Background: Bri2 is a substrate for intramembrane proteolysis by SPPL2b.
Results: Reducing the α-helical content of the Bri2 transmembrane domain increases its intramembrane cleavage by SPPL2b. Destabilization of the Bri2 transmembrane domain is predominantly mediated by Gly-60.
Conclusion: A single helix-destabilizing glycine residue of the transmembrane domain of Bri2 is sufficient for efficient intramembrane proteolysis by SPPL2b.
Significance: Understanding substrate requirements of intramembrane-cleaving proteases.
Keywords: Alzheimer Disease, Aspartic Protease, Intramembrane Proteolysis, Protease, Tumor Necrosis Factor (TNF), Bri2 (itm2b), Familial British Dementia, GXGD-type Aspartyl Proteases, Signal Peptide Peptidase (SPP), Signal Peptide Peptidase-like Protease (SPPL)
Abstract
Regulated intramembrane proteolysis is a widely accepted concept describing the processing of various transmembrane proteins via ectodomain shedding followed by an intramembrane cleavage. The resulting cleavage products can be involved in reverse signaling. Presenilins, which constitute the active center of the γ-secretase complex, signal peptide peptidase (SPP), and its homologues, the SPP-like (SPPL) proteases are members of the family of intramembrane-cleaving aspartyl proteases of the GXGD-type. We recently demonstrated that Bri2 (itm2b) is a substrate for regulated intramembrane proteolysis by SPPL2a and SPPL2b. Intramembrane cleavage of Bri2 is triggered by an initial shedding event catalyzed by A Disintegrin and Metalloprotease 10 (ADAM10). Additionally primary sequence determinants within the intracellular domain, the transmembrane domain and the luminal juxtamembrane domain are required for efficient cleavage of Bri2 by SPPL2b. Using mutagenesis and circular dichroism spectroscopy we now demonstrate that a high α-helical content of the Bri2 transmembrane domain (TMD) reduces cleavage efficiency of Bri2 by SPPL2b, while the presence of a GXXXG dimerization motif influences the intramembrane cleavage only to a minor extent. Surprisingly, only one of the four conserved intramembrane glycine residues significantly affects the secondary structure of the Bri2 TMD and thereby its intramembrane cleavage. Other glycine residues do not influence the α-helical content of the transmembrane domain nor its intramembrane processing.
Introduction
Intramembrane-cleaving proteases (I-CLiPs)3 are involved in a number of pivotal physiological and pathological processes, such as malaria, Alzheimer disease, cholesterol metabolism, Notch-signaling, removal of signal peptides, immune surveillance, and processing of the hepatitis C viral core protein (1). I-CLiPs trigger a variety of cellular pathways leading either to degradation of transmembrane domains or to the liberation of peptides, which in some cases are required for nuclear signaling (2). In most, but not all cases, I-CLiPs are part of a two-step proteolytic cascade known as regulated intramembrane proteolysis (RIP). The first step of RIP, commonly known as shedding, removes a large part of the substrates' ectodomain, which is secreted. In a subsequent step the remaining membrane-bound substrate is cleaved within its transmembrane domain (TMD) to produce an intracellular domain (ICD) and low molecular weight secreted peptides (2). Currently three classes of I-CLiPs have been identified, the intramembrane metalloproteases, represented by the site-2-protease (S2P), intramembrane serine proteases, represented by the large family of rhomboid proteases and finally the GXGD aspartyl proteases (2–4). The latter includes the presenilins (PS1 and PS2), which form the catalytic subunit of the γ-secretase complex (5), signal peptide peptidase (SPP) and its homologues, the SPP-like proteases (SPPLs) (6, 7). They all share the conserved GXGD-motif in TMD 7, which together with a YD-motif in TMD 6 and a PAL-motif in TMD 9, forms the catalytic center of these proteases (8–12). While γ-secretase exclusively accepts type-1-oriented proteins as substrates, SPP and SPPLs only cleave type-2 transmembrane proteins (6, 7, 13).
Numerous substrates for almost all known I-CLiP classes have now been described (1, 2). These I-CLiP substrates are generally TMDs of membrane proteins, which tend to adopt an α-helical conformation to minimize the energetically unfavorable exposure of their amino acid side chains to the hydrophobic core of lipid bilayers (14). Because their peptide bonds are hardly accessible to proteases, α-helical TMDs are expected to be poor protease substrates. To make TMDs susceptible to intramembrane proteolysis their helical content may be reduced by helix-destabilizing amino acids (15, 16). Amino acids with potential helix destabilizing properties in TMDs are in particular proline, glycine, asparagine, and serine (17). Consistent with this hypothesis, the substrates of S2P, rhomboids, and SPP have been shown to contain critical helix-destabilizing moieties within their TMDs (18–21). However, for rhomboid proteases it has been demonstrated recently, that additional primary structure elements within the juxtamembrane domain of the substrate determine the cleavage site, and are more strictly required than TMD helix-destabilizing residues (22). γ-Secretase substrates in contrast apparently do not contain obvious primary sequence elements required for their recognition (13) and γ-secretase is therefore believed to be a rather unselective “membrane proteasome” (23). However, for the β-amyloid precursor protein (APP), a major γ-secretase substrate, it has been shown that the luminal juxtamembrane domain (24) as well as a GXXXG dimerization motif within the TMD (25) affect substrate recognition and cleavage by γ-secretase (26). In line with these findings so far unknown determinants within a short luminal juxtamembrane sequence of the Bri2 protein as well as in the intracellular domain and the TMD are required for efficient intramembrane proteolysis by signal peptide peptidase-like 2b (SPPL2b) (27). To specify these determinants in more detail, we now investigated whether helix destabilizing amino acids in the TMD of Bri2 affect the efficiency of intramembrane proteolysis. We demonstrate that a single glycine residue in the N-terminal part of the Bri2 TMD (G60) is essential for the destabilization of the α-helical structure of the TMD and thus for its cleavage by SPPL2b. In contrast, the GXXXG motif affects intramembrane cleavage of Bri2 only to a minor extent. Consistent with these findings, Gly-60 is a major determinant of the secondary structure of the Bri2 TMD in lipid vesicles, while other potentially helix destabilizing residues within the TMD of Bri2 do not influence secondary structure and cleavage efficiency of Bri2.
