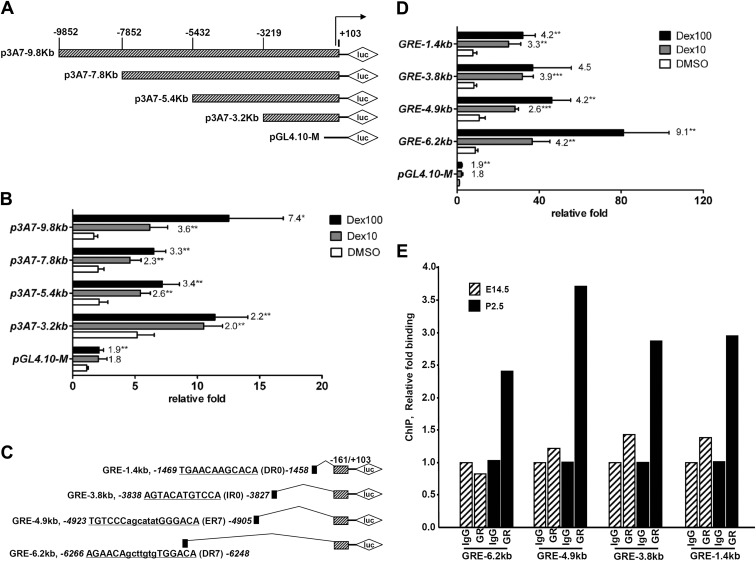
Fig. 5.
Induction of CYP3A7 via the far upstream region mediated by the GR. A–D, Luciferase constructs with various lengths of CYP3A7 5′-flanking region (A) were subjected to luciferase reporter assays (B) or isolated GRE located in the upstream region of CYP3A7 (C) were transiently transfected into hepatoblasts from E18.5 Tg3A4/7-hPXR (D). After culture of hepatoblasts for 24 h, cells were cotransfected with 100 ng human GR, 300 ng pGL4.10-M basic plasmid or various luciferase constructs, and 30 ng internal control plasmid phRL-SV40. At 6 h after transfection, the cells were treated with 0.1% DMSO, 10 nm Dex, or 100 nm Dex. After 24 h treatment, the cells were harvested for the dual luciferase assay. E, Age-related GR binding in predicted GRE binding sites determined by ChIP assays with E14.5 and P2.5 mouse livers; shown is the qPCR analysis of the immunoprecipitated DNA. Histograms of the PCR products resolved on a gel, done in a separate experiment, are shown in Supplemental Fig. 2. The relative fold increases of luciferase activity are expressed as mean ± sd; n = 4. *, P < 0.05; **, P < 0.01 compared with DMSO-treated cells. Numbers on the top of columns denote the fold differences between the Dex-treated group and DMSO-treated group in each construct.