Abstract
Biochemical and genetic evidence establishes a central role of the amyloid precursor protein (APP) in Alzheimer disease (AD) pathogenesis. Biochemically, deposition of the β-amyloid (Aβ) peptides produced from proteolytic processing of APP forms the defining pathological hallmark of AD; genetically, both point mutations and duplications of wild-type APP are linked to a subset of early onset of familial AD (FAD) and cerebral amyloid angiopathy. As such, the biological functions of APP and its processing products have been the subject of intense investigation, and the past 20+ years of research have met with both excitement and challenges. This article will review the current understanding of the physiological functions of APP in the context of APP family members.
A pathological hallmark of Alzheimer disease is the deposition of β-amyloid (Aβ) peptides from amyloid precursor protein (APP). APP undergoes complex proteolytic processing and is part of a gene family whose members have partially overlapping functions.
Synaptic dysfunction, cognitive decline, and plaque deposition of the β-amyloid peptide Aβ, derived from the β-amyloid precursor protein APP, are hallmark features of Alzheimer disease (AD). Since the molecular cloning of APP, more than 20 years ago (Goldgaber et al. 1987; Kang et al. 1987; Tanzi et al. 1987), a large body of biochemical and genetic evidence has accumulated that identified Aβ as a central trigger for AD pathogenesis. Despite this, the physiological role of APP and the question of whether a loss of its functions contributes to AD are still unclear. The secretases involved in APP processing and Aβ generation have been cloned (see De Strooper et al. 2011; Haass et al. 2011) and have since become major therapeutic targets. Understanding the physiological function of APP is also of immediate relevance for AD pathogenesis. As Aβ is generated as part of normal APP processing (Haass et al. 1992), deregulation of Aβ production (either during pathogenesis or as a consequence of secretase inhibitors) is expected to simultaneously affect other APP processing products and may thus compromise physiologically important signaling pathways. Two major obstacles complicate the analysis of functions of APP in vivo: (1) APP is subject to complex proteolytical processing that generates several polypeptides each of which likely performs specific functions, and (2) APP is part of a gene family with partially overlapping functions.
CELL BIOLOGY AND EXPRESSION
APP Processing
APP is an integral type I transmembrane protein with a single transmembrane domain, a large extracellular ectodomain, and a short cytoplasmic tail (Fig. 1). Processing is initiated either by cleavage of APP by α-secretase within the Aβ region, or by cleavage by β-secretase (BACE) at the amino terminus of Aβ, leading to the secretion of large soluble ectodomains, termed APPsα and APPsβ, respectively. Subsequent processing of the carboxy-terminal fragments (CTFβ or CTFα) by γ-secretase results in the production of Aβ, p3, and the APP intracellular domain (AICD). More recently, a novel amino-terminal fragment (N-APP286) derived from APPsβ was identified as a ligand for death receptor 6 (DR6), a member of the TNFR gene family (Nikolaev et al. 2009). Whereas in fibroblasts and nonneuronal cell lines (e.g., HEK293 cells) α-secretase processing is the dominant pathway, primary neuronal cultures express high levels of BACE and thus generate considerable amounts of APPsβ and Aβ (Simons et al. 1996; Kuhn et al. 2010). In adult mouse brain, secreted total APPs constitutes at least 50% of all APP isoforms and in vivo studies using cycloheximide injections revealed a half-life of 4–5 h for both APPsα and APPsβ, whereas APP-FL is turned over much more rapidly (half-life of ca. 1 h) (Morales-Corraliza et al. 2009).
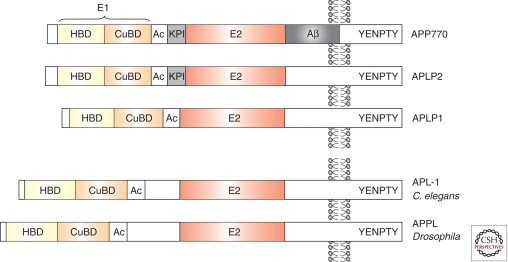
Figure 1.
Schematic overview of domain structure of APP family proteins. All APP family members share conserved E1 and E2 extracellular domains, an acidic domain (Ac) and the YENPTY motif in the carboxyl terminus. Note that Aβ is unique for APP. HBD, Heparin binding domain; CuBD, Copper binding domain; KPI, Kunitz-type protease inhibitor domain.
APP Gene Family and Structure
APP is a member of an evolutionary conserved gene family including APL-1 in Caenorhabditis elegans (Daigle and Li 1993), APPL in Drosophila (Rosen et al. 1989; Luo et al. 1990), appa and appb in zebrafish (Musa et al. 2001), and in mammals besides APP the two amyloid precursor-like proteins, APLP1 and APLP2 (Wasco et al. 1992, 1993; Slunt et al. 1994). APP family proteins share conserved regions within the ectodomain, in particular the E1 and E2 domains and the intracellular tail that shows the largest sequence identity (Fig. 1). Interestingly, the extracellular juxtamembrane regions are highly divergent with the Aβ sequence being unique for APP. The E1 domain can be further subdivided into a heparin-binding/growth factor-like domain and a metal (copper and zinc) binding domain. The E1 domain is followed by an acidic region and a Kunitz-type protease inhibitor (KPI) domain (that is subject to alternative splicing in both APP and APLP2). The E2 region contains a second heparin binding domain and a RERMS motif implicated in trophic functions (Ninomiya et al. 1993; Roch et al. 1994). APP family proteins are posttranslationally modified including N- and O-glycosylation, sialylation, and CS GAG modification of the ectodomain and are phosphorylated at multiple sites within the intracellular carboxy-terminal domain (reviewed in Suzuki and Nakaya 2008; Jacobsen and Iverfeldt 2009). Crystal structures of several subdomains (reviewed in Reinhard et al. 2005; Gralle and Ferreira 2007), including the recently determined complete E1 structure (Dahms et al. 2010) and AICD bound to the adaptor protein Fe65 are available (Radzimanowski et al. 2010). Membrane bound APP/APLP holoproteins resemble cell surface receptors and have been shown to bind to extracellular matrix components (see below), but also interact with cell surface proteins including Alcadein (Araki et al. 2003), F-spondin (Ho and Südhof 2004), Reelin (Hoe et al. 2009b), LRP1 (Pietrzik et al. 2004), sorL1/LR11 (Schmidt et al. 2007), Nogo-66 receptor (Park et al. 2006), Notch2 (Chen et al. 2006), and Netrin (Lourenco et al. 2009). Although several of these interactions regulate APP processing, the physiological relevance of these interactions is poorly understood. Interaction screens have led to the identification of multiple intracellular binding partners (reviewed in Jacobsen and Iverfeldt 2009). Notably, the YENPTY motif that is conserved from C. elegans to mammalian APP/APLPs, confers clathrin mediated endocytosis, modulates Aβ generation (Perez et al. 1999; Ring et al. 2007), and binds several kinases, as well as adaptor proteins including mDab1, JIP, Shc, Grb2, Numb, X11/mint family, and Fe65 family proteins. Although in vitro studies have shown that these interactions may not only modulate APP processing but may also mediate cell signaling, the in vivo relevance is only starting to be revealed.
Although APLP1 and APLP2 lack the Aβ region they are similarly processed. Both APLPs undergo ectodoamin shedding and soluble APLPs have been detected in conditioned medium of transfected cell lines or human cerebrospinal fluid (Slunt et al. 1994; Webster et al. 1995; Paliga et al. 1997). Likewise, p3/Aβ-like fragments (Eggert et al. 2004; Minogue et al. 2009), as well as APLP1 and APLP2 intracellular fragments (termed ALIDs) are generated in a γ-secretase dependent manner (Scheinfeld et al. 2002; Walsh et al. 2003). Whereas there has been robust evidence indicating that APLP2 is processed by α- and β-secretase (Eggert et al. 2004; Pastorino et al. 2004; Endres et al. 2005), APLP1 shedding appeared to be independent of BACE activity as it was not affected by BACE inhibitors (Eggert et al. 2004; Minogue et al. 2009). A recent study using BACE-KO and overexpressing mice showed, however, that BACE deficiency substantially reduces brain APLP1s levels and that ICDs of APP family members are released in the absence of BACE (Frigerio et al. 2010).
Expression, Subcellular Localization, and Axonal Transport
APP and APLP2 are expressed ubiquitously, with particularly high expression in neurons, in largely overlapping patterns during embryonic development and in adult tissue (Slunt et al. 1994; Lorent et al. 1995; Thinakaran et al. 1995). In contrast, APLP1 is found primarily in the nervous system (Lorent et al. 1995). Regarding their subcellular localization, APP/APLPs are found both in somata and dendrites as well as in axons (Yamazaki et al. 1995; Back et al. 2007; Hoe et al. 2009a). APP/APLP expression is up-regulated during neuronal maturation and differentiation, undergoes rapid anterograde transport, and is targeted in vesicles distinct from synaptophysin transport vesicles to synaptic sites (Koo et al. 1990; Sisodia et al. 1993; Kaether et al. 2000; Szodorai et al. 2009). The initial hypothesis that APP anchors these vesicles via its carboxyl terminus to kinesin (Kamal et al. 2001), has been broadly questioned (Tienari et al. 1996; Lazarov et al. 2005; Back et al. 2007). Using time-lapse microscopy, Szodorai recently showed unaltered velocity of APPΔCT-GFP transport and a requirement for Rab3A GTPase activity for vesicle assembly (Szodorai et al. 2009).