EXPERIMENTAL PROCEDURES
Cell Culture, cDNAs, and Transfection
HEK293EBNA or HEK293 TR cells (Invitrogen GmbH) were cultured in DMEM with Glutamax (Invitrogen GmbH) supplemented with 10% fetal calf serum (Invitrogen GmbH) and 1% penicillin/streptomycin (Invitrogen GmbH) to the culture medium of HEK293 TR cells.
5 μg/ml Blasticidin (Invitrogen GmbH) was added. The cell lines expressing SPPL2b wt or the proteolytically inactive SPPL2b D421A (28) containing a C-terminal hemagglutinin (HA) tag (AYPYDVPDYA) as well as the Bri2ΔE construct (27) containing a N-terminal Flag tag (DYKDDDK) and a C-terminal V5 tag (GKPIPNPLLGLDST) to enable the detection of the Bri2 C-Peptide have been described before. Bri2 wt reflects Bri2ΔFC, which has been described before (27). Bri2 wt lacks the propeptide and carries an N-terminal Flag tag (DYKDDDK) and a C-terminal V5 tag (GKPIPNPLLGLDST) to detect Bri2 ICD species and the BRICHOS domain, respectively. The indicated mutations have been introduced using site-directed mutagenesis. Sequences of the respective oligonucleotides are available upon request. All cDNA constructs were sequenced for verification. Transient transfections of cells were carried out using Lipofectamine 2000 (Invitrogen GmbH) according to the manufacturer's instructions. To induce expression of the SPPL2b constructs, cells were incubated with 1 μg/ml doxycycline (BD Biosciences, San Jose) added to the cell culture medium for at least 48 h.
Antibodies, Immunoprecipitation, and Immunoblotting
The anti-HA-peroxidase coupled 3F10 antibody was obtained from Roche Diagnostics GmbH (Mannheim, Germany). The monoclonal anti-Flag M2 and the polyclonal HA 6908 antibody were obtained from Sigma, the poly- and monoclonal V5 antibodies were purchased from Chemicon (Schwalbach, Germany) and Invitrogen GmbH, respectively. Anti-mouse and anti-rabbit secondary antibodies were purchased from Promega. Immunoprecipitation assays were carried out as described previously (29). To detect Bri2 species in the cell lysate or BRICHOS in the conditioned media samples were separated on Tris-Tricine gels or 12% sodium dodecyl sulfate-polyacrylamide gels, respectively. Proteins were subsequently transferred to a polyvinylidene difluoride (PVDF) membrane (Immobilon, Millipore Corporation, Eschborn, Germany) and detected by using an enhanced chemiluminescence technique (ECL) Western blotting Detection Reagent (GE Healthcare, Little Chalfont, UK). For quantification, proteins were detected using the enhanced chemiluminescence technique (ECL) Plus Western blotting Detection System (GE Healthcare). The chemiluminescence signals of at least six independent experiments were measured with a CCD camera-based imaging system (ImageQuant LAS-4000 Fuji Film). Intensities of individual gels were normalized to one control experiment. Mean and S.D. of all individual experiments were determined. The relative signal intensity of ICD compared with Bri2 + Bri2 ICD of the respective sample was measured for the constructs indicated and set to 100% for Bri2 wt. Statistical significance was determined using Student's t test. Statistical significant p values <0.05, <0.005, or <0.0005 are represented by *, ** or ***, respectively.
Immunocytochemistry and Confocal Imaging
HEK293-EBNA cells were grown on polylysine-coated glass coverslips to 50–80% confluence. After transient transfection with the indicated Bri2 constructs, cells were fixed for 20 min with 3.7% paraformaldehyde in phosphate-buffered saline (PBS), permeabilized for 20 min with 0.2% Triton X-100 in PBS (were indicated) and blocked with 1% bovine serum albumin in PBS. Incubations with the monoclonal V5 antibody were performed for 1 h at room temperature in PBS followed by incubation with Alexa 488-coupled secondary antibodies (Molecular Probes). Mounted samples were analyzed with a Zeiss 510Meta confocal laser scanning microscope system equipped with a 100/1.3 objective described previously (30).
Vesicle Preparation and CD Spectrometry
The synthetic peptides covering the following sequences have been obtained from Peptide Specialty Laboratories GmbH, Heidelberg.
Bri2 wt: VGQRRAWCWCMCFGLAFMLAGVILGGAYLYKYFALQPDD (MW = 4432 Da); Bri2 G60A: VGQRRAWCWCMCFALAFMLAGVILGGAYLYKYFALQPDD (MW = 4446 Da); Bri2 G71A: VGQRRAWCWCMCFGLAFMLAGVILAGAYLYKYFALQPDD (MW = 4446 Da); Bri2 G72A: VGQRRAWCWCMCFGLAFMLAGVILGAAYLYKYFALQPDD (MW =4446 Da); Bri2 GallA: VGQRRAWCWCMCFALAFMLAAVILAAAYLYKYFALQPDD (MW = 4488 Da).
