Summary
Riboswitches are structured noncoding RNA domains that selectively bind metabolites and control gene expression (Mandal and Breaker 2004a; Coppins et al. 2007; Roth and Breaker 2009). Nearly all examples of the known riboswitches reside in noncoding regions of messenger RNAs where they control transcription or translation. Newfound classes of riboswitches are being reported at a rate of about three per year (Ames and Breaker 2009), and these have been shown to selectively respond to fundamental metabolites including coenzymes, nucleobases or their derivatives, amino acids, and other small molecule ligands.
The characteristics of some riboswitches suggest they could be modern descendents of an ancient sensory and regulatory system that likely functioned before the emergence of enzymes and genetic factors made of protein (Nahvi et al. 2002; Vitreschak et al. 2004; Breaker 2006). If true, then some of the riboswitch structures and functions that serve modern cells so well may accurately reflect the capabilities of RNA sensors and switches that existed in the RNA World. This article will address some of the characteristics of modern riboswitches that may be relevant to ancient versions of these metabolite-sensing RNAs.
Noncoding sequences in mRNA regulate transcription and translation by directly binding metabolites. These riboswitches may be descendants of an ancient sensory system that existed before proteins took over.
1. RIBOSWITCHES AND THEIR MOVING PARTS
The term riboswitch was established to define RNAs that control gene expression by binding metabolites without the need for protein factors. More recently, the name has begun to be used for riboswitch-like RNAs that respond to changes in temperature (Johansson 2009; Klinkert and Narberhaus 2009), tRNA binding (Gutiérrez-Preciado et al. 2009), or metal ion binding (Cromie et al. 2006; Dann et al. 2007). Although the functions of the RNAs encompassed by this expanded definition certainly would have been useful in an RNA World, the discussion hereafter will be focused on riboswitches that have evolved to respond to small organic compounds.
Riboswitches need to form molecular architectures with sufficient complexity to carry out two main functions: molecular recognition and conformational switching. Simple riboswitches each carry one aptamer that senses a single ligand and one expression platform that usually controls gene expression via a single mechanism. Because only four types of monomers are used by RNA to form selective binding pockets for target metabolites, aptamer sequences and structures tend to be strikingly well conserved over great evolutionary distances (e.g., see Grundy and Henkin 1998; Gelfand et al. 1999; Sudarsan et al. 2003; Nahvi et al. 2004). This sequence and structure conservation serves as the basis for assigning riboswitch representatives to specific classes (Fig. 1). Aspects of the tertiary structure folds used by riboswitch aptamers are presented in greater detail in Garst et al. (2010).
Figure 1.
Classes of validated and candidate riboswitches plotted in descending order relative to their frequency in the genomes of ∼700 bacterial species. Classes with at least some biochemical or genetic validation (filled bars) are named for the ligand that is most tightly bound. Multiple compound names indicate that binding site variants exist with altered ligand specificity. Candidate riboswitches whose ligand-binding functions have not been validated (open bars) are named for genes that are commonly associated with the motif.
The expression platform of each bacterial riboswitch usually is located downstream of the aptamer, where it assesses the ligand binding status of the RNA and regulates gene expression accordingly (Mandal and Breaker 2004a; Barrick and Breaker 2007). Alternately folding structures are common for RNA, and folding differences can be harnessed to influence several different processes that contribute to gene expression efficiency (Fig. 2). Therefore, expression platforms tend to be far less conserved through evolution compared to aptamer domains.
Figure 2.
Established or predicted mechanisms of riboswitch-mediated gene regulation. The most common mechanisms are (A) transcription termination, (B) translation initiation, and (C) splicing control (in eukaryotes). More rare mechanisms observed or predicted in some bacterial species include (D) transcription interference or possibly antisense action, (E) dual transcription and translation control, and (F) ligand-dependent self-cleaving ribozyme action. The numbers in a identify steps that are important for kinetically driven riboswitches (Wickiser et al. 2005a,b; Gilbert et al. 2006). Numbers represent (1) folding of the aptamer, (2) docking of the ligand, (3) folding of the expression platform, and (4) speed of RNA polymerase (RNAP). In B, Rho represents the transcriptional terminator protein (Skordalakes and Berger 2003).
Members of all experimentally validated riboswitch classes can bind their target ligand without the need for protein factors. Also, at least some representatives of each class have predicted or experimentally proven RNA structures residing in expression platforms that could independently control gene expression. In other words, the RNA alone is sufficient to carry out molecular recognition and gene control actions without the obligate assistance of protein factors. As with ancient ribozymes, any RNA World riboswitches would have probably had to function in an environment devoid of genetically encoded proteins. However, proteins could be directly involved in the folding and action of some modern riboswitches, much like protein factors assist large ribozymes such as RNase P (Sharin et al. 2005) and self-splicing ribozymes (Halls et al. 2007).
Aptamers can be structurally preorganized to various extents in the absence of ligand, but ligand binding induces at least some structural reorganization or stabilization of aptamer substructures that consequently influences the folding and function of the adjoining expression platform. It is this metabolite-dependent interplay between aptamer and expression platform that is harnessed by the riboswitch to modulate gene expression through various mechanisms.
2. MODERN MECHANISMS FOR RIBOSWITCH-MEDIATED GENE CONTROL
One of the most common mechanisms used by bacterial riboswitches involves the modulation of transcription termination (Fig. 2A) (Barrick and Breaker 2007). The formation of a strong stem followed by a run of uridine residues constitutes an intrinsic transcription terminator (Gusarov and Nudler 1999; Yarnell and Roberts 1999), which causes RNA polymerase to stall transcription and eventually to release the DNA template and nascent RNA product. Ligand binding to the aptamer controls the formation of the terminator stem usually by regulating the formation of a competing secondary structure or anti-terminator. Similarly, mutually-exclusive base-paired structures are exploited by riboswitches to control ribosome access to the ribosome binding site or Shine-Dalgarno (SD) sequence, thereby regulating translation initiation (Fig. 2B). This mechanism could be used to regulate translation from full-length mRNAs, but perhaps the transcription terminator protein Rho (Skordalakes and Berger 2003) will recognize nascent mRNAs that are not being actively translated because of SD access control by a riboswitch, thus causing transcription termination as well.
The most widespread riboswitch class discovered to date responds to the coenzyme thiamin pyrophosphate (TPP) (Mironov et al. 2002; Winkler et al. 2002b; Barrick and Breaker 2007). This riboswitch is quite common in plants and fungi (Sudarsan et al. 2003) where representatives have been shown to control splicing (Fig. 2C) (Kubodera et al. 2003; Cheah et al. 2007; Croft et al. 2007; Bocobza et al. 2007; Wachter et al. 2007). Additional data is needed to validate the proposed existence of a eukaryotic riboswitch class responsive to arginine (Borsuk et al. 2007), but it seems likely that more metabolite-binding riboswitches will be discovered in eukaryotes. Because eukaryotic TPP riboswitches appear to most commonly control splicing, introns may be excellent vehicles for riboswitch expression and regulatory function, and therefore may yield future riboswitch discoveries.
