Abstract
Hepatic stellate cells (HSCs) are recognized as a major player in liver fibrogenesis. Upon liver injury, HSCs differentiate into myofibroblasts and participate in progression of fibrosis and cirrhosis. Additional cell types such as resident liver fibroblasts/myofibroblasts or bone marrow cells are also known to generate myofibroblasts. One of the major obstacles to understanding the mechanism of liver fibrogenesis is the lack of knowledge regarding the developmental origin of HSCs and other liver mesenchymal cells. Recent cell lineage analyses demonstrate that HSCs are derived from mesoderm during liver development. MesP1-expressing mesoderm gives rise to the septum transversum mesenchyme before liver formation and then to the liver mesothelium and mesenchymal cells, including HSCs and perivascular mesenchymal cells around the veins during liver development. During the growth of embryonic liver, the mesothelium, consisting of mesothelial cells and submesothelial cells, migrates inward from the liver surface and gives rise to HSCs and perivascular mesenchymal cells, including portal fibroblasts, smooth muscle cells around the portal vein, and fibroblasts around the central vein. Cell lineage analyses indicate that mesothelial cells are HSC progenitor cells capable of differentiating into HSCs and other liver mesenchymal cells during liver development.
Keywords: Cell lineage analysis, mesoderm, mesothelial cells, submesothelial cells
Hepatic stellate cells and liver fibrosis
Hepatic stellate cells (HSCs) reside in the space of Disse between hepatocytes and sinusoidal endothelial cells in the liver and extend their dendritic processes along the wall of the sinusoid.1–3 One of the unique features of HSCs is that they store vitamin A lipids in their cytoplasm. Upon liver injury, quiescent HSCs radically change their phenotype into an activated state. They lose their stored vitamin A lipids, begin expressing α-smooth muscle actin (SMA) and synthesizing both proinflammatory cytokines and extracellular matrix proteins. It is generally believed that activated HSCs become myofibroblasts in injured liver and participate in fibrogenesis. Therefore, the activation step of HSCs has been considered as a possible therapeutic target for suppression of fibrosis and its progression to cirrhosis.4,5 In fact, many studies have been aimed at understanding the mechanism of HSC activation. For example, transforming growth factor-β (TGFβ) is known to induce activation of HSCs and suppression of TGFβ signaling was shown to ameliorate liver fibrosis.6
Presence of different fibrogenic cell types in the liver
Although HSCs are recognized as a major source of myofibroblasts, other cell types are also known to differentiate into myofibroblasts during fibrogenesis.7–10 Electron microscopy has been used to classify different types of fibrogenic cells in the liver.7 Portal fibroblasts around the portal vein participate in biliary fibrosis. Around the central vein, fibroblastic cells, so-called second-layered cells, exist.7 The liver surface is covered with mesothelium, which is composed of mesothelial cells and fibroblastic cells called capsular fibroblasts. These fibroblastic cells around the veins and beneath the mesothelial cells have been suggested to differentiate into myofibroblasts and to participate in fibrogenesis in rat liver caused by porcine serum injection.7 Due to the lack of molecular markers, however, the lineage relationship between the fibroblastic cells and HSCs and how they participate in liver fibrogenesis remains to be determined. Among these liver fibrogenic cells, portal fibroblasts have been best-characterized in biliary fibrosis.9,11,12 Upon injury to the bile duct, portal fibroblasts proliferate and differentiate into SMA-expressing myofibroblasts and synthesize extracellular matrix proteins, similar to activated HSCs. Interestingly, unlike activated HSCs, portal fibroblasts do not respond to platelet derived growth factor (PDGF) and their growth is rather inhibited by TGF-β.13 Knittel et al. reported that liver myofibroblasts and activated HSCs are found in different populations within fibrotic livers in rats.8 Isolated myofibroblasts are resistant to spontaneous apoptosis in vitro.14 Thus, it will be desirable to consider targeting a particular myofibroblast type or activated HSCs for effective anti-fibrotic therapy. To this end, we must better understand the molecular mechanisms of the myofibroblastic conversion from different cell types.
