and identify 62 sites of recombination throughout the adult brain, including several nuclei implicated in energy homeostasis regulation. To compare the relationship between acute Pomc promoter activity and Pomc-Cre-mediated recombination at the single cell level, we crossed Pomc-enhanced green fluorescent protein (eGFP) and Pomc-Cre;ROSA-tdTomato lines. We detect the highest concentration of Pomc-eGFP+ cells in the arcuate nucleus of the hypothalamus and dentate gyrus but also observe smaller populations of labeled cells in the nucleus of the solitary tract, periventricular zone of the third ventricle, and cerebellum. Consistent with the dynamic nature of Pomc expression in the embryo, the vast majority of neurons marked with the tdTomato reporter do not express eGFP in the adult. Thus, recombination in off-target sites could contribute to physiological phenotypes using Pomc-Cre transgenics. For example, we find that approximately 83% of the cells in the arcuate nucleus of the hypothalamus immunoreactive for leptin-induced phosphorylated signal transducer and activator of transcription 3 are marked with Pomc-Cre;ROSA-tdTomato; only 13% of these are eGFP+ POMC neurons.
Severe obesity in mice lacking the Proopioimelanocortin (Pomc) or Melanocortin receptor 4 (Mc4r) genes led to the widely held belief that POMC neurons play a central role in regulating body weight (1, 2). POMC and Agouti-related peptide (AgRP) neurons located in the arcuate nucleus of the hypothalamus (ARH) are thought to be the principal source of MC4R ligands in the brain; however, the contribution of signals from POMC neurons in the nucleus of the solitary tract (NTS) has not been well defined. A cleavage product of the POMC peptide, α-MSH, and AgRP induce opposite effects on signaling via MC4R (3, 4). Observations that leptin signaling induces the expression of the POMC-derived agonist and inhibits the expression of the AgRP antagonist led to the hypothesis that melanocortin circuits mediate leptin's effects on energy homeostasis (5–9). Because these early seminal studies relied on pharmacological administration of leptin and/or melanocortin antagonists and agonists, they could not distinguish between the contributions of discrete neuronal populations to energy homeostasis.
Two types of genetically engineered mouse strains were generated to specifically target and interrogate the role of POMC neurons in leptin-responsive circuits. Enhanced green fluorescent protein (eGFP) reporters driven by Pomc promoter elements (Pomc-eGFP) (8, 10) have been used to identify POMC neurons for electrophysiological and immunohistochemical analyses. In addition, a Pomc-Cre BAC transgenic strain was developed to restrict genetic manipulations to POMC neurons and thus evaluate their function in vivo (11). These seminal studies using Pomc transgenic reagents serve as the basis for prevailing models of hypothalamic regulation of body weight and energy homeostasis.
Previously, we demonstrated that the Pomc-Cre transgene activity during gestation directs recombination of floxed alleles in many ARH neurons that do not express POMC in adulthood (12). In follow-up studies, we observed that Pomc is broadly expressed throughout the central nervous system (CNS) during development. These observations raise the possibility that off-target effects in diverse populations in the CNS could contribute to physiological outcomes of genetic manipulations intended for POMC neurons. To assist in interpreting experiments using Pomc-Cre mice, we provide a comprehensive map of brain nuclei marked by Pomc-Cre-mediated recombination of ROSA26-(loxSTOPlox)-LacZ (R26R-LacZ) or ROSA26-(loxSTOPlox)-tdTOMATO (R26R-TOM) reporter strains (13, 14). Next, we compare the relationship between endogenous Pomc transcription, expression of a Pomc-eGFP reporter, and Pomc-Cre reporters at the single cell level in Pomc-eGFP; Pomc-Cre;R26R-TOM mice. Because melanocortin and leptin pathways likely interact to regulate energy homeostasis, we characterize the relationship between leptin-induced phosphorylated signal transducer and activator of transcription 3 (P-STAT3) immunoreactivity, neurons marked by a Pomc-Cre reporter, and those that express Pomc or Neuropeptide Y (Npy). Temporal and spatial patterns of Pomc-eGFP and Pomc-Cre reporter expression should inform the analysis and design of functional studies using these reagents.
Materials and Methods
Animals
Animals were housed in temperature-controlled rooms at 21 C and subject to a 12-h light, 12-h dark cycle. Mice had ad libitum access to standard chow diet (PicoLab Rodent Diet 5053; LabDiet, Henderson, CO) and water. ROSA(26R)-LacZ (Jackson 003309) (13) and C57BL/6, ROSA(26R)-TOM (Jackson 012567) (14) mice were obtained from The Jackson Laboratory (Bar Harbor, ME) and bred at the Russ Berrie Animal Facility. Pomc-Cre (11) and Pomc-eGFP (8) transgenic animals were generously provided by Joel Elmquist/Bradford Lowell and Malcolm Low, respectively. Note, the BAC transgenic Pomc-Cre incorporates at least 45 kb of 5′ and 70 kb of 3′ Pomc flanking sequences, and the Pomc-eGFP transgenic incorporates 13 kb of 5′ and 2 kb of 3′ Pomc flanking sequences. All procedures were performed in accordance with the guidelines of the Institutional Animal Care and Use Committee at the Columbia University Health Sciences Division.
Genotyping
Genotyping at the ROSA26 locus for LacZ was performed using the following three-primer set: oIMR 0316 [wild-type (wt)-forward] 5′-GGAGCGGGAGAAATGGATATG-3′, oIMR 0883 (wt-reverse) 5′-AAAGTCGCTCTGAGTTGTTAT-3′, and oIMR 0315 (mutant-LacZ) 5′-GCGAAGAGTTTGTCCTCAACC-3′ (13). Genotyping at the ROSA26 locus for TOM was performed using a three-primer set including both wt primers (0316 and 0883) above and oIMR 9105 (mutant-TOM) 5′-CTGTTCCTGTACGGCATGG-3′ (14). The Cre transgene was assessed with: 5′-GCGGTCTGGCAGTAAAAACTATC-3′ and 5′-GTGAAACAGCATTGCTGTCACTT-3′.
Tissue processing
P9 and adult mice were anesthetized and transcardially perfused with 4% paraformaldehyde fixative. Embryos were dissected in cold PBS and fixed at 4 C overnight. NeutrAvidin-amplified in situ hybridization (ISH) was performed on fresh frozen tissue. Tissue used for immunohistochemistry (IHC) and/or fluorescent ISH (FISH) was embedded in O.C.T. (Tissue Tek, Torrance, CA) and frozen at −80 C; 10-μm-thick coronal sections were collected on slides.
X-Gal staining
Immediately after fixation, tissue was permeabilized using 2 mm MgCl2, 0.001% sodium deoxycholate, and 0.02% Nonidet P-40 buffer. The tissue was then incubated in: 5 mm potassium ferricyanide, 5 mm potassium ferrocyanide, 20 mm Tris, and 1 mg/ml X-Gal, overnight at room temperature. Whole-mount embryos were cleared in glycerol and imaged. Adult brains were vibrabome sectioned (200 μm) and slide mounted. Images were acquired with a Zeiss LSM5 Pascal Confocal (Zeiss, Oberkochen, Germany) or a Nikon Eclipse 80i (Nikon, Melville, NY) equipped with a Retiga EXi camera and Q Capture Pro imaging software (QImaging, Surrey, Canada).
Scoring
The density of positive X-Gal staining in each brain nucleus, expressed as a percentage of total nuclear area, was used to determine the scores assigned to the nuclei as follows: 0–2% scored ±, 2–8% +, 8–14% ++, 14–30% +++, 30–50% ++++, and more than 50% +++++.
Dot rendering
Stain density in each designated nucleus (according to Allen Reference Atlas, http://www.brain-map.org) (15) was quantified and rendered into dots using Adobe Photoshop. Within each nucleus, dot number is proportional to the stained area, and placement reflects the spatial distribution of stained cells. Note, dots were not assigned to nuclei with less than 2% total stain.