In a few instances in bacteria, a riboswitch aptamer resides upstream of an intrinsic transcription terminator stem that is formed using SD nucleotides (Fig. 2D). This arrangement should allow metabolite-controlled transcription termination as well as allow control of translation initiation for any mRNAs whose transcription progress extends into the coding region.
Also, riboswitches do not necessarily function to exclusively control the expression of an adjoining ORF, but could act in a bimolecular fashion on other RNAs, even after they may have modulated expression of a local gene. One possible example of this is a SAM-I riboswitch identified by bioinformatics (Rodionov et al. 2004) that is transcribed in the opposite direction of the genes it controls. Experimental analysis of this riboswitch (André et al. 2008) confirms the production of antisense RNA in response to changing sulfur levels. However the true mechanism of gene control may not be because of base pairing between sense and antisense transcripts. Rather, competition between RNA polymerases initiating at convergent promoters for the transcription of the mRNA versus the riboswitch-linked RNA may be the dominant control mechanism (Fig. 2E).
One of the most interesting mechanisms for riboswitch-mediated gene control integrates ligand binding and ribozyme activities (Fig. 2F). Representatives of the glmS riboswitch class function as metabolite-responsive self-cleaving ribozymes (Winkler et al. 2004). The ligand for this ribozyme-riboswitch is glucosamine-6-phosphate (GlcN6P), which is the metabolic product of the protein whose expression is controlled by the RNA. A glmS ribozyme construct from Bacillus subtilis cleaves with an GlcN6P-induced rate constant of nearly 100/min (Brooks and Hampel 2009), which is an enhancement of more than 1 billion-fold above the rate constant for spontaneous RNA cleavage by phosphoester transfer. Ribozyme self-cleavage initiates rapid destruction of its associated mRNA coding region via selective nuclease degradation (Collins et al. 2007).
3. POSSIBLE MECHANISMS FOR RIBOSWITCH CONTROL OF RIBOZYMES
It seems reasonable to speculate that metabolite-regulated ribozymes may have been prevalent in RNA World organisms. However, some features of the riboswitch mechanisms described above would not be applicable in a hypothetical RNA World cell. Perhaps a mechanism involving intrinsic transcription termination hairpins existed, but these hairpins would have needed to control transcription by an RNA polymerase ribozyme instead of protein-based polymerase enzymes. Of course, expression platforms that control ribosome access to the SD sequence could not have been relevant until at least a primitive translation apparatus was established.
RNA World organisms could have easily made use of numerous allosteric ribozymes with architectural features that were similar to those created in the laboratory by fusing aptamers to ribozymes (Breaker 2002; Silverman 2003). Such allosteric ribozymes could have emerged through RNA genome rearrangements that placed aptamers near ribozymes, so that ligand-mediated structure modulation affects a neighboring structure critical for ribozyme activity. Alternatively, the ligand could have functioned as a cofactor to directly participate in accelerating the chemical step of the ribozyme, rather than functioning as an allosteric effector. A modern example of cofactor participation is seen with glmS ribozymes, wherein GlcN6P actively participates in glmS ribozyme self-cleavage (Cochrane et al. 2009).
Self-splicing ribozymes also may have had functional homologs in the RNA World that were regulated by small molecule ligands. Grafting of an aptamer to a structurally sensitive region of a group I ribozyme (Hougland et al. 2006) has been shown to yield ligand-dependent splicing and gene control in cells (Thompson et al. 2002). Given that group I ribozymes use guanosine or one of its phosphorylated derivatives to initiate the first step of splicing, perhaps at least some representatives of these RNAs serve as metabolite-responsive riboswitches in modern cells. This function would be consistent with the fact that many group I ribozymes are associated with similar genes, which implies a functional linkage like that seen between riboswitches and the genes they control. If true, group I ribozymes may be one of the oldest and most widespread of all the known riboswitch classes.
4. LIGAND BINDING AFFINITY AND KINETICS OF RIBOSWITCH FUNCTION
Riboswitch aptamers can tightly bind their target ligands with values for dissociation constants (KD) ranging from the mid micromolar as measured for GlcN6P binding by glmS ribozymes (Winkler et al. 2004) to the mid picomolar range as observed for some riboswitch representatives that bind TPP (Welz and Breaker 2007), FMN (Lee et al. 2009) and c-di-GMP (Sudarsan et al. 2008). For Escherichia coli, the presence of a single molecule per cell corresponds to a concentration in the low nanomolar range. Therefore, picomolar KD values for aptamers are at least two orders of magnitude better than required for a riboswitch to detect compounds as rare as one per cell!
There are several possible explanations for this KD and metabolite concentration paradox (Ames and Breaker 2009). Perhaps, the conditions used to estimate ligand affinities are not a good approximation of cellular conditions, and the actual KD values may be much poorer. It is also known that nucleotides flanking the aptamer domains can diminish measured KD values, presumably by forming alternative folds that compete with the ligand-receptive structure. Thus the affinities of riboswitch aptamers may be poorer for nascent transcripts as they emerge from RNA polymerase. Another hypothesis that would resolve this contradiction is that some riboswitches may not reach equilibrium with their target metabolite, but they rely on the kinetics of RNA folding and ligand binding to properly modulate gene expression. In other words, the speed of ligand association, rather than the equilibrium constant reflecting ligand affinity, may be the critical determinant for the concentration of ligand needed to trigger riboswitch action. Unless the ligand binds before RNA polymerase passes beyond the terminator stem, transcription will progress to completion even if ligand binding eventually occurs.
Evidence for the importance of ligand-binding kinetics has emerged from an analysis of an FMN riboswitch from the ribD operon of B. subtilis (Wickiser et al. 2005b). This riboswitch operates via transcription termination whereby ligand binding permits formation of a terminator stem (Fig. 3A). The study revealed that the concentration required to trigger efficient transcription termination is more than 10-fold higher than the KD value measured for the minimal aptamer domain. Furthermore, conducting transcription reactions under conditions that accelerate the speed of RNA polymerization (e.g., increasing NTP concentrations or using mutations to remove RNA polymerase pause sites) creates a demand for even higher FMN concentration to trigger termination.
Figure 3.