Characterization of fetal HSCs in mice
HSCs in the adult liver can be isolated by discontinuous gradient ultracentrifugation, based on their high buoyancy, which is attributable to the presence of stored vitamin A lipids.15 Storage of vitamin A lipids in the liver begins after birth,16 making it impossible to isolate these cells from fetuses using the same method used to isolate adult HSCs. Similar to adult HSCs, fetal HSCs express desmin in embryonic livers.17–19 In addition, fetal HSCs express transcription factors, Foxf1, Lhx2, and Hlx.20–22 Mice with knockouts of each of these transcription factors show abnormalities in liver development. For example, Lhx2 knockout mice show increased expression of SMA in HSCs and fibrosis in embryonic liver,22 yet the mechanism of the causality of this phenotype remains to be clarified.
Presence of different liver mesenchymal cell types in embryonic livers
To characterize fetal HSCs, several groups, including us, have attempted to identify cell surface markers. Kubota et al. purified fetal HSCs based on expression of VCAM-1 and Integrin β3 from rat embryonic livers by fluorescence activated cell sorting (FACS).23 Hoppo et al. isolated a Thy1+ liver mesenchymal cell population from mouse embryonic livers.24 Suzuki et al. made a specific antibody for fetal HSCs and identified its antigen as p75 neurotrophin receptor (p75NTR).25 Our group also identified markers for liver mesenchymal cell populations in mouse embryos.26 We used Msx2-promoter-driven lacZ transgenic mice which express lacZ reporter in desmin+ fetal HSCs and mesenchymal cells around the vein in stage E12.5 embryonic liver. The bile duct is not yet formed at this stage, and therefore it is impossible to morphologically distinguish portal or central veins. Thus, we termed the desmin+lacZ+ mesenchymal cells around the vein as “perivascular mesenchymal cells (PMCs)” in embryonic livers (Fig. 1).26 We also found expression of lacZ in mesothelial cells covering developing liver and submesothelial cells associated with mesothelial cells (Fig. 1).26 Mesothelial cells and submesothelial cells are separated by a basal lamina composed of type IV collagen.
Figure 1.
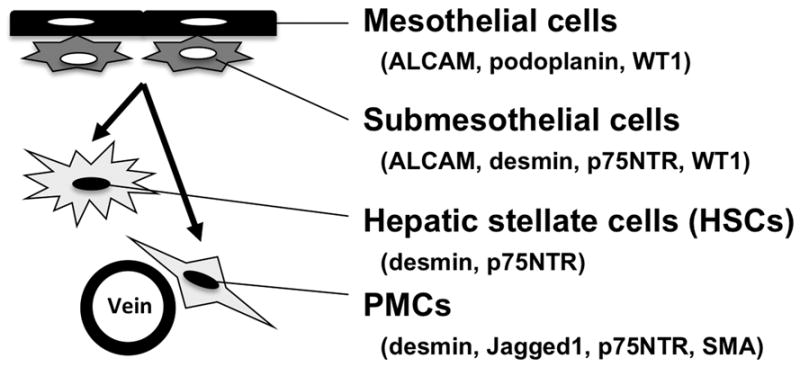
Classification of liver mesenchymal cell types in mouse developing livers. HSCs associated with hepatoblasts express desmin and p75NTR. PMCs around the vein express the HSC markers, Jagged1, and SMA. Mesothelial cells covering liver surface express ALCAM, Podoplanin, and WT1. Submesothelial cells associated with mesothelial cells show an intermediate phenotype between mesothelial cells and HSCs. The mesothelium consisting of mesothelial cells and submesothelial cells gives rise to HSCs and PMCs during mouse liver development (arrows).
Following isolation of these lacZ+ mesenchymal cells by FACS using a fluorescence substrate for lacZ, we surveyed gene expression in these cells by microarray analysis.26 Foxf1, Lhx2, and Hlx are strongly expressed in the lacZ+ cells. In addition to these markers, we found high expression of activated leukocyte cell adhesion molecule (ALCAM/CD166), type I collagen, type IV collagen, desmin, Jagged1, p75Ntr, PDGFRα, podoplanin, SMA, and Wilms tumor 1 homolog (WT1).26 Using immunohistochemistry, we examined marker expression and classified the lacZ+ cells as follows: 1) fetal HSCs inside the liver (desmin, p75NTR); 2) PMCs around the vein (desmin, Jagged1, p75NTR, SMA); 3) submesothelial cells beneath mesothelial cells (ALCAM, desmin, p75NTR, PDGFRα, WT1); 4) mesothelial cells covering the liver surface (ALCAM, podoplanin, WT1).26 Submesothelial cells are of particular interest, because they show an intermediate phenotype between mesothelial cells expressing ALCAM and WT and fetal HSCs expressing desmin and p75NTR. Immunohistochemical observation suggests that ALCAM+WT1+ mesothelial cells and submesothelial cells migrate inward from the liver surface and give rise to HSCs during mouse liver development.26 Similar observations were also made in human fetal livers.27
The neural crest is not the origin of HSCs
In the adult liver, HSCs express mesenchymal cell markers such as desmin, type I collagen, and vimentin, as well as neural cell markers such as glial fibrillary acidic protein (GFAP), nestin, and p75Ntr.1 Based on expression profiles, the neural crest was believed to be the origin of HSCs. To test this notion, Cassiman et al. used Wnt1Cre and Rosa26 reporter mice that are generally used to trace neural crest descendants during embryogenesis.28 Using this method, they failed to detect neural crest-derived HSCs in developing liver.