Immunohistochemistry
Primary antibody incubation was performed using either guinea pig anti-green fluorescent protein (GFP) (1:2000; Molecular Probes, Eugene, OR) or rabbit anti-P-STAT3 (1:800; Cell Signaling, Beverly, MA) at 4 C overnight. Tissue was then incubated in with secondary using either goat antiguinea pig Alexa Fluor 488 (1:500; Invitrogen, Carlsbad, CA) or goat antirabbit cy5 (1:100; Jackson ImmunoResearch, West Grove, PA), for 1 h at room temperature. Note, for P-STAT3, IHC unmasking was performed before primary antibody incubation by the following sequential pretreatment: 1% NaOH with 1% H2O2 solution, 0.3% glycine, and 0.3% sodium dodecyl sulfate. Combined IHC and FISH was performed according to previously published methods (12).
FISH and NeutrAvidin-amplified ISH
Frozen sections were processed as described in the tyramide-based amplification system (TSA) Plus Cy3 System manual (PerkinElmer, Waltham, MA). Antisense digoxigenin- or fluorescein-labeled riboprobes were generated from plasmids containing PCR fragments of Npy and Pomc using the following primers sets: NPY, 5′-TGCTAGGTAACAAGCGAATGG-3′/5′-CAACAACAACAAGGGAAATGG-3 and POMC, 5′- GTTAAGAGCAGTGACTAAGAGAGGC-3′/5′-CCTAACACAGGTAACTCTAAGAGGC-3′. NeutrAvidin-amplified ISH was achieved using biotinylated-tyramide (PerkinElmer) in conjunction with NeutrAvidin Protein, Alkaline Phosphatase Conjugated (Thermo Scientific, Auburn, AL) followed by nitro-blue tetrazolium chloride and 5-bromo-4-chloro-3′-indolyphosphate p-toluidine salt staining.
Results
Dynamic Pomc expression in the embryonic CNS
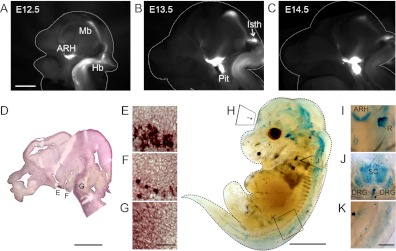
Off-target sites of Pomc-Cre-mediated recombination could result from 1) early transient expression, 2) subthreshold expression in the adult, or 3) ectopic expression in cells that do not express endogenous Pomc transcripts. Distinguishing between these possibilities is critical to developing reagents and strategies that accurately target POMC neurons. Initially, we characterized Pomc expression in the developing embryo using a Pomc-eGFP reporter (8). In two independent transgenic lines, Pomc-eGFP reporter expression accurately reflects endogenous Pomc transcription in ARH neurons (8, 10, 12). At embryonic day (E)12.5, we observe robust GFP signals in the presumptive ARH, retina, optic tract, midbrain, and in a broad domain in the ventral hindbrain and spinal cord (Fig. 1A). We first detect expression in the isthmus and pituitary at E13.5 (Fig. 1B). We confirmed that these sites of GFP fluorescence correlate with Pomc expression, as visualized by ISH (Fig. 1, D–G). Prominent domains of GFP expression persist through E14.5 in the optic tract, ventral hypothalamus, and isthmus; however, the fluorescent signal is dramatically decreased in the midbrain and hindbrain (Fig. 1C). Pomc expression in the developing pituitary steadily increases throughout gestation, in parallel with the maturation of this organ (Fig. 1, B and C).
Fig. 1.
Pomc is broadly distributed and transiently expressed in the embryonic CNS and periphery. Pomc-eGFP reporter fluorescence in hemisected embryonic brains at (A) E12.5, (B) E13.5, and (C) E14.5. D, Images captured from the ventricular surface (magnification, ×1.6) of the ventricular surface. D–G, Pomc ISH of a sagittal section at E13.5. E–G, Magnified views of D indicated by boxes, including: ARH (E), pituitary (F), and hindbrain (G). H–K, E16.5 Pomc-Cre;R26R-LacZ whole mount embryo stained with X-Gal. I–K, Magnified views of the retina from the anterior surface (I), cross-sectional coronal plane of the spinal cord (J), and the duodenum from the lateral surface (K). I, To visualize X-Gal stain in the retina, an albino embryo of the same genotype was processed. I, Representative of the region I denoted in H. All images are presented rostral, left. DRG, Dorsal root ganglia; Hb, hindbrain; Isth, isthmus; Mb, midbrain; Pit, pituitary; R, retina; SC, spinal cord. Arrows, developing vertebrae (J) and duodenal melanocyte (K). Scale bars, 1 mm (A–C), 1 mm (D), 2 mm (H), and 100 μm (G and K).
Distribution of Pomc-Cre activity
We next examined whether transient Pomc expression in the embryo is sufficient to drive Cre-mediated recombination of a floxed reporter allele. At E16.5, we detect X-Gal-labeled cells in the hypothalamus, thalamus, midbrain, hindbrain, retina, olfactory bulb, spinal cord, and dorsal root ganglia in Pomc-Cre;R26R-LacZ embryos (Fig. 1, H–K). In the periphery, we observe X-Gal+ cells in the pituitary and duodenum, consistent with previously identified melanocyte and corticotrophic reservoirs (Fig. 1, H and K) (16, 17). Despite a dramatic down-regulation of Pomc expression in caudal brain regions at E14.5 (Fig. 1C), we detect extensive X-Gal staining at E16.5, indicating that the transient burst of expression is sufficient to drive recombination of floxed alleles in many brain regions.
To capture the cumulative distribution of cells that expressed Pomc at any point during development, we performed X-Gal staining on serial sections of Pomc-Cre;R26R-LacZ adult brains in both sagittal and coronal planes. A comprehensive map of the density and distribution of X-Gal+ neurons is presented in Table 1. The lowest score, ±, denotes regions with very few cells (significantly above background), whereas the highest score of +++++ represents regions with a majority of cells stained in a particular area (Table 1). Analyses of the density and distribution of stained neurons are rendered into dot plot diagrams in eight rostrocaudal and two mediolateral planes of section, as described in Materials and Methods (Fig. 2).
Table 1.
Brain sites of Pomc-Cre-mediated recombination
| Score | |
|---|---|
| Hypothalamus (layers) | |
| Anterior hypothalamus | + |
| ARH | +++++ |
| Dorsal medial hypothalamus | ± |
| Lateral preoptic area | + |
| Medial mammillary nucleus | + |
| Medial preoptic area | + |
| Median preoptic nucleus | ++++ |
| Lateral hypothalamic area | ± |
| PVH | ++ |
| Periventricular (posterior) | +++++ |
| Posterior hypothalamus | + |
| Retrochiasmatic area | ++ |
| SCN | +++ |
| Supraoptic nucleus | ± |
| Tuberal nucleus | +++ |
| VMH (ventromedial) | ++ |
| Ventral premammilary nucleus | + |
| Zona incerta | ± |
| Thalamus | |
| Central medial nucleus | + |
| Lateral dorsal nucleus | + |
| Medial habenula | ± |
| Mediodorsal nucleus | ± |
| Nucleus of reunions | + |
| PVH | + |
| Hippocampus | |
| CA3 (pyramidal cell layer) | + |
| DG (granular cell) | +++++ |
| Entorhinal area (2a,2b,3) | + |
| Parsubiculum | + |
| Postsubiculum | ± |
| Presubiculum | +++ |
| Subiculum (1,2,3) | +++ |
| Olfactory bulb | |
| Cortical amygdalar area (1,2) | ++ |
| Piriform area (2,3) | ++ |
| Taenia tecta | + |
| Septum | |
| Lateral septal nucleus (rostroventral) | + |
| Cerebral cortex | |
| Amygdalar nucleus, basomedial | + |
| BNST | ++ |
| Cerebral cortex (layer 4) | + |
| Amygdalar nucleus, medial | ++ |
| Primary motor area (1,2,3) | ± |
| Primary somatosensory area (3,4) | + |
| Primary somatosensory barrel field (2,3,4) | ± |
| Midbrain | |
| Inferior colliculus (dorsal/external) | ++ |
| Interpeduncular fossa | +++ |
| Medial prectectal area | |
| Parabigeminal | ++ |
| Periaqueductal gray | ± |
| Superior colliculus | ± |
| Ventral tegmental area | ± |
| Pons | |
| LB, external lateral | ++ |
| Pontine central gray | + |
| Pontine reticular nucleus | ± |
| Principal sensory nucleus of the trigeminal | ± |
| Medulla | |
| AP | +++ |
| Cochlear nucleus | ++++ |
| Facial motor nucleus | ++ |
| Intermediate reticular nucleus | ± |
| NTS, medial | ++ |
| NTS, total | + |
| Parvicellular reticular nucleus | ± |
| Spinal nucleus of the trigeminal | + |
| Vestibular nucleus | ± |
Fig. 2.
