Background: The FANCA (Fanconi anemia complementation group A) gene is mutated in ∼60% of Fanconi anemia patients, but no biochemical activity has been identified.
Results: Purified FANCA binds to nucleic acids of a certain length through its C terminus.
Conclusion: FANCA has an intrinsic affinity for nucleic acids with a strong preference for single-stranded forms.
Significance: Unveiling the biochemical activity of FANCA is critical for understanding its functions in DNA repair.
Keywords: DNA Damage, DNA Enzymes, DNA Repair, DNA Replication, Protein-DNA Interaction, FANCA, Fanconi Anemia, Interstrand Cross-link
Abstract
The Fanconi anemia complementation group A (FANCA) gene is one of 15 disease-causing genes and has been found to be mutated in ∼60% of Fanconi anemia patients. Using purified protein, we report that human FANCA has intrinsic affinity for nucleic acids. FANCA binds to both single-stranded (ssDNA) and double-stranded (dsDNA) DNAs; however, its affinity for ssDNA is significantly higher than for dsDNA in an electrophoretic mobility shift assay. FANCA also binds to RNA with an intriguingly higher affinity than its DNA counterpart. FANCA requires a certain length of nucleic acids for optimal binding. Using DNA and RNA ladders, we determined that the minimum number of nucleotides required for FANCA recognition is ∼30 for both DNA and RNA. By testing the affinity between FANCA and a variety of DNA structures, we found that a 5′-flap or 5′-tail on DNA facilitates its interaction with FANCA. A patient-derived FANCA truncation mutant (Q772X) has diminished affinity for both DNA and RNA. In contrast, the complementing C-terminal fragment of Q772X, C772–1455, retains the differentiated nucleic acid-binding activity (RNA > ssDNA > dsDNA), indicating that the nucleic acid-binding domain of FANCA is located primarily at its C terminus, where most disease-causing mutations are found.
Introduction
Fanconi anemia (FA)3 is an autosomal recessive or X-linked disorder characterized by bone marrow failure, developmental abnormalities, predisposition to cancer, and hypersensitivity to cross-linking agents (1–10). Thus far, 15 distinct genes have been identified to cause the deadly disease (6, 11–14). Eight of them are components of the FA core complex (FANCA, FANCB, FANCC, FANCE, FANCF, FANCG, FANCL, and FANCM) that monoubiquitinates both FANCD2 and FANCI, a key event that initiates interstrand cross-link (ICL) repair (6, 8, 9, 15). Downstream of the FANCD2 and FANCI monoubiquitination are double-strand break repair proteins (i.e. FANCD1/BRCA2, FANCJ/BRIP1, and FANCN/PALB2, FANCO/RAD51C, and FANCP/SLX4) that are critical to achieving successful ICL repair (6, 11–14, 16, 17). Although a deficiency in each FA gene shows similar clinical and cellular phenotypes, ∼85% of FA patients present defective FANCA (∼60%), FANCC (∼15%), or FANCG (∼10%) genes, and ∼15% of them have defects in the other FA genes (8, 9, 18).
FANCA physically interacts with FANCG and the transcription factor HES1 within the FA core complex (19–25), which has been found to be localized to chromatin (26, 27). FANCA was also found to be involved in psoralen ICL-induced mutagenesis and in spontaneous and UV light-induced base substitution mutagenesis in human cells, implying its involvement in the mutagenic translesion synthesis of DNA damage (28, 29). Additionally, FANCA was shown to be required for recruiting FANCO/RAD51 and FANCD1/BRCA2 into mitomycin C-induced nuclear foci, indicating its role in the homologous recombination repair of ICLs (12, 30, 31). These data imply that FANCA may play multiple roles in DNA metabolism and transactions. However, because FANCA is not evolutionarily conserved and lacks identifiable domains/motifs, it remains largely unknown how FANCA is involved in these biological processes.
In this study, we report that purified human FANCA binds to nucleic acids with strong preference for single-stranded forms. This novel property of FANCA supports its role in DNA damage repair and should be helpful in understanding how FANCA and the whole FA core complex contribute to the maintenance of genomic stability.