Contribution of the septum transversum mesenchyme to HSCs
Morphological studies suggest the septum transversum mesenchyme (STM) is the source of HSCs in developing liver. Liver bud forms from the foregut endoderm around embryonic day (E) 9 in mouse embryos. The STM surrounding the foregut endoderm and cardiac mesoderm secretes fibroblast growth factors (FGFs) and bone morphogenetic proteins (BMPs), inducing differentiation of hepatoblasts, progenitor cells for both hepatocytes and biliary epithelial cells, from the endoderm.29 Hepatoblasts budding from the foregut endoderm invade into the surrounding STM and form a liver bud around E9.5 to E10.5. During this process, the cells in the STM seem to be trapped between growing hepatoblasts and eventually become HSCs in the liver bud.30 Due to the lack of markers, however, it remains unclear what the contribution of the STM to HSCs might be.
In mouse embryos, the STM develops along the foregut endoderm near the cardiac mesoderm around E7–8. The portion of the STM nearest the cardiac mesoderm is named the proepicardium. The proepicardial cells in the STM are known to traverse the cardiac cavity toward the heart, cover the heart surface, and give rise to the epicardium during heart development.31 Because the STM develops near the cardiac mesoderm, we hypothesized that, similar to the cardiac mesoderm, the STM is derived from mesoderm during embryogenesis. To test this, we analyzed a mesoderm lineage using MesP1Cre and Rosa26lacZ reporter mice.32,33 MesP1 is a transcription factor which is transiently expressed in nascent mesoderm during gastrulation.33 The MesP1+ mesoderm gives rise to a wide range of mesodermal cells, such as cardiomyocytes and endothelial cells. As we anticipated, the MesP1+ mesoderm gives rise to the STM during mouse embryogenesis before liver formation (Fig. 2).32
Figure 2.
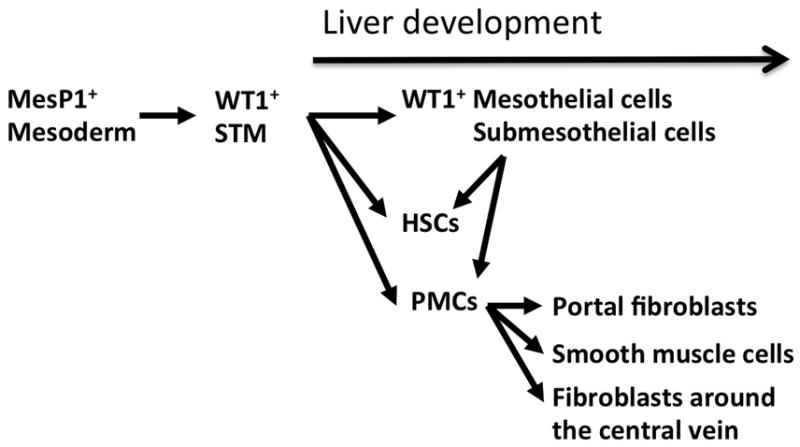
Summary of cell lineage analyses of HSCs during mouse liver development. MesP1+ mesoderm in early embryos gives rise to Wilms tumor 1 (WT1)+ septum transversum mesenchyme (STM). At the onset of liver development, the STM adjacent to the foregut endoderm gives rise to the WT1+ mesothelium and WT1− liver mesenchymal cells including HSCs and PMCs. In later stages, the mesothelium migrates inward from the liver surface and gives rise to HSCs and PMCs including portal fibroblasts and smooth muscle cells around the portal vein and fibroblasts around the central vein. The WT1+ mesothelium does not contribute to hepatoblasts, endothelial cells, or Kupffer cells in liver development.
