Abstract
As extracellular proteins age, they undergo and accumulate nonenzymatic post-translational modifications that cannot be repaired. We hypothesized that these could be used to systemically monitor loss of extracellular matrix due to chronic arthritic diseases such as osteoarthritis (OA). To test this, we predicted sites of deamidation in cartilage oligomeric matrix protein (COMP) and confirmed, by mass spectroscopy, the presence of deamidated (Asp64) and native (Asn64) COMP epitopes (mean 0.95% deamidated COMP (D-COMP) relative to native COMP) in cartilage. An Asp64, D-COMP-specific ELISA was developed using a newly created monoclonal antibody 6-1A12. In a joint replacement study, serum D-COMP (p = 0.017), but not total COMP (p = 0.5), declined significantly after replacement demonstrating a joint tissue source for D-COMP. In analyses of 450 participants from the Johnston County Osteoarthritis Project controlled for age, gender, and race, D-COMP was associated with radiographic hip (p < 0.0001) but not knee (p = 0.95) OA severity. In contrast, total COMP was associated with radiographic knee (p < 0.0001) but not hip (p = 0.47) OA severity. D-COMP was higher in soluble proteins extracted from hip cartilage proximal to OA lesions compared with remote from lesions (p = 0.007) or lesional and remote OA knee (p < 0.01) cartilage. Total COMP in cartilage did not vary by joint site or proximity to the lesion. This study demonstrates the presence of D-COMP in articular cartilage and the systemic circulation, and to our knowledge, it is the first biomarker to show specificity for a particular joint site. We believe that enrichment of deamidated epitope in hip OA cartilage indicates a lesser repair response of hip OA compared with knee OA cartilage.
Keywords: Aging, Aspartate, Extracellular Matrix, Extracellular Matrix Proteins, Protein Deamidation, COMP, Cartilage Biology, Osteoarthritis, Protein Turnover
Introduction
As proteins age they undergo nonenzymatic post-translational modifications. When aged proteins are intracellular, they can be repaired or the protein replaced (1); however, in the extracellular milieu, where no repair mechanisms exist, nonenzymatic modifications can accumulate in a time-dependent manner in proteins whose turnover is slow. Accumulation of these modifications in long lived proteins can potentially alter both their structural and functional properties and provide insights into turnover rates at which proteins are replaced through net synthesis and degradation processes.
One form of nonenzymatic protein modification, deamidation, is believed to be a mechanism of amino acid damage and aging in numerous proteins (2) and a variety of tissues (2, 3). Protein deamidation has been demonstrated to interfere with protein function of interleukin-1β (4), soluble CD4 (5), angiogenin (6), and Bcl-xL (4–7) and may also incite autoimmunity (8). Nonenzymatic deamidation involves the conversion of Asn to Asp or the conversion of Gln to Glu in a spontaneous manner without enzymes (3). During nonenzymatic Asn deamidation, the side chain amine group is lost, causing formation of a succinimide ring. The succinimide ring is unstable and susceptible to hydrolysis at the imide to form Asp in either an α-Asp or β-Asp (isomerized) form depending upon which side of the imide hydrolysis occurs. Things are further complicated as the α-carbon atom in the succinimide ring can undergo racemization allowing the formation of both levorotatory and dextrorotatory optical isomers after hydrolysis. Similarly, Gln deamidation involves formation of a ring structure (glutarimide), where loss of the functional amine group leads to the formation of the acidic residue Glu in again both the levorotatory and dextrorotatory forms of α- or β-Glu. Asn deamidation occurs roughly twice as fast as Gln deamidation because of the less favorable 6-membered glutarimide ring structure. Particular hot spots for deamidation are predicted to exist based upon factors such as steric hindrance and protein context (3), as well as peptide sequence (for instance, dipeptides that deamidate more readily than others include Gly-Asn, Asn-Gly, and Gln-Gly (3)).
One of the tissues in the body most susceptible to accumulation of nonenzymatic protein modifications is cartilage. This is due to the slow turnover rate of many cartilage proteins; for instance, cartilage aggrecan has a predicted half-life of 25 years (9), whereas cartilage collagens have predicted half-lives of at least 120 years (10, 11). Nothing at present is known of the biological effects of these amino acid changes in cartilage, thus representing a significant knowledge gap. We hypothesized that the nonenzymatic modifications that accumulate with cartilage aging could be used to systemically monitor onset and degree of extracellular matrix loss during osteoarthritis (OA).2 To test this hypothesis, we identified a novel protein modification due to deamidation in cartilage oligomeric matrix protein (COMP). COMP is a noncollagenous glycoprotein and a member of the thrombospondin family of extracellular calcium-binding proteins that was initially isolated from cartilage (12). Although primarily expressed in cartilage, COMP is also expressed in tendons, meniscus, and synovial membranes. The carboxyl-terminal globular domain of COMP binds to collagens I and II (13). The amino terminus of COMP oligomerizes to form a pentamer of five identical subunits of 110 kDa (14), creating a pore that is believed to bind chloride and vitamin D3 (15).
We raised novel monoclonal antibodies (mAb) and developed an ELISA that specifically distinguished and quantified the deamidated form of the epitope from the total amount of the epitope (amidated and deamidated). We further demonstrated the presence of deamidated COMP (D-COMP) in articular cartilage and the systemic circulation, an enrichment of this epitope specifically in hip articular cartilage, and a correlation of this epitope in the systemic circulation with hip OA severity.
EXPERIMENTAL PROCEDURES
Patient Samples
The Biomarker and Joint Arthroplasty (BAJA) cohort provided serum samples drawn before or 6 months after either total hip or knee arthroplasty. A total of 14 subjects (9 male and 5 female) underwent total knee replacement (n = 10, six male and four female) or total hip replacement (n = 4, three male and one female) (Table 1). The overall mean age was 63.4 ± 13.2 years (59.6 ± 13.8 years for men, 70.2 ± 9.8 for women). The population was overweight with a mean body mass index of 26.8 ± 5.8 (27.8 ± 5.6 for men and 25.0 ± 6.6 for women). Samples were collected under approval of the Institutional Review Board of Duke University.
TABLE 1.
Cohort demographics
JoCo OA indicates a subset of 450 subjects from the Johnston County Osteoarthritis project.
| BAJA | JoCo OA | Human cartilage | |
|---|---|---|---|
| N (% female) | 14 (36%) | 450 (54%) | 26 (42%) |
| Age mean ± S.D. years (range) | 63.4 ± 13.2 (44–81) | 58.88 ± 9.62 (44–86) | 71.3 ± 10.0 (52–90+) |
| No OA (N) | 0 | 164 | 11 |
| Hip OA without knee OA (N) | 4 | 143 | 11 |
| Knee OA without hip OA (N) | 10 | 79 | 15 |
| Hip and knee OA (N) | 0 | 64 | N/A |
The Johnston County OA Project (JoCo OA) provided selected serum samples from 450 individuals at baseline evaluation. The JoCo OA is an ongoing, community-based study of knee and hip OA in African American and Caucasian residents in a rural county in North Carolina. Details of this study have been reported previously (16), and demographics are summarized in Table 1. Briefly, this study involved civilian, noninstitutionalized adults aged 45 years and older who resided in six townships in Johnston County. Participants were recruited by probability sampling, with oversampling of African Americans. A total of 3,187 individuals completed a baseline clinical evaluation from 1991 to 1997. Serum was collected for all participants at baseline. To allow for analyses of biomarkers in a sample balanced for gender and age, 450 participants were selected with complete radiographic data at baseline to represent roughly equal proportions of women (54%) and men (46%) across a range of ages (see Table 1). A total of 39% of participants were African American. Individuals having radiographic evidence of rheumatoid arthritis or other inflammatory arthropathies in the knees or hips were not included in the subsample. Participants completed bilateral anteroposterior weight-bearing radiography of the knees with foot mat placement and supine anteroposterior hip radiographs. Radiographs were read, without knowledge of participant clinical or biomarker status by a single musculoskeletal radiologist (J. B. R.), for overall radiographic severity by Kellgren-Lawrence (17) (KL, score 0–4) of the knees and hips and for knee osteophyte (score 0–3) and joint space narrowing (score 0–3) based on the standardized Burnett atlas (18). Inter-rater reliability (comparison of radiograph readings between J. B. R. and another radiologist) and intra-rater reliability (comparison of radiograph readings completed by J. B. R. at two separate times) were high (weighted κ for inter-rater reliability 0.9; κ for intra-rater reliability 0.9) (19). The study was conducted under approval of the Institutional Review Boards of the University of North Carolina, Chapel Hill, and the Centers for Disease Control and Prevention.
Waste articular cartilage specimens were obtained from randomly selected total knee (n = 15, mean age 66.5 ± 8.9, 54–88 years) and total hip (n = 11, mean age 77.3 ± 11.3, 57–90+ years) arthroplasties performed at Duke University Medical Center to alleviate symptoms of OA. Patient characteristics are summarized in Table 1. From each arthritic joint, cartilage was harvested from around the lesion (lesion cartilage) and for comparison cartilage remote from the lesion (remote cartilage). Nonarthritic control samples (n = 11) were obtained from National Disease Research Interchange (Philadelphia) or at the time of reconstructive surgery for trauma from patients without evidence of OA as determined by the surgeon and macroscopic inspection of the specimens. Samples were collected under Duke Institutional Review Board approval as waste surgical specimens.