EXPERIMENTAL PROCEDURES
Expression and Purification of Human FANCA Proteins
cDNA for human FANCA was obtained by PCR amplification from a universal cDNA pool (BioChain Institute, Inc.). The full-length open reading frame of FANCA was sequenced and found to exactly match NCBI Reference Sequence NM_000135. Overexpression of hexahistidine-tagged FANCA was achieved in insect High Five cells using the Bac-to-Bac expression system (Invitrogen). Truncation mutants were produced through a PCR-based method (32). Expression of FANCA and its mutants was confirmed by Western blot analysis using a Pierce ECL kit. Antibodies against FANCA were kindly provided by the Fanconi Anemia Research Fund. Monoclonal antibody THE against the His6 tag (GenScript, Piscataway, NJ) was also used to confirm expression and subsequent purification. Upon expression of the recombinant proteins in insect cells, the cells were homogenized using a Dounce homogenizer to prepare extracts. Wild-type FANCA and various truncation mutants were purified using a HiTrap Q-Sepharose Fast Flow column; a 5-ml HiTrap Blue column; a Mono S, Mono Q, and/or Superdex 200 gel filtration column (GE Healthcare); and/or a 2-ml high-resolution hydroxylapatite column (Calbiochem) and by tracing FANCA protein through SDS-PAGE and Western blotting. Protein concentration was determined using the Coomassie (Bradford) protein assay reagent (Pierce). The purified proteins were stored at −80 °C in aliquots. Purified replication protein A (RPA) was prepared as described previously (33).
EMSA
Oligonucleotides that were used to create single-stranded DNA (ssDNA; 61-mer), double-stranded DNA (dsDNA; 61 bp), the 5′-tail (30-mer for the single-stranded part and 31 bp for the double-stranded part), the 3′-tail (30 bp for the double-stranded part and 31-mer for the single-stranded part), the splayed arm (30 bp for the double-stranded part and 31-mer for the single-stranded part), the 5′-flap (with a 31-mer flap), the 3′-flap (with a 31-mer flap), the static fork (all arms are 30 bp), and the static Holliday junction (all four arms are symmetrically 30 bp) were adopted from a design by Gari et al. (34) with the same sequences. It should be noted that there is a 1-base 5′-overhang, originally designed to label the 3′-end of the substrates, on the double-stranded area of the 5′- and 3′-tails, splayed arm, and 5′- and 3′-flaps and on one arm of the static fork and Holliday junctions. Annealing was carried out in a water bath within ∼5 h by slowly cooling from 85 °C to 20 °C. The quality of annealing was monitored by native gel electrophoresis. Proper annealing was verified by the mobility of a corresponding substrate, e.g. the static Holliday junction moves slowest because of its largest size. RNA was chemically synthesized by Integrated DNA Technologies, Inc. using the same sequence as the 61-mer ssDNA.
DNA binding EMSA analysis was performed as described previously (35) in a 10-μl reaction containing 25 mm Tris-HCl (pH 7.5), 100 mm NaCl, 5 mm EDTA, 1 mm DTT, 6% glycerol, 1 nm 5′-32P-labeled oligonucleotide substrates, and the indicated amounts of protein. The reactions were incubated at 18 °C for 45 min, followed by the addition of 4 μl of 50% (w/v) sucrose. The reaction mixtures were resolved by electrophoresis through a 4% nondenaturing polyacrylamide gel in 40 mm Tris acetate (pH 7.6) and 10 mm EDTA with 6% glycerol using the Owl P9DS electrophoresis system (Thermo Scientific). The setting was 100 V (∼1.5 watts/gel) for 40 or 90 min as indicated. DNA substrates and shifted bands were visualized by autoradiography. Quantitation of the bands was performed using NIH ImageJ software.
Determination of Dissociation Constant of FANCA for Nucleic Acids
At steady state (equilibrium), Kd can be determined through the following equation: Kd = [A][B]/[AB], where [A], [B], and [AB] are the concentrations of FANCA, nucleic acids, and the FANCA-nucleic acid complex, respectively. Because the concentration of nucleic acids ([B]) was very low in our EMSA experiments (1 nm), the FANCA protein concentration ([A]) that shifted 50% of nucleic acids ([AB] = [B], thus [B]/[AB] = 1) was used to estimate Kd.