Kinetic function of an FMN riboswitch. (A) Sequence and secondary structure models for the ribD FMN riboswitch from B. subtilis (Winkler et al. 2002a). The RNA functions as a genetic “OFF” switch wherein FMN binding stabilizes P1 formation, precludes the formation of an antiterminator stem, and permits the formation of a terminator stem that represses gene expression. (B) Simplified kinetic scheme for the function of the ribD FMN riboswitch depicted in A. Steps represented by black and gray arrows lead to termination and full-length mRNA production, respectively. See elsewhere for details (Wickiser et al. 2005b).
This and related data (Wickiser et al. 2005a; Gilbert et al. 2006) indicate that at least some riboswitches are not thermodynamically driven, but rely on the kinetics of transcription, RNA folding and metabolite binding to tune the concentration of ligand needed to trigger genetic control. Interestingly, this characteristic may give riboswitches an advantage over protein genetic factors in some instances (Ames and Breaker 2009). A kinetically driven riboswitch can be tuned to respond to a different concentration of metabolite by accruing mutations in the aptamer that change the rate constant for ligand association. Perhaps more likely to occur are mutations in the span of nucleotides linking aptamer to expression platform that change the time needed for RNA polymerase to reach the terminator stem. Kinetically driven riboswitches could lower or raise the concentration needed to trigger function by simply inserting or deleting nucleotides to this linker, respectively. RNA World riboswitches may have exploited similar characteristics to experience a more smooth evolutionary landscape where functional tuning could occur by mutations outside the binding and catalytic sites of receptors and ribozymes.
5. HOW MANY RIBOSWITCH CLASSES CURRENTLY EXIST?
Precisely defining what constitutes a distinctive riboswitch class can be difficult because of variation in aptamer sequence, structure, and function. For example, one or a few mutations to the core of guanine riboswitch aptamers can change ligand specificity to adenine (Mandal and Breaker 2004b) or 2′-deoxyguanosine (Kim et al. 2007), respectively. These mutations result in changes to the binding pocket, but the global architecture of the RNA remains largely unchanged (Serganov et al. 2004; Edwards and Batey 2009). Alternatively, some riboswitch aptamers carry substantial differences in sequence and structural elements that are distal from their near-identical ligand binding cores (see SAM riboswitch discussion later). Comparative sequence analysis algorithms cluster these RNAs into different groups despite the fact that they use similar binding sites to bind the same ligand (Weinberg et al. 2008).
A total of 17 riboswitch classes with at least some experimental validation have been identified based on a conservative approach to classification, wherein representatives are grouped either by similar global architecture or by similar binding site architecture (Fig. 1). At least seven candidate riboswitch classes also have been identified, although evidence for metabolite binding has not been reported for these candidates. Analysis of the numbers of representatives in each class can be conducted to determine if nearly all the riboswitch classes have been reported, or whether there are many more left to be discovered (Ames and Breaker 2009).
There are several reasons why it is important to estimate how many riboswitch classes still may await discovery in modern cells. From an RNA World vantage point, each new riboswitch discovery provides additional insights into the structural potential that primitive organisms may have harnessed long ago. Of course it may be difficult, or even impossible, to prove that a given riboswitch class is a direct descendant of one that existed in a ribo-organism. Regardless, these discoveries also provide a more complete understanding of the role that metabolite-sensing RNAs serve in modern cells. Locating riboswitch sequences in the genomes of organisms reveals new genetic regulons for compounds that are sensed by RNA (e.g., see Sudarsan et al. 2008). Associating genes of unknown function to specific metabolites via riboswitch regulation networks provides clues that are useful to establish protein function. Moreover, if some of these riboswitches control key metabolic processes in pathogenic organisms, then they may serve as new targets for the development of antibacterial compounds (Blount and Breaker 2006; Breaker 2009).
A plot of the riboswitch classes in descending order of frequency of incidence (Fig. 1) reveals that the numbers of representatives for these classes do not sort randomly. Assuming bioinformatics search algorithms used to identify new classes are revealing most common examples in sequence databases, then there naturally appears to be an increasing number of classes that have ever fewer representatives. This distribution is similar to that expected for phenomena that follow a power law relationship (Ames and Breaker 2009). If this relationship holds, then we speculate that more than 100 novel riboswitch classes may exist even in the ∼700 bacterial species whose genomes have been fully sequenced. The actual number of bacterial riboswitch classes that ultimately will be discovered may be far greater because of the vast amount of novel genomic DNA information that remains to be sequenced.
6. SOMETHING SPECIAL ABOUT SAM?
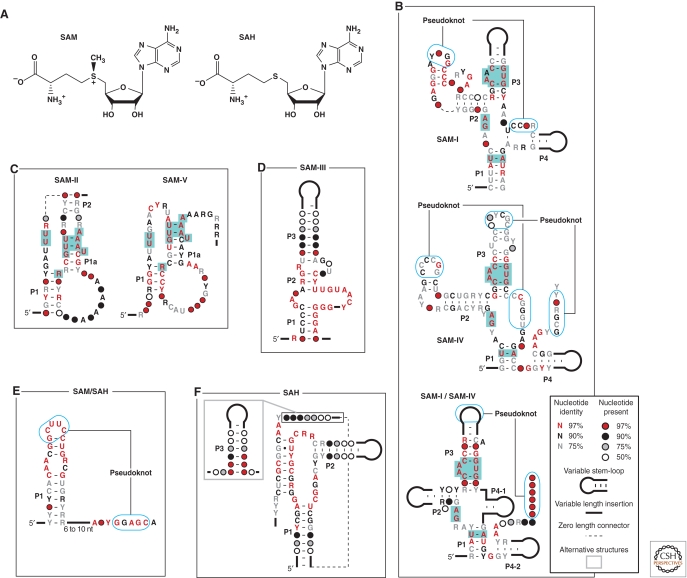
S-adenosylmethionine (SAM; Fig. 4A) is a coenzyme made by fusing the amino acid l-methionine with ATP such that the triphosphate moiety of ATP is removed. Compounds like SAM and other nucleotide-like coenzymes have been proposed to be relics from an RNA World (White III 1976). RNA World organisms would therefore have needed ribozymes to synthesize this compound, probably used aptamers to sense this compound, and used ribozymes that used the compound as a methylating agent. When SAM is used as a coenzyme for methylation reactions, the resulting byproduct after methyl group transfer is S-adenosylhomocysteine (SAH; Fig. 4A). This compound is similar in structure to SAM, which creates a considerable molecular recognition challenge for enzymes and receptors that need to selectively bind only SAM or only SAH. However, this precise molecular discrimination is important, because SAH is toxic to cells and needs to be selectively recycled to regenerate SAM.
Figure 4.