Distribution of neurons labeled with the Pomc-Cre;R26R-LacZ reporter in the adult brain. Dot plots rendered from X-Gal-stained Pomc-Cre;R26R-LacZ brains (200-μm-thick sections in the coronal and sagittal planes) and labeled with atlas coordinates (15). The density of dots directly corresponds to the density of X-Gal staining in each brain region. See Table 1 for complete listing and scoring.
Hypothalamus and thalamus
In the hypothalamus, we detect the highest density of X-Gal staining in the ARH and the posterior periventricular nucleus. In the ARH, X-Gal+ cells are distributed throughout the nucleus; with notable saturation along the ventral and lateral borders. We observe moderate staining density in the suprachiasmatic nucleus (SCN) and tuberal area and low density staining in the ventromedial division of the ventral medial nucleus (VMH), retrochiasmatic nucleus, and paraventricular nucleus (PVH). There are a few stained cells in the anterior hypothalamus, lateral and medial preoptic areas, medial mammillary nucleus, ventral premammilary nucleus, and posterior hypothalamus. We consistently find sparse X-Gal staining in the zona incerta, supraoptic nucleus, dorsal medial hypothalamus, and lateral hypothalamic area (Fig. 2, bregma 0 to −1.4, and Table 1).
In addition to the ARH, other hypothalamic nuclei that contain a substantial number of X-Gal+ cells also contribute to energy homeostasis. The SCN is the master regulator of circadian rhythms; disruption of circadian inputs leads to hyperphagia, obesity, and insulin resistance (18). The VMH is thought to integrate metabolic and reproductive functions and to modulate physiological responses to high-fat diet (19, 20). Homeostatic, circadian, and hedonic inputs from the hypothalamus and brain stem are integrated in the PVH and relayed to the periphery via neuroendocrine signals and efferent projections to the sympathetic nervous system (21).
In the rostral thalamus, we observe moderate X-Gal staining in the central medial and lateral dorsal nuclei, along with the nucleus of the reunions and PVH. We detect very low levels of expression in the medial habenula and mediodorsal nucleus (Fig. 2, bregma −0.1, and Table 1).
Hippocampus, olfactory bulb, and septum
Outside of the ARH, we find the densest region of X-Gal staining in the granule cell layer of the dentate gyrus (DG), particularly in the medial and lateral blades. We observe moderate staining density in the presubiculum and in layers 1, 2, and 3 of the subiculum and a few X-Gal+ cells in the entorhinal area and in the parsubiculum. In the rostral hippocampus, we detect a few X-Gal+ cells in the CA3 region. We observe sparse staining in the postsubiculum (Fig. 2, bregma −0.1, −1.4, −4.5 and lateral 0.2, 2.6, and Table 1). Processing of cognitive factors by the hippocampus is reported to influence feeding behavior and body weight, in part through effects on learning and memories related to food. For example, hippocampal ablations result in increased food intake and body weight, whereas leptin injections into the ventral hippocampus are reported to suppress food intake and processing of food-related memories (22–24).
In the olfactory bulb, we observe low levels of X-Gal stain in layers 1 and 2 of the cortical amygdalar area and also in layers 2 and 3 of the piriform area. We also detect staining in the taenia tecta and in the rostroventral aspect of the lateral septal nucleus (Fig. 2, bregma 1.4, 0, and Table 1). Olfactory circuits modulate appetite and food intake in response to select odors (25), and in turn, the sensitivity of odor detection is influenced by prandial state and leptin signaling (26, 27).
Cerebral cortex
We observe moderate levels of expression in the bed nucleus of the stria terminalis (BNST) and the medial amygdalar nucleus. The BNST receives afferent input from the parabrachial nucleus (PB), which is reported to elicit potent orexigenic effects on food intake (28), and also from NPY/AgRP neurons in the ARH (29, 30). The medial amygdala plays a critical role in regulating behavioral stress response, and lesions of the medial amygdala significantly attenuate chronic stress-induced weight gain (31). In addition, we detect a few X-Gal+ cells in the basomedial amygdalar nucleus, the primary somatosensory area (layers 3 and 4), and in layer 4 of the cerebral cortex. We find sparse staining in the primary motor area and in the primary somatosensory barrel field (Fig. 2, bregma −0.1, −1.4, and Table 1).
Midbrain, pons, and medulla
In the midbrain, we find a moderate density of X-Gal staining in the limbic interpeduncular fossa. We also observe a few stained cells in the inferior colliculus, parabigeminal, medial basal reticular nucleus, and the superior colliculus. The periaqueductal gray and the ventral tegmental area contain sparse staining (Fig. 2, bregma −5.0, −5.7, and Table 1).
In the pons, we detect a low density of X-Gal staining in the external lateral division of the lateral PB (lPB). The PB nucleus mediates conditioned taste aversion and more recently has been shown to receive inhibitory inputs from ARH neurons that influence feeding behavior (28). We detect a few X-Gal+ cells in the pontine central gray, with sparse staining in the pontine reticular nucleus and the principal sensory nucleus of the trigeminal (Fig. 2, bregma −4.5, −5.0, −5.7, and Table 1).
In the medulla, we observe strong staining in the cochlear nucleus and moderate staining in the area postrema (AP). The facial motor nucleus, the medial aspect of the NTS, and the spinal nucleus of the trigeminal have low levels of X-Gal+ staining. We find sparse staining in the parvicellular reticular and vestibular nuclei (Fig. 2, bregma −7.8 and lateral, 0.2, 2.6, and Table 1). The caudal brain stem integrates hypothalamic and vagal inputs with circulating gut and satiety signals and influences processes related to food intake and energy expenditure in the periphery via the autonomic nervous system (32–34). Lesions of the AP and NTS independently result in hypophagia accompanied by substantial reductions in body weight (35, 36).
Cerebellum
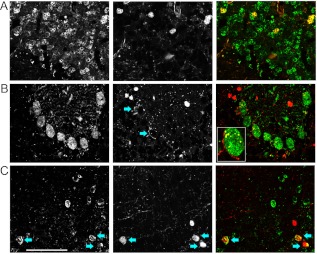
We detect X-Gal+ cells in the nuclear and molecular layers of the cerebellum (Fig. 3). We used IHC for NeuN, parvalbumin (PV), and calbindin to identify granule, stellate, and Purkinjie cells, respectively (37). After confirming that the distribution of cells labeled in Pomc-Cre;R26R-TOM mice recapitulates the pattern observed in Pomc-Cre;R26R-LacZ mice, we assessed TOM reporter expression in conjunction with IHC for the markers listed above. TOM+ cells in the nuclear layer coexpress the granule cell marker NeuN (Fig. 3A), whereas many TOM+ cells in the molecular layer coexpress PV (Fig. 3, B and C). Although Purkinje cells are not marked with the lineage trace, TOM+ fibers are observed in close proximity to their large soma (Fig. 3B, arrows).
