Background: Protein phosphorylation is an important posttranslational modification.
Results: Both in planta and when expressed in yeast, the P-type proton pump is phosphorylated at multiple new positions at its terminal regulatory domains.
Conclusion: Multiple methods for phosphopeptide enrichment are required for complete phosphosite mapping.
Significance: This work provides a surprising example of functional conservation of protein kinase action between plants and yeast.
Keywords: Membrane Proteins, Phosphoproteomics, Phosphorylation, Plasma Membrane Proton Pumps, Protein Kinases
Abstract
Phosphorylation is an important posttranslational modification of proteins in living cells and primarily serves regulatory purposes. Several methods were employed for isolating phosphopeptides from proteolytically digested plasma membranes of Arabidopsis thaliana. After a mass spectrometric analysis of the resulting peptides we could identify 10 different phosphorylation sites in plasma membrane H+-ATPases AHA1, AHA2, AHA3, and AHA4/11, five of which have not been reported before, bringing the total number of phosphosites up to 11, which is substantially higher than reported so far for any other P-type ATPase. Phosphosites were almost exclusively (9 of 10) in the terminal regulatory domains of the pumps. The AHA2 isoform was subsequently expressed in the yeast Saccharomyces cerevisiae. The plant protein was phosphorylated at multiple sites in yeast, and surprisingly, seven of nine of the phosphosites identified in AHA2 were identical in the plant and fungal systems even though none of the target sequences in AHA2 show homology to proteins of the fungal host. These findings suggest an unexpected accessibility of the terminal regulatory domain of plasma membrane H+-ATPase to protein kinase action.
Introduction
Post-translational modifications play important roles in a wide range of cellular functions. It is estimated that about one-third of all proteins in eukaryotic cells are phosphorylated at any given time (1). Reversible phosphorylation regulates the activity, stability, and spatial organization of large number of proteins (2). Post-translational regulation by phosphorylation of proteins has a key role in signal transduction cascades in cells (1, 3, 4). Furthermore, protein kinases influence protein-protein binding properties by regulating the phosphorylation-dependent binding of target motifs to modular protein domains (5, 6). Examples of domains that specifically bind to phosphorylated targets are the Src homology 2 (SH2) domain (7), the BRCA1 C-terminal (BRCT) domain (8), and the 14-3-3 protein (9).
Proteins are phosphorylated by protein kinases, a large family of highly related enzymes (10). In the yeast Saccharomyces cerevisiae, 122 protein kinases are present (11), humans have 518 putative protein kinases (12), and 836 and 1386 protein kinases have been identified in the genomes of the plants Arabidopsis thaliana and Oryza sativa (rice), respectively (13).
Characteristic for the catalytic function of protein kinases is that they recognize and phosphorylate linear motifs (11, 14–16). Linear motifs are short regions of proteins (typically of less than 10 residues) that often reside in regions without an ordered structure (17, 18). The short length of protein kinase target sequences and often poorly conserved sequences makes them difficult to predict with certainty (16). Furthermore, phosphorylation motifs are likely to arise/disappear spontaneously via mutations, for which reason they are evolutionarily labile. Unlike protein domains, which are conserved over long evolutionary distances, phosphorylation motifs often reside in fast-evolving regions (15, 17, 19, 20). These properties render phosphorylation sites difficult to align and trace evolutionarily (21–25). Given the presence of short linear and poorly conserved phosphorylation motifs, the number of potentially different phosphorylation sites becomes relatively small. With a large number of protein kinases it is, therefore, not surprising that target sequences often can be phosphorylated by more than one protein kinase, and single protein sequences can have several phosphorylation sites (26).
Quantitative mass spectrometry (MS) measurements of phosphorylation networks and their dynamics are now rapidly unraveling thousands of cellular phosphorylation sites (e.g. Refs. 27–38)). Even though such large scale phosphoproteomics studies do not capture all phosphorylated peptides under a given condition, large advances in enrichment strategies and mass spectrometry techniques have been made in the past few years (24, 39–41).
There are 11 isoforms of plasma membrane proton pumps in A. thaliana that are expressed throughout the plant. The closely related isoforms AHA13 and AHA2 together are essential for plant growth (42, 43). Plasma membrane proton pumps are phosphorylated at multiple sites in vivo (44), and six phosphosites have been identified so far. By combining several methods for extraction and enrichment of phosphopeptides, we could confirm the presence of five of these phosphosites and identified five new phosphosites. A commonly used tool for studying protein phosphorylation is expression of proteins in heterologous hosts, this being bacteria, yeast, or mammalian cell lines. We compared the phosphorylation profile of AHA2 in its natural environment with that of the plant pump expressed heterologously in the fungal host S. cerevisiae. The plant proton pump was phosphorylated to a large extent in yeast and, to our surprise, at several of the same sites as in Arabidopsis even though phosphorylated sequences did not share homology to any predicted yeast gene product.
EXPERIMENTAL PROCEDURES
Purification of A. thaliana Plasma Membranes
A. thaliana ecotype Columbia (Col-0) plantlets were grown in liquid cultures. Growth conditions involved 16 h light, 21 °C, 200 microeinsteins in ½ Murashige and Skoog medium including 30 mm sucrose for 8 days. After 8 days the medium was changed to ½ Murashige and Skoog for 24 h. Seedlings were homogenized in buffer that contained 50 mm MOPS-KOH, pH 7.5, 330 mm sucrose, 5 mm sodium ascorbate, 5 mm EDTA, and phosphatase inhibitors: 25 mm NaF, 50 mm sodium pyrophosphate (Na4O7P2), and 1 mm sodium molybdate (Na2MoO4). Plasma membrane vesicles were purified from the microsomal membrane fraction (10 000 × g) by two-phase partitioning at 4 °C (44, 45). The final plasma membrane pellet was suspended in sucrose 330 mm, 100 mm Tris-HCl, 1 mm EDTA, and 1 mm DTT.
Expression of AHA2 in Yeast
The S. cerevisiae strain RS-72 (46) was transformed and cultured essentially as described previously (47). In RS-72 (MATa ade1-100 his4-519 leu2-3,112), the natural constitutive promoter of the endogenous yeast plasma membrane H+-ATPase PMA1 was replaced by the galactose-dependent promoter of GAL1. A centromeric yeast expression vector (pMP1733) was used for expression of different versions of the AHA2 gene placed under control of the PMA1 promoter. Depending on the experiment, wild-type AHA2 (pMP 1625 or 1745).
Site-directed Mutagenesis
The construction of the wild -type H+-ATPase vector for heterologous expression in the yeast S. cerevisiae has been described (48). Mutants were constructed by site-directed mutagenesis using an overlap extension polymerase chain reaction. All mutated sequences were verified by DNA sequencing.
Yeast Complementation Assays
The yeast strain RS-72 was employed for functional complementation growth analysis. Plasmid-borne plant H+-ATPases carrying point mutations were tested for their ability to rescue a pma1 mutant on glucose medium. Each experiment was replicated independently three times, each time with cells from independent transformation events.
Isolation of Membrane from Transformed Yeast Cells
Cells were harvested, and membranes were isolated as described (49, 50) with the following modifications. Yeast cells were homogenized in the presence of phosphatase inhibitors: 50 mm Tris HCl, pH 7.5, 10% glycerol, 1 mm EDTA, 1 mm DTT, 25 mm NaF, 25 mm sodium pyrophosphate (Na4O7P2), and 10 mm sodium molybdate (Na2MoO4). Isolated membranes from transformed yeast cells were routinely subjected to Western blotting using anti-AHA2 antibodies to confirm equal expression of wild-type AHA2 and mutants derived from this protein. Protein concentration was determined by Bradford assay (51) using bovine serum albumin as reference
In Solution Digestion of Proteins
For determination of H+-ATPase phosphorylation in planta, plant plasma membrane vesicles (100 μg protein) were inverted by Brij58 (0.01%) or treated with urea (7 m) and thiourea (2 m) and sonicated for 5 min. The plasma membrane proteins were digested with trypsin overnight at 37 °C or with Lys-C for 4 h at room temperature followed with trypsin for overnight at 37 °C. Yeast microsomal membrane proteins containing recombinant AHA2 or His-tag purified AHA2-His6 were subjected to reduction by DTT and alkylation by IAM and subsequently digested individually with three proteases (trypsin, Lys-C, and Glu-C).