0.3–0.4 mg of the respective peptide were mixed with 3.12 mg 1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-choline (POPC) in 1 ml tert-butanol (Sigma). The mixture was chilled in liquid nitrogen and subsequently lyophilized. The phospholipid-peptide mixtures were hydrated in buffer (100 mm KCl, 20 mm sodium phosphate, pH 7.2) for 15 min at 30 °C resulting in a final peptide concentration of ∼80 μm. Small unilamellar vesicles (SUV) were prepared by sonication of the phospholipid-peptide suspensions for ∼20 min under argon at temperatures of ∼5 °C above the respective transition temperature tm of the lipids. A Branson sonifier equipped with a microtip was employed, using a duty cycle of 30%. CD spectra were measured with a Jasco J-810 Spectropolarimeter (Jasco, Gross-Umstadt, Germany) using a cuvette with a path length of 0.1 cm. Spectra of 11 scans were averaged. CD data are expressed as the mean residue ellipticity (Θ) in deg cm2 dmol−1 at the respective wave length in nm. The mean residue ellipticity was calculated from the measured ellipticity (θ, in mdeg) by dividing the molar ellipticity (θ × 100/([Protein] × d), d is the cuvette path length in cm) by the amount of residues (n = 39 for all Bri2 peptides). The molecular weights of the peptides are indicated above.
RESULTS
Contribution of Glycine Residues within the TMD of BRi2 to Its Intramembrane Cleavage
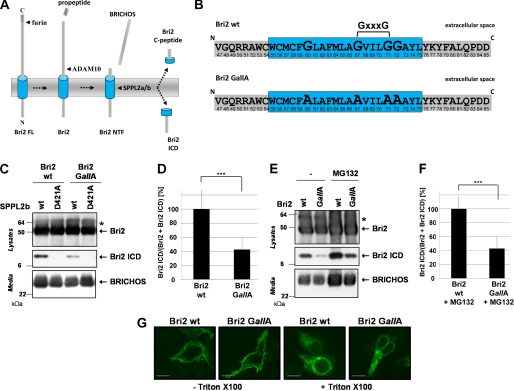
Bri2 (itm2b), the protein genetically involved in familial British dementia (31) was recently identified as a substrate for SPPL2b (27). After the release of a 4 kDa C-terminal propeptide (32, 33) Bri2 undergoes a shedding event catalyzed by ADAM10, which removes the bulk of the Bri2 ectodomain containing the BRICHOS domain (Refs. 27, 34 and Fig. 1A). The shedding event significantly facilitates subsequent intramembrane cleavage by SPPL2b (35), which generates a low molecular weight intracellular peptide (Bri2 ICD) and a secreted C-terminal peptide (Bri2 C-peptide; (Ref. 27 and Fig. 1A)). The efficiency of Bri2 intramembrane cleavage depends on so far unknown determinants within the intracellular domain, the TMD, and in the luminal juxtamembrane domain of Bri2 (35). To further specify these determinants we first investigated, whether mutation of potential helix-destabilizing residues within the TMD of Bri2 affect its intramembrane cleavage by SPPL2b, as it has been suggested for S2P, rhomboid, and SPP substrates (18–21). For this purpose a Bri2 variant lacking all four glycine residues in its TMD (Bri2 GallA) was generated (Fig. 1B). Because no other potential helix-destabilizing residues are present in the Bri2 TMD, Bri2 GallA reflects a mutant lacking all potential helix destabilizers. Compared with wt Bri2 ICD production was strongly reduced when Bri2 GallA was co-expressed with SPPL2b (Fig. 1C). As expected ICD production from both Bri2 variants was fully blocked in cells expressing the catalytically inactive SPPL2b D421A (Fig. 1C). Quantification of ICD production relative to total Bri2 revealed a significant reduction to 42.4% ± 17.3% in Bri2 GallA-expressing cells (Fig. 1D). To ensure that the reduced ICD level is neither due to an enhanced proteosomal degradation nor to missorting of the Bri2 GallA substrate we treated cells co-expressing SPPL2b and either Bri2 wt or Bri2 GallA with 10 μm MG132 (Fig. 1E) and determined the subcellular localization of Bri2 by immunofluorescence (Fig. 1G), respectively. Inhibition of proteosomal degradation stabilized all Bri2 species, including Bri2 ICDs to a similar extent regardless whether Bri2 wt or Bri2 GallA was expressed (Fig. 1E). Moreover, ICD levels generated from Bri2 GallA in cells treated with MG132 were reduced to a similar extent as in nontreated cells (42.9% ± 16.3%; Fig. 1F). Cell surface transport of the Bri2 protein was not affected by the removal of glycine residues from the TMD as shown by immunofluorescence staining of nonpermeabilized cells (Fig. 1G). These findings therefore demonstrate that removal of all potential helix destabilizing residues from the TMD of Bri2 significantly reduces its intramembrane cleavage by SPPL2b.
FIGURE 1.
Glycine residues within the TMD of BRi2 greatly facilitate its intramembrane cleavage by SPPL2b. A, model depicting the proteolytic processing of Bri2 by furin, shedding, and subsequent intramembrane proteolysis. B, amino acid sequence of the Bri2 TMD and the flanking regions. The potential helix-destabilizing residues are enlarged and the corresponding alanine mutations are indicated below. C, Bri2 GallA is less efficiently processed by SPPL2b than Bri2 wt. Co-expression of Bri2 wt or Bri2 GallA with SPPL2b wt or the catalytically inactive SPPL2b D421A mutant shows reduced ICD generation for Bri2 GallA. Bri2 and Bri2 ICD were detected using the anti-FLAG antibody. The corresponding BRICHOS domain was isolated from conditioned media using the polyclonal V5 antibody. D, quantitative analysis of the experiment shown in C. Data represent the means ± S.D. of twelve independent experiments. SPPL2b-dependent ICD generation is significantly reduced to 42.4 ± 17.3% for Bri2 GallA (p < 0.000004). E, proteasomal degradation of Bri2 GallA is not increased. Cells co-expressing SPPL2b wt and either Bri2 wt or Bri2 GallA were treated with 10 μm MG132 or the respective carrier for 24h. Bri2 and Bri2 ICD were detected using the anti-FLAG antibody. The corresponding BRICHOS domain was isolated from conditioned media using the polyclonal V5 antibody. F, quantitative analysis of the experiment shown in E. Data represent the means ± S.D. of six independent experiments. G, cellular localization of Bri2 GallA is similar to Bri2 wt. Immunohistochemistry of HEK293 TR cells transfected with Bri2 wt or Bri2 GallA reveals similar localization of both proteins at the plasma membrane (staining without Triton X-100) and within the secretory pathway (staining with Triton X-100). Bri2 wt and Bri2 GallA were detected using an anti-V5 antibody. Scale bar, 10 μm.