Monoclonal Antibody Generation and Screening
To generate novel specific mAbs to investigate the predicted deamidated epitopes in COMP, a COMP peptide, TFLKD64TVMEC, specific for the deamidated epitope was used to immunize mice (A&G Pharmaceutical, Columbia, MD). Antibody specificity was confirmed using a direct ELISA. Briefly, 96-well ELISA plates were coated with COMP purified from hip cartilage (a gift from Dr. V. Vilim) or BSA conjugated to the deamidation-specific COMP peptide (EITFLKD64TVMEC) or the nondeamidated native COMP peptide (EITFLKN64TVMEC). For later screening experiments, we synthesized two further constructs with different peptides coupled to BSA as follows: one BSA-peptide construct contained the native CELQETN42AALQ sequence, and the other coupled the deamidated CELQETD42AALQ peptide to BSA. The COMP peptides (procured from AnaSpec, Fremont, CA), or COMP isolated from cartilage, were coated in 0.1 m sodium carbonate/bicarbonate coating buffer, pH 9.6, overnight at 4 °C. Plates were blocked overnight with 5% w/v BSA in phosphate-buffered saline (PBS), pH 7.4, at 4 °C before excess blocking buffer was discarded, and the plates were washed with 0.05% v/v PBS wash buffer. Washed wells were incubated with undiluted hybridoma supernatants overnight at 4 °C. Unbound mAb was discarded and washed with 0.05% v/v Tween 20 in PBS, pH 7.4 (PBS/Tween), before addition of alkaline phosphatase-conjugated anti-mouse antibody (Promega, Madison, WI) and development with o-phenylenediamine dihydrochloride (OPD) substrate and detection at 450 nm.
Protein G Antibody Purification
The 6-1A12 mAb was produced in-house and purified using protein G-agarose (Thermo Scientific, Rockford, IL) per the manufacturer's instructions. Briefly, the 6-1A12 hybridoma was grown in roller bottle culture in serum-free hybridoma medium (Sigma). The supernatants were collected, filtered through a 0.2-μm filter, and concentrated 20-fold using Amicon stirred ultrafiltration cells with a 100-kDa molecular mass cutoff (Millipore, Billerica, MA). Concentrated hybridoma medium was diluted 1:1 with 20 mm sodium phosphate binding buffer, pH 7.0, before incubating with protein G-agarose (1 ml of resin to 10 ml of diluted hybridoma medium) overnight at 4 °C with gentle mixing. After incubation, the protein G bead/hybridoma medium was passed into a column, and unbound proteins were washed from the column with a further 5 ml of binding buffer; this wash was collected in 1-ml aliquots, and a Bradford protein assay (Thermo Scientific) was performed to confirm all unbound protein was removed from the column. The 6-1A12 mAb was eluted from the column with 0.1 m glycine buffer, pH 2.7, and 0.5-ml aliquots were collected. Each aliquot was neutralized with 1 m Tris buffer, pH 7.4, to prevent mAb degradation. Fractions containing mAb, as determined by a Bradford protein assay, were pooled and dialyzed against PBS. A final mAb concentration was determined by Bradford protein assay (Thermo Scientific), and the purified mAbs were aliquoted and stored at −80 °C until needed.
Extraction of Soluble Cartilage Matrix Proteins
Cartilage was pulverized under liquid nitrogen and extracted in 4 m guanidine HCl (Gdn-HCl) in sodium acetate buffer, pH 4.0, with protease inhibitor mixture (Sigma) at a ratio of 2 ml of Gdn-HCl extraction buffer per 1 g of wet weight cartilage. Cartilage was extracted for 24 h at 4 °C with gentle mixing, followed by centrifugation and collection of the supernatant. The remaining cartilage was extracted a second time with Gdn-HCl extraction buffer for a further 24 h. Both the first and second Gdn-HCl extractions were combined and stored at −80 °C until needed. A 200-μl aliquot of the extract was buffer-exchanged on an AKTA purifier 10 (GE Healthcare) using a HiTrap desalting column (GE Healthcare) into PBS and stored at −80 °C. Proteoglycan content was determined using the dimethylmethylene blue assay as described previously (20). Protein concentrations were determined using the Bradford protein assay (Thermo Scientific) to allow normalization of values.
To confirm the efficiency of our Gdn-HCl extraction protocol, we performed five sequential Gdn-HCl extractions in triplicate with brief water washes between two specimens, a hip (non-OA, female, 69 years) and a knee (non-OA, male, 77 years). These analyses showed that the method we used routinely, that of two serial Gdn-HCl extractions, was highly efficient and extracted between 99.3% (as estimated by glycosaminoglycan) and 100.0% (as estimated by protein) of the soluble proteins from cartilage (supplemental Table 1).
Mass Spectroscopic Identification of Deamidated COMP
To prevent interference with downstream liquid chromatography-mass spectroscopy (LC-MS/MS), residual Gdn-HCl was removed by buffer exchanges into 50 mm ammonium bicarbonate, pH 7.8, using a HiTrap desalting column on an AKTA HPLC system (GE Healthcare) followed by trypsin digestion. The insoluble residue left after Gdn-HCl extraction was prepared for LC-MS/MS by extensive washing of the cartilage with 50 mm ammonium bicarbonate, pH 7.8, for 3 days with five buffer changes per day. After these extensive washes, the residual cartilage was weighed before direct trypsin digestion. After trypsin digestion, all samples were subjected to shotgun proteomics using LC-MS/MS as described previously (21). Briefly, 0.1% v/v rapigest (Waters) was added before reduction with 10 mm dithiothreitol at 80 °C for 15 min, alkylated with 20 mm iodoacetamide for 30 min at room temperature, and digested with proteomics grade trypsin (Promega, Madison, WI) overnight at 37 °C. Prior to LC-MS/MS analysis, all samples were resuspended in 20 μl of 2% acetonitrile, 0.1% formic acid, pH 3.0. Chromatographic separation of peptide mixtures was performed on a Waters NanoAcquity UPLC equipped with a 1.7-μm BEH130 C18 75-μm inner diameter × 250-mm reversed-phase column. The mobile phase was 0.1% formic acid in water (buffer A) and 0.1% formic acid in acetonitrile (buffer B). MS analysis was performed on a Waters Synapt G2 mass spectrometer, and label-free quantification and integration of qualitative peptide identifications were performed using Rosetta Elucidator (Version 3.3, Rosetta Inpharmatics, Seattle). All raw LC-MS/MS data were subjected to chromatographic retention time alignment using the PeakTeller® algorithm. Quantification of all signals in the precursor MS spectra was performed by Elucidator by calculating peak volume (area under curve). Data were searched with Mascot (Matrix Science, Boston) against a human protein data base downloaded from SwissProt appended with yeast alcohol dehydrogenase.
D-COMP Competitive ELISA
A competitive ELISA was developed to determine whether the D-COMP epitope could be measured using our mAbs in biological samples. Briefly, coating concentrations were optimized (data not shown), and binding plates were prepared by coating 96-well ELISA plates with 6.5 μg/ml D-COMP-specific BSA-peptide (BSA-EITFLKD64TVMEC) in 0.02 m sodium carbonate coating buffer, pH 9.6, overnight at 4 °C. Plates were blocked for at least 2 h with 5% w/v BSA in PBS, pH 7.4, at 37 °C prior to use. Protein G-purified 6-1A12 was diluted to 2 μg/ml in protein diluents (0.1% w/v BSA in PBS, pH 7.4) before addition of 80 μl of mAb to 80 μl of either sample or standard in a low protein binding mixing plate for 30 min at room temperature with gentle mixing. The BSA-EITFLKD64TVMEC coupled construct was used as a standard for the competitive assay and diluted for a standard curve with a range of 0.1–10 μg/ml. The binding reaction was transferred to the coated capture plate, 60 μl per well, and incubated at room temperature for 1 h with gentle mixing to allow free mAb binding to the plate. Subsequent steps were the same as for the direct ELISA described above.
D-COMP and Total COMP Sandwich ELISAs
The D-COMP sandwich ELISA was based upon capture with the D-COMP-specific mAb 6-1A12 and detection with 17-C10, a mAb to total COMP (specificity for COMP irrespective of deamidation state) (22). The method was the same as for the competitive ELISA above with a few exceptions as follows: 96-well plates were coated with the 6-1A12 D-COMP-specific mAb; samples or standard was diluted in 0.1% w/v BSA in PBS as required prior to incubation of 50 μl of sample or standard overnight at 4 °C; and unbound sample was discarded and the plate washed with PBS/Tween wash buffer before incubation with the biotinylated 17-C10 total COMP detection mAb (a gift from Dr. V. Vilim). Unbound 17-C10 was discarded, and the plate was washed. Because of the lower levels of D-COMP, the D-COMP ELISA sensitivity was increased using avidin poly-horseradish peroxidase (poly-HRP, Thermo Scientific), added for 30 min at room temperature with gentle mixing. Excess avidin-HRP was discarded; the plate was washed, and signal was detected using tetramethylbenzidine (Sigma) reagent after stopping with 2 m HCl and detection at 450 nm. Total COMP was measured using the 16-F12 and 17-C10 mAbs as described previously (22) with several modifications. For this project, we used avidin-HRP and tetramethylbenzidine as the detection system so that the total COMP and D-COMP methods were as similar as possible.