Phosphopeptide Enrichment by TiO2 Microcolumn
Peptide mixtures from 100 μg of plant plasma membrane proteins and 100–200 μg of yeast microsomal membrane proteins containing recombinant AHA2 were desalted with a Poros R3 column as described (52), and phosphopeptide enrichment was performed using titanium dioxide (TiO2) microcolumns as described (53).
Phosphopeptide Enrichment by IMAC
Peptide mixtures from 100 μg of plant plasma membrane proteins were desalted with a Poros R3 column, and phosphopeptide enrichment was performed using IMAC as described (54).
Phosphopeptide Enrichment by Calcium Phosphate Precipitation
Peptide mixtures from 100 μg of plant plasma membrane proteins were subjected to phosphopeptide enrichment by calcium phosphate precipitation as described (55). The pellet was dissolved with 5% FA, the peptide mixture was desalted with a Poros R3 column, and the bound peptides were eluted with TiO2 loading buffer (1 m glycolic acid in 80% acetonitrile and 5% TFA). Further phosphopeptide enrichment was performed by TiO2 microcolumn.
Mass Spectrometry
Samples were analyzed by an EASY nanoflow LC system (Proxeon Biosystems)-coupled LTQ-Orbitrap XL mass spectrometer (Thermo Fisher Scientific, Bremen, Germany). Samples resuspended in Solvent A (0.5% FA) were loaded onto a custom-made 15-cm analytical column (100-μm inner diameter, 375-μm outer diameter, packed with Reprosil C18, 3-μm reversed-phase particles (Dr. Maisch GmbH, Germany)) at a high flow rate of 550 nl/min. The peptides bound to the reversed phase material were eluted with a 50-min gradient of 0–34% Solvent B (90% acetonitrile, 0.5% FA). The instrument was operated in a data-dependent mode. The peptides were detected in the Orbitrap, and up to five of the most intense peptides were selected and subjected to fragmentation using Multistage Activation (MSA) method in the linear ion trap.
Data Analysis
Raw data from LTQ-Orbitrap MS were processed using Proteome Discoverer 1.0 (Thermo) and searched in an in-house Mascot server (Version 2.2.04, Matrix Science Ltd., London, UK). The NCBI nr data base was used as the searching data base, and A. thaliana and S. cerevisiae was used as taxons when appropriate.
Searching parameters were set as: tryptic (or Lys-C and Glu-C when appropriate) peptides with up to two missed cleavages sites; carbamidomethyl cysteine as a fixed modification; protein N-acetylation-oxidized methionine and phospho_STY, permitted as variable modifications. The results were searched with a peptide mass tolerance of 5 ppm and a fragment mass tolerance of ±0.6 Da. A decoy data base search was performed. Only peptides that identified as peptide rank 1 and with an expected value lower than 0.05 were considered as candidates. All phosphopeptides and phosphorylation sites presented in this work were validated manually.
RESULTS
Attempts to Get Complete Coverage of Phosphosites in Membrane Protein
By employing a systematic approach involving several different methods for enrichment of phosphopeptides, we succeeded in identifying five new in vivo phosphosites in the plant plasma membrane H+-ATPase (Fig. 1; Table 1). Our results demonstrate that a number of complementary methods are required to get a close to complete coverage of phosphosites present in the membrane-bound transporter.
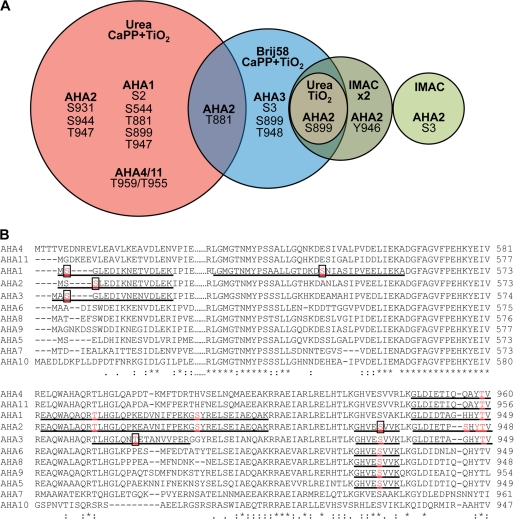
FIGURE 1.
Phosphosites identified in plasma membrane H+-ATPases homologously expressed in A thaliana membranes. A, phosphosites were determined in A. thaliana plasma membrane H+-ATPases using different approaches for phosphopeptide enrichment. Plasma membranes purified from A. thaliana seedlings were exposed for trypsin digestion directly, after treatment with Brij58, or after solubilization by urea/thiourea. IMAC, TiO2, and calcium phosphate precipitation followed by TiO2 (CaPP+TiO2) were used for phosphopeptide enrichment. Phosphorylation sites in the phosphopeptides were validated manually and are unique for the isoform of AHA mentioned in this figure with following exceptions: Site Ser-931 of AHA2 was determined in the peptide GHVEpSVVK, which can derive also from AHA2, AHA3, AHA5, AHA6, AHA8, AHA9, and Thr-959/955 in the peptide GLDIETIQQAYpTV that is shared by AHA4 and AHA11 isoforms. All the phosphopeptides determined are presented in the supplemental material. B, shown is ClustalW analysis of parts of the N terminus (N), P domain (P), and C terminus (C) of the ATPase isoforms of Arabidopsis. The identified phosphopeptides are underlined, and phosphorylated residues printed in bold and red. Novel residues are boxed.
TABLE 1.
Phosphorylation sites in AHAs expressed homologously in plant membranes
| Phosphorylation sites detected in AHA isoforms | Residue in AHA2a | Cytosolic domain | Conservation between AHAsb | Reference |
|---|---|---|---|---|
| Ser-2-AHA1 | Ser-2 | N terminus | This work | |
| Ser-3-AHA2 | Ser-3 | N terminus | This work | |
| Ser-3-AHA3 | ||||
| Ser-544-AHA1 | Ala-544 | P domain | 2/11 | This work |
| Thr-881-AHA1 | Thr-881 | C terminus | 10 (11)/11; Ser or Thr | (77, 78) and this work |
| Thr-881-AHA2 | This work | |||
| Thr-889-AHA3 | Lys-888 | C terminus | 1/11; | This work |
| Ser-899-AHA1 | Ser-899 | C terminus | 5 (7)/11; Ser or Thr | (57, 77) and this work |
| Ser-899-AHA2 | This work | |||
| Not detected in this work | Ser-904 | C terminus | 9/11 | (57) |
| Ser-931-AHA2/3/5/6/8/9c | Ser-931 | C terminus | 11/11 | This work |
| Ser-944-AHA2 | Ser-944 | C terminus | 1/11 | (79) and this work |
| Tyr-946-AHA2 | Tyr-946 | C terminus | 10/11 | (56) and this work |
| Thr-948-AHA1 | Thr-947 | C terminus | 11/11 | (44, 56, 57, 77, 78, 80) and this work |
| Thr-947-AHA2 | ||||
| Thr-948-AHA3 | ||||
| Thr-959/955-AHA4/11c |
a Homologous residues in A. thaliana AHA2.
b Number of sequences in which the residue is conserved out of the 11 AHAs encoded for in the genome of A. thaliana (81).
c The identified phosphopeptide cannot be distinguished between several isoforms (AHA2/3/5/6/8 and AHA4/11).