Fig. 3.
Pomc-Cre;R26R-TOM reporter is observed in the molecular and nuclear layers of the adult cerebellum. A–C, Direct TOM fluorescence is visualized in conjunction with IHC for: NeuN (A), calbindin (B), and PV (C). IHC staining (left panels) appears green in the merged image (right panels), whereas TOM fluorescence (center panels) appears red in merged images (right panels). B, Arrows indicate TOM+ fibers, one indicated cell is shown at higher magnification in the bottom left. C, Arrows indicate TOM/PV coexpression. Scale bar, 50 μm.
Comparison of Pomc-eGFP vs. Pomc-Cre reporters
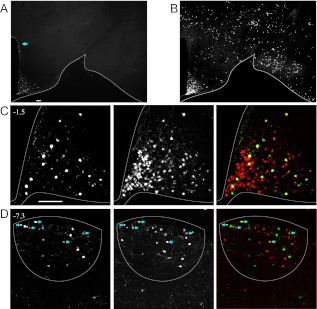
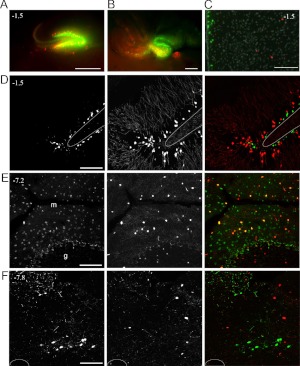
Because both Pomc-eGFP and Pomc-Cre transgenic lines have been used as the basis for studies of POMC neuronal activity, connectivity, and function, we directly compared sites of Pomc promoter activity with those marked by Pomc-Cre-mediated recombination in Pomc-eGFP;Pomc-Cre;R26R-TOM mice. Confirming previous reports, we observe Pomc-eGFP+ cells in the ARH, DG, AP, and NTS (Figs. 4 and 5) (38–40). In the ARH, all GFP+ cells coexpress the Pomc-Cre;R26R-TOM reporter. Consistent with our previous findings, we observe that many TOM+ cells do not express the Pomc-eGFP reporter (Fig. 4, A–C) (12). We also observe a discrete population of Pomc-eGFP+ cells in dorsal periventricular zone at the level of the ARH (Fig. 4A, arrow). However, cells in this region are not marked with the Pomc-Cre;R26R-TOM reporter (Figs. 4B and 5C).
Fig. 4.
POMC neurons represent a minority of the cells labeled with a Pomc-Cre;R26R-TOM reporter in adulthood. A–D, Confocal images of direct TOM fluorescence in conjunction with IHC for GFP in 10-μm sections from adult Pomc-eGFP;Pomc-Cre;R26R-TOM animals. A and B, Low-magnification (×4) images of GFP+ (A) and TOM+ (B) populations in the hypothalamus at the level of the ARH (A–C). D, Expression in the AP of the hindbrain. A, Arrow designates the periventricular zone of the dorsal hypothalamus. C and D, Higher-magnification (×20) images of the ARH (C) and AP (D). For each section, GFP IHC is in the left panel, direct TOM fluorescence is in the center panel, and merged composites are in the right panels. Atlas coordinates are indicated. D, Arrows indicate double positive GFP/TOM cells. Scale bars, 100 μm.
Fig. 5.
Expression of the Pomc-eGFP reporter in the absence of a Pomc-Cre;R26R-TOM reporter is observed in several brain regions in Pomc-eGFP;Pomc-Cre;R26R-TOM mice. A and B, ×2 images of direct GFP and TOM fluorescence visualized from the ventricular surface in hemisected brains from P9. A, Hippocampus. B, Cerebellum. C, Third ventricle of an adult hypothalamus, dorsal to the ARH; 4′,6-diamidino-2-phenylindole staining is presented in gray. D–F, ×20 of GFP IHC (left, green in merge), direct TOM fluorescence (center, red in merge), and the merged image (right) in 10-μm sections from adult animals. D, DG. E, Cerebellum. F, NTS. Coronal atlas coordinates are indicated. Scale bars, 500 μm (A and B) and 100 μm (C–F).
In addition to the periventricular hypothalamus, we observe Pomc-eGFP+ cells that do not express TOM in the DG, cerebellum, and caudal brain stem. In the DG and CA3, we find that GFP and TOM reporter expression is nearly mutually exclusive, with GFP+ cells in the subgranular zone and TOM+ cells in the adjacent granular layer at both P9 and in adulthood (Fig. 5, A and D). Transient Pomc-eGFP expression has been reported in newly born immature neurons derived from the neurogenic niche in the adult DG (40).
We detect Pomc-eGFP+ cells in both the AP and NTS. We find that a minority of GFP+ cells in the AP coexpress TOM (25% of GFP+ are TOM+) (Fig. 4D, arrows), whereas in the NTS, we observe that TOM and GFP expression do not overlap (Fig. 5F). In the cerebellum, we also detect Pomc-eGFP+ cells that do not express the Pomc-Cre reporter. These GFP+ cells are localized to the medial and caudal cerebellum at P9, a period of external granule layer proliferation (9). However, in the adult, GFP+ cells are distributed throughout the rostro-caudal extent of the molecular cell layer (Fig. 5, B and E).
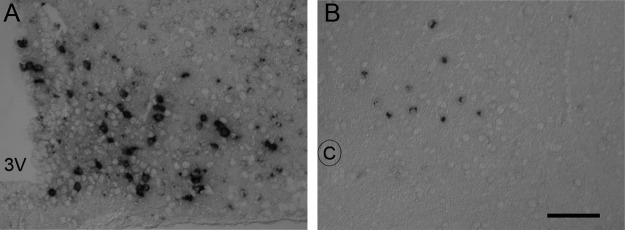
In contrast to the ARH, many GFP+ cells in the hypothalamic periventricular zone, DG, NTS, AP, and lPB are not labeled with a Pomc-Cre reporter. This observation raises the possibility that some of the GFP signal results from ectopic expression of the transgene and not endogenous Pomc transcription. To ascertain whether Pomc is expressed in any of these sites, we first used the TSA system that we routinely use to mark POMC neurons in the ARH (12). Although we do not detect a Pomc signal in the NTS, AP, DG, or lPB with the TSA system, when we use a more sensitive NeutrAvidin-based amplification system (15), we observe Pomc expression in the NTS but not in the other brain sites listed above (Fig. 6).
Fig. 6.
Pomc transcript can be detected in the NTS using NeutrAvidin-amplified ISH. A and B, ISH stain of Pomc transcript: ARH (A) and NTS (B). 3V, Third ventricle; C, central canal. Scale bar, 100 μm.
Peptidergic identities of leptin-sensing neurons in the ARH and caudal brain stem
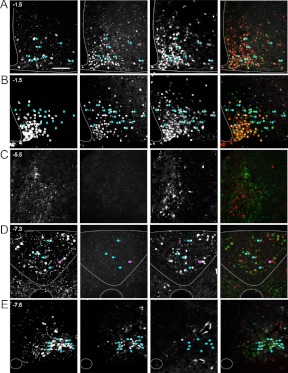
Pomc-expressing populations in both the ARH and NTS have been implicated in the regulation of energy balance, in part, through transduction of leptin signals. To determine whether cells expressing Pomc-eGFP and/or Pomc-Cre reporter cells are leptin-sensing, we assessed P-STAT3 immunoreactivity in conjunction with GFP and TOM in adult brains after a peripheral leptin injection. Consistent with published analyses (34, 41), we find that approximately 40% of mature POMC neurons in the ARH are P-STAT3+ after leptin treatment (Fig. 7A and Supplemental Table 1, published on The Endocrine Society's Journals Online web site at http://endo.endojournals.org). We also observe that approximately half of the TOM+ cells that do not coexpress GFP in the ARH are leptin sensing (of 90 ± 7 TOM only, 47 ± 2 are P-STAT3+) (Fig. 7A); 64% of these non-POMC TOM+/P-STAT3+ cells are NPY neurons (of 47 ± 2 TOM only/P-STAT3+, 30 ± 6 also express Npy) (Fig. 7B). Although we observe Pomc-Cre-mediated recombination in 83% of the leptin-sensing population in the ARH, only 13% of these leptin-sensing cells are POMC neurons.