IN VITRO AND EX VIVO STUDIES OF APP
Cell and Synaptic Adhesion
Investigations of conserved domains support an adhesion property for all members of the APP family. The extracellular sequence of APP has been found to interact with various extracellular matrix components, such as heparin (Clarris et al. 1997; Mok et al. 1997), collagen type I (Beher et al. 1996), and laminin (Kibbey et al. 1993), indicating a role of APP in cell-matrix adhesion. Structural and functional studies also implicate a role of the APP extracellular domains in facilitating cell–cell adhesion through transcellular interactions. Of interest, X-ray analysis revealed that the E2 domain of APP could form antiparallel dimers (Wang and Ha 2004). Both Dahms et al. (2010) and Gralle et al. (2006) reported that heparin binding to the extracellular E1 or E2 domain induces APP/APP dimerization. Cell culture studies revealed that APP family members form homo- or heterotypic cis-dimers, mainly via the E1 domain and the GxxxG motif in the transmembrane domain (Kaden et al. 2008), and that cis-dimerization modulates γ-secretase cleavage (Richter et al. 2010). Trans-dimerization of APP family members can promote cell–cell adhesion (Soba et al. 2005). Using a primary neuron/HEK293 mixed culture assay, Wang et al. (2009) reported that transcellular APP/APP interaction induces presynaptic specializations in cocultured neurons. These studies identify APP proteins as a novel class of synaptic adhesion molecules (SAM) with shared biochemical properties as neurexins (NX)/neuroligins (NL), SynCAMs, and leucine-rich repeat transmembrane neuronal proteins (LRRTM) (Scheiffele et al. 2000; Biederer et al. 2002; Graf et al. 2004; Sara et al. 2005; Fogel et al. 2007; Linhoff et al. 2009). Like NX/NL and SynCAM-mediated synaptic adhesion in which extracellular sequences engage transsynaptic interactions and the intracellular domains recruit pre- or postsynaptic complexes (reviewed in Dalva et al. 2007), both the extracellular and intracellular domains of APP are required to mediate the synaptogenic activity. Consistent with Soba et al. (2005), the E1 domain plays a more active role in synaptic adhesion. Interestingly, the highly conserved GYENPTY sequence of the APP intracellular domain could form a tripartite complex with Munc 18 interacting protein (Mint/X11) and calcium/calmodulin-dependent serine protein kinase (CASK) similar to that of neurexin and SynCAM (Hata et al. 1996; Biederer and Südhof 2000; Biederer et al. 2002), and the SynCAM carboxy-terminal sequence could functionally replace the corresponding APP domain in the coculture assay (Wang et al. 2009), suggesting that the Mint/CASK complexes may be the common mediators for the different classes of synaptic adhesion proteins. Thus, the precise role of APP-mediated synaptic adhesion in central synapses, whether it involves interaction with other SAMs, and the relationship between APP-mediated synaptogenesis and synaptic dysfunction occurring in AD are interesting questions that warrant further investigation.
Besides a direct role of APP/APP interaction in cell and synaptic adhesion, APP has been shown to colocalize with integrins on the surface of axons and at the sites of adhesion (Storey et al. 1996; Yamazaki et al. 1997; Young-Pearse et al. 2008). It has also been reported to interact with other cell adhesion molecules including NCAM (Ashley et al. 2005), NgCAM (Osterfield et al. 2008), and TAG 1 (Ma et al. 2008). As such, APP may play a modulatory role through interacting with these cell adhesion molecules.
Neural and Synapto-Trophic Functions
A large body of evidence supports a trophic function of APP in neurons and synapses. Consistent with its expression pattern, deletion or reduction of APP is associated with impaired neuronal viability in vitro and reduced synaptic activity in vivo (Allinquant et al. 1995; Perez et al. 1997; Hérard et al. 2006). Hippocampal neurons deficient for APP (or APLPs) show initially reduced neurite outgrowth, whereas, after prolonged culture axons are elongated and neurite branching is reduced (Perez et al. 1997; Young-Pearse et al. 2008). However, it should be noted that studies using neuronal cultures derived from various APP/APLP1/APLP2 knockout combinations or obtained by differentiation of embryonic stem cells lacking APP family members failed to detect a requirement of APP proteins in either neuronal differentiation or survival (Heber et al. 2000; Bergmans et al. 2010).
The trophic activity of APP can be mediated by the full-length protein and likely involves the APP adhesion properties discussed above. In particular, binding of APP to extracellular proteoglycans has been suggested to play a role in inducing neurite outgrowth, and a peptide homologous to the APP heparin-binding domain blocked this effect (Small et al. 1994, 1999). Qiu et al. found that when APP-transfected CHO cells were used as a substrate for the growth of primary rat hippocampal neurons, increased surface APP expression stimulated short-term neuronal adhesion and longer-term neurite outgrowth (Qiu et al. 1995). Nevertheless, ample literature points to a potent role of the α-secretase processed soluble fragment (APPsα) in the growth promoting and neurotrophic activities. One of the earliest indications came from the observation that secreted APPs, through the “RERMS” motif in the E2 domain, promoted fibroblast proliferation (Saitoh et al. 1989; Ninomiya et al. 1993a; Jin et al. 1994). Moderate overexpression of APP in transgenic mice, infusion of APPsα or the RERMS pentapeptide into the ventricle, or an indirect increase of APPsα levels because of overexpression of α-secretase, has been shown to increase synaptic density (Mucke et al. 1994; Roch et al. 1994; Meziane et al. 1998; Bell et al. 2008). Moreover, gain- or loss-of-function studies with either intraventricular APPsα infusion, down-regulation by antibody infusion or pharmacological inhibition of α-secretase coherently showed a function for APPsα in spatial memory and for LTP (Turner et al. 2003; Taylor et al. 2008). Caille et al. provided evidence that APPsα and APLP2s act as cofactors for epidermal growth factor (EGF) to stimulate the proliferation of neurosphere cultures in vitro and neural stem cells in the subventricular zone of adult rodent brain in vivo (Caille et al. 2004). Gakhar-Koppole et al. (2008) and Rohe et al. (2008) also reported that APPs stimulated neurogenesis and neurite outgrowth, but suggested that it is mediated through enhanced ERK phosphorylation and may be dependent on membrane-bound APP. Han et al. (2005) offered yet a different mechanism that the growth promoting property is mediated by the ability of APPsα to down-regulate CDK5 and inhibit τ hyperphosphorylation. Of direct physiological relevance, growth and neuronal phenotypes reported in APP deficient mice were shown to be fully restored by expressing only APPsα (Ring et al. 2007), and the lethality of the C. elegans apl-1 null mutant can be rescued by expressing only the APL-1 extracellular domain (Hornsten et al. 2007; Wiese et al. 2010).
Axon Pruning and Degeneration
APPsα has shown synaptotrophic and neuroprotective functions, whereas APPsβ was reported to be much less active or may even be toxic (reviewed in Turner et al. 2003). Recently, employing organotypic slice cultures, Copanaki et al. showed that APPsα (and not APPsβ) antagonizes dendritic degeneration and neuron death triggered by proteasomal stress (Copanaki et al. 2010). The most striking difference came from the study of Nikolaev et al. (2009), which reported that soluble APPsβ, but not APPsα, undergoes further cleavage to produce an amino-terminal ∼35 kDa APP derivative (N-APP), which in turn binds to the death receptor DR6 and mediates axon pruning and degeneration under trophic withdrawal conditions. The investigators attempted to link this pathway to both axonal pruning during normal development and axon- and neurodegeneration in AD. The APPsβ isoform specific cleavage and the differential, or opposite activities between APPsα and APPsβ, are intriguing as there is only 17 amino acids differences between the two isoforms.
Intracellular Signaling
Besides the γ-secretase cleavage that yields Aβ40 and Aβ42, PS-dependent proteolysis also occurs at the ε-site of the membrane-intracellular boundary to generate AICD (Sastre et al. 2001; Weidemann et al. 2002; Zhao et al. 2005). This cleavage is highly reminiscent of the PS-mediated release of the Notch intracellular domain (NICD) obligatory for Notch signaling (reviewed in Selkoe and Kopan 2003). Accordingly, AICD has been shown to translocate to the nucleus (Cupers et al. 2001; Gao and Pimplikar 2001; Kimberly et al. 2001). AICD is labile but can be stabilized by Fe65 (Kimberly et al. 2001). Using a heterologous reporter system, AICD was shown to form a transcriptionally active complex with Fe65 and the chromatin-remodeling factor Tip60 (Cao and Südhof 2001; Gao and Pimplikar 2001). However, subsequent analyses painted a more complex picture: (1) Follow-up studies by Cao et al. provided a modified model, whereby Fe65 is first recruited to the membrane-anchored APP where it is activated through an unknown mechanism. γ-secretase cleavage then releases Fe65 together with AICD, thereby allowing Fe65 to enter the nucleus and to interact with Tip60 (Cao and Südhof 2004); (2) Hass and Yankner revealed that PS-dependent AICD production is not required for the APP signaling activity as it proceeds normally in PS null cells and on PS inhibitor treatment (Hass and Yankner 2005). Instead, the investigators provided an alternative pathway involving Tip60 phosphorylation; (3) a later report documented that the proposed signaling activity is, in fact, executed by Fe65 independently of APP (Yang et al. 2006). Last, the link of Fe65 to chromatin remodeling instead of transcription suggests that APP may not act on specific genes, but rather modulates the overall transcriptional state of a cell (Giliberto et al. 2008).
Regardless of the molecular mechanisms, a trans-activating role of the APP/Fe65/Tip60 complex has been consistently documented, at least in overexpression systems using artificial reporter constructs. Accordingly, effort has been taken to identify the downstream targets, which reportedly include KAI (Baek et al. 2002), GSK3β (Kim et al. 2003; Ryan and Pimplikar 2005), neprilysin (Pardossi-Piquard et al. 2005), EGFR (Zhang et al. 2007), p53 (Checler et al. 2007), LRP (Liu et al. 2007), APP itself (von Rotz et al. 2004), and genes involved in calcium regulation (Leissring et al. 2002) and cytoskeletal dynamics (Müller et al. 2007). However, the validity of these proposed targets have been either questioned or disputed (Hebert et al. 2006; Yang et al. 2006; Chen and Selkoe 2007; Repetto et al. 2007; Giliberto et al. 2008; Tamboli et al. 2008; Waldron et al. 2008; Aydin et al. 2011). Overall, as attractive as the APP/AICD signaling model is, and regardless of the intense effort devoted to this topic in the past 10 years, neither the molecular pathways nor the downstream targets have been unambiguously established.
Apoptosis
Interestingly, AICD has been shown to be further cleaved by caspases at amino acid 664 of APP (695 numbering) to release two smaller fragments, Jcasp and C31; the latter contains the last 31 amino acids of APP and has been proposed to mediate cytoxicity in a full-length APP dependent manner (Bertrand et al. 2001; Lu et al. 2003; Park et al. 2009). In support of a functional role of this pathway, neuronal cultures generated from AICD transgenic mice are found to be more susceptible to toxic stimuli (Giliberto et al. 2008), and impaired synaptic plasticity and learning and memory seen in APP transgenic models were corrected in a mouse line in which the caspase site was mutated despite the presence of abundant amyloid pathology (Galvan et al. 2006). However, a more recent publication challenged these findings (Harris et al. 2010), and the physiological significance of this cleavage event thus requires further investigation.
IN VIVO LOSS-OF-FUNCTION STUDIES OF APP FAMILY PROTEINS
C. elegans and Drosophila
Drosophila deficient for the single APPL gene are viable, show a defect in fast phototaxis (Luo et al. 1992), and reduced synaptic bouton numbers at the neuromuscular junction NMJ. This activity involves a complex between APPL, the cell adhesion molecule fasciclin and Drosophila Mint/X11 (Torroja et al. 1999; Ashley et al. 2005). Knockout of the C. elegans ortholog APL-1, which is expressed in multiple tissue including neurons and muscle, disrupts molting and morphogenesis and results in laval lethality. Interestingly, this lethality could be rescued by neuronal expression of only the extracellular domain of APL-1, suggesting a key physiological role for this APPsα related fragment (Hornsten et al. 2007).