AHA proteins represented less than 3% of all proteins in the Arabidopsis plasma membranes employed. In previous proteomic studies of H+-ATPase phosphorylation, Brij58 was used to prepare inside-out vesicles before digestion (56). To increase the number of phosphorylated sites detected, we treated the membranes with urea/thiourea and employed different phosphopeptide enrichment methods such as IMAC, TiO2, and CaPP followed by TiO2. These different approaches resulted in identification of the phosphorylation sites presented in Fig. 1 and Table 1.
The phosphopeptides identified belong to four or five different isoforms of the plasma membrane H+-ATPases in Arabidopsis (AHA1, AHA2, AHA3 and AHA4/11; the last two isoforms could not be distinguished). We detected all phosphorylation sites previously identified in the C-terminal cytoplasmic domain (57, 58) with the numbering of AHA2: Thr-881, Ser-899, Ser-904, Ser-931, Ser-944, Tyr-946, and Thr-947, except for one, Ser-904 (Table 1). Furthermore, we identified new phosphorylation sites at the N terminus (Ser-2 and Ser-3), in the P-domain (Ser-544 in AHA1), and in C terminus of (Thr-889 in AHA3 and Ser-931 in AHA2). Interestingly, among the phosphorylation sites identified in planta all but one, Ser-544, were in the N- or C-terminal domains. Phosphorylation of Ser-544 was only observed in AHA1, and this residue is not conserved in other AHAs. As AHA2 is the best characterized plasma membrane H+-ATPase and a crystal structure is available for this pump (59), phosphorylated residues identified in AHA2 were, therefore, analyzed in more detail.
Functional Analysis of Phosphosites in N and C termini of AHA2
A well characterized function of the N- and C-terminal domains of plasma membrane H+-ATPases is to regulate pump function, possibly by restricting domain movements during catalysis and/or blocking access to the H+ entry pathway (60). This would suggest that the identified phosphorylation sites in these termini could play a role in regulation of enzyme activity. A tempting hypothesis is that phosphorylation of at least some of these sites abolishes the negative interaction between the termini and the rest of the pump.
To test whether introduction of a negative charge at these positions could result in an activated pump, we performed site-directed mutagenesis of all Ser (four residues), Thr (five residues), and Tyr (two residues) in the C terminus of AHA2 (Fig. 2A). Furthermore, we mutagenized Ser-2 and Ser-3 in the N-terminal domain of AHA2 (Fig. 2B). All residues were changed to either a negatively charged residue (Asp or Glu), to mimic the charge effect of phosphorylation, or to a neutral residue (Ala) and expressed in the yeast strain RS-72 (46). Yeast is equipped with its own plasma membrane H+-ATPase, Pma1p, but in the yeast strain RS-72 the endogenous PMA1 gene has been put under the control of a galactose dependent promoter. This implies that introduced plant plasma membrane H+-ATPase, the gene of which had been brought under control of the constitutive PMA1 promoter, will be produced and replace Pma1p when the cells are grown on glucose medium.
FIGURE 2.
Analysis of the importance of phosphosites in AHA2 for the activity of the pump by functional complementation of yeast pma1. The yeast strain RS-72 is dependent on the activity of the H+-ATPase expressed when grown on glucose media. When grown on galactose the endogenous yeast H+-ATPase PMA1 is expressed. A, shown is an analysis of pumps mutated in C-terminal phosphosites. B, shown is an analysis of pumps mutated in the N-terminal phosphosites.
Single-point mutations at the C terminus had profound effects. In the complementation test, transformed cells expressing mutants of Thr-924, Ser-931, Thr-942, Ser-944, Tyr-946, and Thr-947 exhibited growth that was considerably impaired compared with the wild-type AHA2 no matter whether they were mutated to Ala or Asp/Glu. This suggests a functional importance of these residues. In planta phosphorylation of Thr-947 is known to create a binding site for 14-3-3 protein that activates the pump after its interaction with the pump C terminus (61). When expressed in yeast, endogenous protein kinase(s) phosphorylates Thr-947, which is then allowed to form a complex with yeast 14-3-3 protein with pump activation as the result (62). This provides an explanation why both the T947A and the T947D mutations reduce the ability of AHA2 to complement pma1; T947A because it cannot get phosphorylated in yeast, and T947D because the introduction of a negative charge at this position is not sufficient to create a 14-3-3 protein binding site.
In the case of five AHA2 residues (Thr-881, Ser-899, Tyr-900, and Ser-904), the Ala substitution resulted in a pump with less ability to complement pma1, whereas Asp or Glu substitutions produced a pump that was far better in supporting yeast growth and even better than the wild-type AHA2. This suggests that phosphorylation of these residues results in an activated pump due to neutralization of the C-terminal constraint. Thr-881 has previously been demonstrated as important for regulation of AHA2 activity (44). It is located in the regulatory region R-I (49) and appears to be homologous to Thr-912 in yeast Pma1p, which is phosphorylated in yeast in response to glucose in the medium (63). Glucose activation of Pma1p results in an activated pump with strict coupling between ATP hydrolysis and proton pumping (64).
Substitution of Ser-899 with an Asp had a clear positive effect on the activity of the pump, but this residue is not conserved in plant plasma membrane H+-ATPases other than AHA1 and AHA2. This could indicate an isoform-specific regulation of AHA1/AHA2. aha2 S904D showed the most pronounced activation compared with wild-type AHA2 and aha2 S904A. Ser-904 has previously been identified as a phosphorylated residue in planta (56). Ser-904 is located in the regulatory region R-II (49), a part of the C terminus that is thought to form intramolecular binding to the core of the H+ pump protein as well as being important for stabilization of 14-3-3 binding (65, 66).
The C-terminal residue Ser-931 was consistently phosphorylated in planta and showed unusual properties. An Ala substitution at this position resulted in a pump more efficient in complementing pma1, whereas an Asp substitution gave rise to a mutant pump that could not support yeast growth in the absence of Pma1p, i.e. the opposite effect of the corresponding Thr-881 and Ser-904 mutations. Phosphorylation at this position by the Arabidopsis protein kinase PKS5/CIPK11/SnRK3.22, which is related to Snf1p in S. cerevisiae, has been shown to impair binding of 14-3-3 protein no matter whether the penultimate Thr is phosphorylated or not (48, 58).
Mutations of Ser-2 and Ser-3 at the N terminus did not affect the ability of the plant pump mutants to complement pma1 (Fig. 2A). A functional role of phosphorylation of these residues is, therefore, difficult to deduce. However, it remains a possibility that these events are important for interaction with regulatory proteins(s) present in the natural hosts but absent in the yeast system. In the future it will be important to separate those phosphorylation events that regulate activity from those that might regulate protein-protein interactions, trafficking, etc. in planta.
Phosphorylated Residues in Recombinant AHA2 Heterologously Expressed in Yeast Membranes
A number of Ala substitutions resulted in a plant pump less effective in supporting yeast growth, and this effect could be reversed by an Asp or Glu substitution (see above and Fig. 2). This suggested to us that the involved residues might already be subject to phosphorylation in the heterologous host, which would produce an activated pump. One such example, phosphorylation of AHA2 Thr-947 in yeast, has previously been reported (62, 67). We, therefore, decided to determine whether other phosphorylation sites could be identified in recombinant AHA2 produced in yeast.
As a starting point, we found that AHA2 protein represented around 5.5–10% of protein in the microsomal membrane fraction obtained from transformed yeast cells. When membrane fractions containing recombinant AHA2 were subjected to mass spectrometry, several phosphorylation sites were identified in the plant pump (Fig. 3). Surprisingly, seven of nine phosphorylation sites of AHA2 that we had identified in planta were also phosphorylated in yeast (Fig. 3 and Table 2: Ser-2, Ser-3, Ser-899, Ser-931, Ser-944, Tyr-946, and Thr-947). The two in planta phosphorylation sites that could not be identified in AHA2 produced in yeast were Ser-544 and Thr-881. In the reverse, in fungus, two phosphorylation sites identified in recombinant AHA2 were not observed in the plant material, namely Thr-511 and Thr-942.