Fig. 7.
Peptidergic identities of leptin-sensing neurons in the ARH and brain stem. Adult Pomc-eGFP;Pomc-Cre;R26R-TOM animals were injected with leptin (4 mg/kg ip) after an overnight fast. ×20 images of GFP IHC (left, green in merged image), P-STAT3 IHC (center left, white in merged image), direct TOM fluorescence (center right, red in merged image), and merged image (right) in 10-μm sections. Exception (B, left) represents Npy transcript by FISH; this technique is combined with IHC for P-STAT3, TOM is detected by direct fluorescence. A and B, ARH. A, Arrows represent leptin-sensing GFP+ cells. In the merged image pink cells (red and white) represent leptin-sensing non-POMC neurons that are labeled with the TOM reporter. B, Arrows represent leptin-sensing NPY cells that are marked with the TOM reporter. C, PB. D, Brain stem at the level of the AP. Blue arrows indicate leptin-sensing POMC neurons in the AP, pink arrow represents a leptin-sensing non-POMC neuron that is labeled with the TOM reporter. E, Caudal brain stem. Blue arrows indicate leptin-sensing POMC neurons in the NTS. Stereotactic coordinates defined according to Allen Brain Atlas are indicated in the top left. Scale bar, 100 μm.
In the AP, we find that 60% of the TOM+ population is leptin-sensing (Fig. 7D and Supplemental Table 1). In the NTS, there are relatively few TOM+ cells compared with GFP+, none of which are P-STAT3+. However, consistent with previously published reports, we find that approximately 50% of the GFP+ population in the medial NTS is leptin-sensing (Fig. 7E and Supplemental Table 1) (42).
Discussion
Pomc-eGFP and Pomc-Cre transgenic mouse lines have been used extensively to investigate the role of melanocortin circuits in regulating energy homeostasis. We demonstrate that transient activation of Pomc elements during embryonic development is sufficient to drive Cre-mediated recombination in diverse populations throughout the CNS (Fig. 1). Because Pomc-Cre-mediated recombination is permanent, overlapping expression of the transgene and a floxed allele in off-target sites during gestation would confound analysis of the contribution of POMC neurons to resulting phenotypes.
Pomc-eGFP reporter as a marker of endogenous Pomc transcription
To assist in the analysis of studies involving the Pomc-Cre transgene, as well as to facilitate the design of future experiments, we systematically characterized the distribution of cells marked with Pomc-Cre and Pomc-eGFP reporters across the murine brain. Consistent with previous studies, we find that nearly all Pomc-eGFP± cells in the embryonic and adult ARH express Pomc transcripts (8, 10, 12). Moreover, we observe a close correspondence between domains of Pomc-eGFP+ cells and Pomc expression in the embryonic pituitary, midbrain and hindbrain, and spinal cord (Fig. 1, A–D). Therefore, the Pomc-eGFP reporter is appropriate for use in neuroanatomical or electrophysiological studies of POMC neurons in the ARH.
In several brain regions outside of the ARH (including the DG, periventricular zone of the dorsal hypothalamus, and cerebellum), we and others have identified Pomc-eGFP+ cells that do not express detectable levels of Pomc transcript (Fig. 5) (40). We considered three possible explanations for these findings: 1) Pomc is expressed below the level of detection for this assay, 2) transient bursts of transcription are preserved by the persistence of GFP protein after transcription is extinguished, and 3) ectopic activation of the transgene. Using highly sensitive ISH methods, such as radioactive and NeutrAvidin-amplified detection systems, we and others detect Pomc transcripts in the NTS (Fig. 6 and Ref. 39). Because the integrity of fluorescent reporter staining is not maintained under the conditions needed for NeutrAvidin-amplified detection, we could not assess the relationship between the Pomc-expressing cells in the NTS and those marked with either Pomc-eGFP or a Pomc-Cre reporter. However, the similarity between the distribution and number of neurons marked in both cases supports the idea that Pomc-eGFP+ cells express low levels of Pomc transcript (Figs. 5 and 6B). Extra-ARH Pomc expression in the adult AP, DG, and cerebellum is also supported by data from the Allen Mouse Brain Atlas (http://mouse.brain-map.org) (15) and PCR amplification from the rat cortex and cerebellum (43). Together, these data support the idea that Pomc-eGFP expression in the NTS and in some extra-ARH brain regions reflect subthreshold or transient Pomc transcription, but we cannot exclude the possibility that some sites of GFP+ cells result from ectopic expression of the transgene.
Interpretation and design of functional studies using Pomc-Cre transgenic lines
Previously, we determined that Pomc-Cre directs recombination to a subset of NPY/AgRP neurons in the ARH (12). Approximately 40% of ARH neurons are labeled with the TOM reporter; many of these do not express Pomc or Npy in adulthood (12). Because distinct ARH neuronal subpopulations are physically intermingled and project to similar downstream targets, the use of Pomc-Cre transgenes as part of a strategy to genetically label cell bodies or projection patterns or to characterize electrophysiological properties of POMC neurons is not advisable. Depending on the experimental design, off-target Pomc-Cre-mediated recombination could complicate the interpretation of conditional gain or loss of gene function studies. The two most important considerations in analyzing phenotypes from genetic manipulations using Pomc-Cre transgenes are: 1) the endogenous expression pattern of the gene of interest in relation to sites listed in Table 1; and 2) the timing of Pomc-Cre expression. When the gene of interest is broadly expressed in the CNS, off-target effects in nuclei with moderate to high X-Gal staining could also contribute to body composition and metabolic outcomes.
The phenotypes reported in studies using Pomc-Cre transgenic mice include effects on lipid metabolism, fertility, sexually dimorphic effects on glucose homeostasis and body weight phenotypes, food intake, locomotor activity, energy expenditure, and alterations in soma size (11, 44–49). We find that not only does this Cre line direct recombination to more cells than expected in the ARH, it is also present in other regions of the brain implicated in energy homeostasis, including PVH, VMH, subiculum, medial amygdala, lPB, and AP (Table 1 and Fig. 2). Considering that little is known about the precise physiological functions of many subpopulations affected by the Pomc-Cre transgene, and that circuits regulating energy balance exhibit strong developmental compensation (50), the extent to which off-target recombination events contribute to phenotypes observed with conditional knockouts cannot be easily parsed.
Another feature of Pomc-Cre transgenics that could complicate analyses of physiological phenotypes is variability in the extent of recombination, as evidenced by differences in the absolute number of neurons labeled when crossed to different reporter lines. For example, we find that 25% of NPY/AgRP neurons express the R26R-eGFP label, whereas 73% express R26R-LacZ and/or R26R-tdTOM reporters (Fig 7B and Ref. 12). The discrepancy between labeled neurons using different reporter strains likely reflects differences in recombination efficiency, although this is difficult to assess directly. Variations in recombination efficiency are likely to produce more pronounced effects when the Cre driver is expressed at low levels and/or transiently, as in the case with the Pomc-Cre, but is applicable for all Cre transgenics.