APP/APLP Single Knockout Mice
Three APP mouse mutants, one carrying a hypomorphic mutation of APP (APPΔ) (Müller et al. 1994) and two with complete deficiencies of APP (Zheng et al. 1995; Li et al. 1996) have been generated and revealed comparable phenotypes (Anliker and Müller 2006). APP-KO mice are viable and fertile, showing reduced body weight (about 15%–20% smaller) and brain weight (about 10% less) that was associated with reduced size of forebrain commissures and agenesis of the corpus callosum, consistent with a role of APP for neurite outgrowth and/or axonal pathfinding (Zheng et al. 1995; Magara et al. 1999). APP-KO mice also showed increased brain levels of copper (White et al. 1999), cholesterol and sphingolipid (Grimm et al. 2005). In addition, APP-KO animals showed hypersensitivity to kainate-induced seizures (Steinbach et al. 1998), suggesting a role of APP for neuronal excitation/inhibition balance. Behavioral studies revealed reduced locomotor and exploratory activity, altered circadian activity (Müller et al. 1994; Zheng et al. 1995; Ring et al. 2007), and a deficit in grip strength (Zheng et al. 1995; Ring et al. 2007), indicating compromised neuronal or muscular function (see also NMJ phenotype of double knockouts below). In the Morris water maze, APP-KO mice show impairments, both in learning and spatial memory, that are associated with a defect in long-term potentiation (LTP) (Dawson et al. 1999; Phinney et al. 1999; Seabrook et al. 1999; Ring et al. 2007). However, these impairments are not caused by a gross loss of neurons or synapses, as stereological quantification revealed normal neuron and synaptic bouton counts in the hippocampus of aged APP null mice (Phinney et al. 1999). Surprisingly, a recent study showed that APP deficiency leads to an increase in spine density in apical dendrites of cortical (layers 3 and 5) neurons (Bittner et al. 2009). The same group had previously reported an increase in synapse density in low-density cultures of self-innervating (autaptic) hippocampal neurons (Priller et al. 2006), but normal synaptic density in adult APP-KO mice (Priller et al. 2006). Thus, adaptive mechanisms (e.g., activity-dependent synaptic elimination) likely counteract early developmental changes. It remains to be seen whether alterations in spine density are also present in other brain areas of APP family KOs, which signaling pathways are involved, and how this may relate to functional changes.
Although basal glutamatergic synaptic transmission and paired pulse facilitation was unaffected in hippocampal slice recordings of APP-KO mice, a deficit in paired pulse depression of GABAergic IPSCs may contribute to the LTP defect of APP-KO mice (Seabrook et al. 1999). This may involve, as hypothesized (Seabrook et al. 1999), a reduction in feedback suppression mediated by presynaptic GABAB autoreceptors (but see below Yang et al. 2009). Although the molecular mechanisms of these alterations remain to be determined, these studies indicate that defects in Ca2+-handling, synaptic plasticity and/or neuronal network properties, rather than gross structural changes, cause functional impairments of APP knockout mice. Indeed, recently it was shown that APP is involved in the regulation of L-type Ca channel LTCCs level (Yang et al. 2009). APP-KO mice showed increased levels of CaV1.2 channels in the striatum that lead to alterations in GABAergic short term plasticity in striatal and hippocampal neurons, such as reduced GABAergic paired pulse inhibition and increased GABAergic posttetanic potentiation (Yang et al. 2009). Moreover, there is recent evidence from overexpression and APP knockdown studies in hippocampal neurons indicating an Aβ independent role of APP for the regulation of Ca2+-oscillations (Santos et al. 2009).
Combined Knockouts of APP Family Members
To test whether APLPs may functionally compensate for APP deficiency, mice lacking individual or all possible combinations of APP family proteins have been generated (reviewed in Anliker and Müller 2006; Zheng and Koo 2006). APLP1-KO mice revealed a somatic growth deficit as the only abnormality (Heber et al. 2000a), whereas, to date, no abnormalities have been found for APLP2-KO mice (von Koch et al. 1997). It should be kept in mind, however, that APLP deficient mice have not been examined in comparable detail as APP-KOs. In contrast to the subtle phenotypes of single mutants, double knockout mice (DKO) carrying APLP2/APLP1 and APLP2/APP-deficiencies proved lethal shortly after birth (von Koch et al. 1997; Heber et al. 2000). Surprisingly, APLP1/APP-deficient mice turned out to be viable, fertile, and without any additional abnormalities (Heber et al. 2000). These data indicated redundancy between APLP2 and both other family members, and corroborate a key physiological role for APLP2. None of the lethal double mutants, however, displayed obvious histopathological abnormalities (examined at the light microscopic level) in the brain. So far, the postnatal lethality of the APP/APLP2-DKO precluded the analysis of APP/APLP2 mediated functions in the postnatal and adult nervous system. However, organotypic hippocampal slice cultures can be studied in case of early postnatal lethality. Of note, using this technique, APP/APLP2-DKO mutants revealed defects in basal glutamatergic synaptic transmission that were absent in single mutants (Schrenk-Siemens et al. 2008). Thus, a more complete picture of APP/APLP function in the CNS will await the generation of brain specific conditional mutants.
In the peripheral nervous system, APP and APLP2 play a redundant and essential role for neuromuscular synapse formation and function, as diaphragm preparations from newborn APP/APLP2-DKO mice show excessive nerve growth, a widened endplate pattern, reduced apposition of pre- and postsynaptic components, and severely impaired (spontaneous and evoked) neurotransmission (Wang et al. 2005). Moreover, submandibular ganglia of APP/APLP2-DKO mice showed a reduction in active zone size, synaptic vesicle density, and number of docked vesicles (Yang et al. 2005) pointing to primarily presynaptic defects (but see conditional mutants). Thus, impaired function of the NMJ likely causes early postnatal lethality of combined mutants and defects in grip strength in APP single KOs. Indeed, subsequent analysis of neuromuscular transmission of APP-KO mice showed reduced paired pulse facilitation that was associated with an increase in asynchronous presynaptic transmitter release mediated by N- and L-type Ca2+ channels (Yang et al. 2007).
Triple KO mice lacking all three APP family members die shortly after birth. Unlike the DKO mutants, which did not display histological alterations in the brain, 80% of all triple knockouts showed cranial abnormalities (Herms et al. 2004). The majority of animals showed focal dysplasia resembling human type II lissencephaly and a partial loss of cortical Cajal-Retzius cells (Herms et al. 2004). Within affected areas, neuronal cells from the cortical plate migrated beyond their normal positions and protruded into the marginal zone and the subarachnoid space, indicating a critical role for APP family members in neuronal adhesion and/or positioning (Herms et al. 2004). Interestingly, a very similar phenotype was detected in mice lacking the APP interactors Fe65 and Fe65L1 (Guénette et al. 2006). These data suggest that APP family proteins may mediate some of their function(s) via an APP/Fe65 signaling complex. A role of APP family members in neuronal positioning/migration is further supported by acute in utero knockdown of APP (Young-Pearse et al. 2007) in rats using shRNA electroporation. In summary, these data corroborate an essential role of the APP gene family for normal brain development.
APP CONDITIONAL KNOCKOUT
Germline deletion of APP and APLP2 in mice results in a general impairment in pre- and postsynaptic patterning and a specific defect in presynaptic targeting of CHT (Wang et al. 2005, 2007). Conditional alleles of APP and APLP2 have been generated (Wang et al. 2009; Mallm et al. 2010). Consistent with the synaptic adhesion property of APP, deletion of APP (on a global APLP2-KO background) in either presynaptic motor neurons or in postsynaptic muscle was shown to lead to similar neuromuscular synapse defects (Wang et al. 2009). Interestingly, postsynaptic APP expression is required to mediate presynaptic CHT targeting and synaptic transmission, suggesting that transsynaptic APP/APP interaction is necessary in recruiting the presynaptic APP/CHT complex and cholinergic synaptic function. Whether APP modulates other synaptic processes through similar recruitment of synaptic proteins is an interesting question requiring further investigation.
In Vivo Defined Genetic Modifications of APP Proteins
The above knockout animals provide important information concerning the physiological functions of APP proteins, which may be executed either as a full-length protein or as various processing products. The creation of knockin alleles expressing defined proteolytic fragments of APP offers a powerful system to delineate the APP functional domains in vivo. In this regard, knockin mice that express α- or β-secretase processed soluble APP (APPsα or APPsβ) or membrane anchored APP containing mutations of the highly conserved carboxy-terminal sequences have been generated. These alleles are summarized in Figure 2 and will be discussed in this section.
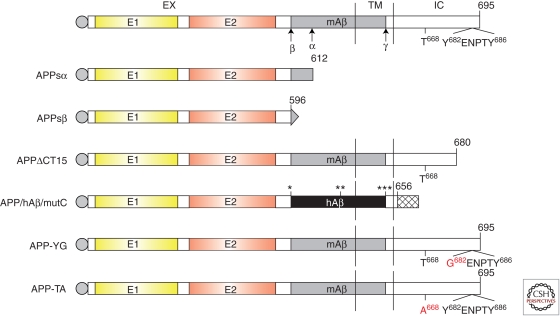
Figure 2.
Schematic representation of APP and its knock-in constructs (not drawn to scale). EX, TM, and IC stand for extracellular, transmembrane, and intracellular region, respectively. E1 and E2 domains are marked in yellow and orange, respectively. mAβ and hAβ represent mouse and human Aβ, respectively. β, α, and γ indicate the cleavage sites by β-, α-, and γ-secretase, respectively. *** represents signal peptide. *** symbolizes the FLAG tag. Residue T668, and YENPTY motif are labeled to illustrate the corresponding point mutations in APP-YG knock-in and APP-TA knock-in mice. *, Swedish mutation (K595M596-N595L596); **, Arctic mutation (E618-G618); ***, London mutation (V642-I618), which are introduced in the APP/hAβ/mutC knock-in allele. All residues are numbered according to APP695 numbering.
APPsα and APPsβ Knockin
Ring et al. (2007) created a strain of APPsα knockin mice by introducing a stop codon immediately after the α-secretase cleavage site. Interestingly, all of the phenotypes reported in APP deficient mice including body and brain weight deficits, grip strength deficits, alterations in locomotor activity, and impaired spatial learning and LTP have been shown to be fully restored by expressing only APPsα (Ring et al. 2007). Consistently, Taylor et al. (2008) showed a requirement for APPsα for in vivo LTP employing infusion of α-secretase inhibitor or recombinant APPsα, respectively. This crucial function of APPsα for synaptic plasticity and cognition is also of relevance for AD, as reduced CSF levels of APPsα and α-secretase ADAM10 are prominent features of sporadic AD cases (Lannfelt et al. 1995; Sennvik et al. 2000; Colciaghi et al. 2002; Tyler et al. 2002).