The GXXXG Dimerization Motif Is Not the Major Determinant for the Intramembrane Cleavage of Bri2
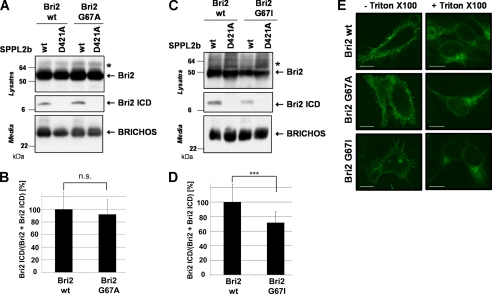
Because the removal of all glycine residues also disrupts the GXXXG dimerization motif in the TMD of Bri2, we next investigated whether a single point mutation in the GXXXG motif would be sufficient to copy the effect of the Bri2 GallA mutation on Bri2 ICD production. An alanine mutation of the first glycine residue in the GXXXG motif (G67A) did not significantly affect generation of the Bri2 ICD (92.1 ± 23.4%; p < 0.54; Fig. 2, A and B), suggesting that the effect on intramembrane proteolysis observed in the Bri2 GallA mutation (Fig. 1) is not caused by the potential disruption of the GXXXG motif.
FIGURE 2.
The GXXXG dimerization motif is not a major determinant for the intramembrane cleavage of Bri2. A, GXXXG to AXXXG mutation does not change Bri2 ICD generation. Co-expression of Bri2 wt or Bri2 G67A with SPPL2b wt or the inactive SPPL2b D421A revealed similar ICD production from both Bri2 variants. All proteolytic products were detected as described in Fig. 1C. B, quantitative analysis of the experiment shown in A. Data represent the means ± S.D. of eight independent experiments. C, GXXXG to IXXXG mutation slightly reduced SPPL2b dependent Bri2 ICD generation. Co-expression of Bri2 wt or Bri2 G67I with SPPL2b wt or the inactive SPPL2b D421A revealed a slightly reduced ICD production from Bri2 G67I. All proteolytic products were detected as described in Fig. 1C. D, quantitative analysis of the experiment shown in C. Data represent the means ± S.D. of eighteen independent experiments. E, mutations in GXXXG motif did not affect subcellular localization of Bri2. Immunohistochemistry of HEK293 TR cells transfected with Bri2 wt, Bri2 G67A, or Bri2 G67I reveals similar localization of all Bri2 protein variants at the plasma membrane (staining without Triton X-100) and within the secretory pathway (staining with Triton X-100). Scale bar, 10 μm.
Because it has been shown that glycine to isoleucine mutations in the GXXXG motif of APP affect its cleavage by γ-secretase to a much greater extent than glycine to alanine mutations (26), we also introduced a G67I mutation to further investigate whether the GXXXG motif has an essential role in the intramembrane proteolysis of Bri2 by SPPL2b. Although the G67I mutation reduced Bri2 ICD generation, (71.8 ± 14.9% (p < 0.0003); Fig. 2, C and D) the reduction was significantly less as compared with the Bri2 GallA mutation (42.4 ± 17.3% (p < 0.000004); Fig. 1D). Mutation of Gly-67 did not affect cell surface transport of the Bri2 protein as shown by immunofluorescence staining of nonpermeabilized cells (Fig. 2E).