FIGURE 3.
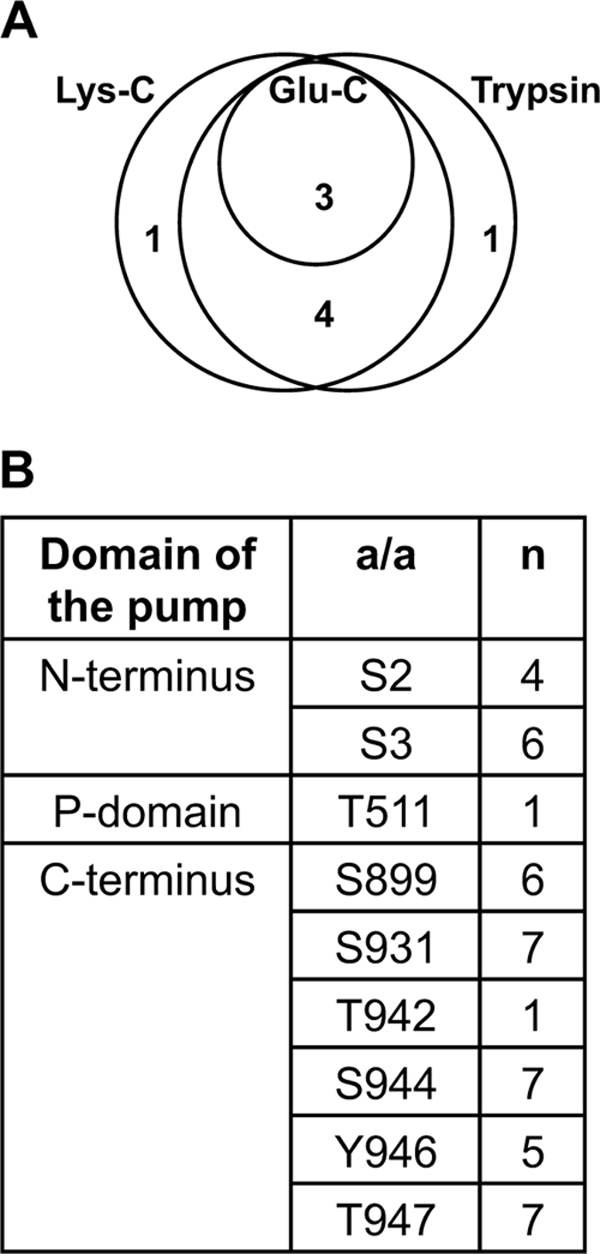
Phosphorylation of recombinant autoinhibited plasma membrane H+-ATPase 2 (AHA2) heterologously expressed in yeast membranes. A, shown is a Euler diagram of the number of phosphosites determined by digesting of AHA2 in a mixture of solubilized membrane proteins with trypsin, Lys-C, and Glu-C. Trypsin and Lys-C were of the same effectiveness and gave one unique site each. B, shown is a summary table of phosphosites in AHA2 determined using complementary digestion by three enzymes, their distribution within the H+-ATPase domains, and number of cases (n) when phosphosites were determined within a total number of 7 samples analyzed. a/a, amino acid. Almost all the phosphosites determined are in the N- and C-terminal ends of the pump.
TABLE 2.
Comparison of phosphorylation sites in AHAs expressed homologously in plant membranes with those of AHA2 expressed heterologously in yeast membranes
| Cytosolic domain | Phosphorylated in planta |
Phosphorylated in fungus |
|||
|---|---|---|---|---|---|
| AHA1 | AHA2 | AHA3 | AHA2 | Pma1p | |
| N-terminal domain | Ser-2 | Ser-2 | |||
| Ser-3 | Ser-3 | Ser-3 | |||
| Ser-11 | |||||
| Ser-12 | |||||
| Ser-14 | |||||
| Ser-52 | |||||
| Ser-61 | |||||
| Central part | Ser-464 | ||||
| Thr-511 | |||||
| Ser-544 | |||||
| Thr-647 | |||||
| C-terminal domain | Thr-881 | Thr-881 | |||
| Ser-899 | Ser-899 | Ser-899 | |||
| Thr-889 | |||||
| Ser-931 | Ser-931 | ||||
| Ser-942 | |||||
| Ser-944 | Ser-944 | ||||
| Tyr-946 | Tyr-946 | ||||
| Thr-948 | Thr-947 | Thr-948 | Thr-947 | ||
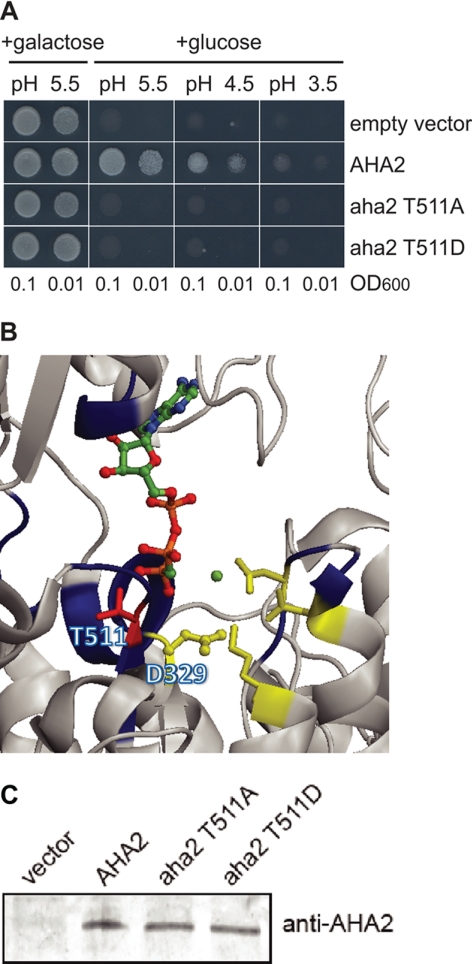
Thr-511 is situated in the cytoplasmic P-domain and was the only residue in yeast expressed AHA2 that was phosphorylated outside the N and C termini. The corresponding phosphopeptide was only observed once in yeast, and phosphorylation of this residue could not be detected in planta. Nevertheless, Thr-511 was analyzed in more detail (Fig. 4), as it is a highly conservative residue in P-type ATPases (68) and is situated in the conserved motif MXTGD and close to the Asp-329, which plays an essential role in the catalytic mechanism of the pump (Fig. 4B). Both Ala and Asp mutants completely abolished yeast growth (Fig. 4A) even though they were expressed at the same levels as wild-type AHA2 (Fig. 4C). This suggests a crucial role of this residue in the functioning of the enzyme. Obviously, in planta phosphorylation at this position, should it occur, would result in effective inhibition of pump activity.
FIGURE 4.
Phosphorylation of highly conservative Thr-551 situated in the catalytic center inhibits pump activity. A, shown is abolishment of yeast growth complementation when mutant in Thr-511 AHA2 was expressed. B, in the crystal structure of AHA2, Thr-511 is situated close to Asp-329, the conserved Asp phosphorylated during catalysis (59). C, shown is a Western blot analysis of expression level of the mutant AHA2 in yeast cells.
DISCUSSION
New in Planta Phosphosites Identified in Arabidopsis Plasma Membrane H+-ATPases
By employing complementary phosphopeptide enrichment methods, we have in this work identified 10 different in vivo phosphosites in different isoforms of the plasma membrane H+-ATPase (Table 1), five of which have not been reported before, bringing the total number of phosphosites in this pump up to 11, which is substantially higher than reported for any other P-type ATPase.