Next, we tested whether the STM gives rise to HSCs during liver development. We found specific expression of WT1 in the STM before liver development. In later stages of liver morphogenesis, WT1 is only expressed in the mesothelium consisting of mesothelial cells and submesothelial cells. Thus, WT1 is an ideal marker for tracing the WT1+ STM, as well as the mesothelium.32 Using Wt1CreERT2 and Rosa26 reporter mice, we first conditionally traced the STM lineage. In Wt1-expressing cells, the Wt1CreERT2 mouse expresses a fusion protein of Cre DNA recombinase and modified estrogen receptor (ERT2). Upon treatment with tamoxifen, a synthetic ligand for ERT2, the CreERT2 protein translocates to the nuclei and activates lacZ reporter gene expression from the Rosa26 gene locus.31 Thus, by tamoxifen injection to the pregnant mouse, we are able to specifically label the STM as lacZ+ cells and trace their lineage thereafter. This conditional cell lineage analysis revealed that the lacZ+ STM gives rise to mesothelial cells, submesothelial cells, HSCs, and PMCs during early mouse liver development (Fig. 2).32 These results demonstrate that the MesP1+ mesoderm gives rise to the STM and then differentiates into the each of the liver mesenchymal cell types, including HSCs.
Contribution of the mesothelium to HSCs
As described above, in mouse embryos, the cardiac mesoderm and STM induce liver bud formation from the foregut endoderm via FGF and BMP signaling.29 Similar to mice, the FGF and BMP signals from mesoderm are also essential for liver development in chicken.34 In contrast to mammals, however, in chick embryos, the liver bud develops along the ductus venosus. In chick embryos, the liver mesothelium has been suggested to contribute to the development of the HSCs and sinusoidal endothelial cells by fluorescence dye labeling.35 Ijpenberg et al. observed a similar differentiation pathway from mesothelial cells to HSCs in mouse embryos using the Wt1lacZ mouse.36 We also noted that ALCAM+WT1+ mesothelial cells and submesothelial cells seem to migrate inward from the liver surface and give rise to Alcam− WT− HSCs in mouse embryos.26 To rigorously demonstrate this migration, we analyzed the development of WT1CreERT2/Rosa26 reporter mice and found that WT1+ mesothelial cells and submesothelial cells do migrate inward and give rise to WT1− HSCs and PMCs in mouse embryogenesis around E10–E13 (Fig. 2).32
Common origin for HSCs and other liver mesenchymal cell types
To determine the contribution of the liver mesothelium to different liver mesenchymal cells in detail, we labeled the WT1+ mesothelium at E10.5 and then analyzed E18.5 embryonic liver. In E18.5 liver, the bile duct has formed and the portal and central veins are morphologically identifiable. Conditional cell lineage analysis using the WT1CreERT2/Rosa26 reporter mouse revealed that the WT1+ mesothelium contributes to desmin+ HSCs in the sinusoid, Jagged1+ portal fibroblasts adjacent to the bile duct, SMA+ vascular smooth muscle cells in the wall of the portal vein, and desmin+ fibroblasts along the wall of the central vein (Fig. 2).32 Thus, our data indicate a common origin for HSCs, portal fibroblasts, fibroblasts around the central vein, and vascular smooth muscle cells in the liver.
In human fetuses, endoderm was suggested to be the origin of HSCs based on the expression of CD34 and cytokeratin 7/8.37 Chagraoui et al. reported a possible precursor for both hepatoblasts and mesenchymal cells in embryonic livers.38 Our cell lineage analysis, in mice, failed to detect hepatoblast differentiation from either MesP1+ mesoderm or WT1+ STM.26,32 In addition, our cell lineages indicate that the WT1+ mesothelium does not contribute to sinusoidal endothelial cells or Kupffer cells. Therefore, a HSC lineage is distinct from that of hepatoblasts, sinusoidal endothelial cells, and Kupffer cells in mouse liver development.
Contribution of the mesothelium in the heart, lung, and intestine
Our findings indicate the contribution of the mesothelium to liver mesenchymal cells in developing liver. Similar to observations made in the liver, recently, the mesothelium has been shown to contribute to mesenchymal cells in the developing heart, lung, and gut.31,39,40 This suggests that migration and differentiation of mesothelial cells to mesenchymal cells, such as fibroblasts, is a common mechanism for the formation of organs during embryogenesis.