Gly-60 in the N-terminal Part of the Bri2 TMD Is Required for Efficient Cleavage by SPPL2b
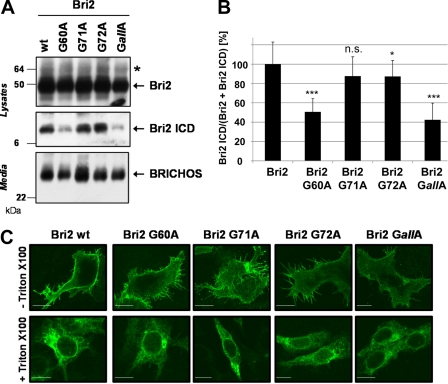
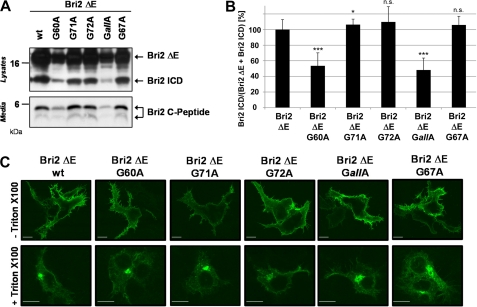
To test whether the interplay of all glycine residues in the TMD of Bri2 is necessary for efficient intramembrane proteolysis or whether a single glycine residue is sufficient to allow efficient cleavage by SPPL2b, we generated individual glycine mutations. Interestingly, mutagenesis of Gly-60 was sufficient to reduce Bri2 ICD generation to a similar extent (50.6 ± 13.8%; p < 4·10−13) as mutagenesis of all glycine residues (GallA; 42.4 ± 17.3%; p < 0.000004). In contrast SPPL2b-dependent ICD generation was hardly changed for Bri2 G71A (87.6 ± 20.0%; p < 0.15) and for Bri2 G72A (87.3 ± 16.5%; p < 0.03) (Fig. 3, A and B). Cell surface transport of the Bri2 protein was not affected by any of the point mutations (Fig. 3C). To further ensure that the G60A mutation in Bri2 directly affects intramembrane cleavage and not shedding, we co-expressed SPPL2b and Bri2ΔE cDNA constructs, which lack the entire Bri2 ectodomain and have been previously shown to be solely processed by SPPL2a and SPPL2b independent of a preceding shedding event (27). Because of a V5-tag added to the C terminus, these constructs furthermore allowed us to detect the C-terminal counterpart (Bri2 C-Peptide) of the Bri2 ICD in the conditioned media (27). In line with the data obtained upon expression of the Bri2 constructs ICD generation in cells co-expressing SPPL2b and either Bri2ΔE G60A (53.5 ± 16.6%; p < 1 × 10−10) or Bri2ΔE GallA (48.2 ± 15.3%; p < 8 × 10−9) was significantly reduced, while SPPL2b-dependent ICD generation was hardly changed for Bri2ΔE G71A (106.3 ± 7.1%; p < 0.04), Bri2 ΔE G72A (109.7 ± 19.6%; p < 0.14) and Bri2 ΔE G67A (105.9 ± 11.2%; p < 0.10) (Fig. 4, A and B). Consistent with these findings levels of Bri2 C-peptides in conditioned media from cells expressing Bri2ΔE G67A, Bri2ΔE G71A, or Bri2ΔE G72A were hardly changed compared with Bri2ΔE wt, while those of Bri2ΔE G60A and Bri2ΔE GallA-expressing cells were markedly reduced (Fig. 4A). Thus, these findings independently confirm the conclusions drawn from the expression of the full-length constructs and demonstrate that G60 strongly affects Bri2 cleavage by SPPL2b. As demonstrated for the Bri2 variants, all Bri2 ΔE variants localized to the cell surface (Fig. 4C).
FIGURE 3.
Gly-60 in the N-terminal part of the Bri2 TMD enables efficient cleavage by SPPL2b. A, G60A mutation in the TMD of Bri2 strongly reduces intramembrane cleavage. Co-expression of SPPL2b wt and the indicated Bri2 mutants revealed a strongly reduced Bri2 ICD generation only in cells expressing the G60A or the GallA mutant. All proteolytic products were detected as described in Fig. 1C. B, quantification of the experiment shown in A. Data represent the means ± S.D. of at least twelve independent experiments. C, glycine mutations in the Bri2 TMD do not affect subcellular localization of Bri2. Immunohistochemistry of HEK293 TR cells transfected with the indicated Bri2 variants reveals similar localization of all protein variants at the plasma membrane (staining without Triton X-100) and within the secretory pathway (staining with Triton X-100). Scale bar, 10 μm.
FIGURE 4.
Gly-60 directly affects the intramembrane cleavage of Bri2 by SPPL2b and not shedding of Bri2. A, Bri2 ΔE variants lacking the BRICHOS domain affect Bri2 ICD production similar to the respective full-length constructs. Co-expression of SPPL2b wt and the indicated Bri2ΔE mutants revealed a significantly reduced Bri2 ICD generation only in cells expressing Bri2ΔE G60A or Bri2ΔE GallA. Bri ΔE and its corresponding ICDs were detected using the anti-FLAG antibody. The corresponding Bri2 C-peptides were isolated from conditioned media after 4 h using the polyclonal V5 antibody. B, quantification of the experiment shown in A. Data represent the means ± S.D. of at least twelve independent experiments. C, Bri2 ΔE variants all share a similar subcellular localization. Immunohistochemistry of HEK293 TR cells transfected with the indicated Bri2 ΔE variants reveals similar localization of all protein variants at the plasma membrane (staining without Triton X-100 and within the secretory pathway (staining with Triton X-100). Scale bar, 10 μm.
Gly-60 Selectively Disrupts the α-Helical Structure of the Bri2 TMD
Because TMDs tend to adopt an α-helical conformation in the hydrophobic core of lipid bilayers (14), it was proposed that helix-destabilizing residues are required to facilitate intramembrane proteolysis (19, 20). We therefore asked whether the glycine residues in the TMD of Bri2 affect its secondary structure. To address this question, synthetic peptides reflecting the Bri2 TMD and the flanking regions (Fig. 1B) were generated. The lyophilized peptides were reconstituted in small unilamellar vesicles (SUVs) consisting of 1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-choline (POPC), and circular dichrosim (CD) spectra were collected. As indicated by the single minimum of the ellipticity between 220 and 230 nm, the peptide reflecting the Bri2 wt TMD is mainly in a β-strand like conformation (Fig. 5). In contrast, the peptides mimicking the G60A or the Bri2 GallA mutation revealed spectra with a smaller ellipticity at 210 nm to 215 nm. In particular the Bri2 GallA peptide revealed a minimum of the ellipticity at 210 nm, suggesting a predominantly α-helical conformation (Fig. 5). In contrast, peptides carrying the G71A or the G72A mutation behaved highly similar to that of the Bri2 wt peptide. The secondary structure of a peptide carrying the G67A mutation, which is according to the biochemical data described above presumably in a similar conformation as the wt peptide, could not be determined, because the synthesis of the peptide was impossible. These data indicate that the non α-helical conformation of the Bri2 TMD is mainly caused by Gly-60 in the N-terminal part of the TMD, while potential helix-destabilizing residues in the C-terminal part of the TMD do not influence the secondary structure of the Bri2 TMD in lipid vesicles. Thus the biophysical in vitro data are in line with our findings in living cells.