Riboswitch aptamers that respond to the coenzyme SAM or its metabolic derivative SAH. (A) The chemical structures of SAM and SAH. (B) Three variations of the SAM-I aptamer class. Despite substantial differences in sequence and structural subdomains, SAM-I (Grundy and Henkin 1998), SAM-IV (Weinberg et al. 2008), and SAM-I/SAM-IV (Weinberg et al. 2010) aptamer families carry a common ligand binding core (green-shaded nucleotides), including the identities of nucleotides that directly contact SAM (red bold letters). (C) A second superfamily of SAM-specific riboswitches use either the SAM-II (Corbino et al. 2005) or SAM-V (Poiata et al. 2009) aptamer families, which also carry a common ligand binding core. Annotations are as described for B. (D) Sequence and structural features of SAM-III aptamers (Fuchs et al. 2006), representing the third class of SAM-specific riboswitches. (E) Sequence and structural features of SAM/SAH aptamers (Weinberg et al. 2010), which genetically track like SAM riboswitches but bind SAH more tightly. (F) A distinct class of aptamers that genetically track as SAH riboswitches and selectively bind SAH while strongly discriminating against SAM (Wang et al. 2008).
Interestingly, at least five distinct classes of riboswitches exist that bind SAM or its derivative SAH (Wang and Breaker 2008). SAM-I riboswitches (Fig. 4B) strongly discriminate between SAM and SAH by using carbonyl oxygen atoms (O2) of two U residues to create a partial negative-charged surface that attracts the distinguishing positive charge present on the sulfur of SAM (Montange and Batey 2006). The conserved nucleotides that form the binding pocket of SAM-I aptamers are also present in two additional structural families SAM-IV (Weinberg et al. 2008) and SAM-I/SAM-IV (Weinberg et al. 2010). Although these three structural families were discovered at different times because they are clustered separately by bioinformatics algorithms, they appear to use different accessory structures to form ligand-binding cores that are identical.
A similar situation is observed for SAM-II and SAM-V riboswitches (Fig. 4C). SAM-II riboswitch aptamers (Corbino et al. 2005) form a binding pocket that selectively recognizes SAM in part by exploiting the carbonyl oxygen atoms (O4) to bind the positive charge on sulfur (Gilbert et al. 2008). SAM-V aptamers (Poiata et al. 2009) represent a different structural family member but representatives carry distinctive sequences that flank a putative ligand-binding core that appears to be identical to that formed by SAM-II. It is not clear whether members of the structural superfamilies most commonly represented by SAM-I and SAM-II riboswitches have emerged via divergent or convergent evolution, but their existence helps showcase the diversity of structures that can be used by RNA to support the formation of identical metabolite binding pockets.
SAM-III or SMK riboswitches (Fuchs et al. 2006) represent a third distinct class of SAM-binding RNAs. Yet again, SAM is bound more tightly than SAH in large part by exploiting the positive charge on sulfur, which is recognized by a carbonyl oxygen (O4) of a U residue (Lu et al. 2008). The ligand binding core is otherwise distinct from the two classes discussed previously. It is interesting to note that riboswitch aptamers examined in detail so far rely only on carbonyl oxygen atoms from U residues to interact with the positive charge on sulfur. Phosphate oxygen atoms appear ideally suited to associate with positive-charged ligands, and yet no SAM riboswitches exploiting this possible interaction have been discovered to date.
A fourth distinct SAM riboswitch class exists, but representatives of this class do not strongly discriminate between SAM and SAH. The SAM/SAH aptamers (Weinberg et al. 2010) are exceptionally small for natural metabolite-binding RNAs (as short as 43 nucleotides), and actually bind SAH at least fivefold more tightly than SAM. However, the motif is always genetically associated with genes for SAM biosynthesis, suggesting that SAM is the biologically relevant ligand. Most likely, SAH does not attain a concentration needed to inappropriately trigger riboswitch function.
Much greater discrimination in favor of SAH and against SAM is achieved by a fifth distinct riboswitch class in this series (Wang et al. 2008). SAH riboswitches are genetically associated with genes for enzymes that degrade SAH to prevent its toxic build-up and that recycle components for the regeneration of SAM. Given that SAM concentrations are likely to be higher that SAH concentrations, SAH riboswitches must strongly discriminate against SAM. Indeed, a representative SAH riboswitch appears to discriminate against SAM by approximately 1000 fold. This discrimination conceivably could be achieved by forming an SAH binding pocket that creates a steric block of the extra methyl group on SAM.
Based on current data, there does not appear to be anything that is particularly special about SAM as a riboswitch ligand. The existence of a diversity of riboswitch aptamers for SAM and SAH appears to be because of the importance that cells place on sensing these ligands, and perhaps due more to the rich structural potential that RNA can rely on to form selective binding pockets for metabolites. As evidence of this structural potential, we are discovering that multiple riboswitch classes have evolved to sense other metabolites, including preQ1 (Roth et al. 2007; Meyer et al. 2008). This same structural potential of course would also have been available to organisms from the RNA World.
7. INCREASING CAPABILITIES BY STACKING RIBOSWITCH COMPONENTS
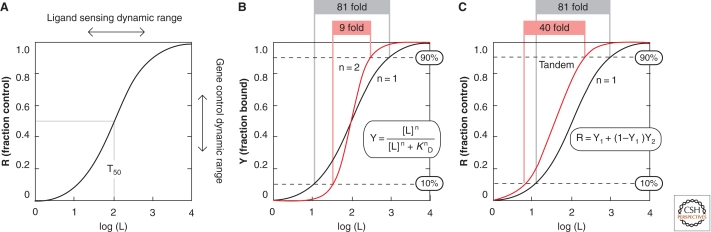
The architecture of simple riboswitches composed of a single aptamer and a single expression platform has very limited functional capabilities. Most obvious is the fact that the riboswitch will respond only to its target ligand, or perhaps also to a close chemical analog that may also be present in a cell. Furthermore, the dose-response curve for a simple riboswitch can do no better than conform to the functional optimum for a one-to-one interaction between a receptor and its ligand (Fig. 5). Specifically, a simple riboswitch that functions to perfection will require an 81-fold change in ligand concentration to progress from 10% to 90% gene modulation (Fig. 5B). If some of the riboswitch RNAs being made fail to function properly, because of folding problems for example, then the dynamic range for gene control will be reduced and the dynamic range for ligand sensing may be expanded.
Figure 5.