Determination of Minimum Length of Nucleic Acids Required for FANCA Binding Using DNA and RNA Ladders
DNA ladders (10-, 20-, 30-, 40-, 50-, 61-, 71-, and 99-mers) were created by mixing oligonucleotides with different sequences and labeling with 32P. RNA ladders (10-, 20-, 30-, 40-, 50-, 60-, 70-, 80-, 90-, and 100-mers) was created as recommended by the manufacturer (Ambion) and labeled with 32P. 10-mer, 15-mer, and 30-mer oligonucleotides were the 3′-truncated forms of a 61-mer oligonucleotide: GACGCTGCCGAATTCTACCAGTGCCTTGCTAGGACATCTTTGCCCACCTGCAGGTTCACCC. Corresponding dsDNAs were prepared by annealing with the complementing oligonucleotides.
RESULTS
Purified Human FANCA Efficiently Interacts with Nucleic Acids
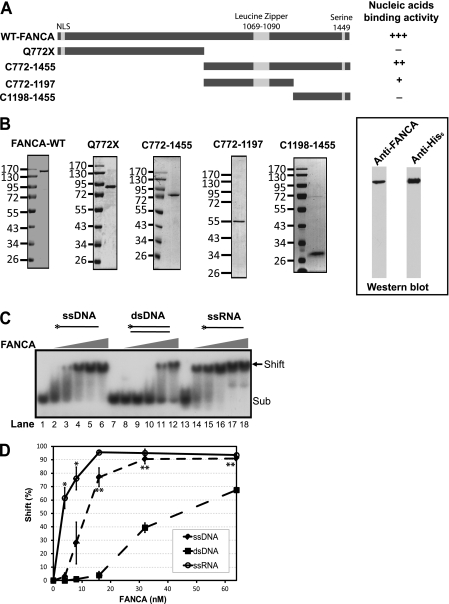
To study its biochemical properties, we overexpressed wild-type human FANCA protein in insect cells using the Bac-to-Bac expression system. As shown in Fig. 1B, we purified WT FANCA to near homogeneity. Purified WT FANCA migrated to a position corresponding to its calculated molecular mass of 164 kDa on SDS-polyacrylamide gel, indicating that it was the full-length protein. The identity of FANCA was further confirmed by Western blotting using a FANCA-specific antibody and an anti-His6 antibody (Fig. 1B).
FIGURE 1.
Purification of FANCA and its affinity for nucleic acids. A, schematic diagram of predicted FANCA motifs and strategy of mutagenesis. The range of numbers indicates how FANCA was truncated. NLS, predicted nuclear localization signal (amino acids 18–34 or 19–35). The leucine zipper (amino acids 1069–1090) and a phosphorylation site (serine) are also indicated. B, SDS-PAGE of the purified proteins stained with Coomassie Brilliant Blue R-250 and Western blotting. Q772X is a C-terminal truncation mutant derived from an FA patient. C772–1455 is C-terminal residues 772–1455 of FANCA. All peptides shown were tagged with hexahistidine at their N termini. Protein markers in kilodaltons are indicated. C, EMSA titration of WT FANCA for nucleic acid-binding activity. Diagrams of the nucleic acid substrates are shown on top of each set of reactions. The 32P-labeled 5′-end is indicated by an asterisk. The concentrations of FANCA indicated are 0, 4, 8, 16, 32, and 64 nm. The 61-mer nucleic acid substrate concentration was 1 nm. The reactions were resolved in 40-min nondenaturing gel electrophoresis. The protein-nucleic acid complex is indicated by an arrow. Sub, nucleic acid substrates. D, quantitation of three independent EMSA experiments with His6-tagged FANCA in C. Shift (%) is the ratio of the shifted band versus substrate band. Error bars represent S.D. A paired t test was performed to determine the statistical significance between ssDNA and dsDNA and between ssDNA and ssRNA. **, significant difference (p < 0.05) between ssDNA and dsDNA; *, significant difference (p < 0.05) between ssDNA and ssRNA.
Because FANCA has been shown to be involved in many steps of DNA repair, we reasoned that FANCA is likely to interact directly with DNA. Indeed, EMSA analysis by incubating increasing amounts of purified FANCA with 32P-labeled ssDNA or its dsDNA counterpart showed that FANCA bound to both ssDNA and dsDNA in a concentration-dependent manner (Fig. 1C). Intriguingly, FANCA had significantly greater affinity for ssDNA than for dsDNA. Using the paired t test, we determined the statistical significance of shifts between ssDNA and dsDNA (Fig. 1D). The p values for 16, 32, and 64 nm FANCA between ssDNA and dsDNA were 0.013, 0.031, and 0.014, respectively, which indicates a significant difference (p < 0.05). Furthermore, the Kd determined by protein titration showed that FANCA bound to ssDNA ∼4-fold better than to dsDNA (11.1 nm for ssDNA and 42.3 nm for dsDNA) (Table 1).