Knockout of WT1 is embryonic lethal due to abnormal bleeding in the pericardiac cavity and these embryos show malformation of the heart, diaphragm, and liver.41 In particular, WT1-null liver shows reduced size and abnormal expression of SMA in the HSCs.36,42 Similar to the Wt1-knockout livers, β-catenin deletion in liver mesenchymal cells also resulted in a small liver size and abnormal expression of SMA in HSCs.43 Wt1 and β-catenin may both regulate the phenotypes of HSCs.
Role of liver mesenchymal cells in liver development
In developing liver, similar to regenerating liver, HSCs support proliferation of hepatoblasts by secretion of growth factors, such as hepatocyte growth factor and pleiotrophin.18 WT1-null mesothelial cells demonstrate decreased expression of pleiotrophin and decreased proliferation of hepatoblasts.42 Berg et al. found expression of Fgf10 in fetal HSCs in developing liver.44 A loss of Fgf10 ligand or its receptor in mice reduced the size of the developing liver, suggesting that FGF10 promotes liver development. In addition to the cytokine signals, cell-cell interaction between Thy1+ liver mesenchymal cells and hepatoblasts was shown to be important for maturation of hepatoblasts.24
Possible HSC progenitors in adult liver
It remains unclear whether there are progenitor cells for HSCs in the adult liver. Kordes et al. showed that CD133+ cells separated from the HSC population express stem cell markers such as Oct3/4 and Nanog and are capable of differentiating into endothelial cells and hepatocytes in vitro.45 Using GFAPCre and Rosa26 reporter mice, Yang et al. have suggested a contribution of HSCs to oval cells, hepatic progenitor cells, in injured liver.46 Further studies using different Cre lines are necessary to validate whether mesodermal HSCs can differentiate into other cell lineages in injured livers.
Conclusions and perspectives
Our studies indicate that the mesoderm-derived mesothelium is a major source of HSCs and other liver mesenchymal cells including portal fibroblasts. In the adult liver, bone marrow-derived cells are also thought to contribute to HSCs or myofibroblasts, although several reports indicate that this contribution is negligible.47–50 During embryogenesis, hematopoiesis takes place in the liver and hematopoietic stem cells migrate to the bone marrow before birth. Thus, during embryogenesis, the STM-derived mesothelium, rather than bone marrow cells is likely to be the major source for HSCs. Our cell lineage analyses suggest a common origin for HSCs and other liver fibrogenic cells. It will be important, as we move forward, to determine the mechanisms of differentiation of the mesothelium to vitamin A-storing HSCs and other fibrogenic cells during liver development. Continuing to study the intricate developmental biology of the liver will provide interesting insight into the roles of the liver mesenchymal cells in liver fibrogenesis and regeneration and will potentially lead to the development of treatments for fibrosis and cirrhosis of the liver.
Acknowledgments
This work was supported by NIH grants R01-AA020753, R24-AA12885 Non-parenchymal Liver Cell Core, pilot project funding from P50-AA011999 Southern California Research Center for ALPD and Cirrhosis, and pilot project funding from P30-DK048522 USC Research Center for Liver Disease.
Footnotes
Conflict of Interest
No conflict of interest has been declared by the author.
References
- 1.Geerts A. History, heterogeneity, developmental biology, and functions of quiescent hepatic stellate cells. Semin Liver Dis. 2001;21:311–335. doi: 10.1055/s-2001-17550. [DOI] [PubMed] [Google Scholar]
- 2.Sato M, Suzuki S, Senoo H. Hepatic stellate cells: unique characteristics in cell biology and phenotype. Cell Struct Funct. 2003;28:105–112. doi: 10.1247/csf.28.105. [DOI] [PubMed] [Google Scholar]