Li et al. (2010a) generated an APPsβ knockin allele that allows investigation of the stability and possible cleavage of APPsβ in the absence of APPsα. Contrary to Nikolaev (2009), the APPsβ protein was shown to be highly stable in vivo and does not undergo further cleavage under regular cell culture conditions in vitro. Crossing the APPsβ allele to APLP2 null background revealed that APPsβ failed to rescue the nerve sprouting phenotype of the APP/APLP2 null neuromuscular junction or early postnatal lethality (Li et al. 2010a). These data support the view that APPsβ exists as a stable protein and that the neuromuscular synapse defects present in APP/APLP2 null mice is not caused by the lack of APPsβ and, by extension, a defective APPsβ/DR6 pathway. However, when crossing the APPsα knockin allele (Ring et al. 2007) to an APLP2 null background, most of the combined mutants survived into adulthood (Weyer et al. 2011). These data suggest a distinct functional role of secreted APPsα sufficient to partially rescue the lethality of APP/APLP2-DKO mice, and revealed a synergistic role of both APP and APLP2 for hippocampal function and synaptic plasticity (Weyer et al. 2011).
Deletion or Mutation of the APP Intracellular Domain
Two APP carboxy-terminal deletion knockin mice have been reported. One deletes the last 15 amino acids of the APP sequence (APPΔCT15) (Ring et al. 2007); the other replaces mouse Aβ with the human Aβ sequence containing the Swedish, Arctic, and London FAD mutations and simultaneously deletes the last 39 residues of the APP sequence (APP/hAβ/mutC) (Li et al. 2010b). Crossing the APP/hAβ/mutC allele to APLP2 null background resulted in similar neuromuscular synapse defects and early postnatal lethality as in mice doubly deficient in APP and APLP2, supporting a functional role of the APP carboxy-terminal domain in these development activities. Nevertheless, Aβ production and amyloid pathology could proceed without the carboxy-terminal sequences (Li et al. 2010b). An essential role of the APP carboxy-terminal domain, specifically the YENPTY motif, in development was shown by the creation of APP knockin mice in which the Tyr682 residue of the Y682ENPTY sequence was changed to Gly (APPYG). Crossing the homozygous knockin mice on APLP2-KO background showed that the APPYG/YG/APLP2−/− mice show neuromuscular synapse deficits and early lethality similar to APP/APLP2 double KO mice (Barbagallo et al. 2011a). In sharp contrast, similar analysis of the knockin mice with mutation of the highly conserved Thr668 residue (APPTA) showed that this site is dispensable for the APP-mediated development function (Barbagallo et al. 2011b).
CONCLUDING REMARKS
Because of the central role of APP in AD pathogenesis, a great deal of effort has been devoted to understanding the biological functions of APP since its cloning in 1988. In vitro and in vivo studies have shown important activities of APP in various neuronal and synaptic processes, which can be executed either as a full-length protein or as one of the processing products. However, the underlying mechanisms remain largely undefined and often controversial. Key questions regarding whether APP is a receptor or a ligand or both, whether APP is by itself a signaling molecule or rather plays a secondary role in gene regulation, how APP function is coordinated between its full-length form and the proteolytic cleavage products and by its many intracellular binding partners awaits further investigation. The creation of the comprehensive panel of APP mouse strains including global inactivation, tissue-specific knockout and defined genetic modifications, combined with modern biological tools such as powerful large-scale experimentation and exciting neuroimaging technology, place us in an excellent position to address these questions.
ACKNOWLEDGMENTS
We thank Edith Gibson for editorial support and Hongmei Li for graphic assistance. The investigators’ work cited in this review was supported by grants from NIH (AG032051 and AG033467 to H.Z.), the American Health and Assistance Foundation (A2008-052 to H.Z.), the Deutsche Forschungsgemeinschaft (MU 1457/5-1 and MU 1457/8-1 to U.C.M.), and NGFNplus (01GS08128 to U.C.M.).
Footnotes
Editors: Dennis J. Selkoe, Eckhard Mandelkow, and David M. Holtzman
Additional Perspectives on The Biology of Alzheimer Disease available at www.perspectivesinmedicine.org
REFERENCES
*Reference is also in this collection.
- Allinquant B, Hantraye P, Mailleux P, Moya K, Bouillot C, Prochiantz A 1995. Downregulation of amyloid precursor protein inhibits neurite outgrowth in vitro. J Cell Biol 128: 919–927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anliker B, Müller U 2006. The functions of mammalian amyloid precursor protein and related amyloid precursor-like proteins. Neurodegener Dis 3: 239–246 [DOI] [PubMed] [Google Scholar]
- Araki Y, Tomita S, Yamaguchi H, Miyagi N, Sumioka A, Kirino Y, Suzuki T 2003. Novel cadherin-related membrane proteins, Alcadeins, enhance the X11-like protein-mediated stabilization of amyloid β-protein precursor metabolism. J Biol Chem 278: 49448–49458 [DOI] [PubMed] [Google Scholar]
- Ashley J, Packard M, Ataman B, Budnik V 2005. Fasciclin II signals new synapse formation through amyloid precursor protein and the scaffolding protein dX11/Mint. J Neurosci 25: 5943–5955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aydin D, Filippov MA, Tschäpe JA, Gretz N, Prinz M, Eils R, Brors B, Müller UC 2011. Comparative transcriptome profiling of amyloid precursor protein family members in the adult cortex. BMC Genomics 12: 160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Back S, Haas P, Tschape JA, Gruebl T, Kirsch J, Müller U, Beyreuther K, Kins S 2007. β-amyloid precursor protein can be transported independent of any sorting signal to the axonal and dendritic compartment. J Neurosci Res 85: 2580–2590 [DOI] [PubMed] [Google Scholar]
- Baek SH, Ohgi KA, Rose DW, Koo EH, Glass CK, Rosenfeld MG 2002. Exchange of N-CoR corepressor and Tip60 coactivator complexes links gene expression by NF-κB and β-amyloid precursor protein. Cell 110: 55–67 [DOI] [PubMed] [Google Scholar]
- Barbagallo APM, Wang Z, Zheng H, D’Adamio L 2011a. A single tyrosine residue in the amyloid precursor protein intracellular domain is essential for developmental function. J Biol Chem (in press) (JBC/2011/219837) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbagallo APM, Wang Z, Zheng H, D’Adamio L 2011b. The intracellular threonine of amyloid precursor protein that is essential for docking of Pin1 is dispensable for developmental function. PLoS ONE (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beher D, Hesse L, Masters CL, Multhaup G 1996. Regulation of amyloid protein precursor (APP) binding to collagen and mapping of the binding sites on APP and collagen type I. J Biol Chem 271: 1613–1620 [DOI] [PubMed] [Google Scholar]
- Bell KF, Zheng L, Fahrenholz F, Cuello AC 2008. ADAM-10 over-expression increases cortical synaptogenesis. Neurobiol Aging 29: 554–565 [DOI] [PubMed] [Google Scholar]
- Bergmans BA, Shariati SA, Habets RL, Verstreken P, Schoonjans L, Müller U, Dotti CG, De Strooper B 2010. Neurons generated from APP/APLP1/APLP2 triple knockout embryonic stem cells behave normally in vitro and in vivo: Lack of evidence for a cell autonomous role of the amyloid precursor protein in neuronal differentiation. Stem Cells 28: 399–406 [DOI] [PubMed] [Google Scholar]
- Bertrand E, Brouillet E, Caille I, Bouillot C, Cole GM, Prochiantz A, Allinquant B 2001. A short cytoplasmic domain of the amyloid precursor protein induces apoptosis in vitro and in vivo. Mol Cell Neurosci 18: 503–511 [DOI] [PubMed] [Google Scholar]
- Biederer T, Südhof TC 2000. Mints as adaptors. Direct binding to neurexins and recruitment of munc18. J Biol Chem 275: 39803–39806 [DOI] [PubMed] [Google Scholar]
- Biederer T, Sara Y, Mozhayeva M, Atasoy D, Liu X, Kavalali ET, Südhof TC 2002. SynCAM, a synaptic adhesion molecule that drives synapse assembly. Science 297: 1525–1531 [DOI] [PubMed] [Google Scholar]
- Bittner T, Fuhrmann M, Burgold S, Jung CK, Volbracht C, Steiner H, Mitteregger G, Kretzschmar HA, Haass C, Herms J 2009. γ-Secretase inhibition reduces spine density in vivo via an amyloid precursor protein-dependent pathway. J Neurosci 29: 10405–10409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caille I, Allinquant B, Dupont E, Bouillot C, Langer A, Müller U, Prochiantz A 2004. Soluble form of amyloid precursor protein regulates proliferation of progenitors in the adult subventricular zone. Development 131: 2173–2181 Epub 2004 Apr 8 [DOI] [PubMed] [Google Scholar]
- Cao X, Südhof TC 2001. A transcriptionally [correction of transcriptively] active complex of APP with Fe65 and histone acetyltransferase Tip60. Science 293: 115–120 [DOI] [PubMed] [Google Scholar]
- Cao X, Südhof TC 2004. Dissection of amyloid-β precursor protein-dependent transcriptional transactivation. J Biol Chem 279: 24601–24611 Epub 22004 Mar 24 [DOI] [PubMed] [Google Scholar]
- Checler F, Sunyach C, Pardossi-Piquard R, Sevalle J, Vincent B, Kawarai T, Girardot N, St George-Hyslop P, da Costa CA 2007. The γ/ε-secretase-derived APP intracellular domain fragments regulate 53. Curr Alzheimer Res 4: 423–426 [DOI] [PubMed] [Google Scholar]
- Chen AC, Selkoe DJ 2007. Response to: Pardossi-Piquard et al., “Presenilin-dependent transcriptional control of the Aβ-degrading enzyme neprilysin by intracellular domains of βAPP and APLP.” Neuron 46, 541–554. Neuron 53: 479–483 [DOI] [PubMed] [Google Scholar]
- Chen F, Hasegawa H, Schmitt-Ulms G, Kawarai T, Bohm C, Katayama T, Gu Y, Sanjo N, Glista M, Rogaeva E, et al. 2006. TMP21 is a presenilin complex component that modulates γ-secretase but not ε-secretase activity. Nature 440: 1208–1212 [DOI] [PubMed] [Google Scholar]