FIGURE 5.
Gly-60 significantly disturbs the α-helical structure of the Bri2 TMD. CD spectroscopy of synthetic Bri2 peptides covering the TMD and its flanking regions. The indicated peptides were incorporated in lipid vesicles and CD spectra were recorded at 20 °C. As indicated by the single minimum of the ellipticity between 220 nm and 230 nm the Bri2 wt peptide is mainly in a β-strand like conformation. Similarly, peptides with the G71A or the G72A mutation are predominantly in a β-strand like conformation. Peptides mimicking the G60A or the Bri2 GallA mutation revealed spectra with a smaller ellipticity at 210 nm to 215 nm suggesting a α-helical conformation.
DISCUSSION
Over the past years, intramembrane proteases have attracted a lot of attention, because their substrates are involved in a variety of diseases such as Alzheimer disease, Morbus Chron, psoriasis, hepatitis C, and malaria (4). In contrast to commonly known proteases, intramembrane proteases are capable of cleaving their substrate within the hydrophobic environment of a lipid bilayer (36), which raises the question of how substrate recognition and cleavage under these exceptional conditions is mediated. For the vast majority of classical proteases, which cleave their substrates in a hydrophilic environment, it has been shown that they almost exclusively process substrates, which present themselves in an extended β-strand conformation, while α-helically folded regions of proteins are highly resistant to proteolysis (16). The results of our study indicate that the intramembrane cleavage of Bri2 by SPPL2b, a member of the GXGD-type intramembrane aspartyl proteases, is also only efficiently catalyzed if the TMD of Bri2 adopts a β-strand-like conformation, while TMDs adopting a predominantly α-helical conformation are poor substrates. Similar data have been obtained for SPPL2a, a close homologue of SPPL2b (data not shown).
Our findings are consistent with studies on other intramembrane proteases such as S2P (18), SPP (20), and rhomboids (19, 21), which all suggest that the respective substrates of these intramembrane proteases are only efficiently processed if potentially helix-destabilizing residues are present in their TMDs. Helix-destabilizing residues like proline, glycine, asparagines, and serine have been suggested to disrupt the α-helical conformation which TMDs tend to adopt in the hydrophobic environment of cellular membranes (17) and therefore facilitate their cleavage by intramembrane proteases.
Using CD spectroscopy, we demonstrate that Gly-60 in the TMD of Bri2 is by itself able to stabilize the Bri2 TMD in a β-strand-like conformation when incorporated in lipid vesicles. Unexpectedly, two other glycine residues in the C-terminal part of the Bri2 TMD did not affect the secondary structure of the Bri2 wt TMD, demonstrating that potential helix-destabilizing residues in the context of a TMD may not generally disturb its favored α-helical conformation.
Recently, a study on rhomboids, a family of intramembrane serine proteases, demonstrated that a specific recognition motif surrounding the cleavage site is more strictly required for efficient substrate turnover than helical instability of the substrate TMD (22). However, the same study suggests that rhomboid substrates with their recognition motif located within or near their TMD require the presence of helix-destabilizing residues in the TMD to be efficiently cleaved (22). Assuming that substrate recognition and cleavage by rhomboid proteases would be governed by a similar mechanism as by GXGD intramembrane aspartyl proteases, one would expect an additional recognition motif close to either membrane border or in one of the juxtamembrane domains of Bri2. Although as yet no consistent recognition motif has been identified in the known SPPL2b substrates, our data do not exclude the existence of such a motif. In particular because swapping experiments using the juxtamembrane domains of Bri3, a close homologue of Bri2 but not a substrate for SPPL2b, have demonstrated that both juxtamembrane domains of Bri2 contain as yet unknown determinants that trigger intramembrane cleavage by SPPL2b (35). However, whether these determinants are specific sequence motifs or rather secondary structure requirements remains to be elucidated.
In line with this, presenilin, another member of the GXGD intramembrane aspartyl protease family, also requires so far incompletely characterized determinants in the TMD, the cytosolic domain (13), and the luminal juxtamembrane domain (24) of its substrates.
In addition it has been shown that dimerization of APP via a GXXXG dimerization motif in its TMD strongly reduces its intramembrane cleavage by γ-secretase (37). APP is a well-characterized γ-secretase substrate responsible for the generation of amyloid-β (Aβ), which forms pathologic aggregates in the brains of AD patients (38). Whether dimerization of APP reduces the total amount of secreted Aβ (37) or specifically reduces the pathogenic Aβ42 by modulating cleavage specificity (26, 39) is controversially discussed. Although in contrast to APP (25), it has not yet been formally proven that the GXXXG motif in the Bri2 TMD indeed is capable of triggering Bri2 dimerization, our present data suggest that a potential dimerization of Bri2 via a GXXXG in the TMD affects its intramembrane cleavage by SPPL2b only to a minor extent compared with the robust effect that was observed for APP (26, 37, 39).
The Bri2 TMD, including Gly-60 and the GXXXG motif, is highly conserved among species. In addition Gly-60 of Bri2 is also conserved in the TMD of human Bri3, which is a very poor substrate for SPPL2b (35). These observations may indicate that Gly-60 is only able to successfully destabilize an α-helix of a TMD if embedded in a certain context. However, because we failed to synthesize the peptide reflecting the Bri3 TMD, we could not finally prove this hypothesis in this study.