- 3.Friedman SL. Hepatic stellate cells: protean, multifunctional, and enigmatic cells of the liver. Physiol Rev. 2008;88:125–172. doi: 10.1152/physrev.00013.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bataller R, Brenner DA. Liver fibrosis. J Clin Invest. 2005;115:209–218. doi: 10.1172/JCI24282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Popov Y, Schuppan D. Targeting liver fibrosis: strategies for development and validation of antifibrotic therapies. Hepatology. 2009;50:1294–1306. doi: 10.1002/hep.23123. [DOI] [PubMed] [Google Scholar]
- 6.George J, Roulot D, Koteliansky VE, Bissell DM. In vivo inhibition of rat stellate cell activation by soluble transforming growth factor β type II receptor: a potential new therapy for hepatic fibrosis. Proc Natl Acad Sci U S A. 1999;96:12719–12724. doi: 10.1073/pnas.96.22.12719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bhunchet E, Wake K. Role of mesenchymal cell populations in porcine serum-induced rat liver fibrosis. Hepatology. 1992;16:1452–1473. doi: 10.1002/hep.1840160623. [DOI] [PubMed] [Google Scholar]
- 8.Knittel T, Kobold D, Saile B, et al. Rat liver myofibroblasts and hepatic stellate cells: different cell populations of the fibroblast lineage with fibrogenic potential. Gastroenterology. 1999;117:1205–1221. doi: 10.1016/s0016-5085(99)70407-5. [DOI] [PubMed] [Google Scholar]
- 9.Kinnman N, Francoz C, Barbu V, et al. The myofibroblastic conversion of peribiliary fibrogenic cells distinct from hepatic stellate cells is stimulated by platelet-derived growth factor during liver fibrogenesis. Lab Invest. 2003;83:163–173. doi: 10.1097/01.lab.0000054178.01162.e4. [DOI] [PubMed] [Google Scholar]
- 10.Guyot C, Lepreux S, Combe C, et al. Hepatic fibrosis and cirrhosis: the (myo)fibroblastic cell subpopulations involved. Int J Biochem Cell Biol. 2006;38:135–151. doi: 10.1016/j.biocel.2005.08.021. [DOI] [PubMed] [Google Scholar]
- 11.Uchio K, Tuchweber B, Manabe N, Gabbiani G, Rosenbaum J, Desmoulière A. Cellular retinol-binding protein-1 expression and modulation during in vivo and in vitro myofibroblastic differentiation of rat hepatic stellate cells and portal fibroblasts. Lab Invest. 2002;82:619–628. doi: 10.1038/labinvest.3780456. [DOI] [PubMed] [Google Scholar]
- 12.Kruglov EA, Nathanson RA, Nguyen T, et al. Secretion of MCP-1/CCL2 by bile duct epithelia induces myofibroblastic transdifferentiation of portal fibroblasts. Am J Physiol Gastrointest Liver Physiol. 2006;290:G765–G771. doi: 10.1152/ajpgi.00308.2005. [DOI] [PubMed] [Google Scholar]
- 13.Wells RG, Kruglov E, Dranoff JA. Autocrine release of TGF-β by portal fibroblasts regulates cell growth. FEBS Lett. 2004;559:107–110. doi: 10.1016/S0014-5793(04)00037-7. [DOI] [PubMed] [Google Scholar]
- 14.Saile B, Matthes N, Neubauer K, et al. Rat liver myofibroblasts and hepatic stellate cells differ in CD95-mediated apoptosis and response to TNF-α. Am J Physiol Gastrointest Liver Physiol. 2002;283:G435–444. doi: 10.1152/ajpgi.00441.2001. [DOI] [PubMed] [Google Scholar]
- 15.Friedman SL, Roll FJ. Isolation and culture of hepatic lipocytes, Kupffer cells, and sinusoidal endothelial cells by density gradient centrifugation with Stractan. Anal Biochem. 1987;161:207–218. doi: 10.1016/0003-2697(87)90673-7. [DOI] [PubMed] [Google Scholar]
- 16.Matsumoto E, Hirosawa K, Abe K, Naka S. Development of the vitamin A-storing cell in mouse liver during late fetal and neonatal periods. Anat Embryol. 1984;169:249–259. doi: 10.1007/BF00315630. [DOI] [PubMed] [Google Scholar]
- 17.Kiassov AP, Van Eyken P, van Pelt JF, et al. Desmin expressing nonhematopoietic liver cells during rat liver development: an immunohistochemical and morphometric study. Differentiation. 1995;59:253–258. doi: 10.1046/j.1432-0436.1995.5940253.x. [DOI] [PubMed] [Google Scholar]