The phosphopeptides identified belong to four or five different isoforms (AHA1, AHA2, AHA3, and AHA4/11; the last two isoforms cannot be distinguished) of Arabidopsis plasma membrane H+-ATPases. We detected previously known sites, and in addition we have reported new in planta phosphosites at the N terminus (Ser-2, Ser-3), in the P-domain (Ser-544), and in the C terminus (Thr-889, Ser-931) of this essential plant pump. In a specific isoform, AHA2, we detected seven phosphosites in vivo. The seven sites include almost all known sites (except for Ser-904; see Ref. 57) and in addition two new sites (Ser-3 and Ser-931). Phosphorylation of Ser-931 was previously only demonstrated in tobacco (58) and not in any of the systematic screens performed on material from Arabidopsis, indicating the importance of complementary methods.
Comparison of Phosphosites in Homologously and Heterologously Produced Protein
Our data demonstrate that a heterologously expressed protein can undergo phosphorylation to a large extent. The phosphorylation patterns of AHA2 expressed in planta and in yeast membranes are strikingly similar (Table 2). Phosphosites covering Thr-881 and Ser-544 were the only exceptions being observed in material derived from plants only and not in recombinant AHA2 produced in yeast. Common for phosphorylation sites observed in AHA2 whether expressed in planta or in fungus is that they are situated in the N- and C-terminal domain, which in AHA2 play regulatory roles (60). These extensions of the plant plasma membrane H+-ATPase are not conserved in its fungal counterpart Pma1p (supplemental Figs. 1 and 2). For this reason it was unexpected to find a similar phosphorylation pattern between AHA2 expressed in planta and in yeast (Fig. 5B).
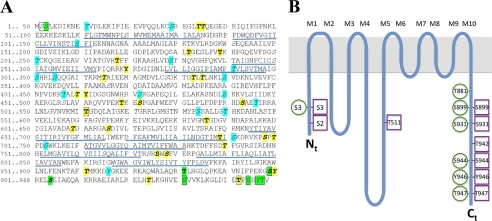
FIGURE 5.
Overview of phosphosites in AHA2. A, shown is the amino acid sequence of AHA2. Green color, phospho-residues identified in AHA2 expressed homologously in planta (this study and previous studies). Boxed, phosphosites identified in recombinant AHA2 expressed heterologously in yeast. Italics (and cyan when not phosphorylated in AHA2 in planta), Phosphosites were predicted by the NetPhosK server. Bold (and yellow when not phosphorylated in AHA2 in planta), phosphosites predicted by the PhosPhAt 3.0 server; underlined, membrane spanning domains in the crystal structure of AHA2 (59). B, shown is a spaghetti model of AHA2 with the phosphosites indicated. Green circles, phosphosites when expressed in planta. Magenta squares, phosphosites when expressed in yeast.
The phosphorylation patterns of AHA2, whether it be in planta or in fungus, could not have been predicted using common phosphophosite prediction servers such as NetPhosK and PhosPhAt 3.0. Phosphorylation of Ser-931, Ser-944, and Thr-947, which were phosphorylated in both biological systems, was not predicted by any server (Fig. 5). Furthermore, Thr-34, Thr-35, Thr-315, Thr-343, Thr-345, Thr-410, Ser-472, Ser-493, Thr-609, Ser-616, Ser-698, Thr-700, Thr-740, Ser-778, Thr-861, Tyr-900, and Thr-934 were predicted as protein kinase targets by both servers (labeled both in bold and italic in Fig. 5A) but were not phosphorylated in any of the two systems. Among these residues, none are in transmembrane segments or loops exposed to the extra-cytoplasmic side, and at least Thr-34, Thr-35, Thr-410, Ser-472, Thr-609, and Thr-700 are surface-exposed to various degrees in the crystal structure of AHA2 (supplemental Fig. 3; Ref. 59).
Several reasons for the observed high similarity between phosphorylation sites in the two distantly related organisms can be proposed. The most obvious possibility is perhaps that the structures of the terminal domains of plasma membrane proton pumps are in extended loose or helical forms that make them extremely accessible to protein kinase action. An extended structure is not unlikely given the fact that at least the C-terminal domain is predicted to make interactions with residues in several cytoplasmic domains (42).
More speculative models could be the following: (a) specific protein kinases phosphorylating plant plasma membrane H+-ATPases are conserved in yeast; (b) both organisms express nonspecific protein kinases that phosphorylate targets at all surface-accessible phosphorylation sites; (c) both organisms have their own palette of protein kinase(s) recognizing essentially the same phosphorylation sites.
Model (a) is unlikely be correct, as there is no sequence similarity between the terminal domains of AHA2 and its yeast counterpart Pma1p (supplemental Fig. 1), and there is no homology between the phosphorylation motifs of AHA2 and any other yeast gene product (supplemental Fig. 2). Still, we cannot entirely rule out the possibility that protein kinases in the two organisms have the same structure and/or substrate specificities even though not all possible substrates are present in each organism.
Model (b) above is not likely to be true as in vivo phosphosites are exclusively located in the terminal domains, whereas Ser and Thr residues, which according to the crystal structure of AHA2 (supplemental Fig. 3; Ref. 59) are exposed in cytosolic domains, fail to become phosphorylated. However, we cannot rule out the possibility that other conformations of AHA2 associated with transport or regulation allow for nonspecific kinases to phosphorylate the cytosolic domains.
According to the Model (c), protein kinases do not evolve according to their protein targets but rather as a group express a variety of potential binding preferences. If this is true, protein targets may preferentially evolve to become the substrate of preexisting protein kinases, which even they have the same specificity, need not be phylogenetically related.
All protein kinases have a very conserved active site that catalyzes phosphoryl transfer from ATP to a protein substrate (69, 70). Flexibility is achieved by separating the region of catalysis from that of molecular recognition, the docking groove (71–73). However, this separation is not absolute, and docking grooves of protein kinases are still intimately connected to the active sites, for which reason protein kinase docking grooves show a limited degree of modularity and evolvability (74–76). The fact that protein kinase recognition motifs can easily be copied by new targets, whereas docking grooves in protein kinases show less flexibility, provides a molecular rationale for Model (c) above.
AHA2 is so far the first example of a protein being targeted by yeast protein kinases in a similar manner as by protein kinases in its natural host. Whether this is a unique feature of AHA2 or a common phenomenon across kingdoms still needs to be demonstrated.
Conclusion
In this work we have demonstrated the power of combining different methods for enrichment of phosphopeptides when mapping phosphosites in a membrane protein. We identified five new phosphosites in the plant plasma membrane H+-ATPase, bringing the total number of phosphosites mapped in this pump up to 11. Most of the phosphosites are in terminal regulatory domains. When the AHA2 pump isoform is expressed in yeast it is phosphorylated at seven of these sites and at two more not observed in planta. The fact that phosphosites are almost identical in these two homologous and heterologous systems is surprising and could indicate that the terminal domains of proton pumps are unusually accessible for protein kinases.
Supplementary Material
Acknowledgment
We thank Mette Niemann for excellent technical support.
This work was supported by a Danish Research Council for Technology and Production grant (to M. G. P. and O. N. J.) and a Danish Basic Research Foundation grant (to M. G. P.).

This article contains supplemental Figs. 1–3.
- AHA
- autoinhibited H+-ATPase
- IMAC
- immobilized metal ion affinity chromatography
- FA
- formic acid.