TABLE 1.
Dissociation constant (Kd) of FANCA for nucleic acids
Kd was calculated based on the following equation: Kd = [A][B]/[AB] (see “Experimental Procedures”). Different nucleic acids are listed in order based on their Kd values.
| Nucleic acid | Kd |
|---|---|
| nm | |
| ssRNA | 2.8 ± 0.49 |
| Splayed arm | 10.2 ± 0.07 |
| ssDNA | 11.1 ± 1.77 |
| 5′-Flap | 12.3 ± 0.49 |
| 5′-Tail | 12.6 ± 0.50 |
| 3′-Tail | 16.9 ± 4.38 |
| 3′-Flap | 17.2 ± 1.63 |
| Holliday junction | 27.0 ± 6.29 |
| Static fork | 35.2 ± 2.33 |
| dsDNA | 42.3 ± 3.89 |
Because FA is also a developmental disease and FANCA is involved in the regulation of gene expression (24, 25, 36), it is conceivable that FANCA could somehow be involved in RNA transactions, e.g. RNA stability, transcription, or translation, to carry out its functions. When FANCA was incubated with a single-stranded RNA (ssRNA) oligonucleotide with the same sequence as the ssDNA, we indeed observed that FANCA possessed affinity for RNA. In fact, its affinity for ssRNA determined by Kd was significantly better than for ssDNA (2.8 nm for ssRNA and 11.1 nm for ssDNA) (Table 1). The p values determined by paired t test for 4 and 8 nm FANCA between ssDNA and ssRNA showed a significant difference (0.041 and 0.033 for 4 and 8 nm FANCA, respectively) (Fig. 1D).
FANCA Requires Large Area of Nucleic Acids for Optimal Binding
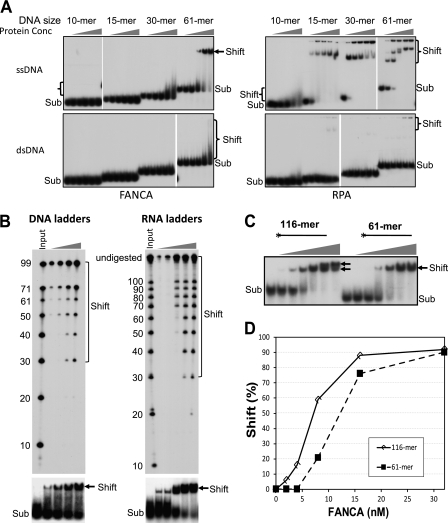
The preferential binding of FANCA to ssDNA resembles that of RPA, a well known ssDNA-binding protein that is involved in DNA replication, damage signaling, recombination, and repair (37–39). EMSA analysis of purified RPA (33) with ssDNA and dsDNA of different lengths indicated that RPA is very specific for ssDNA and forms protein-DNA filaments with increasing amounts of RPA and increasing sizes of ssDNA (multiple shifted bands in Fig. 2A, right panel). A high concentration of RPA could shift 10-nucleotide ssDNA (10-mer ssDNA in Fig. 2A, right panel); however, it did not efficiently interact with dsDNA, even when it was a 61-mer.
FIGURE 2.
FANCA requires large area of nucleic acids for optimal affinity. A, EMSA assays were performed by titration of FANCA and RPA with ssDNA and dsDNA of the indicated lengths. The concentrations of both FANCA and RPA were 0, 8, 16, 32, and 64 nm as indicated. The substrate concentration was 1 nm. The reactions were resolved in 90-min nondenaturing gel electrophoresis. The length of the DNA substrates is shown on top of each set of reactions. The protein-DNA complex is indicated by a bracket or an arrow. Sub, nucleic acid substrates. B, EMSA using DNA and RNA ladders (3 nm). The shifted bands (arrows) were recovered and resolved on a denaturing sequencing gel (upper panels). For DNA ladders, the concentrations of FANCA were 8, 16, 32, and 64 nm as indicated; for RNA ladders, the concentrations of FANCA were 2, 4, 8, 16, and 32 nm as indicated. Brackets indicated DNA or RNA species shifted by FANCA. DNA and RNA inputs with the indicated sizes in nucleotides are shown on the left of each gel. C, EMSA using 116- and 61-mer ssDNA oligonucleotides. The 32P-labeled 5′-end is indicated by an asterisk. The concentrations of FANCA were 0, 2, 4, 8, 16, 32, and 64 nm as indicated. Arrows indicate two different species of shifted bands. D, quantitation of EMSA experiments in C. Shift (%) is the ratio of the shifted band versus substrate band.