D-COMP Standard for ELISA
D-COMP concentrations by sandwich ELISA were determined using a COMP standard derived by pooling Gdn-HCl extracts from hip cartilage as follows: extracts from a 76-year-old normal hip specimen, and extracts from lesioned and remote cartilage regions of 75-year-old knee OA and 72-year-old hip OA specimens (23). The mean quantity of D-COMP in these samples was estimated as 0.95% the quantity of total COMP based on the relative mean abundance of D-COMP and total COMP within the same mass spectrometry trace for each sample (% D-COMP in standard = Asp64 peak intensity/Asn64 peak intensity·100).
Western Blot Protocol
PBS-dialyzed Gdn-HCl cartilage extract and precision plus prestained protein standards (5 μg/well, Bio-Rad) were separated under reducing conditions on a 10% SDS-PAGE. Proteins were transferred to a nitrocellulose membrane. After transfer, the membrane was blocked with 3% w/v milk proteins for 2 h at room temperature. The blocked membrane was incubated with 6-1A12 or 17-C10 mAbs in PBS 1% w/v milk proteins overnight at 4 °C. Excess mAb was discarded, and the membrane was washed with 0.1% v/v PBS/Tween before incubation with anti-mouse HRP-conjugated secondary antibody (Promega, Madison, WI) in 1% w/v milk proteins for 30 min. Excess antibody was discarded, and the membrane was washed before signal development with ECL Plus reagent (GE Healthcare) using Hyperfilm ECL x-ray film (GE Healthcare).
Statistical Methods
Because of the small sample size, the BAJA study data were analyzed using the Wilcoxon signed rank nonparametric paired t test using GraphPad Prism Version 5 (La Jolla, CA). The COMP concentrations from the JoCo OA cohort were log transformed to achieve a normal distribution before analysis using a generalized linear model, the cumulative logit model, controlling for age, gender, and race. As radiographic data from the left and right joints of a person were likely to correlate, the intra-individual correlation was estimated in the model as a variance component. The JoCo OA data represented in Fig. 4 were log transformed and analyzed by one-way analysis of variance with a Bonferroni correction for multiple comparisons. The cartilage extract data were analyzed either with the Wilcoxon signed rank nonparametric paired t test for paired data or using the nonparametric Mann-Whitney U test for unpaired data.
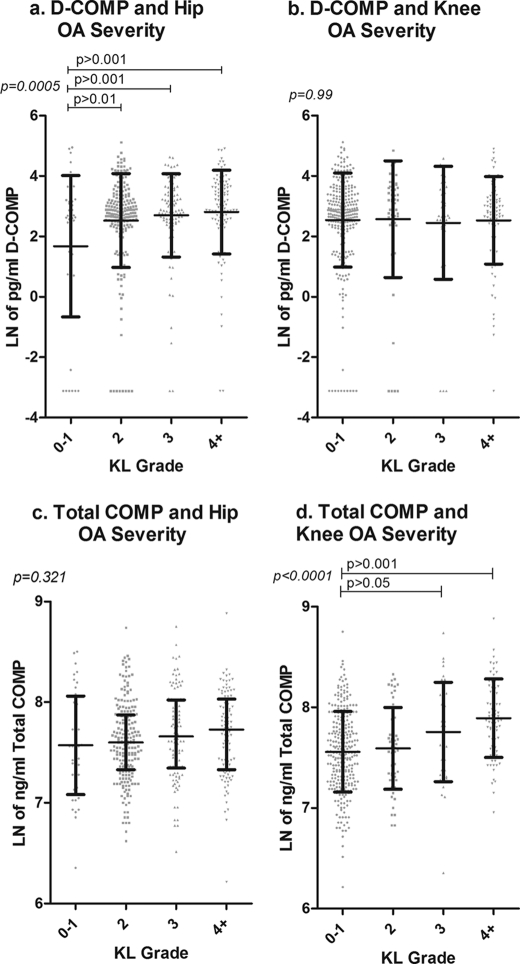
FIGURE 4.
Association of serum D-COMP with hip OA severity and total COMP with knee OA severity in the JoCo OA cohort samples. Sera and x-rays (of both hips and knees) were obtained for 450 subjects from the JoCo OA cohort. X-rays were read for OA severity status as defined by KL grade (0–4 scale) with OA being defined as KL grade ≤2. For the purposes of this analysis, the KL scores were summed for both hips or both knees (total possible range of 0–8). Serum levels for both D-COMP and total COMP were determined, and the data natural log transformed for statistical purposes. The data are represented as scatter plots showing the mean ± S.D. ln D-COMP and ln total COMP for non-OA subjects (KL0–1), early OA (KL2 and KL3), and for advanced OA (KL4+). a demonstrates significant increase in D-COMP with hip OA severity, and b demonstrates no significant change in D-COMP with knee OA severity. c demonstrates no significant change in total COMP with OA hip severity, and d demonstrates significant increase in total COMP with knee OA severity. For each KL group (0–1, 2, 3, 4+) the frequencies were n = 48, 200, 99, and 102 for hip OA, and n = 46, 199, 99, and 102 for knee OA. Subjects were excluded when COMP was not quantifiable as follows: one subject for D-COMP and four subjects for total COMP. Significance was determined using a one-way analysis of variance with a Bonferroni correction for multiple comparisons.
RESULTS
Epitope Selection
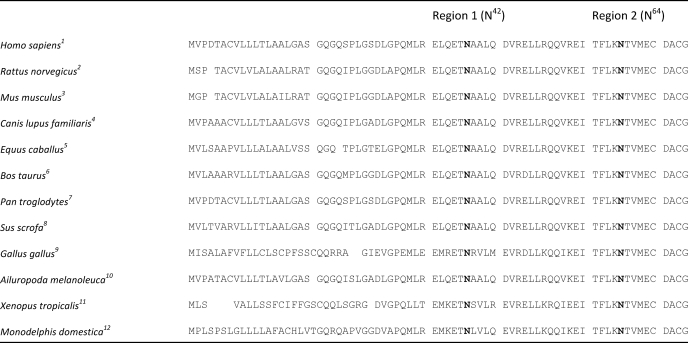
We identified putative deamidation hot spots within COMP using the algorithm developed by Robinson and Robinson (3). This algorithm used inputted crystal structure and amino acid composition of COMP to predict Asn residues susceptible to deamidation and gave an estimate of the deamidation half-life for each individual Asn residue (CD value × 100 days) and half-life for net deamidation value (ID value × 100 days). As the crystal structure for human COMP was not available at the time of this project, we used the two partial rat Protein Data Bank structural files 1fbm (24) and 1vdf (25). Both crystal structures span the amino-terminal region of rat COMP between Gly27 and Gly72. Using the two rat terminal structural files, we identified two Asn residues (amino acids 41 and 63) susceptible to deamidation, in human COMP designated Asn42 and Asn64, respectively (Table 2). Neither of these residues has a particularly fast deamidation rate, with Asn41 predicted to have a deamidation half-life between 21 and 26 years and Asn63 between 39 and 50 years, making them ideal markers for monitoring degradation of a long lived tissue such as cartilage matrix degradation. We compared the sequence alignment for a range of different species available in the NCBI data base (www.ncbi.nlm.nih) and confirmed that the predicted Asn deamidation hot spots were contained in human COMP and conserved in a wide range of other animals (Table 2).
TABLE 2.
Species conservation of predicted deamidation hotspots in COMP
1Human; 2Rat; 3Mouse; 4Dog; 5Horse; 6Cattle; 7Chimpanzee; 8Pig; 9Red jungle fowl; 10Giant panda; 11Western clawed frog; 12Gray short-tailed opossum; sequence numbering based on human COMP.
To determine whether the predicted deamidated sequences occur in vivo, we performed LC-MS/MS analysis of Gdn-HCl extracts from five articular cartilage specimens from three subjects. We were unable to confirm the presence of the predicted deamidation event at Asn42 in these samples. However, we were able to confirm the presence of the native Asn64 tryptic peptide (64NTVMECDACGMQQSVCR) and the deamidated Asp64 peptide (64DTVMECDACGMQQSVCR) in normal hip, OA hip, and OA knee cartilage (both remote and lesioned regions). As we were able to determine peak intensities for both the Asp64 and Asn64 peptides within the same MS run, we calculated within sample ratios of D-COMP to native COMP. In all five specimens, the native Asn64 peptide was more abundant than the Asp64 deamidated peptide (by 83–330-fold) based on intensities of mass spectroscopic traces, with a mean (S.D.) intensity of 145,178 ± 86,144 for the Asn64 peptide compared with an intensity of 1,382 ± 1,092 for the Asp64 deamidated peptide (representing a mean 0.95% relative level of D-COMP to native COMP).