REFERENCES
- 1. Cohen P. (2000) The regulation of protein function by multisite phosphorylation. A 25-year update. Trends Biochem. Sci. 25, 596–601 [DOI] [PubMed] [Google Scholar]
- 2. Ptacek J., Snyder M. (2006) Charging it up. Global analysis of protein phosphorylation. Trends Genet. 22, 545–554 [DOI] [PubMed] [Google Scholar]
- 3. Mann M., Jensen O. N. (2003) Proteomic analysis of post-translational modifications. Nat. Biotechnol. 21, 255–261 [DOI] [PubMed] [Google Scholar]
- 4. Ozlu N., Akten B., Timm W., Haseley N., Steen H., Steen J. A. (2010) Phosphoproteomics. Wiley Interdiscip. Rev. Syst. Biol. Med. 2, 255–276 [DOI] [PubMed] [Google Scholar]
- 5. Pawson T., Nash P. (2000) Protein-protein interactions define specificity in signal transduction. Genes Dev. 14, 1027–1047 [PubMed] [Google Scholar]
- 6. de Lichtenberg U., Jensen L. J., Brunak S., Bork P. (2005) Dynamic complex formation during the yeast cell cycle. Science 307, 724–727 [DOI] [PubMed] [Google Scholar]
- 7. Koch C. A., Anderson D., Moran M. F., Ellis C., Pawson T. (1991) SH2 and SH3 domains. Elements that control interactions of cytoplasmic signaling proteins. Science 252, 668–674 [DOI] [PubMed] [Google Scholar]
- 8. Glover J. N., Williams R. S., Lee M. S. (2004) Interactions between BRCT repeats and phosphoproteins. Tangled up in two. Trends Biochem. Sci. 29, 579–585 [DOI] [PubMed] [Google Scholar]
- 9. Morrison D. K. (2009) The 14-3-3 proteins. Integrators of diverse signaling cues that impact cell fate and cancer development. Trends Cell Biol. 19, 16–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Manning G., Plowman G. D., Hunter T., Sudarsanam S. (2002) Evolution of protein kinase signaling from yeast to man. Trends Biochem. Sci. 27, 514–520 [DOI] [PubMed] [Google Scholar]
- 11. Mok J., Kim P. M., Lam H. Y., Piccirillo S., Zhou X., Jeschke G. R., Sheridan D. L., Parker S. A., Desai V., Jwa M., Cameroni E., Niu H., Good M., Remenyi A., Ma J. L., Sheu Y. J., Sassi H. E., Sopko R., Chan C. S., De Virgilio C., Hollingsworth N. M., Lim W. A., Stern D. F., Stillman B., Andrews B. J., Gerstein M. B., Snyder M., Turk B. E. (2010) Deciphering protein kinase specificity through large scale analysis of yeast phosphorylation site motifs. Sci. Signal. 3, ra12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Manning G., Whyte D. B., Martinez R., Hunter T., Sudarsanam S. (2002) The protein kinase complement of the human genome. Science 298, 1912–1934 [DOI] [PubMed] [Google Scholar]
- 13. Krupa A., Anamika, Srinivasan N. (2006) Genome-wide comparative analyses of domain organization of repertoires of protein kinases of Arabidopsis thaliana and Oryza sativa. Gene 380, 1–13 [DOI] [PubMed] [Google Scholar]
- 14. Kemp B. E., Pearson R. B. (1990) Protein kinase recognition sequence motifs. Trends Biochem. Sci. 15, 342–346 [DOI] [PubMed] [Google Scholar]
- 15. Neduva V., Russell R. B. (2005) Linear motifs. Evolutionary interaction switches. FEBS Lett. 579, 3342–3345 [DOI] [PubMed] [Google Scholar]
- 16. Miller M. L., Jensen L. J., Diella F., Jørgensen C., Tinti M., Li L., Hsiung M., Parker S. A., Bordeaux J., Sicheritz-Ponten T., Olhovsky M., Pasculescu A., Alexander J., Knapp S., Blom N., Bork P., Li S., Cesareni G., Pawson T., Turk B. E., Yaffe M. B., Brunak S., Linding R. (2008) Linear motif atlas for phosphorylation-dependent signaling. Sci. Signal 1, ra2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Linding R., Russell R. B., Neduva V., Gibson T. J. (2003) GlobPlot. Exploring protein sequences for globularity and disorder. Nucleic Acids Res. 31, 3701–3708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gould C. M., Diella F., Via A., Puntervoll P., Gemünd C., Chabanis-Davidson S., Michael S., Sayadi A., Bryne J. C., Chica C., Seiler M., Davey N. E., Haslam N., Weatheritt R. J., Budd A., Hughes T., Pas J., Rychlewski L., Travé G., Aasland R., Helmer-Citterich M., Linding R., Gibson T. J. (2010) ELM: the status of the 2010 eukaryotic linear motif resource. Nucleic Acids Res. 38, D167–D180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Puntervoll P., Linding R., Gemünd C., Chabanis-Davidson S., Mattingsdal M., Cameron S., Martin D. M., Ausiello G., Brannetti B., Costantini A., Ferrè F., Maselli V., Via A., Cesareni G., Diella F., Superti-Furga G., Wyrwicz L., Ramu C., McGuigan C., Gudavalli R., Letunic I., Bork P., Rychlewski L., Küster B., Helmer-Citterich M., Hunter W. N., Aasland R., Gibson T. J. (2003) ELM server. A new resource for investigating short functional sites in modular eukaryotic proteins. Nucleic Acids Res. 31, 3625–3630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Moses A. M., Liku M. E., Li J. J., Durbin R. (2007) Regulatory evolution in proteins by turnover and lineage-specific changes of cyclin-dependent kinase consensus sites. Proc. Natl. Acad. Sci. U.S.A. 104, 17713–17718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Jiménez J. L., Hegemann B., Hutchins J. R., Peters J. M., Durbin R. (2007) A systematic comparative and structural analysis of protein phosphorylation sites based on the mtcPTM database. Genome Biol. 8, R90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Malik R., Nigg E. A., Körner R. (2008) Comparative conservation analysis of the human mitotic phosphoproteome. Bioinformatics 24, 1426–1432 [DOI] [PubMed] [Google Scholar]
- 23. Macek B., Gnad F., Soufi B., Kumar C., Olsen J. V., Mijakovic I., Mann M. (2008) Phosphoproteome analysis of E. coli reveals evolutionary conservation of bacterial Ser/Thr/Tyr phosphorylation. Mol. Cell. Proteomics 7, 299–307 [DOI] [PubMed] [Google Scholar]
- 24. Boekhorst J., van Breukelen B., Heck A., Jr., Snel B. (2008) Comparative phosphoproteomics reveals evolutionary and functional conservation of phosphorylation across eukaryotes. Genome Biol. 9, R144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Jensen L. J., Jensen T. S., de Lichtenberg U., Brunak S., Bork P. (2006) Co-evolution of transcriptional and post-translational cell-cycle regulation. Nature 443, 594–597 [DOI] [PubMed] [Google Scholar]
- 26. Landry C. R., Levy E. D., Michnick S. W. (2009) Weak functional constraints on phosphoproteomes. Trends Genet. 25, 193–197 [DOI] [PubMed] [Google Scholar]
- 27. Gruhler A., Olsen J. V., Mohammed S., Mortensen P., Faergeman N. J., Mann M., Jensen O. N. (2005) Quantitative phosphoproteomics applied to the yeast pheromone signaling pathway. Mol. Cell. Proteomics 4, 310–327 [DOI] [PubMed] [Google Scholar]
- 28. Collins M. O., Yu L., Coba M. P., Husi H., Campuzano I., Blackstock W. P., Choudhary J. S., Grant S. G. (2005) Proteomic analysis of in vivo phosphorylated synaptic proteins. J. Biol. Chem. 280, 5972–5982 [DOI] [PubMed] [Google Scholar]