- 18.Asahina K, Sato H, Yamasaki C, et al. Pleiotrophin/heparin-binding growth-associated molecule as a mitogen of rat hepatocytes and its role in regeneration and development of liver. Am J Pathol. 2002;160:2191–2205. doi: 10.1016/S0002-9440(10)61167-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nitou M, Ishikawa K, Shiojiri N. Immunohistochemical analysis of development of desmin-positive hepatic stellate cells in mouse liver. J Anat. 2000;197:635–646. doi: 10.1046/j.1469-7580.2000.19740635.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hentsch B, Lyons I, Li R, et al. Hlx homeo box gene is essential for an inductive tissue interaction that drives expansion of embryonic liver and gut. Genes Dev. 1996;10:70–79. doi: 10.1101/gad.10.1.70. [DOI] [PubMed] [Google Scholar]
- 21.Kalinichenko VV, Bhattacharyya D, Zhou Y, et al. Foxf1+/− mice exhibit defective stellate cell activation and abnormal liver regeneration following CCl4 injury. Hepatology. 2003;37:107–117. doi: 10.1053/jhep.2003.50005. [DOI] [PubMed] [Google Scholar]
- 22.Wandzioch E, Kolterud Å, Jacobsson M, Friedman SL, Carlsson L. Lhx2−/− mice develop liver fibrosis. Proc Natl Acad Sci U S A. 2004;101:16549–16554. doi: 10.1073/pnas.0404678101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kubota H, Yao HL, Reid LM. Identification and characterization of vitamin A-storing cells in fetal liver: implications for functional importance of hepatic stellate cells in liver development and hematopoiesis. Stem Cells. 2007;25:2339–2349. doi: 10.1634/stemcells.2006-0316. [DOI] [PubMed] [Google Scholar]
- 24.Hoppo T, Fujii H, Hirose T, et al. Thy1-positive mesenchymal cells promote the maturation of CD49f-positive hepatic progenitor cells in the mouse fetal liver. Hepatology. 2004;39:1362–1370. doi: 10.1002/hep.20180. [DOI] [PubMed] [Google Scholar]
- 25.Suzuki K, Tanaka M, Watanabe N, et al. p75 Neurotrophin receptor is a marker for precursors of stellate cells and portal fibroblasts in mouse fetal liver. Gastroenterology. 2008;135:270–281. doi: 10.1053/j.gastro.2008.03.075. [DOI] [PubMed] [Google Scholar]
- 26.Asahina K, Tsai SY, Li P, et al. Mesenchymal origin of hepatic stellate cells, submesothelial cells, and perivascular mesenchymal cells during mouse liver development. Hepatology. 2009;49:998–1011. doi: 10.1002/hep.22721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Loo CK, Wu XJ. Origin of stellate cells from submesothelial cells in a developing human liver. Liver Int. 2008;28:1437–1445. doi: 10.1111/j.1478-3231.2008.01788.x. [DOI] [PubMed] [Google Scholar]
- 28.Cassiman D, Barlow A, Vander Borght S, Libbrecht L, Pachnis V. Hepatic stellate cells do not derive from the neural crest. J Hepatol. 2006;44:1098–1104. doi: 10.1016/j.jhep.2005.09.023. [DOI] [PubMed] [Google Scholar]
- 29.Zaret KS. Regulatory phases of early liver development: paradigms of organogenesis. Nat Rev Genet. 2002;3:499–512. doi: 10.1038/nrg837. [DOI] [PubMed] [Google Scholar]
- 30.Enzan H, Himeno H, Hiroi M, et al. Development of hepatic sinusoidal structure with special reference to the Ito cells. Microsc Res Tech. 1997;39:336–349. doi: 10.1002/(SICI)1097-0029(19971115)39:4<336::AID-JEMT4>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 31.Zhou B, Ma Q, Rajagopal S, et al. Epicardial progenitors contribute to the cardiomyocyte lineage in the developing heart. Nature. 2008;454:109–113. doi: 10.1038/nature07060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Asahina K, Zhou B, Pu WT, Tsukamoto H. Septum transversum-derived mesothelium gives rise to hepatic stellate cells and perivascular mesenchymal cells in developing mouse liver. Hepatology. 2011;53:983–995. doi: 10.1002/hep.24119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Saga Y, Miyagawa-Tomita S, Takagi A, Kitajima S, Miyazaki J, Inoue T. MesP1 is expressed in the heart precursor cells and required for the formation of a single heart tube. Development. 1999;126:3437–3447. doi: 10.1242/dev.126.15.3437. [DOI] [PubMed] [Google Scholar]