Taken together, our data suggest that the Bri2 TMD domain is only efficiently cleaved by SPPL2b if it adopts a non α-helical structure. Gly-60 has a high impact in destabilizing the α-helical character of the Bri2 TMD. Whether the secondary structure of the TMD is solely determined by Gly-60 or may additionally be affected by other primary structure elements and whether the lipid composition of certain cellular membranes may affect the secondary structure of the Bri2 TMD remains, however, to be elucidated.
Acknowledgments
We thank Drs. Harald Steiner and Frits Kamp for helpful discussions and Dr. Rackwitz for providing the synthetic peptides.
This work was supported in part by DFG Grant HA1737-11 (to C. H. and R. F.).
- I-CLiP
- intramembrane-cleaving protease
- SPPL2b
- signal peptide peptidase-like 2b
- RIP
- regulated intramembrane proteolysis
- TMD
- transmembrane domain
- ICD
- intracellular domain
- APP
- β-amyloid precursor protein
- Aβ
- amyloid-β
- AD
- Alzheimer disease
- SUV
- small unilamellar vesicles.
REFERENCES
- 1. Wolfe M. S. (2009) Intramembrane-cleaving proteases. J. Biol. Chem. 284, 13969–13973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Fluhrer R., Steiner H., Haass C. (2009) Intramembrane proteolysis by signal peptide peptidases: a comparative discussion of GXGD-type aspartyl proteases. J. Biol. Chem. 284, 13975–13979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Brown M. S., Ye J., Rawson R. B., Goldstein J. L. (2000) Regulated intramembrane proteolysis: a control mechanism conserved from bacteria to humans. Cell 100, 391–398 [DOI] [PubMed] [Google Scholar]
- 4. Lemberg M. K., Freeman M. (2007) Cutting proteins within lipid bilayers: rhomboid structure and mechanism. Mol. Cell 28, 930–940 [DOI] [PubMed] [Google Scholar]
- 5. Steiner H., Fluhrer R., Haass C. (2008) Intramembrane proteolysis by gamma-secretase. J. Biol. Chem. 283, 29627–29631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Weihofen A., Binns K., Lemberg M. K., Ashman K., Martoglio B. (2002) Identification of signal peptide peptidase, a presenilin-type aspartic protease. Science 296, 2215–2218 [DOI] [PubMed] [Google Scholar]
- 7. Friedmann E., Lemberg M. K., Weihofen A., Dev K. K., Dengler U., Rovelli G., Martoglio B. (2004) Consensus analysis of signal peptide peptidase and homologous human aspartic proteases reveals opposite topology of catalytic domains compared with presenilins. J. Biol. Chem. 279, 50790–50798 [DOI] [PubMed] [Google Scholar]
- 8. Wolfe M. S., Xia W., Ostaszewski B. L., Diehl T. S., Kimberly W. T., Selkoe D. J. (1999) Two transmembrane aspartates in presenilin-1 required for presenilin endoproteolysis and γ-secretase activity. Nature 398, 513–517 [DOI] [PubMed] [Google Scholar]
- 9. Steiner H., Kostka M., Romig H., Basset G., Pesold B., Hardy J., Capell A., Meyn L., Grim M. L., Baumeister R., Fechteler K., Haass C. (2000) Glycine 384 is required for presenilin-1 function and is conserved in bacterial polytopic aspartyl proteases. Nat. Cell Biol. 2, 848–851 [DOI] [PubMed] [Google Scholar]
- 10. Wang J., Beher D., Nyborg A. C., Shearman M. S., Golde T. E., Goate A. (2006) C-terminal PAL motif of presenilin and presenilin homologues required for normal active site conformation. J. Neurochem. 96, 218–227 [DOI] [PubMed] [Google Scholar]
- 11. Tolia A., Horré K., De Strooper B. (2008) Transmembrane domain 9 of presenilin determines the dynamic conformation of the catalytic site of γ-secretase. J. Biol. Chem. 283, 19793–19803 [DOI] [PubMed] [Google Scholar]
- 12. Sato C., Takagi S., Tomita T., Iwatsubo T. (2008) The C-terminal PAL motif and transmembrane domain 9 of presenilin 1 are involved in the formation of the catalytic pore of the γ-secretase. J. Neurosci. 28, 6264–6271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hemming M. L., Elias J. E., Gygi S. P., Selkoe D. J. (2008) Proteomic profiling of γ-secretase substrates and mapping of substrate requirements. PLoS Biol. 6, e257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Popot J. L., Engelman D. M. (2000) Helical membrane protein folding, stability, and evolution. Annu. Rev. Biochem. 69, 881–922 [DOI] [PubMed] [Google Scholar]
- 15. Hubbard S. J. (1998) The structural aspects of limited proteolysis of native proteins. Biochim Biophys Acta 1382, 191–206 [DOI] [PubMed] [Google Scholar]
- 16. Madala P. K., Tyndall J. D., Nall T., Fairlie D. P. (2010) Update 1 of Proteases universally recognize beta-strands in their active sites. Chemical Reviews 110, PR1–PR31 [DOI] [PubMed] [Google Scholar]
- 17. Li S. C., Deber C. M. (1994) A measure of helical propensity for amino acids in membrane environments. Nat Struct Biol 1, 558. [DOI] [PubMed] [Google Scholar]
- 18. Ye J., Davé U. P., Grishin N. V., Goldstein J. L., Brown M. S. (2000) Asparagine-proline sequence within membrane-spanning segment of SREBP triggers intramembrane cleavage by site-2 protease. Proc. Natl. Acad. Sci. U.S.A. 97, 5123–5128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Urban S., Freeman M. (2003) Substrate specificity of rhomboid intramembrane proteases is governed by helix-breaking residues in the substrate transmembrane domain. Mol. Cell 11, 1425–1434 [DOI] [PubMed] [Google Scholar]