- Clarris HJ, Cappai R, Heffernan D, Beyreuther K, Masters CL, Small DH 1997. Identification of heparin-binding domains in the amyloid precursor protein of Alzheimer’s disease by deletion mutagenesis and peptide mapping. J Neurochem 68: 1164–1172 [DOI] [PubMed] [Google Scholar]
- Colciaghi F, Borroni B, Pastorino L, Marcello E, Zimmermann M, Cattabeni F, Padovani A, Di Luca M 2002. [α]-Secretase ADAM10 as well as [α]APPs is reduced in platelets and CSF of Alzheimer disease patients. Mol Med 8: 67–74 [PMC free article] [PubMed] [Google Scholar]
- Copanaki E, Chang S, Vlachos A, Tschape JA, Müller UC, Kogel D, Deller T 2010. sAPPα antagonizes dendritic degeneration and neuron death triggered by proteasomal stress. Mol Cell Neurosci 44: 386–393 [DOI] [PubMed] [Google Scholar]
- Cupers P, Orlans I, Craessaerts K, Annaert W, De Strooper B 2001. The amyloid precursor protein (APP)-cytoplasmic fragment generated by γ-secretase is rapidly degraded but distributes partially in a nuclear fraction of neurones in culture. J Neurochem 78: 1168–1178 [DOI] [PubMed] [Google Scholar]
- Dahms SO, Hoefgen S, Roeser D, Schlott B, Guhrs KH, Than ME 2010. Structure and biochemical analysis of the heparin-induced E1 dimer of the amyloid precursor protein. Proc Natl Acad Sci 107: 5381–5386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daigle I, Li C 1993. apl-1, a Caenorhabditis elegans gene encoding a protein related to the human β-amyloid protein precursor. Proc Natl Acad Sci 90: 12045–12049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalva MB, McClelland AC, Kayser MS 2007. Cell adhesion molecules: Signalling functions at the synapse. Nat Rev Neurosci 8: 206–220 Epub 2007 Feb 14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson GR, Seabrook GR, Zheng H, Smith DW, Graham S, O’Dowd G, Bowery BJ, Boyce S, Trumbauer ME, Chen HY, et al. 1999. Age-related cognitive deficits, impaired long-term potentiation and reduction in synaptic marker density in mice lacking the β-amyloid precursor protein. Neuroscience 90: 1–13 [DOI] [PubMed] [Google Scholar]
- *.De Strooper B, Iwatsubo T, Wolfe MS 2011. Presenilins and γ-secretase: Structure, function and role in Alzheimer disease. Cold Spring Harb Perspect Med 10.1101/cshperspect.a006304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eggert S, Paliga K, Soba P, Evin G, Masters CL, Weidemann A, Beyreuther K 2004. The proteolytic processing of the amyloid precursor protein gene family members APLP-1 and APLP-2 involves α-, β-, γ-, and ε-like cleavages: Modulation of APLP-1 processing by n-glycosylation. J Biol Chem 279: 18146–18156 [DOI] [PubMed] [Google Scholar]
- Endres K, Postina R, Schroeder A, Mueller U, Fahrenholz F 2005. Shedding of the amyloid precursor protein-like protein APLP2 by disintegrin-metalloproteinases. FEBS J 272: 5808–5820 [DOI] [PubMed] [Google Scholar]
- Fogel AI, Akins MR, Krupp AJ, Stagi M, Stein V, Biederer T 2007. SynCAMs organize synapses through heterophilic adhesion. J Neurosci 27: 12516–12530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frigerio CS, Fadeeva JV, Minogue AM, Citron M, Van Leuven F, Staufenbiel M, Paganetti P, Selkoe DJ, Walsh DM 2010. β-Secretase cleavage is not required for generation of the intracellular C-terminal domain of the amyloid precursor family of proteins. FEBS J 277: 1503–1518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gakhar-Koppole N, Hundeshagen P, Mandl C, Weyer SW, Allinquant B, Müller U, Ciccolini F 2008. Activity requires soluble amyloid precursor protein α to promote neurite outgrowth in neural stem cell-derived neurons via activation of the MAPK pathway. Eur J Neurosci 28: 871–882 [DOI] [PubMed] [Google Scholar]
- Galvan V, Gorostiza OF, Banwait S, Ataie M, Logvinova AV, Sitaraman S, Carlson E, Sagi SA, Chevallier N, Jin K, et al. 2006. Reversal of Alzheimer’s-like pathology and behavior in human APP transgenic mice by mutation of Asp664. Proc Natl Acad Sci 103: 7130–7135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y, Pimplikar SW 2001. The γ-secretase-cleaved C-terminal fragment of amyloid precursor protein mediates signaling to the nucleus. Proc Natl Acad Sci 98: 14979–14984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giliberto L, Zhou D, Weldon R, Tamagno E, De Luca P, Tabaton M, D’Adamio L 2008. Evidence that the amyloid β precursor protein-intracellular domain lowers the stress threshold of neurons and has a “regulated” transcriptional role. Mol Neurodegener 3: 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldgaber D, Lerman MI, McBride OW, Saffiotti U, Gajdusek DC 1987. Characterization and chromosomal localization of a cDNA encoding brain amyloid of Alzheimer’s disease. Science 235: 877–880 [DOI] [PubMed] [Google Scholar]
- Graf ER, Zhang X, Jin SX, Linhoff MW, Craig AM 2004. Neurexins induce differentiation of GABA and glutamate postsynaptic specializations via neuroligins. Cell 119: 1013–1026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gralle M, Ferreira ST 2007. Structure and functions of the human amyloid precursor protein: The whole is more than the sum of its parts. Prog Neurobiol 82: 11–32 [DOI] [PubMed] [Google Scholar]
- Gralle M, Oliveira CL, Guerreiro LH, McKinstry WJ, Galatis D, Masters CL, Cappai R, Parker MW, Ramos CH, Torriani I, et al. 2006. Solution conformation and heparin-induced dimerization of the full-length extracellular domain of the human amyloid precursor protein. J Mol Biol 357: 493–508 [DOI] [PubMed] [Google Scholar]
- Grimm MO, Grimm HS, Patzold AJ, Zinser EG, Halonen R, Duering M, Tschape JA, De Strooper B, Müller U, Shen J, et al. 2005. Regulation of cholesterol and sphingomyelin metabolism by amyloid-β and presenilin. Nat Cell Biol 7: 1118–1123 [DOI] [PubMed] [Google Scholar]
- Guénette S, Chang Y, Hiesberger T, Richardson JA, Eckman CB, Eckman EA, Hammer RE, Herz J 2006. Essential roles for the FE65 amyloid precursor protein-interacting proteins in brain development. EMBO J 25: 420–431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haass C, Schlossmacher MG, Hung AY, Vigo-Pelfrey C, Mellon A, Ostaszewski BL, Lieberburg I, Koo EH, Schenk D, Teplow DB, et al. 1992. Amyloid β-peptide is produced by cultured cells during normal metabolism. Nature 359: 322–325 [DOI] [PubMed] [Google Scholar]
- Han P, Dou F, Li F, Zhang X, Zhang YW, Zheng H, Lipton SA, Xu H, Liao FF 2005. Suppression of cyclin-dependent kinase 5 activation by amyloid precursor protein: A novel excitoprotective mechanism involving modulation of τ phosphorylation. J Neurosci 25: 11542–11552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris J, Devidze N, Halabisky B, Lo I, Thwin MT, Yu GQ, Bredesen DE, Masliah E, Mucke L 2010. Many neuronal and behavioral impairments in transgenic mouse models of Alzheimer’s disease are independent of caspase cleavage of the amyloid precursor protein. J Neurosci 30: 372–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hass MR, Yankner BA 2005. A {γ}-secretase-independent mechanism of signal transduction by the amyloid precursor protein. J Biol Chem 280: 36895–36904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- *.Haass C 2011. Trafficking and proteolytic processing of APP. Cold Spring Harb Perspect Med 10.1101/cshperspect.a006270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hata Y, Butz S, Südhof TC 1996. CASK: A novel dlg/PSD95 homolog with an N-terminal calmodulin-dependent protein kinase domain identified by interaction with neurexins. J Neurosci 16: 2488–2494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heber S, Herms J, Gajic V, Hainfellner J, Aguzzi A, Rulicke T, von Kretzschmar H, von Koch C, Sisodia S, Tremml P, et al. 2000. Mice with combined gene knock-outs reveal essential and partially redundant functions of amyloid precursor protein family members. J Neurosci 20: 7951–7963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebert SS, Serneels L, Tolia A, Craessaerts K, Derks C, Filippov MA, Müller U, De Strooper B 2006. Regulated intramembrane proteolysis of amyloid precursor protein and regulation of expression of putative target genes. EMBO Rep 7: 739–745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hérard AS, Besret L, Dubois A, Dauguet J, Delzescaux T, Hantraye P, Bonvento G, Moya KL 2006. siRNA targeted against amyloid precursor protein impairs synaptic activity in vivo. Neurobiol Aging 27: 1740–1750 [DOI] [PubMed] [Google Scholar]
- Herms J, Anliker B, Heber S, Ring S, Fuhrmann M, Kretzschmar H, Sisodia S, Müller U 2004. Cortical dysplasia resembling human type 2 lissencephaly in mice lacking all three APP family members. Embo J 23: 4106–4115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho A, Südhof TC 2004. Binding of F-spondin to amyloid-β precursor protein: A candidate amyloid-β precursor protein ligand that modulates amyloid-β precursor protein cleavage. Proc Natl Acad Sci 101: 2548–2553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoe HS, Fu Z, Makarova A, Lee JY, Lu C, Feng L, Pajoohesh-Ganji A, Matsuoka Y, Hyman BT, Ehlers MD, et al. 2009a. The effects of amyloid precursor protein on postsynaptic composition and activity. J Biol Chem 284: 8495–8506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoe HS, Lee KJ, Carney RS, Lee J, Markova A, Lee JY, Howell BW, Hyman BT, Pak DT, Bu G, et al. 2009b. Interaction of reelin with amyloid precursor protein promotes neurite outgrowth. J Neurosci 29: 7459–7473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornsten A, Lieberthal J, Fadia S, Malins R, Ha L, Xu X, Daigle I, Markowitz M, O’Connor G, Plasterk R, et al. 2007. APL-1, a Caenorhabditis elegans protein related to the human β-amyloid precursor protein, is essential for viability. Proc Natl Acad Sci 104: 1971–1976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobsen KT, Iverfeldt K 2009. Amyloid precursor protein and its homologues: A family of proteolysis-dependent receptors. Cell Mol Life Sci 66: 2299–2318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin LW, Ninomiya H, Roch JM, Schubert D, Masliah E, Otero DA, Saitoh T 1994. Peptides containing the RERMS sequence of amyloid β/A4 protein precursor bind cell surface and promote neurite extension. J Neurosci 14: 5461–5470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaden D, Munter LM, Joshi M, Treiber C, Weise C, Bethge T, Voigt P, Schaefer M, Beyermann M, Reif B, et al. 2008. Homophilic interactions of the amyloid precursor protein (APP) ectodomain are regulated by the loop region and affect β-secretase cleavage of APP. J Biol Chem 283: 7271–7279 [DOI] [PubMed] [Google Scholar]