Binding kinetics for simple riboswitches, or complex riboswitches that use cooperative binding or a tandem architecture using riboswitches of from the same class. (A) Dose-response curve for a typical riboswitch carrying a single aptamer that functions perfectly. Note that the plot represents the performance of a population of individual riboswitch molecules where [R] represents the fraction of riboswitches causing gene expression change on ligand binding. Ligand concentration [L] is in arbitrary units, and T50 (Wickiser et al. 2005b) represents the concentration of ligand needed to half-maximally modulate gene expression. (B) Comparison of the dose-response curve for a simple riboswitch (one aptamer and one expression platform) versus a cooperative riboswitch (two aptamers and one expression platform). The curve for the cooperative riboswitch reflects perfect cooperativity between the aptamers and a Hill coefficient (n) of two. Note that [Y], the fraction of riboswitches bound by ligand, is equivalent to [R] if ligand binding always triggers a change in gene expression. (C) Comparison of the dose-response curve for a simple riboswitch versus a tandem arrangement of independently functioning riboswitches of the same class and near identical T50 values. Other annotations are as described in A and B. (Adapted, with permission, from Welz and Breaker 2007.)
These biochemical certainties for simple riboswitches place severe limitations on the performance characteristics of metabolite-sensing RNAs. However, modern organisms (and likely their RNA World ancestors) found ways to overcome the biochemical limitations of simple riboswitches. For example, riboswitches for the amino acid glycine commonly carry two aptamers and only a single expression platform (Fig. 6A). These aptamers function cooperatively, such that glycine binding by one aptamer increases the affinity for glycine binding to the adjacent aptamer. This cooperative binding function allows the riboswitch to show a far narrower ligand sensing dynamic range, which yields a genetic switch that has more digital character (Fig. 5B).
Figure 6.
Architectures and functions of tandem riboswitch arrangements. (A) Cooperative glycine riboswitch system that yields a more digital genetic response in numerous Gram positive bacteria including in the 5′ UTR of the B. subtilis gcvT gene (Mandal et al. 2004). (B) Tandem TPP riboswitches from the 5′ UTR of the thiamin metabolism gene tenA from Bacillus anthracis (Welz and Breaker 2007). (C) Tandem SAM-II and SAM-V riboswitches identified in ocean bacteria such as “Cand. P. ubique” (Poiata et al. 2009). (D) A two-input Boolean NOR logic gate composed of tandem riboswitches for SAM and AdoCbl located in the metE gene from Bacillus clausii (Sudarsan et al. 2006).
Ligand binding by a glycine riboswitch from Vibrio cholerae that includes two cooperative aptamers shows a Hill coefficient of 1.64 (Mandal et al. 2004). This value does not attain perfection, but must give cells that carry cooperative aptamers an evolutionary advantage. Many glycine riboswitches control expression of genes whose protein products form the glycine cleavage system, which allows cells to use excess glycine as an energy source. Apparently some cells take advantage of the cooperative riboswitch to more quickly produce proteins for glycine cleavage when the amino acid is in slight excess, and then turn off production of the glycine cleavage system before glycine concentrations drop to a point that may jeopardize synthesis of proteins.
Other tandem-arranged riboswitch configurations also exist, and these further expand the functional characteristics of metabolite-binding RNAs. For example, a type of tandem riboswitch architecture has been observed that can produce a more digital genetic response without cooperative ligand binding. Two independently functioning riboswitches from the same class (or that sense the same ligand) can occur in the same mRNA to produce a dose-response curve that is steeper than that for a single riboswitch (Fig. 6b). An example of this type of gene control system in Bacillus anthracis is composed of two TPP riboswitches (Sudarsan et al. 2006; Welz and Breaker 2007). If the concentrations of ligand needed to half-maximally modulate gene expression (T50 values) are near identical, then only a 40-fold change in ligand concentration is needed to progress from 10% to 90% gene modulation.
A very similar type of tandem riboswitch architecture has recently been observed in Candidatus pelagibacter ubique (Poiata et al. 2009). This arrangement is composed of representatives of SAM-II and SAM-V riboswitch classes (Fig. 6C). The SAM-II riboswitch occurs first in the nascent mRNA, and has an expression platform that includes an intrinsic terminator stem. The SAM-V riboswitch occurs second, and likely controls gene expression by regulating access to the ribosome binding site. This arrangement could yield a more digital dose-response curve like the tandem TPP riboswitches described above. In addition, the two distinct expression platforms that control either transcription or translation provide a particularly economical genetic switch. If SAM concentrations are adequate, newly initiated transcripts are terminated by the SAM-II riboswitch before the coding region is produced. If SAM concentrations become adequate only after the full mRNA is produced, then the SAM-V riboswitch can repress translation without requiring that the mRNA be degraded. This capability could be particularly advantageous for organisms that live in environments that are constantly limited for nutrients.
Although simple riboswitches only respond to one ligand type, this restriction in signaling complexity can be overcome by stacking tandem riboswitches from different ligand-binding classes such that gene expression is responsive to more than one chemical signal (Fig. 6D). Indeed, a natural example of such a two-input Boolean logic gate has been observed in the metE mRNA from the bacterium Bacillus clausii (Sudarsan et al. 2006). A SAM-I riboswitch aptamer precedes an aptamer for AdoCbl, and each aptamer is associated with its own intrinsic terminator stem. The binding of either SAM or AdoCbl causes transcription termination before the coding region of the mRNA is made, and thus the tandem arrangement functions as Boolean NOR gate.
The examples described above are undoubtedly just a small sample of the ways that riboswitches can be conjoined or combined with other functional domains to create even more complex RNA devices. Many types of RNA switches are being created (or recreated) in laboratories by molecular engineers (Breaker 2004; Silverman 2003). For example, integration of multiple aptamers into a structurally sensitive site of a hammerhead ribozyme produced a cooperative allosteric ribozyme that functions as a Boolean AND gate (Jose et al. 2001). This RNA construct incorporates several features of natural riboswitches with ribozyme function to provide just one example of the incredible complexity of function that could be achieved by RNA. Although there may be little demand in modern cells for controlling ribozyme functions using aptamers as allosteric binding sites, RNA World organisms could have made extensive use of ribozymes whose reaction kinetics were controlled by metabolite binding aptamers integrated into their structures. Of course riboswitches from the RNA World would not necessarily have carried expression platforms like those from modern cells, but could have used other ligand-mediated structural changes to regulate early metabolic processes.
8. THE ORIGIN OF RIBOSWITCHES
The fact that riboswitches are entrusted by some modern cells to sense many compounds and regulate key metabolic genes strongly validates the hypothesis that RNA World organisms could have used similar RNA-based sensors and switches. However, we cannot presume that all modern riboswitches are direct descendants from homologous riboswitches from the RNA World. Biological innovation with RNA polymers would not have ceased with the emergence of proteins, and so new or reinvented riboswitch classes are probably continuing to appear.