Antibody Generation and Validation
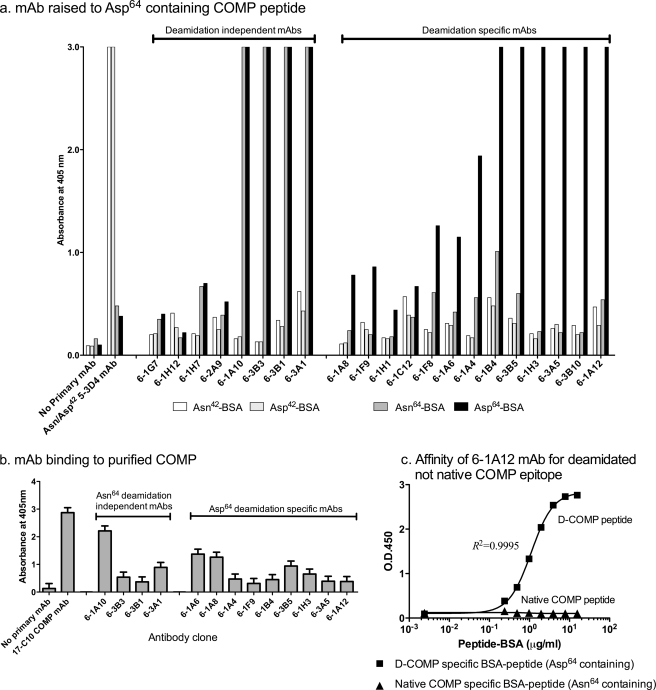
To generate mAbs to deamidated epitopes in COMP, mice were immunized with a peptide containing Asp64 (TFLKD64TVMEC) found in deamidated COMP. Using this approach, we expected to generate two classes of mAbs, those that recognized the deamidated Asp64 residue and mAbs that were deamidation-independent, recognizing both the amidated and deamidated sequence. We identified 21 mAb clones that recognized the immunogen. To determine the specificities of these 21 clones, we compared their immunoreactivity to four different BSA-conjugated COMP peptides as follows: the deamidated Asp64 (TFLKD64TVMEC) immunogen, native Asn64 (TFLKN64TVMEC), deamidated Asp42 (CELQETD42AALQ), and native Asn42 (CELQETN42AALQ) (Fig. 1a). As anticipated, we identified the following two types of mAbs using this approach: mAbs that appeared to be deamidation-independent as they recognized both the Asn64 and Asp64 containing BSA constructs (clones 6-1A10, 6-3B3, 6-3B1, 6-3A1, 6-1G7, 6-1H12, 6-1H7, and 6-2A9), and deamidation-specific clones (clones 6-1A12, 6-1A6, 6-1A8, 6-1A4, 6-1F9, 6-1B4, 6-3B5, 6-1H3, 6-3A5, 6-3B10, 6-1F8, 6-1H1, and 6-1C12). None of the 21 clones raised to the Asp64 peptide bound the BSA-coupled Asn42 or Asp42 control peptides; the immunoreactivity of these control peptides was confirmed by reactivity with the deamidation independent mAb (5-3D4) raised specifically to the Asp42-containing peptide (Fig. 1a).
FIGURE 1.
Monoclonal antibody specificity of mAbs raised to the COMP-deamidated epitope Asp64. a, screening of mAbs against deamidated and native COMP peptides. Two native COMP BSA-peptide constructs, Asn42 (BSA-CELQETN42AALQ) and Asn64 (TFLKN64TVMEC-BSA), and the corresponding two deamidated BSA constructs, Asp42 (BSA-CELQETD42AALQ) and Asp64 (TFLKD64TVMEC-BSA), were coated onto 96-well plates. Hybridoma culture media were incubated with the coated plates for 2 h before addition of goat anti-mouse alkaline phosphatase secondary antibody and ELISA development using OPD substrate. A negative control containing no primary antibody was included, and a mAb raised against CELQETD42AALQ was used to confirm immunoreactivity of the BSA-CELQET(N/D)42AALQ control peptides. Screening yielded a total of eight deamidation-independent (immunoreactivity to Asn64 and Asp64) and 13 deamidation-dependent mAbs (preferential immunoreactivity to Asp64). b, screening of mAbs against purified cartilage COMP. Purified COMP protein (a generous gift from V. Vilim) was coated onto 96-well plates. Hybridoma culture media were incubated with the coated plates for 2 h before addition of goat anti-mouse alkaline phosphatase secondary antibody and ELISA development using OPD substrate. A negative control containing no primary antibody and a positive control using the anti-COMP 17-C10 mAb were included. c, mAb 6-1A12 preferentially reacted against deamidated COMP peptide and not the native COMP sequence. To test the affinity of the 6-1A12 mAb for both native and deamidated COMP, different concentrations of either the D-COMP Asp64 specific TFLKD64TVMEC-BSA or the native COMP-specific Asn64 TFLKN64TVMEC-BSA were coated onto a 96-well plate in a direct ELISA, performed as for a. A standard curve with an appropriate dose response is generated for 6-1A12 and the deamidated COMP Asp64-BSA construct; 6-1A12 did not recognize the native Asn64-BSA construct.
To confirm that the mAbs recognized COMP from human cartilage and not just peptides, a subset of these mAbs was screened by direct ELISA against COMP purified from human hip cartilage pooled from two subjects (Fig. 1b) (generous gift from V. Vilim (see Ref. 26)). All mAbs tested displayed higher than background immunoreactivity to cartilage COMP. Of note, the mAb preparations used in these validation experiments were not purified so the apparent differences in immunoreactivity could reflect mAb concentration rather than mAb affinity. In addition, the fact that the deamidation-specific mAbs reacted to cartilage COMP suggested sufficient sensitivity on the part of these mAbs to identify the presence of deamidated Asp64 in COMP purified from human articular cartilage.
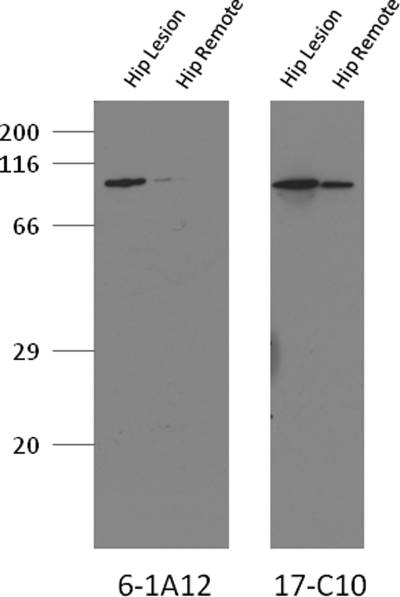
For the further experiments in this study, we selected the Asp64 deamidation-specific 6-1A12 mAb because it grew well in culture and produced a high yield of mAb. The specificity and affinity of the 6-1A12 mAb was confirmed through evaluation of its binding to different coating concentrations of both the native and deamidated BSA peptides (Fig. 1c). The 6-1A12 mAb had no affinity for the native Asn64-containing sequence but instead had a high specificity for only the deamidated Asp64 COMP-containing peptide. To further confirm that the 6-1A12 mAb recognized COMP extracted from cartilage and not just a COMP-specific peptide, we performed a reduced Western blot of OA hip cartilage Gdn-HCl extract (Fig. 2). The deamidation specific 6-1A12 mAb detected a 110-kDa protein that corresponded to a band of similar molecular mass detected by the established anti-COMP mAb 17-C10 (27) and that corresponded to the size of monomeric COMP.
FIGURE 2.
Western blot demonstrating D-COMP mAb binding to COMP from Gdn-HCl extract of hip articular cartilage. Protein extracts were prepared from osteoarthritic hip cartilage harvested from both an OA lesional site and remote from the lesion in remote OA cartilage. Prior to SDS-PAGE separation on a 10% gel, samples were dialyzed into PBS, protein levels determined, and 5 μg of protein loaded per lane. Samples were separated under standard conditions and transferred to nitrocellulose that was blocked with 3% milk proteins prior to mAb incubation and development with ECL reagents. D-COMP was immunolocated with mAb 6-1A12 specific for deamidated COMP (left) and total COMP was immunolocated with mAb 17-C10 (right). For clarity, the membrane immunostained for total COMP was exposed for a shorter time period (30 s), although the membrane immunostained for D-COMP, due to it lower levels, required a longer exposure (5 min).