Unlike RPA, purified FANCA did not bind to ssDNA efficiently when it was shorter than 30 nucleotides (Fig. 2A, left panel). A nitrocellulose filter binding assay using labeled 25-mer ssDNA and RNA also demonstrated negative results for FANCA-nucleic acid interaction. When a 30-mer ssDNA oligonucleotide was used for EMSA, we observed an unstable interaction between FANCA and the ssDNA (smear bands in Fig. 2A, left panel). However, the interaction was dramatically improved when a 61-mer oligonucleotide was used, indicating that FANCA requires a larger area of DNA compared with RPA for optimal interaction. Also apart from RPA, FANCA began to interact with dsDNA when it was a 61-mer (61-mer dsDNA in Fig. 2A, left panel).
To further define the minimum size of nucleic acids for optimal interaction, we first incubated a set of ssDNA or RNA ladders with purified FANCA. After EMSA, we recovered the nucleic acids in the shifted bands and reran them on a denaturing sequencing gel. As shown in Fig. 2B, with increasing amounts of FANCA, more ssDNA and RNA species were identified in the shifted band. However, even with high concentrations of FANCA, ssDNA and RNA oligonucleotides shorter than 30 nucleotides were barely detectable in the shifted band, further supporting that FANCA requires ∼30 nucleotides of nucleic acids for efficient interaction.
On the basis of the molecular masses and migration distances of FANCA-DNA and RPA-DNA complexes on the EMSA gel, we reasoned that only one FANCA is bound to the 61-mer ssDNA oligonucleotide (61-mer ssDNA in Fig. 2A, left panel). Our next question was whether longer ssDNA has the capacity to accommodate more FANCA molecules and to form a FANCA-DNA filament. Using a synthetic 116-mer ssDNA oligonucleotide, which was almost double the 61-mer oligonucleotide, we found two distinct shifted bands with increasing amounts of FANCA (compare the shifted bands of the 116-mer and 61-mer oligonucleotides in Fig. 2C), indicating two FANCA molecules on one 116-mer ssDNA molecule. In contrast, only one shifted band corresponding to the faster band in the 116-mer experiment was observed with the 61-mer oligonucleotide. FANCA had greater overall affinity when the ssDNA was longer (compare the shifted bands between the 116- and 61-mers in Fig. 2, C and D).
FANCA Prefers DNA Structures with 5′-Flap or 5′-Tail
Because FA proteins are involved in maintaining the stability of replication forks (40, 41), we reasoned that the DNA-binding activity of FANCA is likely involved in recognition of branched structures. To test this possibility, we performed EMSA with purified WT FANCA and a variety of DNA structures, including the 5′- and 3′-tails, splayed arm, 5′- and 3′-flaps, static fork, and Holliday junction (Fig. 3) (35). Intriguingly, the affinity between FANCA and different DNA structures could be divided into three groups: Group I, the 5′-tail, 5′-flap, and splayed arm structures showed the highest close-to-ssDNA level affinity (Fig. 3, solid red lines); Group II, the 3′-tail and 3′-flap structures had lower affinity compared with Group I (Fig. 3, dashed green lines); and Group III, the static fork and Holliday junction possessed the lowest affinity for FANCA (Fig. 3, solid blue lines). The apparent Kd constants of FANCA for all structures also support this grouping (Table 1).
FIGURE 3.
FANCA prefers DNA structures with 5′-flap or 5′-tail. EMSA assays were performed by titration of FANCA with different DNA structures. The concentrations of FANCA were 8, 16, 32, and 64 nm. The substrate concentration was 1 nm. The reactions were resolved in 40-min nondenaturing gel electrophoresis, quantitated, and expressed as shift (%), which is the ratio of the shifted band versus substrate band. Diagrams of the DNA structures are shown below. Three groups of DNA structures are color-coded. Red asterisks indicate 32P labeling. RNA, ssDNA, and dsDNA were used as references for comparison. Error bars represent S.D.