- Kaether C, Skehel P, Dotti CG 2000. Axonal membrane proteins are transported in distinct carriers: A two-color video microscopy study in cultured hippocampal neurons. Mol Biol Cell 11: 1213–1224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamal A, Almenar-Queralt A, LeBlanc JF, Roberts EA, Goldstein LS 2001. Kinesin-mediated axonal transport of a membrane compartment containing β-secretase and presenilin-1 requires APP. Nature 414: 643–648 [DOI] [PubMed] [Google Scholar]
- Kang J, Lemaire HG, Unterbeck A, Salbaum JM, Masters CL, Grzeschik KH, Multhaup G, Beyreuther K, Müller-Hill B 1987. The precursor of Alzheimer’s disease amyloid A4 protein resembles a cell-surface receptor. Nature 325: 733–736 [DOI] [PubMed] [Google Scholar]
- Kibbey MC, Jucker M, Weeks BS, Neve RL, Van Nostrand WE, Kleinman HK 1993. β-Amyloid precursor protein binds to the neurite-promoting IKVAV site of laminin. Proc Natl Acad Sci 90: 10150–10153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HS, Kim EM, Lee JP, Park CH, Kim S, Seo JH, Chang KA, Yu E, Jeong SJ, Chong YH, et al. 2003. C-terminal fragments of amyloid precursor protein exert neurotoxicity by inducing glycogen synthase kinase-3β expression. FASEB J 17: 1951–1953 [DOI] [PubMed] [Google Scholar]
- Kimberly WT, Zheng JB, Guenette SY, Selkoe DJ 2001. The intracellular domain of the β-amyloid precursor protein is stabilized by Fe65 and translocates to the nucleus in a notch-like manner. J Biol Chem 276: 40288–40292 [DOI] [PubMed] [Google Scholar]
- Koo EH, Sisodia SS, Archer DR, Martin LJ, Weidemann A, Beyreuther K, Fischer P, Masters CL, Price DL 1990. Precursor of amyloid protein in Alzheimer disease undergoes fast anterograde axonal transport. Proc Natl Acad Sci 87: 1561–1565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn PH, Wang H, Dislich B, Colombo A, Zeitschel U, Ellwart JW, Kremmer E, Rossner S, Lichtenthaler SF 2010. ADAM10 is the physiologically relevant, constitutive α-secretase of the amyloid precursor protein in primary neurons. EMBO J 29: 3020–3032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lannfelt L, Basun H, Vigo-Pelfrey C, Wahlund LO, Winblad B, Lieberburg I, Schenk D 1995. Amyloid β-peptide in cerebrospinal fluid in individuals with the Swedish Alzheimer amyloid precursor protein mutation. Neurosci Lett 199: 203–206 [DOI] [PubMed] [Google Scholar]
- Lazarov O, Morfini GA, Lee EB, Farah MH, Szodorai A, DeBoer SR, Koliatsos VE, Kins S, Lee VM, Wong PC, et al. 2005. Axonal transport, amyloid precursor protein, kinesin-1, and the processing apparatus: Revisited. J Neurosci 25: 2386–2395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leissring MA, Murphy MP, Mead TR, Akbari Y, Sugarman MC, Jannatipour M, Anliker B, Müller U, Saftig P, De Strooper B, et al. 2002. A physiologic signaling role for the γ-secretase-derived intracellular fragment of APP. Proc Natl Acad Sci 99: 4697–4702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li ZW, Stark G, Gotz J, Rulicke T, Gschwind M, Huber G, Müller U, Weissmann C 1996. Generation of mice with a 200-kb amyloid precursor protein gene deletion by Cre recombinase-mediated site-specific recombination in embryonic stem cells. Proc Natl Acad Sci 93: 6158–6162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Wang B, Wang Z, Guo Q, Tabuchi K, Hammer RE, Südhof TC, Wang R, Zheng H 2010a. Soluble APP regulates transthyretin and Klotho gene expression without rescuing the essential function of APP. Proc Natl Acad Sci 107: 17362–17367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Wang Z, Wang B, Guo Q, Dolios G, Tabuchi K, Hammer RE, Südhof TC, Wang R, Zheng H 2010b. Genetic dissection of the amyloid precursor protein in developmental function and amyloid pathogenesis. J Biol Chem 285: 30598–30605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linhoff MW, Lauren J, Cassidy RM, Dobie FA, Takahashi H, Nygaard HB, Airaksinen MS, Strittmatter SM, Craig AM 2009. An unbiased expression screen for synaptogenic proteins identifies the LRRTM protein family as synaptic organizers. Neuron 61: 734–749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Zerbinatti CV, Zhang J, Hoe HS, Wang B, Cole SL, Herz J, Muglia L, Bu G 2007. Amyloid precursor protein regulates brain apolipoprotein E and cholesterol metabolism through lipoprotein receptor LRP1. Neuron 56: 66–78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorent K, Overbergh L, Moechars D, De Strooper B, Van Leuven F, Van den Berghe H 1995. Expression in mouse embryos and in adult mouse brain of three members of the amyloid precursor protein family, of the α-2-macroglobulin receptor/low density lipoprotein receptor-related protein and of its ligands apolipoprotein E, lipoprotein lipase, α-2-macroglobulin and the 40,000 molecular weight receptor-associated protein. Neuroscience 65: 1009–1025 [DOI] [PubMed] [Google Scholar]
- Lourenco FC, Galvan V, Fombonne J, Corset V, Llambi F, Müller U, Bredesen DE, Mehlen P 2009. Netrin-1 interacts with amyloid precursor protein and regulates amyloid-β production. Cell Death Differ 16: 655–663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu DC, Soriano S, Bredesen DE, Koo EH 2003. Caspase cleavage of the amyloid precursor protein modulates amyloid β-protein toxicity. J Neurochem 87: 733–741 [DOI] [PubMed] [Google Scholar]
- Luo LQ, Martin-Morris LE, White K 1990. Identification, secretion, and neural expression of APPL, a Drosophila protein similar to human amyloid protein precursor. J Neurosci 10: 3849–3861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo L, Tully T, White K 1992. Human amyloid precursor protein ameliorates behavioral deficit of flies deleted for Appl gene. Neuron 9: 595–605 [DOI] [PubMed] [Google Scholar]
- Ma QH, Futagawa T, Yang WL, Jiang XD, Zeng L, Takeda Y, Xu RX, Bagnard D, Schachner M, Furley AJ, et al. 2008. A TAG1-APP signalling pathway through Fe65 negatively modulates neurogenesis [see comment]. Nat Cell Biol 10: 283–294 [DOI] [PubMed] [Google Scholar]
- Magara F, Müller U, Li ZW, Lipp HP, Weissmann C, Stagljar M, Wolfer DP 1999. Genetic background changes the pattern of forebrain commissure defects in transgenic mice underexpressing the β-amyloid-precursor protein. Proc Natl Acad Sci 96: 4656–4661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallm JP, Tschape JA, Hick M, Filippov MA, Müller UC 2010. Generation of conditional null alleles for APP and APLP2. Genesis 48: 200–206 [DOI] [PubMed] [Google Scholar]
- Meziane H, Dodart JC, Mathis C, Little S, Clemens J, Paul SM, Ungerer A 1998. Memory-enhancing effects of secreted forms of the β-amyloid precursor protein in normal and amnestic mice. Proc Natl Acad Sci 95: 12683–12688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minogue AM, Stubbs AK, Frigerio CS, Boland B, Fadeeva JV, Tang J, Selkoe DJ, Walsh DM 2009. γ-secretase processing of APLP1 leads to the production of a p3-like peptide that does not aggregate and is not toxic to neurons. Brain Res 1262: 89–99 [DOI] [PubMed] [Google Scholar]
- Mok SS, Sberna G, Heffernan D, Cappai R, Galatis D, Clarris HJ, Sawyer WH, Beyreuther K, Masters CL, Small DH 1997. Expression and analysis of heparin-binding regions of the amyloid precursor protein of Alzheimer’s disease. FEBS Lett 415: 303–307 [DOI] [PubMed] [Google Scholar]
- Morales-Corraliza J, Mazzella MJ, Berger JD, Diaz NS, Choi JH, Levy E, Matsuoka Y, Planel E, Mathews PM 2009. In vivo turnover of τ and APP metabolites in the brains of wild-type and Tg2576 mice: Greater stability of sAPP in the β-amyloid depositing mice. PLoS One 4: e7134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mucke L, Masliah E, Johnson WB, Ruppe MD, Alford M, Rockenstein EM, Forss-Petter S, Pietropaolo M, Mallory M, Abraham CR 1994. Synaptotrophic effects of human amyloid β protein precursors in the cortex of transgenic mice. Brain Res 666: 151–167 [DOI] [PubMed] [Google Scholar]
- Müller U, Cristina N, Li ZW, Wolfer DP, Lipp HP, Rulicke T, Brandner S, Aguzzi A, Weissmann C 1994. Behavioral and anatomical deficits in mice homozygous for a modified β-amyloid precursor protein gene. Cell 79: 755–765 [DOI] [PubMed] [Google Scholar]
- Müller T, Concannon CG, Ward MW, Walsh CM, Tirniceriu AL, Tribl F, Kogel D, Prehn JH, Egensperger R 2007. Modulation of gene expression and cytoskeletal dynamics by the amyloid precursor protein intracellular domain (AICD). Mol Biol Cell 18: 201–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munter LM, Voigt P, Harmeier A, Kaden D, Gottschalk KE, Weise C, Pipkorn R, Schaefer M, Langosch D, Multhaup G 2007. GxxxG motifs within the amyloid precursor protein transmembrane sequence are critical for the etiology of Aβ42. EMBO J 26: 1702–1712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musa A, Lehrach H, Russo VA 2001. Distinct expression patterns of two zebrafish homologues of the human APP gene during embryonic development. Dev Genes Evol 211: 563–567 [DOI] [PubMed] [Google Scholar]
- Nikolaev A, McLaughlin T, O’Leary DDM, Tessier-Lavigne M 2009. APP binds DR6 to trigger axon pruning and neuron death via distinct caspases. Nature 457: 981–990 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Ninomiya H, Roch JM, Sundsmo MP, Otero DA, Saitoh T 1993. Amino acid sequence RERMS represents the active domain of amyloid β/A4 protein precursor that promotes fibroblast growth. J Cell Biol 121: 879–886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osterfield M, Egelund R, Young LM, Flanagan JG 2008. Interaction of amyloid precursor protein with contactins and NgCAM in the retinotectal system. Development 135: 1189–1199 [DOI] [PubMed] [Google Scholar]