Decline in Serum Concentrations of D-COMP but Not Total COMP after Joint Arthroplasty
To determine whether the D-COMP epitope could be measured in biological fluid, we developed a competitive ELISA using the 6-1A12 mAb and the BSA-coupled deamidated COMP-specific (TFLKD64TVMEC) construct as both the coating antigen and the assay standard. In the BAJA study, serum (n = 14) was collected before joint replacement (pre-joint arthroplasty samples) and 6 months after either hip (n = 4) or knee (n = 10) total arthroplasty. We expected to observe a decrease in the serum concentrations of biomarker after joint arthroplasty, thereby supporting in principle a joint tissue source for the measured biomarker in the serum. Six months after joint replacement, serum total COMP concentration (1,513 ± 457 ng/ml) was not significantly changed (p = 0.502) from baseline prior to joint replacement (1,600 ± 655 ng/ml) (Fig. 3a). However, we did observe a small but significant (p = 0.017) decrease in the serum concentrations of D-COMP 6 months after joint replacement (578.6 ± 154 pg/ml) compared with baseline prior to joint replacement (726 ± 158 pg/ml) (Fig. 3b). Stratified by joint site, we observed a small but nonsignificant decreasing trend for D-COMP following knee replacement (13% mean decline from 732 ± 48 pg/ml prior replacement to 638 ± 34 pg/ml 6 months post-replacement) and a larger decreasing trend for D-COMP following hip replacement (39% mean decline from 711 ± 108 pg/ml to 431 ± 83 pg/ml post-replacement). There was minimal change in total COMP stratified by joint site for knee (5% mean decline from 1680 ± 225 to 1593 ± 166 ng/ml post-replacement) or hip (6% mean decline from 1400 ± 260 to 1316 ± 26 ng/ml post-replacement).
FIGURE 3.
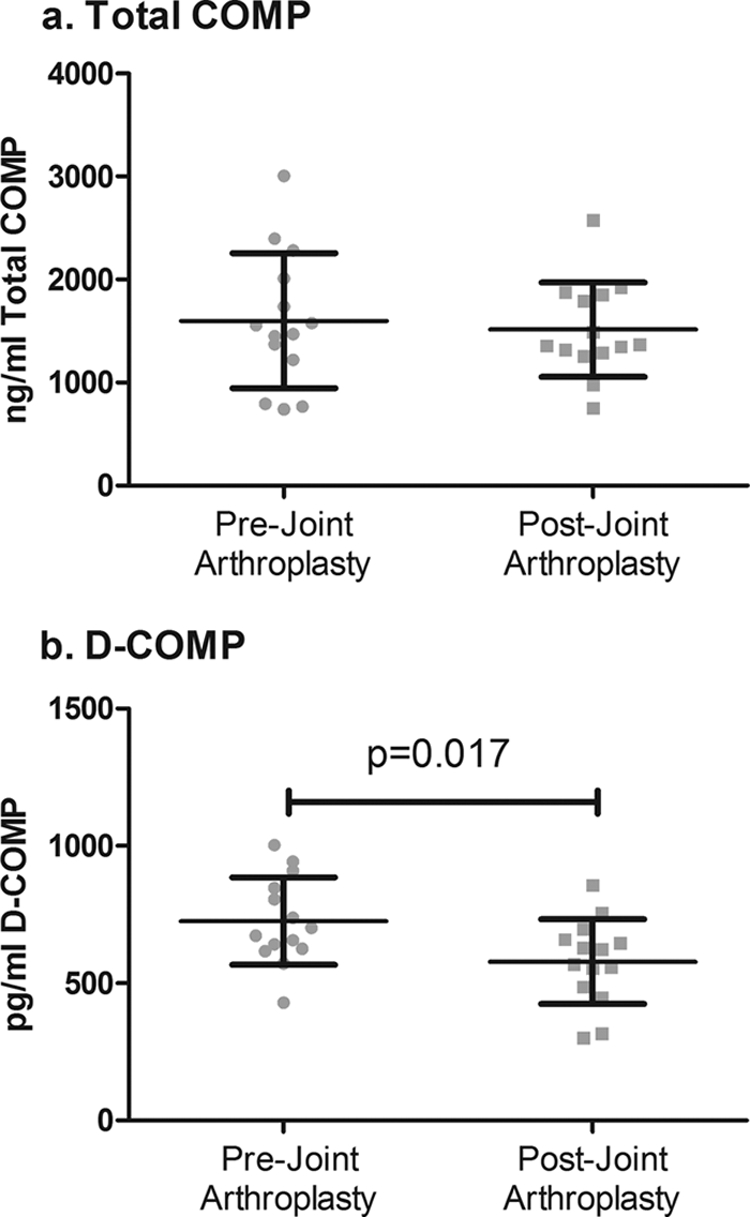
Serum D-COMP but not total COMP concentrations declined following replacement of OA joints. Sera were obtained before and 6 months after joint arthroplasty in the BAJA cohort. Total serum COMP, measured by sandwich ELISA, and serum D-COMP, measured by competition ELISA, were determined pre- and 6 months post-joint replacement for n = 14 patents (nine knee and five hip replacements). mAb 6-1A12 was used for D-COMP competition ELISA; for total COMP sandwich ELISA mAbs 16-F12 captured and biotinylated 17-C10 for detection. Significant differences were determined using the paired nonparametric Wilcoxon signed rank test. Total COMP was unchanged after joint replacement (a), and D-COMP declined significantly (p = 0.017) following joint replacement (b).
Serum concentrations of D-COMP and total COMP were reported on hip and knee OA severity, respectively. To further investigate the utility of D-COMP as a novel biomarker for OA, we measured D-COMP and total COMP in the JoCo OA subsample of patients. This sample subset was derived from the population-based JoCo study (16). The total subsample of 450 individuals included 898 nonreplaced knees and 896 nonreplaced hips, graded for radiographic OA severity by KL grade. For improved sensitivity, we developed and optimized (data not shown) a sandwich ELISA using our deamidation-specific 6-1A12 mAb for capture and the 17-C10 total COMP mAb for detection. Using linear regression analysis, we evaluated for an age association of serum D-COMP and total COMP concentrations in the 158 non-OA subjects defined as KL 0-1 for both hips and knees. This subset ranged in age from 44 to 77 years (mean ± S.D. of 55.9 ± 7.8 years). There was a significant but minimal positive association of total COMP with age (p = 0.007, r2 = 0.045) but no association of D-COMP (p = 0.90, r2<0.001) with age (supplemental Fig. 1).
To determine how serum COMP levels changed with severity of OA, we analyzed the full JoCo 450 subjects using generalized linear models (cumulative logit models) controlling for the intra-individual correlation in the model as a component of variation, as well as age, gender, and race. Radiographic knee OA was present with the following frequencies of KL knee OA severity grades: 535 KL0; 153 KL1; 115 KL2; 81 KL3; 14 KL4, and two knee replacements. Radiographic hip OA occurred with the following frequencies of severity: 73 KL0; 512 KL1; 287 KL2; 20 KL3; 4 KL4, and four hip replacements. Total COMP and D-COMP were associated with age but not gender or race. Independent of age, total COMP was significantly associated with knee OA KL grade (p < 0.0001) but not hip KL grade (p = 0.40). In contrast and independent of age, D-COMP was significantly associated with hip OA KL grade (p < 0.0001) but not with knee OA KL grade (p = 0.87). For knee OA, we also had available the scores for radiographic OA features of joint space narrowing and osteophyte. Total COMP was strongly associated with both knee joint space narrowing (p < 0.0001) and knee osteophyte (p < 0.0001), whereas D-COMP was not. To provide a graphic representation of the results, D-COMP values are shown in scatter plots stratified by knee and hip OA status (Fig. 4). These data strongly suggest that the serum D-COMP assay provides a new biomarker indicative of the presence and severity of hip OA, whereas native nondeamidated or total COMP is an indicator of the presence and severity of knee OA.
Concentrations of D-COMP in Hip Cartilage Exceeded Those of Knee Cartilage
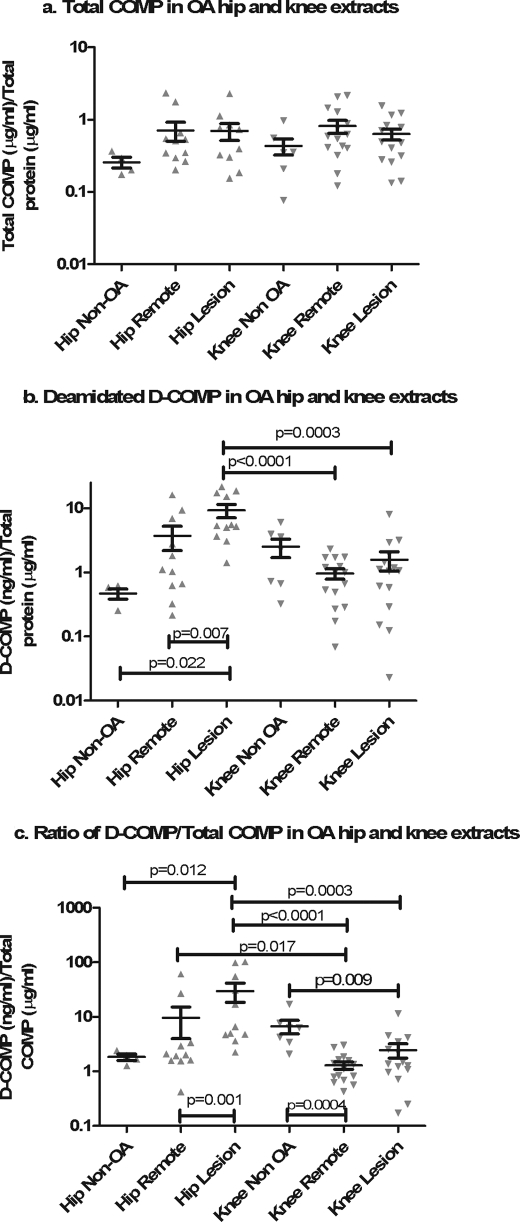
To better understand the results from the JoCo OA cohort and the relationship between D-COMP and hip OA, we investigated the amount of D-COMP and total COMP in Gdn-HCl-extracted soluble protein from OA hip (n = 12) and knee cartilage samples (n = 16) collected as waste surgical tissue at the time of joint arthroplasty.