Some riboswitch classes, particularly those that sense TPP, AdoCbl, and FMN are exceptionally widespread (Barrick and Breaker 2007). Moreover, these riboswitch ligands are proposed to be molecular relics from an RNA World (White 1976, Benner et al. 1989). Such characteristics are precisely what would be expected for riboswitches that have an ancient origin. Furthermore, the most widespread riboswitch classes have some of the most complex aptamer structures, and these information-rich structures are unlikely to emerge frequently during evolution. Rather, widespread complex riboswitches most likely have an early and perhaps RNA World origin.
In contrast, the more narrowly distributed riboswitches that have less-complex structures could represent more recent evolutionary inventions. Even relatively widespread riboswitches with simplistic aptamers, such as those that bind purines (Kim and Breaker 2008) or preQ1 (Roth et al. 2007) could be reinvented rather than directly descend from primordial homologs. Perhaps modern classes of riboswitch aptamers have mixed beginnings; with some having emerged relatively recently to satisfy new regulatory needs whereas others representing an unbroken structural and functional lineage from RNAs that have changed little since the RNA World.
One intriguing hypothesis is that some riboswitch aptamers, particularly those that bind coenzymes, may be descendent from ancient ribozymes that synthesized these coenzymes or used them to catalyze metabolic reactions. A coenzyme-dependent ribozyme and a coenzyme-responsive riboswitch both would need to form a selective binding pocket for the compound, and therefore it is likely that some RNA sequences that function as ribozymes could be very close in sequence-space to those that function as riboswitches (Fig. 7).
Figure 7.
A possible path for the descent of modern riboswitches from RNA World ribozymes. SAM-dependent RNAs were arbitrarily chosen as an example. (Stage I) Emergence of ribozymes that synthesize SAM. Ribozymes might have coupled methionine to a 5′-terminal ATP moiety or coupled methionine to a free ATP substrate. (Stage II) Utilization of SAM by diverse methyltransferase ribozymes. In a complex RNA World, various methyltransferase ribozymes might have been present that used SAM as a prosthetic group or as a diffusible cofactor. (Stage III) Co-opting ribozyme subdomains to create RNA switches. Allosteric systems that controlled ribozyme function or that controlled the biosynthesis of new RNAs could have made use of variant SAM-binding domains of certain methyltransferase ribozymes. (Stage IV) Perseverance of ancient SAM-binding aptamers and their use in modern riboswitches. (Adapted, with permission, from Breaker 2006.)
9. CONCLUSIONS
The existence of so many riboswitch classes that are involved in key regulatory processes provides support for the hypothesis that RNA World organisms could have harnessed the structural and functional potential of aptamers to create sophisticated sensory and regulatory networks without the need for proteins. Also as seen with modern organisms, when more complex biochemical functions are demanded for the regulatory task, multiple riboswitches can be assembled in different architectures to provide more sophisticated RNA devices. Tandem aptamers that yield more digital genetic responses, multi-mechanism gene control, or Boolean logic gate function result from simply grafting the parts from two or more riboswitches together. Because examples of sophisticated riboswitches are successfully competing with proteins for demanding roles in modern cells, there is reason to believe that RNA World organisms could have used similar RNAs to regulate very complex metabolic states before the emergence of proteins.
There also appears to be considerable potential for exploiting riboswitches for practical applications. Existing riboswitches can be fused to coding regions to create ligand-controlled genetic constructs. Reverse engineering of natural riboswitches could help guide the creation of designer riboswitches to expand the tools available for genetics studies (Suess and Weigand 2008). Also, analogs of riboswitch ligands can be used to deregulate key metabolic pathways in bacteria, and thus may serve as new classes of antibiotics (Blount and Breaker 2006; Breaker 2009). Therefore the continued discovery and analysis of riboswitches should help efforts to create molecular tools and drug targets from RNA, as well as provide modern examples of RNAs whose functions may closely mimic those from the RNA World.
ACKNOWLEDGMENTS
RNA research in the Breaker laboratory is supported by the Howard Hughes Medical Institute and by grants from the National Institutes of Health.
Footnotes
Editors: John F. Atkins, Raymond F. Gesteland, and Thomas R. Cech
Additional Perspectives on RNA Worlds available at www.cshperspectives.org
REFERENCES
- André G, et al. 2008. S-box and T-box riboswitches and antisense RNA control a sulfur metabolic operon of Clostridium acetobutylicum. Nucleic Acids Res 36: 5955–5969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ames TD, Breaker RR 2009. Bacterial riboswitch discovery and analysis. In: The chemical biology of nucleic acids, Meyer G., ed., John Wiley & Sons, Ltd; (in press) [Google Scholar]
- Barrick JE, Breaker RR 2007. The distributions, mechanisms, and structures of metabolite-binding riboswitches. Genome Biol 8: R239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benner SA, Ellington AD, Tauer A 1989. Modern metabolism as a palimpsest of the RNA world. Proc Natl Acad Sci USA 86: 7054–7058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blount KF, Breaker RR 2006. Riboswitches as antibacterial drug targets. Nat Biotechnol 24: 1558–1564 [DOI] [PubMed] [Google Scholar]
- Bocobza S, Adato A, Mandel T, Shapira M, Nudler E, Aharoni A 2007. Riboswitch-dependent gene regulation and its evolution in the plant kingdom. Genes Dev 21: 2874–2879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borsuk P, Przykorska A, Blachnio K, Koper M, Pawlowicz JM, Pekala M, Weglenski P 2007. L-arginine influences the structure and function of arginase mRNA in Aspergillus nidulans. Biol Chem 388: 135–144 [DOI] [PubMed] [Google Scholar]
- Breaker RR 2002. Engineered allosteric ribozymes as biosensor components. Curr Opin Biotechnol 13: 31–39 [DOI] [PubMed] [Google Scholar]
- Breaker RR 2004. Natural and engineered nucleic acids as tools to explore biology. Nature 432: 838–845 [DOI] [PubMed] [Google Scholar]