In our analysis of the peptidergic identities of leptin-sensing cells in the ARH, we find that 83% of P-STAT3+ cells are TOM+ (Fig. 7 and Supplemental Table 1). Of the leptin-sensing neurons affected by the Pomc-Cre transgene, 30% express Pomc, 55% express Npy, and the remaining 15% have yet to be defined (of 54 ± 2 P-STAT3+/TOM+, 14 ± 1 are POMC+, 30 ± 6 are NPY+). Simultaneous Cre-mediated recombination in POMC and NPY/AgRP neurons could contribute to the mild phenotypes observed with Pomc-Cre-driven conditional knockouts. In addition, non-POMC/NPY neurons marked with Pomc-Cre could also contribute to physiological outcomes. However, because a floxed locus typically requires both alleles to become inactivated for gene expression to become phenotypically observable, the most relevant assessment of the extent of Cre-mediated recombination is the extent to which the modulation of the expression or function of the targeted gene is achieved.
Many off-target sites of Pomc-Cre reporter activity result from transient Pomc expression during gestation; thus, delaying transgene expression to adulthood should reduce the opportunity for unintended Pomc-Cre-mediated recombination events. For example, after injections of a viral construct expressing a Cre-dependent mCherry reporter into the ARH of adult Pomc-Cre;R26R-yellow fluorescent protein mice, the mCherry label is observed in a subset of the neurons labeled with yellow fluorescent protein (51). Although these data are consistent with reduced off-target recombination, this was not directly assessed. In theory, activation of an inducible Pomc-Cre transgene in adulthood should also reduce off-target recombination. Although strategies to spatially and/or temporally restrict Pomc-Cre expression can limit unintended recombination events, consideration should be given to extra-ARH sites of Pomc-Cre activity in the adult, discussed below.
Pomc-eGFP expression in the absence of Pomc-Cre-mediated recombination
We observe little overlap between cells expressing Pomc-eGFP and Pomc-Cre reporters in the periventricular hypothalamus, DG, and NTS. In the DG, Pomc-eGFP is expressed in an immature population in the subgranular zone, whereas the Pomc-Cre reporter is detected in the adjacent population of cells in the granular layer (Fig. 5D and Ref. 40). The most parsimonious explanation of the pattern of reporter expression we observe in the DG is that the Pomc-eGFP and Pomc-Cre transgenes are transiently expressed in immature neurons in the subgranular layer, but due to delayed Cre-mediated recombination relative to the onset of Cre expression (52), the lineage trace is not produced until the mature cells reach the granular layer. However, we cannot exclude the possibility that the Pomc-eGFP and Pomc-Cre transgenes are ectopically expressed in the subgranular layer and the granular layer of the DG, respectively. In both the periventricular hypothalamus and NTS, Pomc-eGFP+ cells are medially located, whereas a few cells expressing the Pomc-Cre reporter are distributed more laterally and are not interspersed with the GFP+ cells (Fig. 5, C and F). It is not clear whether there is a direct relationship between cells expressing Pomc-eGFP and Pomc-Cre reporters in the adult NTS; the latter could also be the descendants of cells that transiently expressed Pomc in the caudal CNS during embryogenesis (Fig. 1, A–D).
The relationship between cells expressing Pomc-eGFP and Pomc-Cre reporters in the developing cerebellum is different than in the mature structure. During the period of external germinal layer proliferation in the P9 cerebellum (9), Pomc-eGFP and Pomc-Cre reporters are expressed in adjacent populations (Fig. 5B), reminiscent of the expression profile in the neurogenic DG (Fig. 5D). In the adult cerebellum, as well as in the AP, we detect some cells expressing either Pomc-eGFP or Pomc-Cre reporter alone as well as some cells that coexpress both reporters (Figs. 4D and 5E). Because Pomc-eGFP+ cells are scattered throughout the molecular layer and are not preferentially localized to the proposed site of adult neurogenesis in the subpial layer (53, 54), these cells are less likely to represent newly born neurons. Expression of Pomc-Cre reporter in the absence of Pomc-eGFP in the cerebellum and AP could represent: 1) cells that expressed Pomc at an earlier development stage, 2) cells that expressed Pomc-eGFP in adulthood but subsequently extinguished it, or 3) ectopic transgene expression. Additional studies are needed to distinguish between these possibilities.
Pomc is expressed transiently in newly born neurons in both the embryonic ARH and adult DG (12, 40). Because Pomc-eGFP and Pomc-Cre reporters are expressed in adjacent populations during a period of external germinal layer proliferation (Fig. 5B), Pomc may also be transiently expressed in immature cerebellar neurons at P9. These observations raise the possibility that Pomc-eGFP expression may mark a transient stage of neuronal differentiation in several different CNS regions. Given this, it is striking that we also observe Pomc-eGFP+ cells that do not express a Pomc-Cre reporter in two proposed sites of adult neurogenesis in the periventricular zone of the dorsal hypothalamus and the NTS (55, 56). Further studies are needed to explore whether Pomc-eGFP is expressed in newly born neurons in the hypothalamus and/or NTS; if proven true, it could provide a powerful tool to investigate the contribution of adult-born neurons to obesity-related outcomes.
Summary
To date, most efforts to validate Cre-transgenic lines involved ensuring that expression of the floxed gene is indeed disrupted in the intended target population. Our studies highlight the need to ascertain whether recombination occurs in off-target sites as well. Because some genes are expressed more broadly during development and others are not, this determination must be made for each Cre-transgenic line generated. Moreover, as low or transient expression of a Cre transgene could lead to variability in recombination efficiency, validation of models using Cre-based reagents should involve the direct assessment of the desired change in protein expression or function. Although the transient expression of Pomc-Cre transgenes in immature neurons throughout the CNS makes it an extreme case, the lessons learned are applicable to all Cre reagents.
Supplementary Material
Acknowledgments
We thank J. P. Pardere for helping with dot plot renderings; M. Madra for assistance with tissue preparation and ISH staining; L. Yang of the Diabetes and Endocrinology Research Center Pathology Core for cryosectioning (PD30 DK63608); and M. Low (University of Michigan, Ann Arbor, MI), J. Elmquist (University of Texas Southwestern, Dallas, TX), and B. Lowell (Beth Israel Deaconess Medical Center, Boston, MA) for generously providing mouse reagents.
This work was supported by a Naomi Berrie Young Investigator award (L.M.Z.) and NIH Grant F31DK079372 (to S.L.P.).
Disclosure Summary: The authors have nothing to disclose.
For editorial see page 1005
- AgRP
- Agouti-related peptide
- AP
- area postrema
- ARH
- arcuate nucleus of the hypothalamus
- BNST
- bed nucleus of the stria terminalis
- CNS
- central nervous system
- DG
- dentate gyrus
- E
- embryonic day
- eGFP
- enhanced GFP
- FISH
- fluorescent ISH
- GFP
- green fluorescent protein
- IHC
- immunohistochemistry
- ISH
- in situ hybridization
- lPB
- lateral PB
- Mc4r
- Melanocortin receptor 4
- Npy
- Neuropeptide Y
- NTS
- nucleus of the solitary tract
- PB
- parabrachial nucleus
- Pomc
- Proopioimelanocortin
- P-STAT3
- phosphorylated signal transducer and activator of transcription 3
- PV
- parvalbumin
- PVH
- paraventricular nucleus
- SCN
- suprachiasmatic nucleus
- TSA
- tyramide-based amplification system
- VMH
- ventral medial nucleus
- wt
- wild type.