- 34.Zhang W, Yatskievych TA, Baker RK, Antin PB. Regulation of Hex gene expression and initial stages of avian hepatogenesis by Bmp and Fgf signaling. Dev Biol. 2004;268:312–326. doi: 10.1016/j.ydbio.2004.01.019. [DOI] [PubMed] [Google Scholar]
- 35.Pérez-Pomares JM, Carmona R, González-Iriarte M, Macías D, Guadix JA, Muñoz-Chápuli R. Contribution of mesothelium-derived cells to liver sinusoids in avian embryos. Dev Dyn. 2004;229:465–474. doi: 10.1002/dvdy.10455. [DOI] [PubMed] [Google Scholar]
- 36.Ijpenberg A, Pérez-Pomares JM, Guadix JA, et al. Wt1 and retinoic acid signaling are essential for stellate cell development and liver morphogenesis. Dev Biol. 2007;312:157–170. doi: 10.1016/j.ydbio.2007.09.014. [DOI] [PubMed] [Google Scholar]
- 37.Suskind DL, Muench MO. Searching for common stem cells of the hepatic and hematopoietic systems in the human fetal liver: CD34+ cytokeratin 7/8+ cells express markers for stellate cells. J Hepatol. 2004;40:261–268. doi: 10.1016/j.jhep.2003.11.007. [DOI] [PubMed] [Google Scholar]
- 38.Chagraoui J, Lepage-Noll A, Anjo A, Uzan G, Charbord P. Fetal liver stroma consists of cells in epithelial-to-mesenchymal transition. Blood. 2003;101:2973–2982. doi: 10.1182/blood-2002-05-1341. [DOI] [PubMed] [Google Scholar]
- 39.Wilm B, Ipenberg A, Hastie ND, et al. The serosal mesothelium is a major source of smooth muscle cells of the gut vasculature. Development. 2005;132:5317–5328. doi: 10.1242/dev.02141. [DOI] [PubMed] [Google Scholar]
- 40.Que J, Wilm B, Hasegawa H, et al. Mesothelium contributes to vascular smooth muscle and mesenchyme during lung development. Proc Natl Acad Sci U S A. 2008;105:16626–16630. doi: 10.1073/pnas.0808649105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kreidberg JA, Sariola H, Loring JM, et al. WT-1 is required for early kidney development. Cell. 1993;74:679–691. doi: 10.1016/0092-8674(93)90515-r. [DOI] [PubMed] [Google Scholar]
- 42.Onitsuka I, Tanaka M, Miyajima A. Characterization and functional analyses of hepatic mesothelial cells in mouse liver development. Gastroenterology. 2010;138:1525–1535. doi: 10.1053/j.gastro.2009.12.059. [DOI] [PubMed] [Google Scholar]
- 43.Berg T, DeLanghe S, Al Alam D, et al. β-Catenin regulates mesenchymal progenitor cell differentiation during hepatogenesis. J Surg Res. 2010;164:276–285. doi: 10.1016/j.jss.2009.10.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Berg T, Rountree CB, Lee L, et al. Fibroblast growth factor 10 is critical for liver growth during embryogenesis and controls hepatoblast survival via β-catenin activation. Hepatology. 2007;46:1187–1197. doi: 10.1002/hep.21814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kordes C, Sawitza I, Müller-Marbach A, et al. CD133+ hepatic stellate cells are progenitor cells. Biochem Biophys Res Commun. 2007;352:410–417. doi: 10.1016/j.bbrc.2006.11.029. [DOI] [PubMed] [Google Scholar]
- 46.Yang L, Jung Y, Omenetti A, et al. Fate-mapping evidence that hepatic stellate cells are epithelial progenitors in adult mouse livers. Stem Cells. 2008;26:2104–2113. doi: 10.1634/stemcells.2008-0115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Baba S, Fujii H, Hirose T, et al. Commitment of bone marrow cells to hepatic stellate cells in mouse. J Hepatol. 2004;40:255–260. doi: 10.1016/j.jhep.2003.10.012. [DOI] [PubMed] [Google Scholar]
- 48.Miyata E, Masuya M, Yoshida S, et al. Hematopoietic origin of hepatic stellate cells in the adult liver. Blood. 2008;111:2427–2435. doi: 10.1182/blood-2007-07-101261. [DOI] [PubMed] [Google Scholar]
- 49.Kisseleva T, Uchinami H, Feirt N, et al. Bone marrow-derived fibrocytes participate in pathogenesis of liver fibrosis. J Hepatol. 2006;45:429–438. doi: 10.1016/j.jhep.2006.04.014. [DOI] [PubMed] [Google Scholar]
- 50.Higashiyama R, Moro T, Nakao S, et al. Negligible contribution of bone marrow-derived cells to collagen production during hepatic fibrogenesis in mice. Gastroenterology. 2009;137:1459–1466. doi: 10.1053/j.gastro.2009.07.006. [DOI] [PubMed] [Google Scholar]