- Breaker RR 2006. Riboswitches and the RNA world. In: The RNA world, 3rd ed., Gesteland R.F., Cech T.R., Atkins J.F., eds., Cold Spring Harbor Laboratory Press, pp. 89–107 [Google Scholar]
- Breaker RR 2009. Riboswitches: from ancient gene-control systems to modern drug targets. Future Microbiol 4: 771–773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooks KM, Hampel KJ 2009. A rate-limiting conformational step in the catalytic pathway of the glmS ribozyme. Biochemistry 48: 5669–5678 [DOI] [PubMed] [Google Scholar]
- Cheah MT, Wachter A, Sudarsan N, Breaker RR 2007. Control of alternative RNA splicing and gene expression by eukaryotic riboswitches. Nature 447: 497–500 [DOI] [PubMed] [Google Scholar]
- Cochrane JC, Lipchock SV, Smith KD, Strobel SA 2009. Structural and chemical basis for glucosamine-6-phosphate binding and activation of the glmS ribozyme. Biochemistry 48: 3239–3246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins JA, Irnov I, Baker S, Winkler WC 2007. Mechanism of mRNA destabilization by the glmS ribozyme. Genes Dev 21: 3356–3368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coppins RL, Hall KB, Groisman EA 2007. The intricate world of riboswitches. Curr Opin Microbiol 10: 176–181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbino KA, Barrick JE, Lim J, Welz R, Tucker BJ, Puskarz I, Mandal M, Rudnick ND, Breaker RR 2005. Evidence for a second class of S-adenosylmethionine riboswitches and other regulatory RNA motifs in α-proteobacteria. Genome Biol 6: R70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croft MT, Moulin M, Webb ME, Smith AG 2007. Thiamine biosynthesis in algae is regulated by riboswitches. Proc Natl Acad Sci 104: 20770–20775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cromie MJ, Shi Y, Latifi T, Groisman EA 2006. An RNA sensor for intracellular Mg2+. Cell 125: 71–84 [DOI] [PubMed] [Google Scholar]
- Dann CE 3rd, Wakeman CA, Sieling CL, Baker SC, Ironv I, Winkler WC 2007. Structure and mechanism of a metal-sensing regulatory RNA. Cell 130: 878–892 [DOI] [PubMed] [Google Scholar]
- Edwards AL, Batey RT 2009. A structural basis for the recognition of 2′-deoxyguanosine by the purine riboswitch. J Mol Biol 385: 938–948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs RT, Grundy FJ, Henkin TM 2006. The SMK box is a new SAM-binding RNA for translational regulation of SAM synthase. Nat Struct Mol Biol 13: 226–233 [DOI] [PubMed] [Google Scholar]
- Garst AD, Edwards AL, Batey RT 2010. Riboswitches: structures and mechanisms. Cold Spring Harb Perspect Biol [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelfand MS, Mironov AA, Jomantas J, Kozlov YI, Perumov DA 1999. A conserved RNA structure element involved in the regulation of bacterial riboflavin synthesis genes. Trends Genet 15: 439–442 [DOI] [PubMed] [Google Scholar]
- Gilbert SD, Rambo RP, Van Tyne D, Batey RT 2008. Structure of the SAM-II riboswitch bound to S-adenosylmethionine. Nat Struct Mol Biol 15: 177–182 [DOI] [PubMed] [Google Scholar]
- Gilbert SD, Stoddard CD, Wise SJ, Batey RT 2006. Thermodynamic and kinetic characterization of ligand binding to the purine riboswitch aptamer domain. J Mol Biol 359: 754–768 [DOI] [PubMed] [Google Scholar]
- Grundy FJ, Henkin TM 1998. The S box regulon: A new global transcription termination control system for methionine and cysteine biosynthesis genes in gram-positive bacteria. Mol Microbiol 30: 737–749 [DOI] [PubMed] [Google Scholar]
- Gusarov I, Nudler E 1999. The mechanism of intrinsic transcription termination. Mol Cell 3: 495–504 [DOI] [PubMed] [Google Scholar]
- Gutiérrez-Preciado A, Henkin TM, Grundy FJ, Yanofsky C, Merino E 2009. Biochemical features and functional implications of the RNA-based T-box regulatory system. Microbiol Mol Biol Rev 73: 36–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halls C, Mohr S, Del Campo M, Yang Q, Jankowsky E, Lambowitz AM 2007. Involvement of DEAD-box proteins in group I and group II intron splicing. Biochemical characterization of Mss116p, ATP hydrolysis-dependent and -independent mechanisms, and general RNA chaperone activity. J Mol Biol 365: 835–855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hougland JL, Piccirilli JA, Forconi M, Lee J, Herschlag D 2006. How the group I intron works: a case study of RNA structure and function. In: The RNA world, 3rd ed., Gesteland R.F., Cech T.R., Atkins J.F., eds., Cold Spring Harbor Laboratory Press, pp. 133–205 [Google Scholar]
- Johansson J 2009. RNA thermosensors in bacterial pathogens. Contrib Microbiol 16: 150–160 [DOI] [PubMed] [Google Scholar]
- Jose A, Soukup GA, Breaker RR 2001. Cooperative binding of effectors by an allosteric ribozyme. Nucleic Acids Res 29: 1631–1637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JN, Breaker RR 2008. Purine sensing by riboswitches. Biol Cell 100: 1–11 [DOI] [PubMed] [Google Scholar]
- Kim JN, Roth A, Breaker RR 2007. Guanine riboswitch variants from Mesoplasma florum selectively recognize 2′-deoxyguanosine. Proc Natl Acad Sci 104: 16092–16097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klinkert B, Narberhaus F 2009. Microbial thermosensors. Cell Mol Life Sci 66: 2661–2676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubodera T, et al. 2003. Thiamine-regulated gene expression of Aspergillus oryzae thiA requires splicing of the intron containing a riboswitch-like domain in the 5′ UTR. FEBS Lett 555: 516–520 [DOI] [PubMed] [Google Scholar]
- Lee ER, Blount KF, Breaker RR 2009. Roseoflavin is a natural antibacterial compound that binds to FMN riboswitches and regulates gene expression. RNA Biol 6: 187–194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu C, Smith AM, Fuchs RT, Ding F, Rajashankar K, Henkin TM, Ke A 2008. Crystal structures of the SAM-III/SMK riboswitch reveal the SAM-dependent translation inhibition mechanism. Nat Struct Mol Biol 15: 1076–1083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandal M, Breaker RR 2004a. Gene regulation by riboswitches. Nat Rev Mol Cell Biol 5: 451–463 [DOI] [PubMed] [Google Scholar]
- Mandal M, Breaker RR 2004b. Adenine riboswitches and gene activation by disruption of a transcription terminator. Nat Struct Mol Biol 11: 29–35 [DOI] [PubMed] [Google Scholar]
- Mandal M, Lee M, Barrick JE, Weinberg Z, Emilsson GM, Ruzzo WL, Breaker RR 2004. A glycine-dependent riboswitch that uses cooperative binding to control gene expression. Science 306: 275–279 [DOI] [PubMed] [Google Scholar]