References
- 1. Huszar D, Lynch CA, Fairchild-Huntress V, Dunmore JH, Fang Q, Berkemeier LR, Gu W, Kesterson RA, Boston BA, Cone RD, Smith FJ, Campfield LA, Burn P, Lee F. 1997. Targeted disruption of the melanocortin-4 receptor results in obesity in mice. Cell 88:131–141 [DOI] [PubMed] [Google Scholar]
- 2. Yaswen L, Diehl N, Brennan MB, Hochgeschwender U. 1999. Obesity in the mouse model of pro-opiomelanocortin deficiency responds to peripheral melanocortin. Nat Med 5:1066–1070 [DOI] [PubMed] [Google Scholar]
- 3. Fan W, Boston BA, Kesterson RA, Hruby VJ, Cone RD. 1997. Role of melanocortinergic neurons in feeding and the agouti obesity syndrome. Nature 385:165–168 [DOI] [PubMed] [Google Scholar]
- 4. Ollmann MM, Wilson BD, Yang YK, Kerns JA, Chen Y, Gantz I, Barsh GS. 1997. Antagonism of central melanocortin receptors in vitro and in vivo by agouti-related protein. Science 278:135–138 [DOI] [PubMed] [Google Scholar]
- 5. Schwartz MW, Baskin DG, Bukowski TR, Kuijper JL, Foster D, Lasser G, Prunkard DE, Porte D, Jr, Woods SC, Seeley RJ, Weigle DS. 1996. Specificity of leptin action on elevated blood glucose levels and hypothalamic neuropeptide Y gene expression in ob/ob mice. Diabetes 45:531–535 [DOI] [PubMed] [Google Scholar]
- 6. Schwartz MW, Seeley RJ, Woods SC, Weigle DS, Campfield LA, Burn P, Baskin DG. 1997. Leptin increases hypothalamic pro-opiomelanocortin mRNA expression in the rostral arcuate nucleus. Diabetes 46:2119–2123 [DOI] [PubMed] [Google Scholar]
- 7. Mizuno TM, Mobbs CV. 1999. Hypothalamic agouti-related protein messenger ribonucleic acid is inhibited by leptin and stimulated by fasting. Endocrinology 140:814–817 [DOI] [PubMed] [Google Scholar]
- 8. Cowley MA, Smart JL, Rubinstein M, Cerdán MG, Diano S, Horvath TL, Cone RD, Low MJ. 2001. Leptin activates anorexigenic POMC neurons through a neural network in the arcuate nucleus. Nature 411:480–484 [DOI] [PubMed] [Google Scholar]
- 9. Hatten ME, Heintz N. 1995. Mechanisms of neural patterning and specification in the developing cerebellum. Annu Rev Neurosci 18:385–408 [DOI] [PubMed] [Google Scholar]
- 10. Pinto S, Roseberry AG, Liu H, Diano S, Shanabrough M, Cai X, Friedman JM, Horvath TL. 2004. Rapid rewiring of arcuate nucleus feeding circuits by leptin. Science 304:110–115 [DOI] [PubMed] [Google Scholar]
- 11. Balthasar N, Coppari R, McMinn J, Liu SM, Lee CE, Tang V, Kenny CD, McGovern RA, Chua SC, Jr, Elmquist JK, Lowell BB. 2004. Leptin receptor signaling in POMC neurons is required for normal body weight homeostasis. Neuron 42:983–991 [DOI] [PubMed] [Google Scholar]
- 12. Padilla SL, Carmody JS, Zeltser LM. 2010. Pomc-expressing progenitors give rise to antagonistic neuronal populations in hypothalamic feeding circuits. Nat Med 16:403–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Soriano P. 1999. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat Genet 21:70–71 [DOI] [PubMed] [Google Scholar]
- 14. Madisen L, Zwingman TA, Sunkin SM, Oh SW, Zariwala HA, Gu H, Ng LL, Palmiter RD, Hawrylycz MJ, Jones AR, Lein ES, Zeng H. 2010. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat Neurosci 13:133–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lein ES, Hawrylycz MJ, Ao N, Ayres M, Bensinger A, Bernard A, Boe AF, Boguski MS, Brockway KS, Byrnes EJ, Chen L, Chen L, Chen TM, Chin MC, Chong J, Crook BE, Czaplinska A, Dang CN, Datta S, Dee NR, Desaki AL, Desta T, Diep E, Dolbeare TA, Donelan MJ, Dong HW, Dougherty JG, Duncan BJ, Ebbert AJ, Eichele G, Estin LK, Faber C, Facer BA, Fields R, Fischer SR, Fliss TP, Frensley C, Gates SN, Glattfelder KJ, Halverson KR, Hart MR, Hohmann JG, Howell MP, Jeung DP, Johnson RA, Karr PT, Kawal R, Kidney JM, Knapik RH, Kuan CL, Lake JH, Laramee AR, Larsen KD, Lau C, Lemon TA, Liang AJ, Liu Y, Luong LT, Michaels J, Morgan JJ, Morgan RJ, Mortrud MT, Mosqueda NF, Ng LL, Ng R, Orta GJ, Overly CC, Pak TH, Parry SE, Pathak SD, Pearson OC, Puchalski RB, Riley ZL, Rockett HR, Rowland SA, Royall JJ, Ruiz MJ, Sarno NR, Schaffnit K, Shapovalova NV, Sivisay T, Slaughterbeck CR, Smith SC, Smith KA, Smith BI, Sodt AJ, Stewart NN, Stumpf KR, Sunkin SM, Sutram M, Tam A, Teemer CD, Thaller C, Thompson CL, Varnam LR, Visel A, Whitlock RM, Wohnoutka PE, Wolkey CK, Wong VY, Wood M, Yaylaoglu MB, Young RC, Youngstrom BL, Yuan XF, Zhang B, Zwingman TA, Jones AR. 2007. Genome-wide atlas of gene expression in the adult mouse brain. Nature 445:168–176 [DOI] [PubMed] [Google Scholar]
- 16. Wolter HJ. 1984. α-Melanotropin and β-endorphin-like immunoreactivities are contained within neurons and nerve fibers of the rat duodenum. Brain Res 295:378–384 [DOI] [PubMed] [Google Scholar]
- 17. Cohen-Cory S. 2002. The developing synapse: construction and modulation of synaptic structures and circuits. Science 298:770–776 [DOI] [PubMed] [Google Scholar]
- 18. Turek FW, Joshu C, Kohsaka A, Lin E, Ivanova G, McDearmon E, Laposky A, Losee-Olson S, Easton A, Jensen DR, Eckel RH, Takahashi JS, Bass J. 2005. Obesity and metabolic syndrome in circadian Clock mutant mice. Science 308:1043–1045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. King BM. 2006. The rise, fall, and resurrection of the ventromedial hypothalamus in the regulation of feeding behavior and body weight. Physiol Behav 87:221–244 [DOI] [PubMed] [Google Scholar]
- 20. Yi CX, Scherer T, Tschöp MH. 2011. Cajal revisited: does the VMH make us fat? Nat Neurosci 14:806–808 [DOI] [PubMed] [Google Scholar]
- 21. Tolson KP, Gemelli T, Gautron L, Elmquist JK, Zinn AR, Kublaoui BM. 2010. Postnatal Sim1 deficiency causes hyperphagic obesity and reduced Mc4r and oxytocin expression. J Neurosci 30:3803–3812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Forloni G, Fisone G, Guaitani A, Ladinsky H, Consolo S. 1986. Role of the hippocampus in the sex-dependent regulation of eating behavior: studies with kainic acid. Physiol Behav 38:321–326 [DOI] [PubMed] [Google Scholar]
- 23. Davidson TL, Chan K, Jarrard LE, Kanoski SE, Clegg DJ, Benoit SC. 2009. Contributions of the hippocampus and medial prefrontal cortex to energy and body weight regulation. Hippocampus 19:235–252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kanoski SE, Hayes MR, Greenwald HS, Fortin SM, Gianessi CA, Gilbert JR, Grill HJ. 2011. Hippocampal leptin signaling reduces food intake and modulates food-related memory processing. Neuropsychopharmacology 36:1859–1870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Shen J, Niijima A, Tanida M, Horii Y, Maeda K, Nagai K. 2005. Olfactory stimulation with scent of grapefruit oil affects autonomic nerves, lipolysis and appetite in rats. Neurosci Lett 380:289–294 [DOI] [PubMed] [Google Scholar]
- 26. Aimé P, Duchamp-Viret P, Chaput MA, Savigner A, Mahfouz M, Julliard AK. 2007. Fasting increases and satiation decreases olfactory detection for a neutral odor in rats. Behav Brain Res 179:258–264 [DOI] [PubMed] [Google Scholar]