- 29. Wolf-Yadlin A., Hautaniemi S., Lauffenburger D. A., White F. M. (2007) Multiple reaction monitoring for robust quantitative proteomic analysis of cellular signaling networks. Proc. Natl. Acad. Sci. U.S.A. 104, 5860–5865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Beausoleil S. A., Jedrychowski M., Schwartz D., Elias J. E., Villén J., Li J., Cohn M. A., Cantley L. C., Gygi S. P. (2004) Large-scale characterization of HeLa cell nuclear phosphoproteins. Proc. Natl. Acad. Sci. U.S.A. 101, 12130–12135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Olsen J. V., Blagoev B., Gnad F., Macek B., Kumar C., Mortensen P., Mann M. (2006) Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell 127, 635–648 [DOI] [PubMed] [Google Scholar]
- 32. Bodenmiller B., Mueller L. N., Pedrioli P. G., Pflieger D., Jünger M. A., Eng J. K., Aebersold R., Tao W. A. (2007) An integrated chemical, mass spectrometric, and computational strategy for (quantitative) phosphoproteomics. Application to Drosophila melanogaster Kc167 cells. Mol. Biosyst. 3, 275–286 [DOI] [PubMed] [Google Scholar]
- 33. Bodenmiller B., Wanka S., Kraft C., Urban J., Campbell D., Pedrioli P. G., Gerrits B., Picotti P., Lam H., Vitek O., Brusniak M. Y., Roschitzki B., Zhang C., Shokat K. M., Schlapbach R., Colman-Lerner A., Nolan G. P., Nesvizhskii A. I., Peter M., Loewith R., von Mering C., Aebersold R. (2010) Phosphoproteomic analysis reveals interconnected system-wide responses to perturbations of kinases and phosphatases in yeast. Sci. Signal. 3, rs4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Malmström J., Lee H., Aebersold R. (2007) Advances in proteomic workflows for systems biology. Curr. Opin. Biotechnol. 18, 378–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Matsuoka S., Ballif B. A., Smogorzewska A., McDonald E. R., 3rd, Hurov K. E., Luo J., Bakalarski C. E., Zhao Z., Solimini N., Lerenthal Y., Shiloh Y., Gygi S. P., Elledge S. J. (2007) ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science 316, 1160–1166 [DOI] [PubMed] [Google Scholar]
- 36. Rikova K., Guo A., Zeng Q., Possemato A., Yu J., Haack H., Nardone J., Lee K., Reeves C., Li Y., Hu Y., Tan Z., Stokes M., Sullivan L., Mitchell J., Wetzel R., Macneill J., Ren J. M., Yuan J., Bakalarski C. E., Villen J., Kornhauser J. M., Smith B., Li D., Zhou X., Gygi S. P., Gu T. L., Polakiewicz R. D., Rush J., Comb M. J. (2007) Global survey of phosphotyrosine signaling identifies oncogenic kinases in lung cancer. Cell 131, 1190–1203 [DOI] [PubMed] [Google Scholar]
- 37. Huttlin E. L., Jedrychowski M. P., Elias J. E., Goswami T., Rad R., Beausoleil S. A., Villén J., Haas W., Sowa M. E., Gygi S. P. (2010) A tissue-specific atlas of mouse protein phosphorylation and expression. Cell 143, 1174–1189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Rigbolt K. T., Prokhorova T. A., Akimov V., Henningsen J., Johansen P. T., Kratchmarova I., Kassem M., Mann M., Olsen J. V., Blagoev B. (2011) System-wide temporal characterization of the proteome and phosphoproteome of human embryonic stem cell differentiation. Sci. Signal. 4, rs3. [DOI] [PubMed] [Google Scholar]
- 39. Wilson-Grady J. T., Villén J., Gygi S. P. (2008) Phosphoproteome analysis of fission yeast. J. Proteome Res. 7, 1088–1097 [DOI] [PubMed] [Google Scholar]
- 40. Tan C. S., Bodenmiller B., Pasculescu A., Jovanovic M., Hengartner M. O., Jørgensen C., Bader G. D., Aebersold R., Pawson T., Linding R. (2009) Comparative analysis reveals conserved protein phosphorylation networks implicated in multiple diseases. Sci. Signal. 2, ra39. [DOI] [PubMed] [Google Scholar]
- 41. Nakagami H., Sugiyama N., Mochida K., Daudi A., Yoshida Y., Toyoda T., Tomita M., Ishihama Y., Shirasu K. (2010) Large scale comparative phosphoproteomics identifies conserved phosphorylation sites in plants. Plant Physiol. 153, 1161–1174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Palmgren M. G. (2001) Plant plasma membrane H+-ATPases. Powerhouses for nutrient uptake. Annu. Rev. Plant Physiol. Plant Mol. Biol. 52, 817–845 [DOI] [PubMed] [Google Scholar]
- 43. Haruta M., Burch H. L., Nelson R. B., Barrett-Wilt G., Kline K. G., Mohsin S. B., Young J. C., Otegui M. S., Sussman M. R. (2010) Molecular characterization of mutant Arabidopsis plants with reduced plasma membrane proton pump activity. J. Biol. Chem. 285, 17918–17929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Niittylä T., Fuglsang A. T., Palmgren M. G., Frommer W. B., Schulze W. X. (2007) Temporal analysis of sucrose-induced phosphorylation changes in plasma membrane proteins of Arabidopsis. Mol. Cell. Proteomics 6, 1711–1726 [DOI] [PubMed] [Google Scholar]
- 45. Larsson C., Sommarin M., Widell S. (1994) Isolation of highly purified plasma membranes and the separation of inside-out and right-side-out vesicles. Method Enzymol. 228, 451–469 [Google Scholar]
- 46. Cid A., Perona R., Serrano R. (1987) Replacement of the promoter of the yeast plasma membrane ATPase gene by a galactose-dependent promoter and its physiological consequences. Curr. Genet. 12, 105–110 [DOI] [PubMed] [Google Scholar]
- 47. Regenberg B., Villalba J. M., Lanfermeijer F. C., Palmgren M. G. (1995) C-terminal deletion analysis of plant plasma membrane H+-ATPase. Yeast as a model system for solute transport across the plant plasma membrane. Plant Cell 7, 1655–1666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Fuglsang A. T., Guo Y., Cuin T. A., Qiu Q., Song C., Kristiansen K. A., Bych K., Schulz A., Shabala S., Schumaker K. S., Palmgren M. G., Zhu J. K. (2007) Arabidopsis protein kinase PKS5 inhibits the plasma membrane H+-ATPase by preventing interaction with 14–3-3 protein. Plant Cell 19, 1617–1634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Axelsen K. B., Venema K., Jahn T., Baunsgaard L., Palmgren M. G. (1999) Molecular dissection of the C-terminal regulatory domain of the plant plasma membrane H+-ATPase AHA2. Mapping of residues that when altered give rise to an activated enzyme. Biochemistry 38, 7227–7234 [DOI] [PubMed] [Google Scholar]
- 50. Buch-Pedersen M. J., Palmgren M. G. (2003) Conserved Asp-684 in transmembrane segment M6 of the plant plasma membrane P-type proton pump AHA2 is a molecular determinant of proton translocation. J. Biol. Chem. 278, 17845–17851 [DOI] [PubMed] [Google Scholar]
- 51. Bradford M. M. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254 [DOI] [PubMed] [Google Scholar]
- 52. Gobom J., Nordhoff E., Mirgorodskaya E., Ekman R., Roepstorff P. (1999) Sample purification and preparation technique based on nano-scale reversed-phase columns for the sensitive analysis of complex peptide mixtures by matrix-assisted laser desorption/ionization mass spectrometry. J. Mass Spectrom. 34, 105–116 [DOI] [PubMed] [Google Scholar]
- 53. Thingholm T. E., Jørgensen T. J., Jensen O. N., Larsen M. R. (2006) Highly selective enrichment of phosphorylated peptides using titanium dioxide. Nat. Protoc. 1, 1929–1935 [DOI] [PubMed] [Google Scholar]