- Paliga K, Peraus G, Kreger S, Durrwang U, Hesse L, Multhaup G, Masters CL, Beyreuther K, Weidemann A 1997. Human amyloid precursor-like protein 1–cDNA cloning, ectopic expression in COS-7 cells and identification of soluble forms in the cerebrospinal fluid. Eur J Biochem 250: 354–363 [DOI] [PubMed] [Google Scholar]
- Pardossi-Piquard R, Petit A, Kawarai T, Sunyach C, Alves da Costa C, Vincent B, Ring S, D’Adamio L, Shen J, Müller U, et al. 2005. Presenilin-dependent transcriptional control of the Aβ-degrading enzyme neprilysin by intracellular domains of βAPP and APLP. Neuron 46: 541–554 [DOI] [PubMed] [Google Scholar]
- Park JH, Gimbel DA, GrandPre T, Lee JK, Kim JE, Li W, Lee DH, Strittmatter SM 2006. Alzheimer precursor protein interaction with the Nogo-66 receptor reduces amyloid-β plaque deposition. J Neurosci 26: 1386–1395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SA, Shaked GM, Bredesen DE, Koo EH 2009. Mechanism of cytotoxicity mediated by the C31 fragment of the amyloid precursor protein. Biochem Biophys Res Commun 388: 450–455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastorino L, Ikin AF, Lamprianou S, Vacaresse N, Revelli JP, Platt K, Paganetti P, Mathews PM, Harroch S, Buxbaum JD 2004. BACE (β-secretase) modulates the processing of APLP2 in vivo. Mol Cell Neurosci 25: 642–649 [DOI] [PubMed] [Google Scholar]
- Perez RG, Zheng H, Van der Ploeg LH, Koo EH 1997. The β-amyloid precursor protein of Alzheimer’s disease enhances neuron viability and modulates neuronal polarity. J Neurosci 17: 9407–9414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez RG, Soriano S, Hayes JD, Ostaszewski B, Xia W, Selkoe DJ, Chen X, Stokin GB, Koo EH 1999. Mutagenesis identifies new signals for β-amyloid precursor protein endocytosis, turnover, and the generation of secreted fragments, including Aβ42. J Biol Chem 274: 18851–18856 [DOI] [PubMed] [Google Scholar]
- Phinney AL, Calhoun ME, Wolfer DP, Lipp HP, Zheng H, Jucker M 1999. No hippocampal neuron or synaptic bouton loss in learning-impaired aged β-amyloid precursor protein-null mice. Neuroscience 90: 1207–1216 [DOI] [PubMed] [Google Scholar]
- Pietrzik CU, Yoon IS, Jaeger S, Busse T, Weggen S, Koo EH 2004. FE65 constitutes the functional link between the low-density lipoprotein receptor-related protein and the amyloid precursor protein. J Neurosci 24: 4259–4265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Priller C, Bauer T, Mitteregger G, Krebs B, Kretzschmar HA, Herms J 2006. Synapse formation and function is modulated by the amyloid precursor protein. J Neurosci 26: 7212–7221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu WQ, Ferreira A, Miller C, Koo EH, Selkoe DJ 1995. Cell-surface β-amyloid precursor protein stimulates neurite outgrowth of hippocampal neurons in an isoform-dependent manner. J Neurosci 15: 2157–2167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radzimanowski J, Simon B, Sattler M, Beyreuther K, Sinning I, Wild K 2010. Structure of the intracellular domain of the amyloid precursor protein in complex with Fe65-PTB2. EMBO Rep 9: 1136–1140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinhard C, Hebert SS, De Strooper B 2005. The amyloid-β precursor protein: Integrating structure with biological function. EMBO J 24: 3996–4006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Repetto E, Yoon IS, Zheng H, Kang DE 2007. Presenilin 1 regulates epidermal growth factor receptor turnover and signaling in the endosomal-lysosomal pathway. J Biol Chem 282: 31504–31516 [DOI] [PubMed] [Google Scholar]
- Richter L, Munter LM, Ness J, Hildebrand PW, Dasari M, Unterreitmeier S, Bulic B, Beyermann M, Gust R, Reif B, et al. 2010. Amyloid β 42 peptide (Aβ42)-lowering compounds directly bind to Aβ and interfere with amyloid precursor protein (APP) transmembrane dimerization. Proc Natl Acad Sci 107: 14597–14602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ring S, Weyer SW, Kilian SB, Waldron E, Pietrzik CU, Filippov MA, Herms J, Buchholz C, Eckman CB, Korte M, et al. 2007. The secreted β-amyloid precursor protein ectodomain APPs α is sufficient to rescue the anatomical, behavioral, and electrophysiological abnormalities of APP-deficient mice. J Neurosci 27: 7817–7826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roch JM, Masliah E, Roch-Levecq AC, Sundsmo MP, Otero DA, Veinbergs I, Saitoh T 1994. Increase of synaptic density and memory retention by a peptide representing the trophic domain of the amyloid β/A4 protein precursor. Proc Natl Acad Sci 91: 7450–7454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohe M, Carlo AS, Breyhan H, Sporbert A, Militz D, Schmidt V, Wozny C, Harmeier A, Erdmann B, Bales KR, et al. 2008. Sortilin-related receptor with A-type repeats (SORLA) affects the amyloid precursor protein-dependent stimulation of ERK signaling and adult neurogenesis. J Biol Chem 283: 14826–14834 [DOI] [PubMed] [Google Scholar]
- Rosen DR, Martin-Morris L, Luo LQ, White K 1989. A Drosophila gene encoding a protein resembling the human β-amyloid protein precursor. Proc Natl Acad Sci 86: 2478–2482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan KA, Pimplikar SW 2005. Activation of GSK-3 and phosphorylation of CRMP2 in transgenic mice expressing APP intracellular domain. J Cell Biol 171: 327–335 Epub 2005 Oct 17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitoh T, Sundsmo M, Roch JM, Kimura N, Cole G, Schubert D, Oltersdorf T, Schenk DB 1989. Secreted form of amyloid β protein precursor is involved in the growth regulation of fibroblasts. Cell 58: 615–622 [DOI] [PubMed] [Google Scholar]
- Santos SF, Pierrot N, Morel N, Gailly P, Sindic C, Octave JN 2009. Expression of human amyloid precursor protein in rat cortical neurons inhibits calcium oscillations. J Neurosci 29: 4708–4718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sara Y, Biederer T, Atasoy D, Chubykin A, Mozhayeva MG, Südhof TC, Kavalali ET 2005. Selective capability of SynCAM and neuroligin for functional synapse assembly. J Neurosci 25: 260–270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sastre M, Steiner H, Fuchs K, Capell A, Multhaup G, Condron MM, Teplow DB, Haass C 2001. Presenilin-dependent γ-secretase processing of β-amyloid precursor protein at a site corresponding to the S3 cleavage of Notch. EMBO Rep 2: 835–841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheiffele P, Fan J, Choih J, Fetter R, Serafini T 2000. Neuroligin expressed in nonneuronal cells triggers presynaptic development in contacting axons. Cell 101: 657–669 [DOI] [PubMed] [Google Scholar]
- Scheinfeld MH, Ghersi E, Laky K, Fowlkes BJ, D’Adamio L 2002. Processing of β-amyloid precursor-like protein-1 and -2 by γ-secretase regulates transcription. J Biol Chem 277: 44195–44201 [DOI] [PubMed] [Google Scholar]
- Schmidt V, Sporbert A, Rohe M, Reimer T, Rehm A, Andersen OM, Willnow TE 2007. SorLA/LR11 regulates processing of amyloid precursor protein via interaction with adaptors GGA and PACS-1. J Biol Chem 282: 32956–32964 [DOI] [PubMed] [Google Scholar]
- Schrenk-Siemens K, Perez-Alcala S, Richter J, Lacroix E, Rahuel J, Korte M, Müller U, Barde YA, Bibel M 2008. Embryonic stem cell-derived neurons as a cellular system to study gene function: Lack of amyloid precursor proteins APP and APLP2 leads to defective synaptic transmission. Stem Cells 26: 2153–2163 [DOI] [PubMed] [Google Scholar]
- Seabrook GR, Smith DW, Bowery BJ, Easter A, Reynolds T, Fitzjohn SM, Morton RA, Zheng H, Dawson GR, Sirinathsinghji DJ, et al. 1999. Mechanisms contributing to the deficits in hippocampal synaptic plasticity in mice lacking amyloid precursor protein. Neuropharmacology 38: 349–359 [DOI] [PubMed] [Google Scholar]
- Selkoe D, Kopan R 2003. Notch and Presenilin: Regulated intramembrane proteolysis links development and degeneration. Annu Rev Neurosci 26: 565–597 [DOI] [PubMed] [Google Scholar]
- Sennvik K, Fastbom J, Blomberg M, Wahlund LO, Winblad B, Benedikz E 2000. Levels of α- and β-secretase cleaved amyloid precursor protein in the cerebrospinal fluid of Alzheimer’s disease patients. Neurosci Lett 278: 169–172 [DOI] [PubMed] [Google Scholar]
- Simons M, de Strooper B, Multhaup G, Tienari PJ, Dotti CG, Beyreuther K 1996. Amyloidogenic processing of the human amyloid precursor protein in primary cultures of rat hippocampal neurons. J Neurosci 16: 899–908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sisodia SS, Koo EH, Hoffman PN, Perry G, Price DL 1993. Identification and transport of full-length amyloid precursor proteins in rat peripheral nervous system. J Neurosci 13: 3136–3142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slunt HH, Thinakaran G, Von Koch C, Lo AC, Tanzi RE, Sisodia SS 1994. Expression of a ubiquitous, cross-reactive homologue of the mouse β-amyloid precursor protein (APP). J Biol Chem 269: 2637–2644 [PubMed] [Google Scholar]
- Small DH, Nurcombe V, Reed G, Clarris H, Moir R, Beyreuther K, Masters CL 1994. A heparin-binding domain in the amyloid protein precursor of Alzheimer’s disease is involved in the regulation of neurite outgrowth. J Neurosci 14: 2117–2127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Small DH, Clarris HL, Williamson TG, Reed G, Key B, Mok SS, Beyreuther K, Masters CL, Nurcombe V 1999. Neurite-outgrowth regulating functions of the amyloid protein precursor of Alzheimer’s disease. J Alzheimers Dis 1: 275–285 [DOI] [PubMed] [Google Scholar]