We compared D-COMP and total COMP levels from regions of hip and knee cartilages adjacent to OA lesions and from regions of the same joint remote from the OA lesion. We also evaluated the ratio of D-COMP/total COMP as a measure free from confounding by extraction efficiencies and dialysis variations. For comparison, we also included age-matched non-OA hip (n = 4, mean age 73.5 years ±6.9, range 67–82 years) and knee (n = 7, mean age 60.6 years ±14.5, range 52–83 years) cartilage collected as waste trauma or cadaveric tissue.
Total COMP in cartilage did not vary by joint site (lesion or remote) or joint group (knee or hip) (Fig. 5a). In contrast, D-COMP did vary by joint site. D-COMP was significantly higher in hip versus knee lesion cartilage (mean ± S.D., 9.26 ± 7.30 and 1.57 ± 2.01 ng of D-COMP/μg of protein extracted, respectively, p = 0.0003), and a similar but not significant trend was observed for D-COMP in hip remote versus knee remote cartilage (mean ± S.D., 3.72 ± 5.10 and 0.96 ± 0.68 ng of D-COMP/μg of extracted protein, respectively, p = 0.14) (Fig. 5b). D-COMP was significantly higher in the hip lesional cartilage compared with the hip remote cartilage (p = 0.007) and compared with non-OA hip cartilage (p = 0.02) and remote knee cartilage (p < 0.0001) as follows: mean ± S.D. of 9.26 ± 7.3, 3.72 ± 5.06, 0.47 ± 0.17, and 0.96 ± 0.68 ng of D-COMP/μg of extracted protein, respectively. There was no significant difference in D-COMP levels between non-OA hip and knee cartilage samples. These data confirm that the D-COMP epitope is more abundant in OA hip than either OA knee cartilage or non-OA cartilage and is enriched at the site of OA lesions in the hip.
FIGURE 5.
Enrichment of D-COMP in hip cartilage and OA lesion sites. Soluble proteins were extracted from cartilage using 4 m Gdn-HCl before buffer exchange into PBS to allow further analysis. Total COMP (a) and D-COMP (b) were measured by sandwich ELISA and normalized to total protein to correct for variations in the extraction and dialysis efficiency. The ratio of D-COMP/total COMP (c) was calculated as a measure that is independent of technical variation due to variation in protein extraction and dialysis efficiency. Data were plotted on a logarithmic scale for clarity, and statistical differences were determined on nonlogarithmically transformed data using nonparametric statistics. For paired samples (remote and lesion within a joint), the Wilcoxon signed rank test was used; for nonpaired samples (hip and knee joint comparisons) intergroup significance was determined using the nonparametric Mann-Whitney U test. D-COMP and the D-COMP/total COMP ratio were higher in hip than knee cartilages and higher in hip OA lesions than in hip cartilage remote from lesions.
To better understand the turnover of COMP at the different sites, we evaluated the ratio of aged D-COMP/total COMP (Fig. 5c). The D-COMP/total COMP ratio was highest in hip OA lesional cartilage and higher at the hip lesion versus any of the other regions, namely versus the hip remote (p = 0.001), knee remote (p < 0.0001), or knee lesion (p = 0.0003) cartilage (mean ± S.D., 29.96 ± 38.18 hip lesion, 9.58 ± 18.59 hip remote, 1.30 ± 0.78 knee remote, and 2.47 ± 2.84 knee lesion). We also observed a significantly higher D-COMP/total COMP ratio between hip remote and knee remote cartilage (mean ± S.D., 9.58 ± 18.59 and 1.30 ± 0.78 D-COMP/total COMP, respectively, p = 0.017).
Relative to non-OA hip cartilage, hip lesion OA cartilage had significantly higher (p = 0.012) extractable concentrations of D-COMP as a proportion of total COMP (mean ± S.D., 1.84 ± 0.5 and 2.47 ± 2.1 respectively, Fig. 5c). As D-COMP has a long residence time in cartilage, it is conceivable that the differences observed between hip and knee OA D-COMP levels could be due to differential cross-linking of D-COMP to the collagen matrix preventing release from the cartilage in knee OA. To test this, we performed LC-MS/MS analysis of extracts from lesional regions of three age-matched cartilage samples as follows: an OA hip (72 years), an OA knee (75 years), and a normal hip (76 years). Gdn-HCl extraction of the cartilage samples yielded a soluble protein fraction and a paired insoluble fraction that was subjected to trypsin digestion and mass spectroscopic analysis. The tryptic peptide (64NTVMECDACGMQQSVR) corresponding to the native COMP sequence was present in all samples. As expected, more native COMP peptide was found in the Gdn-HCl-soluble fraction than in the insoluble cartilage residua (see supplemental Table 2). However, we were only able to identify the deamidated peptide (64DTVMECDACGMQQSVR), corresponding to the D-COMP epitope, in the Gdn-HCl soluble fraction and not in the insoluble fraction. Although this result does not absolutely exclude its presence in the insoluble fraction, it suggests that the deamidated epitope is not preferentially tethered in cartilage. The ratios of the intensity of the deamidated/native COMP peptide in the soluble protein fractions confirmed the abundance of extractable deamidated COMP from hip compared with knee cartilage (supplemental Table 2).
As we have no evidence to suggest preferential cross-linking in the OA knee cartilage, we believe that these results suggest lower turnover and older proteins in hip OA lesions. Interestingly, we observed the reverse in knee OA. Relative to non-OA knee cartilage (mean ± S.D., 6.73 ± 4.9), knee OA had significantly lower concentrations of D-COMP as a proportion of total COMP at both remote (p = 0.004, 2.47 ± 2.8) and lesion (p = 0.009, 1.30 ± 0.9) sites. These results suggest that the COMP in knee OA cartilage is newer and turning over faster. Non-OA hip and knee cartilages were statistically similar with respect to D-COMP as a proportion of total COMP suggesting that non-OA hip and knee cartilages have similar turnover and that the joint response to OA is quite different by hip versus knee joint site. Taken together, these data are consistent with a model positing lower rates of cartilage COMP synthetic repair in hip OA, and particularly at hip OA lesions, compared with normal hip and non-OA and OA knee.
DISCUSSION
To our knowledge, this is the first demonstration that a cartilage extracellular matrix component, measured systemically, demonstrated specificity for OA severity at a particular joint site. We believe this reflects a fundamental underlying difference between hip and knee cartilage turnover and repair responses in OA. We observed no association between D-COMP serum levels and chronological age in non-OA subjects. This is not surprising as the half-life for generating our epitope is predicted by the Robinson and Robinson algorithm (3) to be between 39 and 50 years. Assuming COMP has a turnover rate similar to the predicted half-life of 25 years for aggrecan (9), then accumulation of D-COMP with age in healthy cartilage would be prevented by normal cartilage turnover.
We demonstrated in our post-joint arthroplasty BAJA study of 14 patients, a significant decrease in the concentrations of our new and novel D-COMP biomarker after the removal of the affected arthritic joints. The greatest decrease in D-COMP was observed for the hip, although the knee showed a much more modest decrease in D-COMP levels. As OA does not usually affect only a single joint, and even normal joint turnover would be expected to contribute to the overall serum D-COMP levels, replacement of a single joint led, as expected, to a decline but not a disappearance of D-COMP from the serum. In contrast, we did not observe any significant decrease in total COMP levels after joint arthroplasty. These results with total COMP are consistent with observations made previously by Sharif et al. (28) who demonstrated that total COMP actually increased, rather than decreased, for at least 6 months following joint replacement. Other reports have demonstrated total COMP production by osteoblasts (29) and the potential for elevated total COMP during the period of reactive bone repair following joint arthroplasty. We believe that D-COMP measured systemically reflects degradation of mature aged cartilage; this is in contrast to native or total (the majority of which is native nondeamidated) COMP epitope in the systemic circulation that we posit reflects, in part, high turnover of newly synthesized tissue, or so-called “frustrated repair” (30), or new COMP production from a reparative bone response occurring after joint replacement.
In the much larger JoCo OA cohort, we were better able to refine our understanding of the utility of D-COMP as a biomarker. In this cohort, we observed that D-COMP was highly significantly correlated with hip OA severity but not with knee OA severity. We also observed that total COMP was strongly associated with knee OA but not with hip OA severity. Of note, in the JOCO cohort sample, the greatest increase in serum D-COMP occurred with the transition from the KL0-1 to the KL2 level of hip OA severity. This would suggest that D-COMP may be of particular value as an early indicator of hip pathology and OA.