- 20. Lemberg M. K., Martoglio B. (2002) Requirements for signal peptide peptidase-catalyzed intramembrane proteolysis. Mol. Cell 10, 735–744 [DOI] [PubMed] [Google Scholar]
- 21. Akiyama Y., Maegawa S. (2007) Sequence features of substrates required for cleavage by GlpG, an Escherichia coli rhomboid protease. Mol. Microbiol. 64, 1028–1037 [DOI] [PubMed] [Google Scholar]
- 22. Strisovsky K., Sharpe H. J., Freeman M. (2009) Sequence-specific intramembrane proteolysis: identification of a recognition motif in rhomboid substrates. Mol. Cell 36, 1048–1059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kopan R., Ilagan M. X. (2004) γ-Secretase: proteasome of the membrane? Nat. Rev. Mol. Cell Biol. 5, 499–504 [DOI] [PubMed] [Google Scholar]
- 24. Ren Z., Schenk D., Basi G. S., Shapiro I. P. (2007) Amyloid β-protein precursor juxtamembrane domain regulates specificity of γ-secretase-dependent cleavages. J. Biol. Chem. 282, 35350–35360 [DOI] [PubMed] [Google Scholar]
- 25. Kaden D., Munter L. M., Joshi M., Treiber C., Weise C., Bethge T., Voigt P., Schaefer M., Beyermann M., Reif B., Multhaup G. (2008) Homophilic interactions of the amyloid precursor protein (APP) ectodomain are regulated by the loop region and affect β-secretase cleavage of APP. J. Biol. Chem. 283, 7271–7279 [DOI] [PubMed] [Google Scholar]
- 26. Munter L. M., Voigt P., Harmeier A., Kaden D., Gottschalk K. E., Weise C., Pipkorn R., Schaefer M., Langosch D., Multhaup G. (2007) GXXXG motifs within the amyloid precursor protein transmembrane sequence are critical for the etiology of Aβ42. EMBO J. 26, 1702–1712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Martin L., Fluhrer R., Reiss K., Kremmer E., Saftig P., Haass C. (2008) Regulated intramembrane proteolysis of Bri2 (Itm2b) by ADAM10 and SPPL2a/SPPL2b. J. Biol. Chem. 283, 1644–1652 [DOI] [PubMed] [Google Scholar]
- 28. Fluhrer R., Fukumori A., Martin L., Grammer G., Haug-Kröper M., Klier B., Winkler E., Kremmer E., Condron M. M., Teplow D. B., Steiner H., Haass C. (2008) Intramembrane proteolysis of GXGD-type aspartyl proteases is slowed by a familial Alzheimer disease-like mutation. J. Biol. Chem. 283, 30121–30128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Fluhrer R., Capell A., Westmeyer G., Willem M., Hartung B., Condron M. M., Teplow D. B., Haass C., Walter J. (2002) A non-amyloidogenic function of BACE-2 in the secretory pathway. J. Neurochem. 81, 1011–1020 [DOI] [PubMed] [Google Scholar]
- 30. Kaether C., Capell A., Edbauer D., Winkler E., Novak B., Steiner H., Haass C. (2004) The presenilin C-terminus is required for ER-retention, nicastrin-binding and γ-secretase activity. EMBO J. 23, 4738–4748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Vidal R., Frangione B., Rostagno A., Mead S., Révész T., Plant G., Ghiso J. (1999) A stop-codon mutation in the BRI gene associated with familial British dementia. Nature 399, 776–781 [DOI] [PubMed] [Google Scholar]
- 32. Kim S. H., Wang R., Gordon D. J., Bass J., Steiner D. F., Lynn D. G., Thinakaran G., Meredith S. C., Sisodia S. S. (1999) Furin mediates enhanced production of fibrillogenic ABri peptides in familial British dementia. Nat. Neurosci. 2, 984–988 [DOI] [PubMed] [Google Scholar]
- 33. Kim S. H., Creemers J. W., Chu S., Thinakaran G., Sisodia S. S. (2002) Proteolytic processing of familial British dementia-associated BRI variants: evidence for enhanced intracellular accumulation of amyloidogenic peptides. J. Biol. Chem. 277, 1872–1877 [DOI] [PubMed] [Google Scholar]
- 34. Sánchez-Pulido L., Devos D., Valencia A. (2002) BRICHOS: a conserved domain in proteins associated with dementia, respiratory distress and cancer. Trends Biochem. Sci 27, 329–332 [DOI] [PubMed] [Google Scholar]
- 35. Martin L., Fluhrer R., Haass C. (2009) Substrate requirements for SPPL2b-dependent regulated intramembrane proteolysis. J. Biol. Chem. 284, 5662–5670 [DOI] [PubMed] [Google Scholar]
- 36. Wolfe M. S. (2010) Structure, mechanism, and inhibition of γ-secretase and presenilin-like proteases. Biol. Chem. 391, 839–847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Eggert S., Midthune B., Cottrell B., Koo E. H. (2009) Induced dimerization of the amyloid precursor protein leads to decreased amyloid-β protein production. J. Biol. Chem. 284, 28943–28952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Selkoe D. J. (2011) Resolving controversies on the path to Alzheimer therapeutics. Nat. Med. 17, 1060–1065 [DOI] [PubMed] [Google Scholar]
- 39. Munter L. M., Botev A., Richter L., Hildebrand P. W., Althoff V., Weise C., Kaden D., Multhaup G. (2010) Aberrant amyloid precursor protein (APP) processing in hereditary forms of Alzheimer disease caused by APP familial Alzheimer disease mutations can be rescued by mutations in the APP GXXXG motif. J. Biol. Chem. 285, 21636–21643 [DOI] [PMC free article] [PubMed] [Google Scholar]