- 27. Prud'homme MJ, Lacroix MC, Badonnel K, Gougis S, Baly C, Salesse R, Caillol M. 2009. Nutritional status modulates behavioural and olfactory bulb Fos responses to isoamyl acetate or food odour in rats: roles of orexins and leptin. Neuroscience 162:1287–1298 [DOI] [PubMed] [Google Scholar]
- 28. Wu Q, Boyle MP, Palmiter RD. 2009. Loss of GABAergic signaling by AgRP neurons to the parabrachial nucleus leads to starvation. Cell 137:1225–1234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Saper CB, Loewy AD. 1980. Efferent connections of the parabrachial nucleus in the rat. Brain Res 197:291–317 [DOI] [PubMed] [Google Scholar]
- 30. Shin JW, Geerling JC, Loewy AD. 2008. Inputs to the ventrolateral bed nucleus of the stria terminalis. J Comp Neurol 511:628–657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Solomon MB, Jones K, Packard BA, Herman JP. 2010. The medial amygdala modulates body weight but not neuroendocrine responses to chronic stress. J Neuroendocrinol 22:13–23 [DOI] [PubMed] [Google Scholar]
- 32. Crawley JN, Schwaber JS. 1983. Nucleus tractus solitarius lesions block the behavioral actions of cholecystokinin. Peptides 4:743–747 [DOI] [PubMed] [Google Scholar]
- 33. Grill HJ, Schwartz MW, Kaplan JM, Foxhall JS, Breininger J, Baskin DG. 2002. Evidence that the caudal brainstem is a target for the inhibitory effect of leptin on food intake. Endocrinology 143:239–246 [DOI] [PubMed] [Google Scholar]
- 34. Münzberg H, Huo L, Nillni EA, Hollenberg AN, Bjørbaek C. 2003. Role of signal transducer and activator of transcription 3 in regulation of hypothalamic proopiomelanocortin gene expression by leptin. Endocrinology 144:2121–2131 [DOI] [PubMed] [Google Scholar]
- 35. Contreras RJ, Fox E, Drugovich ML. 1982. Area postrema lesions produce feeding deficits in the rat: effects of preoperative dieting and 2-deoxy-D-glucose. Physiol Behav 29:875–884 [DOI] [PubMed] [Google Scholar]
- 36. Menani JV, Colombari E, Talman WT, Johnson AK. 1996. Commissural nucleus of the solitary tract lesions reduce food intake and body weight gain in rats. Brain Res 740:102–108 [DOI] [PubMed] [Google Scholar]
- 37. Doyle JP, Dougherty JD, Heiman M, Schmidt EF, Stevens TR, Ma G, Bupp S, Shrestha P, Shah RD, Doughty ML, Gong S, Greengard P, Heintz N. 2008. Application of a translational profiling approach for the comparative analysis of CNS cell types. Cell 135:749–762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Knigge KM, Joseph SA, Nocton J. 1981. Topography of the ACTH-immunoreactive neurons in the basal hypothalamus of the rat brain. Brain Res 216:333–341 [DOI] [PubMed] [Google Scholar]
- 39. Bronstein DM, Schafer MK, Watson SJ, Akil H. 1992. Evidence that β-endorphin is synthesized in cells in the nucleus tractus solitarius: detection of POMC mRNA. Brain Res 587:269–275 [DOI] [PubMed] [Google Scholar]
- 40. Overstreet LS, Hentges ST, Bumaschny VF, de Souza FS, Smart JL, Santangelo AM, Low MJ, Westbrook GL, Rubinstein M. 2004. A transgenic marker for newly born granule cells in dentate gyrus. J Neurosci 24:3251–3259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Williams KW, Margatho LO, Lee CE, Choi M, Lee S, Scott MM, Elias CF, Elmquist JK. 2010. Segregation of acute leptin and insulin effects in distinct populations of arcuate proopiomelanocortin neurons. J Neurosci 30:2472–2479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ellacott KL, Halatchev IG, Cone RD. 2006. Characterization of leptin-responsive neurons in the caudal brainstem. Endocrinology 147:3190–3195 [DOI] [PubMed] [Google Scholar]
- 43. Grauerholz BL, Jacobson JD, Handler MS, Millington WR. 1998. Detection of pro-opiomelanocortin mRNA in human and rat caudal medulla by RT-PCR. Peptides 19:939–948 [DOI] [PubMed] [Google Scholar]
- 44. Hill JW, Elias CF, Fukuda M, Williams KW, Berglund ED, Holland WL, Cho YR, Chuang JC, Xu Y, Choi M, Lauzon D, Lee CE, Coppari R, Richardson JA, Zigman JM, Chua S, Scherer PE, Lowell BB, Brüning JC, Elmquist JK. 2010. Direct insulin and leptin action on pro-opiomelanocortin neurons is required for normal glucose homeostasis and fertility. Cell Metab 11:286–297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Shi H, Sorrell JE, Clegg DJ, Woods SC, Seeley RJ. 2010. The roles of leptin receptors on POMC neurons in the regulation of sex-specific energy homeostasis. Physiol Behav 100:165–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Mori H, Inoki K, Münzberg H, Opland D, Faouzi M, Villanueva EC, Ikenoue T, Kwiatkowski D, MacDougald OA, Myers MG, Jr, Guan KL. 2009. Critical role for hypothalamic mTOR activity in energy balance. Cell Metab 9:362–374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Xu AW, Ste-Marie L, Kaelin CB, Barsh GS. 2007. Inactivation of signal transducer and activator of transcription 3 in proopiomelanocortin (Pomc) neurons causes decreased pomc expression, mild obesity, and defects in compensatory refeeding. Endocrinology 148:72–80 [DOI] [PubMed] [Google Scholar]
- 48. Huo L, Gamber K, Greeley S, Silva J, Huntoon N, Leng XH, Bjørbaek C. 2009. Leptin-dependent control of glucose balance and locomotor activity by POMC neurons. Cell Metab 9:537–547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Banno R, Zimmer D, De Jonghe BC, Atienza M, Rak K, Yang W, Bence KK. 2010. PTP1B and SHP2 in POMC neurons reciprocally regulate energy balance in mice. J Clin Invest 120:720–734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Luquet S, Perez FA, Hnasko TS, Palmiter RD. 2005. NPY/AgRP neurons are essential for feeding in adult mice but can be ablated in neonates. Science 310:683–685 [DOI] [PubMed] [Google Scholar]
- 51. Atasoy D, Aponte Y, Su HH, Sternson SM. 2008. A FLEX switch targets Channelrhodopsin-2 to multiple cell types for imaging and long-range circuit mapping. J Neurosci 28:7025–7030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Branda CS, Dymecki SM. 2004. Talking about a revolution: the impact of site-specific recombinases on genetic analyses in mice. Dev Cell 6:7–28 [DOI] [PubMed] [Google Scholar]
- 53. Ponti G, Peretto P, Bonfanti L. 2006. A subpial, transitory germinal zone forms chains of neuronal precursors in the rabbit cerebellum. Dev Biol 294:168–180 [DOI] [PubMed] [Google Scholar]
- 54. Ponti G, Peretto P, Bonfanti L. 2008. Genesis of neuronal and glial progenitors in the cerebellar cortex of peripuberal and adult rabbits. PLoS One 3:e2366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Xu AW, Kaelin CB, Morton GJ, Ogimoto K, Stanhope K, Graham J, Baskin DG, Havel P, Schwartz MW, Barsh GS. 2005. Effects of hypothalamic neurodegeneration on energy balance. PLoS Biol 3:e415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Bauer S, Hay M, Amilhon B, Jean A, Moyse E. 2005. In vivo neurogenesis in the dorsal vagal complex of the adult rat brainstem. Neuroscience 130:75–90 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.