The association of total COMP with knee OA progression is well established (31–35). A few studies have investigated total COMP in the setting of hip OA (36–39). Three studies observed that higher levels of baseline COMP predicted the development of hip radiographic OA (36, 38, 39). One of these studies suggested that high baseline COMP levels were associated with a reduction in radiographic progression (36). One small study found an association between hip JSN and increasing serum COMP levels over a 1-year follow up period (37), and another larger study found no association of total serum COMP with hip OA but a significant association with knee OA osteophyte (40). We believe the unique ability of our D-COMP biomarker to correlate with hip but not knee OA severity demonstrates the utility and value of studying post-translational modifications in cartilage proteins as biomarkers of OA.
To better understand the underlying biological basis for D-COMP as a hip OA but not knee OA biomarker, we investigated the concentrations of D-COMP in cartilage extracts from OA hips and knees. We hypothesized that D-COMP would be increased in cartilage with a low turnover rate and that biologically older tissues would have a higher D-COMP/total COMP ratio. We observed a significantly higher mean D-COMP/total COMP ratio in OA hip cartilages compared with OA knee cartilages. Total COMP concentrations between hip and knee cartilage were not significantly different confirming that the differences observed in the ratios were due to a higher proportion of D-COMP in hip cartilage. These data strongly suggest that COMP, and by inference the cartilage extracellular matrix in an OA hip joint, is turned over (net of catabolism and anabolism) at a much lower rate than cartilage in an OA knee joint. Conversely, this would indicate that knees are more robust at repairing ongoing degradation than hips.
In the hip, we also observed significantly higher concentrations of D-COMP and D-COMP/total Comp ratio in lesional OA cartilage when compared with remote regions. The elevated concentrations of D-COMP at hip OA lesions further support the serum biomarker observation that D-COMP was associated with hip OA. Moreover, this suggests that lesional cartilage is biologically older than the cartilage remote from the lesion and is consistent with lower rates of COMP synthesis at sites of hip OA lesions. For comparison, we included non-OA and age-matched cartilage collected as cadaveric tissue or shortly after trauma at the time of surgical repair. We found no significant difference between non-OA hip and knee cartilage for either D-COMP or total COMP. However, the D-COMP/total COMP ratio of OA lesional hip cartilage was significantly higher than non-OA cartilage suggesting that cartilage at the lesion is older. In contrast, we observed the opposite in the knee, with significantly lower than normal D-COMP/total COMP ratios in the OA tissue. As we were unable to find any evidence of enhanced cross-linking of older COMP in the knee OA cartilage, we believe these data support the hypothesis that serum D-COMP reflects cartilage turnover and that hip OA tissue is older (low repair response) than non-OA hip, whereas the knee OA tissue is younger (high repair response) than non-OA.
Although higher concentrations of D-COMP at hip lesions could potentially be explained by accelerated D-COMP production, we believe this to be unlikely as there was no evidence for accelerated production of D-COMP at knee OA lesions, which would be expected to be exposed to very similar catabolic conditions during cartilage loss. Radiolabeled proteoglycan studies in cartilage have shown that cartilage matrix turnover is greatest in the superficial zone whereas the matrix becomes older and the protein turnover rate decreases deeper into the cartilage (41). Therefore, we favor a model wherein gradual erosion of the cartilage at hip OA lesions leads, in the relative absence of synthetic repair, to the loss of the superficial and medial cartilage layers, leaving behind the older deeper zones.
In summary, we were able to predict a deamidation event in the cartilage matrix molecule COMP, demonstrate its presence in human cartilage, produce specific mAbs to this modification, and develop an ELISA to study its utility as a biomarker in OA. We identified a D-COMP modification that is not only a novel biomarker in OA but also a biomarker with specificity for hip OA, which, to the best of our knowledge, is the first OA biomarker specific to a particular joint site. Based upon our studies of COMP in cartilage, we believe we can explain the specificity of our D-COMP biomarker through a lower COMP protein synthesis in hip OA when compared with knee OA and higher rate of knee cartilage extracellular matrix turnover. The clear differences in D-COMP concentrations at remote OA hip and knee cartilage are consistent with different biological aging rates in these two large joint sites in OA due in part to less COMP synthesis in hips than knees. Hip OA cartilage lesions, in particular, were differentially enriched for D-COMP. This is compatible both with loss of the more rapidly regenerating superficial cartilage layers and inadequate COMP synthesis, resulting in biologically older remaining deep layers of hip OA cartilage. We believe that D-COMP warrants further study as a marker of hip disease to determine more clearly its utility in a patient setting, although currently we would envision its use as a longitudinal marker to follow disease progression within a subject. Examination is also warranted to determine its ability to detect occult or pre-radiographic hip disease.
Supplementary Material
Acknowledgments
We thank David Quintana for assistance with the BAJA sample collection. We also thank the Duke Proteomics Core Facility (Drs. Erik Soderblom and Arthur Moseley) for their expert assistance and Dr. Vladimir Vilim for the gift of 17-C10 antibody and purified COMP from human cartilage.
This work was supported, in whole or in part, by National Institutes of Health Grant 5P30 AG028716 from NIA (Claude D. Pepper Older Americans Independence Centers), Grant P01 AR050245 from NIAMS, Grant 5-P60-AR49465 from NIAMS (Multidisciplinary Clinical Research Center), and Grant 5-P60-AR-3070 from NIAMS (Multipurpose Arthritis and Musculoskeletal Diseases Center 1). This work was also supported by Centers for Disease Control and Prevention/Association of Schools of Public Health Grants S043 and S3486. We acknowledge use of tissues procured by the National Disease Research Interchange (NDRI) with support from National Institutes of Health Grant 5 U42 RR006042.

This article contains supplemental Fig. 1 and Tables 1 and 2.
- OA
- osteoarthritis
- BAJA
- Biomarkers and Joint Arthroplasty study
- COMP
- cartilage oligomeric matrix protein
- Gdn-HCl
- guanidine HCl
- JoCo OA
- Johnston County Osteoarthritis Project
- KL grade
- Kellgren-Lawrence grade of OA severity
- mAb
- monoclonal antibody
- OPD
- o-phenylenediamine dihydrochloride
- D-COMP
- deamidated COMP.
REFERENCES
- 1. Cloos P. A., Christgau S. (2002) Nonenzymatic covalent modifications of proteins. Mechanisms, physiological consequences, and clinical applications. Matrix Biol. 21, 39–52 [DOI] [PubMed] [Google Scholar]
- 2. McCudden C. R., Kraus V. B. (2006) Biochemistry of amino acid racemization and clinical application to musculoskeletal disease. Clin. Biochem. 39, 1112–1130 [DOI] [PubMed] [Google Scholar]
- 3. Robinson N. E., Robinson A. B. (2004) Molecular Clocks: Deamidation of Asparaginyl and Glutaminyl Residues in Peptides and Proteins, pp. 1–419, Althouse Press, Cave Junction, OR [Google Scholar]
- 4. Daumy G. O., Wilder C. L., Merenda J. M., McColl A. S., Geoghegan K. F., Otterness I. G. (1991) Reduction of biological activity of murine recombinant interleukin-1β by selective deamidation at asparagine 149. FEBS Lett. 278, 98–102 [DOI] [PubMed] [Google Scholar]
- 5. Teshima G., Porter J., Yim K., Ling V., Guzzetta A. (1991) Deamidation of soluble CD4 at asparagine 52 results in reduced binding capacity for the HIV-1 envelope glycoprotein gp120. Biochemistry 30, 3916–3922 [DOI] [PubMed] [Google Scholar]
- 6. Deverman B. E., Cook B. L., Manson S. R., Niederhoff R. A., Langer E. M., Rosová I., Kulans L. A., Fu X., Weinberg J. S., Heinecke J. W., Roth K. A., Weintraub S. J. (2002) Bcl-xL deamidation is a critical switch in the regulation of the response to DNA damage. Cell 111, 51–62 [DOI] [PubMed] [Google Scholar]
- 7. Hallahan T. W., Shapiro R., Strydom D. J., Vallee B. L. (1992) Importance of asparagine 61 and asparagine 109 to the angiogenic activity of human angiogenin. Biochemistry 31, 8022–8029 [DOI] [PubMed] [Google Scholar]
- 8. Moss C. X., Matthews S. P., Lamont D. J., Watts C. (2005) Asparagine deamidation perturbs antigen presentation on class II major histocompatibility complex molecules. J. Biol. Chem. 280, 18498–18503 [DOI] [PubMed] [Google Scholar]