- 54. Ye J., Zhang X., Young C., Zhao X., Hao Q., Cheng L., Jensen O. N. (2010) Optimized IMAC-IMAC protocol for phosphopeptide recovery from complex biological samples. J. Proteome Res. 9, 3561–3573 [DOI] [PubMed] [Google Scholar]
- 55. Zhang X., Ye J., Jensen O. N., Roepstorff P. (2007) Highly efficient phosphopeptide enrichment by calcium phosphate precipitation combined with subsequent IMAC enrichment. Mol. Cell. Proteomics 6, 2032–2042 [DOI] [PubMed] [Google Scholar]
- 56. Nühse T. S., Stensballe A., Jensen O. N., Peck S. C. (2003) Large scale analysis of in vivo phosphorylated membrane proteins by immobilized metal ion affinity chromatography and mass spectrometry. Mol. Cell. Proteomics 2, 1234–1243 [DOI] [PubMed] [Google Scholar]
- 57. Nühse T. S., Stensballe A., Jensen O. N., Peck S. C. (2004) Phosphoproteomics of the Arabidopsis plasma membrane and a new phosphorylation site database. Plant Cell 16, 2394–2405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Duby G., Poreba W., Piotrowiak D., Bobik K., Derua R., Waelkens E., Boutry M. (2009) Activation of plant plasma membrane H+-ATPase by 14-3-3 proteins is negatively controlled by two phosphorylation sites within the H+-ATPase C-terminal region. J. Biol. Chem. 284, 4213–4221 [DOI] [PubMed] [Google Scholar]
- 59. Pedersen B. P., Buch-Pedersen M. J., Morth J. P., Palmgren M. G., Nissen P. (2007) Crystal structure of the plasma membrane proton pump. Nature 450, 1111–1114 [DOI] [PubMed] [Google Scholar]
- 60. Ekberg K., Palmgren M. G., Veierskov B., Buch-Pedersen M. J. (2010) A novel mechanism of P-type ATPase autoinhibition involving both termini of the protein. J. Biol. Chem. 285, 7344–7350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Olsson A., Svennelid F., Ek B., Sommarin M., Larsson C. (1998) A phosphothreonine residue at the C-terminal end of the plasma membrane H+-ATPase is protected by fusicoccin-induced 14-3-3 binding. Plant Physiol. 118, 551–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Jahn T. P., Schulz A., Taipalensuu J., Palmgren M. G. (2002) Post-translational modification of plant plasma membrane H+-ATPase as a requirement for functional complementation of a yeast transport mutant. J. Biol. Chem. 277, 6353–6358 [DOI] [PubMed] [Google Scholar]
- 63. Lecchi S., Nelson C. J., Allen K. E., Swaney D. L., Thompson K. L., Coon J. J., Sussman M. R., Slayman C. W. (2007) Tandem phosphorylation of Ser-911 and Thr-912 at the C terminus of yeast plasma membrane H+-ATPase leads to glucose-dependent activation. J. Biol. Chem. 282, 35471–35481 [DOI] [PubMed] [Google Scholar]
- 64. Venema K., Palmgren M. G. (1995) Metabolic modulation of transport coupling ratio in yeast plasma membrane H+-ATPase. J. Biol. Chem. 270, 19659–19667 [DOI] [PubMed] [Google Scholar]
- 65. Jelich-Ottmann C., Weiler E. W., Oecking C. (2001) Binding of regulatory 14-3-3 proteins to the C terminus of the plant plasma membrane H+-ATPpase involves part of its autoinhibitory region. J. Biol. Chem. 276, 39852–39857 [DOI] [PubMed] [Google Scholar]
- 66. Fuglsang A. T., Borch J., Bych K., Jahn T. P., Roepstorff P., Palmgren M. G. (2003) The binding site for regulatory 14-3-3 protein in plant plasma membrane H+-ATPase. Involvement of a region promoting phosphorylation-independent interaction in addition to the phosphorylation-dependent C-terminal end. J. Biol. Chem. 278, 42266–42272 [DOI] [PubMed] [Google Scholar]
- 67. Fuglsang A. T., Visconti S., Drumm K., Jahn T., Stensballe A., Mattei B., Jensen O. N., Aducci P., Palmgren M. G. (1999) Binding of 14-3-3 protein to the plasma membrane H+-ATPase AHA2 involves the three C-terminal residues Tyr-946—Thr–Val and requires phosphorylation of Thr-947. J. Biol. Chem. 274, 36774–36780 [DOI] [PubMed] [Google Scholar]
- 68. Axelsen K. B., Palmgren M. G. (1998) Evolution of substrate specificities in the P-type ATPase superfamily. J. Mol. Evol. 46, 84–101 [DOI] [PubMed] [Google Scholar]
- 69. Taylor S. S., Kornev A. P. (2011) Protein kinases. Evolution of dynamic regulatory proteins. Trends Biochem. Sci. 36, 65–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Kornev A. P., Taylor S. S. (2010) Defining the conserved internal architecture of a protein kinase. Biochim. Biophys. Acta 1804, 440–444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Holland P. M., Cooper J. A. (1999) Protein modification. Docking sites for kinases. Curr. Biol. 9, R329–R331 [DOI] [PubMed] [Google Scholar]
- 72. Reményi A., Good M. C., Lim W. A. (2006) Docking interactions in protein kinase and phosphatase networks. Curr. Opin Struct. Biol. 16, 676–685 [DOI] [PubMed] [Google Scholar]
- 73. Biondi R. M., Nebreda A. R. (2003) Signaling specificity of Ser/Thr protein kinases through docking site-mediated interactions. Biochem. J. 372, 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Won A. P., Garbarino J. E., Lim W. A. (2011) Recruitment interactions can override catalytic interactions in determining the functional identity of a protein kinase. Proc. Natl. Acad. Sci. U.S.A. 108, 9809–9814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Bhattacharyya R. P., Reményi A., Yeh B. J., Lim W. A. (2006) Domains, motifs, and scaffolds. The role of modular interactions in the evolution and wiring of cell signaling circuits. Annu. Rev. Biochem. 75, 655–680 [DOI] [PubMed] [Google Scholar]
- 76. Reményi A., Good M. C., Bhattacharyya R. P., Lim W. A. (2005) The role of docking interactions in mediating signaling input, output, and discrimination in the yeast MAPK network. Mol. Cell 20, 951–962 [DOI] [PubMed] [Google Scholar]
- 77. Nühse T. S., Bottrill A. R., Jones A. M., Peck S. C. (2007) Quantitative phosphoproteomic analysis of plasma membrane proteins reveals regulatory mechanisms of plant innate immune responses. Plant J. 51, 931–940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Reiland S., Messerli G., Baerenfaller K., Gerrits B., Endler A., Grossmann J., Gruissem W., Baginsky S. (2009) Large scale Arabidopsis phosphoproteome profiling reveals novel chloroplast kinase substrates and phosphorylation networks. Plant Physiol. 150, 889–903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Benschop J. J., Mohammed S., O'Flaherty M., Heck A. J., Slijper M., Menke F. L. (2007) Quantitative phosphoproteomics of early elicitor signaling in Arabidopsis. Mol. Cell. Proteomics 6, 1198–1214 [DOI] [PubMed] [Google Scholar]
- 80. Jones A. M., MacLean D., Studholme D. J., Serna-Sanz A., Andreasson E., Rathjen J. P., Peck S. C. (2009) Phosphoproteomic analysis of nuclei-enriched fractions from Arabidopsis thaliana. J. Proteomics 72, 439–451 [DOI] [PubMed] [Google Scholar]
- 81. Axelsen K. B., Palmgren M. G. (2001) Inventory of the superfamily of P-type ion pumps in Arabidopsis. Plant Physiol 126, 696–706 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.