- Meyer MM, Roth A, Chervin SM, Garcia GA, Breaker RR 2008. Confirmation of a second natural preQ1 aptamer class in Streptococcaceae bacteria. RNA 14: 685–695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mironov AS, Gusarov I, Rafikov R, Lopez LE, Shatalin K, Kreneva RA, Perumov DA, Nudler E 2002. Sensing small molecules by nascent RNA: a mechanism to control transcription in bacteria. Cell 111: 747–756 [DOI] [PubMed] [Google Scholar]
- Montange RK, Batey RT 2006. Structure of the S-adenosylmethionine riboswitch regulatory mRNA element. Nature 441: 1172–1175 [DOI] [PubMed] [Google Scholar]
- Nahvi A, Barrick JE, Breaker RR 2004. Coenzyme B12 riboswitches are widespread genetic control elements in prokaryotes. Nucleic Acids Res 32: 143–150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nahvi A, Sudarsan N, Ebert MS, Zou X, Brown KL, Breaker RR 2002. Genetic control by a metabolite binding mRNA. Chem Biol 9: 1043–1049 [DOI] [PubMed] [Google Scholar]
- Poiata E, Meyer MM, Ames TD, Breaker RR 2009. A variant riboswitch aptamer class for S-adenosylmethionine common in marine bacteria. RNA 15: 2046–2056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodionov DA, Vitreschak AG, Mironov AA, Gelfand MS 2004. Comparative genomics of the methionine metabolism in Gram-positive bacteria: a variety of regulatory systems. Nucleic Acids Res 32: 3340–3353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth A, Breaker RR 2009. The structural and functional diversity of metabolite-binding riboswitches. Annu Rev Biochem 78: 305–334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth A, Winkler WC, Regulski EE, Lee BW, Lim J, Jona I, Barrick JE, Ritwick A, Kim JN, Welz R, et al. 2007. A riboswitch selective for the queuosine precursor preQ1 contains an unusually small aptamer domain. Nat Struct Mol Biol 14: 308–317 [DOI] [PubMed] [Google Scholar]
- Sharin E, Schein A, Mann H, Ben-Asouli Y, Jarrous N 2005. RNase P: Role of distinct protein cofactors in tRNA substrate recognition and RNA-based catalysis. Nucleic Acids Res 33: 5120–5132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman SK 2003. Rube Goldberg goes (ribo)nuclear? Molecular switches and sensors made from RNA. RNA 4: 377–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skordalakes E, Berger JM 2003. Structure of the Rho transcription terminator: Mechanism of mRNA recognition and helicase loading. Cell 114: 135–146 [DOI] [PubMed] [Google Scholar]
- Sudarsan N, Barrick JE, Breaker RR 2003. Metabolite-binding RNA domains are present in the genes of eukaryotes. RNA 9: 644–647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudarsan N, Hammond MC, Block KF, Welz R, Barrick JE, Roth A, Breaker RR 2006. Tandem riboswitch architectures exhibit complex gene control functions. Science 314: 300–304 [DOI] [PubMed] [Google Scholar]
- Sudarsan N, Lee ER, Weinberg Z, Moy RH, Kim JN, Link KH, Breaker RR 2008. Riboswitches in eubacteria sense the second messenger cyclic di-GMP. Science 321: 411–413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suess B, Weigand JE 2008. Engineered riboswitches: overview, problems and trends. RNA Biol 5: 24–29 [DOI] [PubMed] [Google Scholar]
- Serganov A, Yuan YR, Pikovskaya O, Polonskaia A, Malinina L, Phan AT, Hobartner C, Micura R, Breaker RR, Patel DJ 2004. Structural basis for discriminative regulation of gene expression by adenine- and guanine-sensing mRNAs. Chem Biol 11: 1729–1741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson KM, Syrett HA, Knudsen SM, Ellington AD 2002. Group I aptazymes as genetic regulatory switches. BMC Biotechnol 2: 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wachter A, Tunc-Ozdemir M, Grove BC, Green PJ, Shintani DK, Breaker RR 2007. Riboswitch control of gene expression in plants by splicing and alternative 3′ processing of mRNAs. Plant Cell 19: 3437–3450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang JX, Breaker RR 2008. Riboswitches that sense S-adenosylmethionine and S-adenosylhomocysteine. Biochem Cell Biol 86: 157–168 [DOI] [PubMed] [Google Scholar]
- Wang JX, Lee ER, Morales DR, Lim J, Breaker RR 2008. Riboswitches that sense S-adenosylhomocysteine and activate genes involved in coenzyme recycling. Mol Cell 28: 691–702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberg Z, Regulski EE, Hammond MC, Barrick JE, Yao Z, Ruzzo WL, Breaker RR 2008. The aptamer core of SAM-IV riboswitches mimics the ligand-binding site of SAM-I riboswitches. RNA 14: 822–828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberg Z, Wang JX, Bogue J, Yang J, Corbino K, Moy R, Breaker RR 2010. Comparative genomics reveals 104 candidate structured RNAs from bacteria, archaeal and their metagenomes. Genome Biol (submitted). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welz R, Breaker RR 2007. Ligand binding and gene control characteristics of tandem riboswitches in Bacillus anthracis. RNA 13: 573–582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White HB, III. 1976. Coenzymes as fossils of an earlier metabolic state. J Mol Evol 7: 101–104 [DOI] [PubMed] [Google Scholar]
- Wickiser JK, Cheah MT, Breaker RR, Crothers DM 2005a. The kinetics of ligand binding by an adenine-sensing riboswitch. Biochemistry 44: 13404–13414 [DOI] [PubMed] [Google Scholar]
- Wickiser JK, Winkler WC, Breaker RR, Crothers DM 2005b. The speed of RNA transcription and metabolite binding kinetics operate an FMN riboswitch. Mol Cell 18: 49–60 [DOI] [PubMed] [Google Scholar]
- Winkler WC, Cohen-Chalamish S, Breaker RR 2002a. An mRNA structure that controls gene expression by binding FMN. Proc Natl Acad Sci 99: 15908–15913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler W, Nahvi A, Breaker RR 2002b. Thiamine derivatives bind messenger RNAs directly to regulate bacterial gene expression. Nature 419: 952–956 [DOI] [PubMed] [Google Scholar]
- Winkler WC, Nahvi A, Roth A, Collins JA, Breaker RR 2004. Control of gene expression by a natural metabolite-responsive ribozyme. Nature 428: 281–286 [DOI] [PubMed] [Google Scholar]
- Vitreschak AG, Rodionov DA, Mironov AA, Gelfand MS 2004. Riboswitches: The oldest mechanism for the regulation of gene expression? Trends Genet 20: 44–50 [DOI] [PubMed] [Google Scholar]
- Yarnell WS, Roberts JW 1999. Mechanism of intrinsic transcription termination and antitermination. Science 284: 611–615 [DOI] [PubMed] [Google Scholar]