- 9. Maroudas A., Bayliss M. T., Uchitel-Kaushansky N., Schneiderman R., Gilav E. (1998) Aggrecan turnover in human articular cartilage. Use of aspartic acid racemization as a marker of molecular age. Arch. Biochem. Biophys. 350, 61–71 [DOI] [PubMed] [Google Scholar]
- 10. Maroudas A., Palla G., Gilav E. (1992) Racemization of aspartic acid in human articular cartilage. Connect. Tissue Res. 28, 161–169 [DOI] [PubMed] [Google Scholar]
- 11. Verzijl N., DeGroot J., Thorpe S. R., Bank R. A., Shaw J. N., Lyons T. J., Bijlsma J. W., Lafeber F. P., Baynes J. W., TeKoppele J. M. (2000) Effect of collagen turnover on the accumulation of advanced glycation end products. J. Biol. Chem. 275, 39027–39031 [DOI] [PubMed] [Google Scholar]
- 12. Hedbom E., Antonsson P., Hjerpe A., Aeschlimann D., Paulsson M., Rosa-Pimentel E., Sommarin Y., Wendel M., Oldberg A., Heinegård D. (1992) Cartilage matrix proteins. An acidic oligomeric protein (COMP) detected only in cartilage. J. Biol. Chem. 267, 6132–6136 [PubMed] [Google Scholar]
- 13. Rosenberg K., Olsson H., Mörgelin M., Heinegård D. (1998) Cartilage oligomeric matrix protein shows high affinity zinc-dependent interaction with triple helical collagen. J. Biol. Chem. 273, 20397–20403 [DOI] [PubMed] [Google Scholar]
- 14. Mörgelin M., Heinegård D., Engel J., Paulsson M. (1992) Electron microscopy of native cartilage oligomeric matrix protein purified from the Swarm rat chondrosarcoma reveals a five-armed structure. J. Biol. Chem. 267, 6137–6141 [PubMed] [Google Scholar]
- 15. Ozbek S., Engel J., Stetefeld J. (2002) Storage function of cartilage oligomeric matrix protein. The crystal structure of the coiled-coil domain in complex with vitamin D(3). EMBO J. 21, 5960–5968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Jordan J. M., Helmick C. G., Renner J. B., Luta G., Dragomir A. D., Woodard J., Fang F., Schwartz T. A., Abbate L. M., Callahan L. F., Kalsbeek W. D., Hochberg M. C. (2007) Prevalence of knee symptoms and radiographic and symptomatic knee osteoarthritis in African Americans and Caucasians. The Johnston County Osteoarthritis Project. J. Rheumatol. 34, 172–180 [PubMed] [Google Scholar]
- 17. Kellgren J. H., Lawrence J. S. (1957) Radiological assessment of osteo-arthrosis. Ann. Rheum. Dis. 16, 494–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Burnett S., Hart D. J., Cooper C., Spector T. D. (1994) A Radiographic Atlas of Osteoarthritis, pp. 1–45, Springer-Verlag, London [Google Scholar]
- 19. Jordan J. M., Linder G. F., Renner J. B., Fryer J. G. (1995) The impact of arthritis in rural populations. Arthritis Care Res. 8, 242–250 [DOI] [PubMed] [Google Scholar]
- 20. Chandrasekhar S., Esterman M. A., Hoffman H. A. (1987) Microdetermination of proteoglycans and glycosaminoglycans in the presence of guanidine hydrochloride. Anal. Biochem. 161, 103–108 [DOI] [PubMed] [Google Scholar]
- 21. Soderblom E. J., Philipp M., Thompson J. W., Caron M. G., Moseley M. A. (2011) Quantitative label-free phosphoproteomics strategy for multifaceted experimental designs. Anal. Chem. 83, 3758–3764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Vilím V., Vytásek R., Olejárová M., Machácek S., Gatterová J., Procházka B., Kraus V. B., Pavelka K. (2001) Serum cartilage oligomeric matrix protein reflects the presence of clinically diagnosed synovitis in patients with knee osteoarthritis. Osteoarthritis Cartilage 9, 612–618 [DOI] [PubMed] [Google Scholar]
- 23. Vilím V., Olejárová M., Machácek S., Gatterová J., Kraus V. B., Pavelka K. (2002) Serum levels of cartilage oligomeric matrix protein (COMP) correlate with radiographic progression of knee osteoarthritis. Osteoarthritis Cartilage 10, 707–713 [DOI] [PubMed] [Google Scholar]
- 24. Guo Y., Bozic D., Malashkevich V. N., Kammerer R. A., Schulthess T., Engel J. (1998) All-trans-retinol, vitamin D, and other hydrophobic compounds bind in the axial pore of the five-stranded coiled-coil domain of cartilage oligomeric matrix protein. EMBO J. 17, 5265–5272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Malashkevich V. N., Kammerer R. A., Efimov V. P., Schulthess T., Engel J. (1996) The crystal structure of a five-stranded coiled coil in COMP. A prototype ion channel? Science 274, 761–765 [DOI] [PubMed] [Google Scholar]
- 26. Vilím V., Vobùrka Z., Vytásek R., Senolt L., Tchetverikov I., Kraus V. B., Pavelka K. (2003) Monoclonal antibodies to human cartilage oligomeric matrix protein. Epitope mapping and characterization of sandwich ELISA. Clin. Chim. Acta 328, 59–69 [DOI] [PubMed] [Google Scholar]
- 27. Vilim V., Lenz M. E., Vytasek R., Masuda K., Pavelka K., Kuettner K. E., Thonar E. J. (1997) Characterization of monoclonal antibodies recognizing different fragments of cartilage oligomeric matrix protein in human body fluids. Arch. Biochem. Biophys. 341, 8–16 [DOI] [PubMed] [Google Scholar]
- 28. Sharif M., Kirwan J. R., Elson C. J., Granell R., Clarke S. (2004) Suggestion of nonlinear or phasic progression of knee osteoarthritis based on measurements of serum cartilage oligomeric matrix protein levels over five years. Arthritis Rheum. 50, 2479–2488 [DOI] [PubMed] [Google Scholar]
- 29. Leslie M., Fang C., Carlson C., Tulli H., Perris R., DiCesare P. (1999) Expression of cartilage oligomeric matrix protein (COMP) by human embryonic and adult osteoblasts. Trans. Orthop. Res. 24, 581. [DOI] [PubMed] [Google Scholar]
- 30. Sofat N. (2009) Analyzing the role of endogenous matrix molecules in the development of osteoarthritis. Int. J. Exp. Pathol. 90, 463–479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Clark A. G., Jordan J. M., Vilim V., Renner J. B., Dragomir A. D., Luta G., Kraus V. B. (1999) Serum cartilage oligomeric matrix protein reflects osteoarthritis presence and severity. The Johnston County Osteoarthritis Project. Arthritis Rheum. 42, 2356–2364 [DOI] [PubMed] [Google Scholar]
- 32. Golightly Y. M., Marshall S. W., Kraus V. B., Renner J. B., Villaveces A., Casteel C., Jordan J. M. (2011) Biomarkers of incident radiographic knee osteoarthritis. Do they vary by chronic knee symptoms? Arthritis Rheum. 63, 2276–2283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hunter D. J., Li J., LaValley M., Bauer D. C., Nevitt M., DeGroot J., Poole R., Eyre D., Guermazi A., Gale D., Felson D. T. (2007) Cartilage markers and their association with cartilage loss on magnetic resonance imaging in knee osteoarthritis: the Boston Osteoarthritis Knee Study. Arthritis Res. Ther. 9, R108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sowers M. F., Karvonen-Gutierrez C. A., Yosef M., Jannausch M., Jiang Y., Garnero P., Jacobson J. (2009) Longitudinal changes of serum COMP and urinary CTX-II predict X-ray defined knee osteoarthritis severity and stiffness in women. Osteoarthritis Cartilage 17, 1609–1614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Tseng S., Reddi A. H., Di Cesare P. E. (2009) Cartilage Oligomeric Matrix Protein (COMP). A biomarker of arthritis. Biomark Insights 4, 33–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Chaganti R. K., Kelman A., Lui L., Yao W., Javaid M. K., Bauer D., Nevitt M., Lane N. E. (2008) Change in serum measurements of cartilage oligomeric matrix protein and association with the development and worsening of radiographic hip osteoarthritis. Osteoarthritis Cartilage 16, 566–571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Conrozier T., Saxne T., Fan C. S., Mathieu P., Tron A. M., Heinegård D., Vignon E. (1998) Serum concentrations of cartilage oligomeric matrix protein and bone sialoprotein in hip osteoarthritis. A one-year prospective study. Ann. Rheum. Dis. 57, 527–532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kelman A., Lui L., Yao W., Krumme A., Nevitt M., Lane N. E. (2006) Association of higher levels of serum cartilage oligomeric matrix protein and N-telopeptide cross-links with the development of radiographic hip osteoarthritis in elderly women. Arthritis Rheum. 54, 236–243 [DOI] [PubMed] [Google Scholar]
- 39. Dragomir A. D., Kraus V. B., Renner J. B., Luta G., Clark A., Vilim V., Hochberg M. C., Helmick C. G., Jordan J. M. (2002) Serum cartilage oligomeric matrix protein and clinical signs and symptoms of potential pre-radiographic hip and knee pathology. Osteoarthritis Cartilage 10, 687–691 [DOI] [PubMed] [Google Scholar]
- 40. Kraus V. B., Kepler T. B., Stabler T., Renner J., Jordan J. (2010) First qualification study of serum biomarkers as indicators of total body burden of osteoarthritis. PLoS One 5, e9739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Maroudas A. (1975) Glycosaminoglycan turnover in articular cartilage. Philos. Trans. R. Soc. Lond. B Biol. Sci. 271, 293–313 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.