- Soba P, Eggert S, Wagner K, Zentgraf H, Siehl K, Kreger S, Lower A, Langer A, Merdes G, Paro R, et al. 2005. Homo- and heterodimerization of APP family members promotes intercellular adhesion. EMBO J 24: 3624–3634 Epub 2005 Sep 29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinbach JP, Müller U, Leist M, Li ZW, Nicotera P, Aguzzi A 1998. Hypersensitivity to seizures in β-amyloid precursor protein deficient mice. Cell Death Differ 5: 858–866 [DOI] [PubMed] [Google Scholar]
- Storey E, Spurck T, Pickett-Heaps J, Beyreuther K, Masters CL 1996. The amyloid precursor protein of Alzheimer’s disease is found on the surface of static but not activity motile portions of neurites. Brain Res 735: 59–66 [DOI] [PubMed] [Google Scholar]
- Suzuki T, Nakaya T 2008. Regulation of amyloid β-protein precursor by phosphorylation and protein interactions. J Biol Chem 283: 29633–29637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szodorai A, Kuan YH, Hunzelmann S, Engel U, Sakane A, Sasaki T, Takai Y, Kirsch J, Müller U, Beyreuther K, et al. 2009. APP anterograde transport requires Rab3A GTPase activity for assembly of the transport vesicle. J Neurosci 29: 14534–14544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamboli IY, Prager K, Thal DR, Thelen KM, Dewachter I, Pietrzik CU, St George-Hyslop P, Sisodia SS, De Strooper B, Heneka MT, et al. 2008. Loss of γ-secretase function impairs endocytosis of lipoprotein particles and membrane cholesterol homeostasis. J Neurosci 28: 12097–12106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanzi RE, Gusella JF, Watkins PC, Bruns GA, St George-Hyslop P, Van Keuren ML, Patterson D, Pagan S, Kurnit DM, Neve RL 1987. Amyloid β protein gene: cDNA, mRNA distribution, and genetic linkage near the Alzheimer locus. Science 235: 880–884 [DOI] [PubMed] [Google Scholar]
- Taylor CJ, Ireland DR, Ballagh I, Bourne K, Marechal NM, Turner PR, Bilkey DK, Tate WP, Abraham WC 2008. Endogenous secreted amyloid precursor protein-α regulates hippocampal NMDA receptor function, long-term potentiation and spatial memory. Neurobiol Dis 31: 250–260 [DOI] [PubMed] [Google Scholar]
- Thinakaran G, Kitt CA, Roskams AJ, Slunt HH, Masliah E, von Koch C, Ginsberg SD, Ronnett GV, Reed RR, Price DL, et al. 1995. Distribution of an APP homolog, APLP2, in the mouse olfactory system: A potential role for APLP2 in axogenesis. J Neurosci 15: 6314–6326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tienari PJ, De Strooper B, Ikonen E, Simons M, Weidemann A, Czech C, Hartmann T, Ida N, Multhaup G, Masters CL, et al. 1996. The β-amyloid domain is essential for axonal sorting of amyloid precursor protein. EMBO J 15: 5218–5229 [PMC free article] [PubMed] [Google Scholar]
- Torroja L, Packard M, Gorczyca M, White K, Budnik V 1999. The Drosophila β-amyloid precursor protein homolog promotes synapse differentiation at the neuromuscular junction. J Neurosci 19: 7793–7803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner PR, O’Connor K, Tate WP, Abraham WC 2003. Roles of amyloid precursor protein and its fragments in regulating neural activity, plasticity and memory. Prog Neurobiol 70: 1–32 [DOI] [PubMed] [Google Scholar]
- Tyler SJ, Dawbarn D, Wilcock GK, Allen SJ 2002. α- and β-secretase: Profound changes in Alzheimer’s disease. Biochem Biophys Res Commun 299: 373–376 [DOI] [PubMed] [Google Scholar]
- von Koch CS, Zheng H, Chen H, Trumbauer M, Thinakaran G, van der Ploeg LH, Price DL, Sisodia SS 1997. Generation of APLP2 KO mice and early postnatal lethality in APLP2/APP double KO mice. Neurobiol Aging 18: 661–669 [DOI] [PubMed] [Google Scholar]
- von Rotz RC, Kohli BM, Bosset J, Meier M, Suzuki T, Nitsch RM, Konietzko U 2004. The APP intracellular domain forms nuclear multiprotein complexes and regulates the transcription of its own precursor. J Cell Sci 117: 4435–4448 [DOI] [PubMed] [Google Scholar]
- Waldron E, Isbert S, Kern A, Jaeger S, Martin AM, Hebert SS, Behl C, Weggen S, De Strooper B, Pietrzik CU 2008. Increased AICD generation does not result in increased nuclear translocation or activation of target gene transcription. Exp Cell Res 314: 2419–2433 [DOI] [PubMed] [Google Scholar]
- Walsh DM, Fadeeva JV, LaVoie MJ, Paliga K, Eggert S, Kimberly WT, Wasco W, Selkoe DJ 2003. γ-Secretase cleavage and binding to FE65 regulate the nuclear translocation of the intracellular C-terminal domain (ICD) of the APP family of proteins. Biochemistry 42: 6664–6673 [DOI] [PubMed] [Google Scholar]
- Wang Y, Ha Y 2004. The X-ray structure of an antiparallel dimer of the human amyloid precursor protein E2 domain. Mol Cell 15: 343–353 [DOI] [PubMed] [Google Scholar]
- Wang P, Yang G, Mosier DR, Chang P, Zaidi T, Gong YD, Zhao NM, Dominguez B, Lee KF, Gan WB, et al. 2005. Defective neuromuscular synapses in mice lacking amyloid precursor protein (APP) and APP-Like protein 2. J Neurosci 25: 1219–1225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Yang L, Wang Z, Zheng H 2007. Amyolid precursor protein mediates presynaptic localization and activity of the high-affinity choline transporter. Proc Natl Acad Sci 104: 14140–14145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Wang B, Yang L, Guo Q, Aithmitti N, Songyang Z, Zheng H 2009. Presynaptic and postsynaptic interaction of the amyloid precursor protein promotes peripheral and central synaptogenesis. J Neurosci 29: 10788–10801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasco W, Bupp K, Magendantz M, Gusella JF, Tanzi RE, Solomon F 1992. Identification of a mouse brain cDNA that encodes a protein related to the Alzheimer disease-associated amyloid β protein precursor. Proc Natl Acad Sci 89: 10758–10762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasco W, Gurubhagavatula S, Paradis MD, Romano DM, Sisodia SS, Hyman BT, Neve RL, Tanzi RE 1993. Isolation and characterization of APLP2 encoding a homologue of the Alzheimer’s associated amyloid β protein precursor. Nat Genet 5: 95–100 [DOI] [PubMed] [Google Scholar]
- Webster MT, Groome N, Francis PT, Pearce BR, Sherriff FE, Thinakaran G, Felsenstein KM, Wasco W, Tanzi RE, Bowen DM 1995. A novel protein, amyloid precursor-like protein 2, is present in human brain, cerebrospinal fluid and conditioned media. Biochem J 310 (Pt 1): 95–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weidemann A, Eggert S, Reinhard FB, Vogel M, Paliga K, Baier G, Masters CL, Beyreuther K, Evin G 2002. A novel ε-cleavage within the transmembrane domain of the Alzheimer amyloid precursor protein demonstrates homology with Notch processing. Biochemistry 41: 2825–2835 [DOI] [PubMed] [Google Scholar]
- Weyer SA, Klevanski M, Delekate A, Voikar V, Aydin D, Hick M, Filippov MA, Drost N, Schaller KL, Saar M, et al. 2011. APP and APLP2 are essential at PNS and CNS synapses for transmission, spatial learning and LTP. EMBO J 30: 2266–2280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White AR, Reyes R, Mercer JF, Camakaris J, Zheng H, Bush AI, Multhaup G, Beyreuther K, Masters CL, Cappai R 1999. Copper levels are increased in the cerebral cortex and liver of APP and APLP2 knockout mice. Brain Res 842: 439–444 [DOI] [PubMed] [Google Scholar]
- Wiese M, Antebi A, Zheng H 2010. Intracellular trafficking and synaptic function of APL-1 in Caenorhabditis elegans. PLoS ONE 5: e12790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki T, Selkoe DJ, Koo EH 1995. Trafficking of cell surface β-amyloid precursor protein: Retrograde and transcytotic transport in cultured neurons. J Cell Biol 129: 431–442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki T, Koo EH, Selkoe DJ 1997. Cell surface amyloid β-protein precursor colocalizes with β 1 integrins at substrate contact sites in neural cells. J Neurosci 17: 1004–1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang G, Gong YD, Gong K, Jiang WL, Kwon E, Wang P, Zheng H, Zhang XF, Gan WB, Zhao NM 2005. Reduced synaptic vesicle density and active zone size in mice lacking amyloid precursor protein (APP) and APP-like protein 2. Neurosci Lett 384: 66–71 [DOI] [PubMed] [Google Scholar]
- Yang Z, Cool BH, Martin GM, Hu Q 2006. A dominant role for FE65 (APBB1) in nuclear signaling. J Biol Chem 281: 4207–4214 [DOI] [PubMed] [Google Scholar]
- Yang L, Wang B, Long C, Wu G, Zheng H 2007. Increased asynchronous release and aberrant calcium channel activation in amyloid precursor protein deficient neuromuscular synapses. Neuroscience 149: 768–778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Wang Z, Wang B, Justice NJ, Zheng H 2009. Amyloid precursor protein regulates Cav1.2 L-type calcium channel levels and function to influence GABAergic short-term plasticity. J Neurosci 29: 15660–15668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young-Pearse TL, Bai J, Chang R, Zheng JB, LoTurco JJ, Selkoe DJ 2007. A critical function for β-amyloid precursor protein in neuronal migration revealed by in utero RNA interference. J Neurosci 27: 14459–14469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young-Pearse TL, Chen AC, Chang R, Marquez C, Selkoe DJ 2008. Secreted APP regulates the function of full-length APP in neurite outgrowth through interaction with integrin β1. Neural Dev 3: 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang YW, Wang R, Liu Q, Zhang H, Liao FF, Xu H 2007. Presenilin/γ-secretase-dependent processing of β-amyloid precursor protein regulates EGF receptor expression. Proc Natl Acad Sci 104: 10613–10618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao G, Cui MZ, Mao G, Dong Y, Tan J, Sun L, Xu X 2005. {γ}-Cleavage is dependent on {ζ}-cleavage during the proteolytic processing of amyloid precursor protein within its transmembrane domain. J Biol Chem 280: 37689–37697 [DOI] [PubMed] [Google Scholar]
- Zheng H, Koo EH 2006. The amyloid precursor protein: Beyond amyloid. Mol Neurodegener 1: 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng H, Jiang M, Trumbauer ME, Sirinathsinghji DJ, Hopkins R, Smith DW, Heavens RP, Dawson GR, Boyce S, Conner MW, et al. 1995. β-Amyloid precursor protein-deficient mice show reactive gliosis and decreased locomotor activity. Cell 81: 525–531 [DOI] [PubMed] [Google Scholar]