Whereas the common feature of Group I structures is that they contain either a 5′-flap or a 5′-tail (31-mer), Group II DNAs have a 3′-flap or a 3′-tail (31-mer as well), and Group III structures barely have any free ends (only a 1-base 5′-overhang designed for 3′ labeling). On the basis of these observations, we conclude that FANCA prefers DNA structures with a 5′-flap or a 5′-tail.
Nucleic Acid-binding Domain of FANCA Is Located at Its C Terminus
FANCA does not have any identifiable domains for its interaction with nucleic acids. To define the nucleic acid-binding domain of FANCA, we first analyzed the primary structure of FANCA using the PSIPRED protein structure prediction server (University College London) and found that FANCA is predicted to contain extensive α-helices with a few short β-strands. On the basis of this prediction, we first chose a truncation mutant of FANCA (Q772X) derived from FA patients. Q772X is located roughly in the middle of the 1455-amino acid protein and in the middle of a coiled structure. EMSA of the purified truncation mutant (Fig. 1, A and B) indicated that the N-terminal moiety of FANCA had diminished nucleic acid-binding activity (Fig. 4, first panel).
FIGURE 4.
Nucleic acid-binding domain of FANCA. EMSA assays were performed with titration of the indicated FANCA truncation mutants. The concentrations of the mutants were 0, 8, 16, 32, and 64 nm as indicated. The 61-mer substrate concentration was 1 nm. The reactions were resolved in 40-min nondenaturing gel electrophoresis. Diagrams of the nucleic acid substrates are shown on top of each set of reactions. The 32P-labeled 5′-end is indicated by an asterisk. The protein-nucleic complex is indicated by brackets or an arrow. Sub, nucleic acid substrates.
However, the complementing fragment of Q772X, i.e. C-terminal amino acids 772–1455 of FANCA (Fig. 1, A and B), bound to nucleic acids more efficiently and retained the differentiated binding activity in the order of RNA > ssDNA > dsDNA (Fig. 4, second panel). To further define the nucleic acid-binding domain, we created two additional C-terminal truncation mutants (Fig. 1, A and B). EMSA tests indicated that the DNA-binding activity of both mutants was severely compromised (Fig. 4, third and fourth panels), although the leucine zipper-containing C772–1197 fragment retained partially affinity for ssDNA. The nucleic acid binding results are summarized in Fig. 1A. Overall, these data demonstrate that the nucleic acid-binding domain is located primarily at the C terminus of FANCA.
DISCUSSION
FA proteins have been generally believed to be involved in the repair of DNA ICLs that block replication and transcription (1–9). Although there are at least 15 FA disease-causing genes, ∼60% of the disease is caused by a deficiency in FANCA (8, 18). Thus far, the established DNA-interacting components (FANCM, FANCI, FANCD2, FANCJ, FANCO, and FANCP) account for only ∼5% of FA, an observation that does not seem to support the role of FA proteins in DNA repair (9). Discovery of the robust nucleic acid-binding activity of FANCA is important because it explains not only how FANCA localizes to chromatin but also provides solid biochemical support for its role in DNA repair.
An intriguing observation of this study is the preferential binding activity of FANCA for ssDNA over dsDNA. It has been known that recruitment of the FA core complex to chromatin relies strictly on replication (40, 41). Therefore, it is likely that FANCA recognizes the exposed ssDNA in the stalled replication forks, which look like a 5′-flap structure or a splayed arm, and contributes to assembly of the FA core complex on the stalled forks. It would be interesting to further investigate whether and how the DNA-binding activity of FANCA facilitates repair of cross-links through damage recognition, translesion synthesis (29), and homologous recombination events (30).
Thus far, FANCM-FAAP24 is the only identified DNA-binding component in the FA core complex (42, 43). FANCM can remodel stalled replication forks through fork reversal and branch migration, thus stabilizing the stalled replication forks and providing temporal and spatial access for the damage to be repaired (34, 44). FANCM appears to be responsible for recruitment of the FA core complex to chromatin (21, 42, 43, 45–48). The monoubiquitinated FANCI-FANCD2 complex may also be recruited to chromatin through a FANCM-dependent mechanism (15, 49–51). However, unlike other factors in the core complex, FANCM is not required for the formation of the eight-subunit (but not the 10-subunit) core complex (45), and FANCM−/− cells are only partially deficient in damage-induced FANCD2 monoubiquitination (52, 53). Fancm−/− knock-out mice further support that FANCM may have a stimulatory but not essential role in monoubiquitinating FANCD2 (54). Additionally, a direct interacting partner for FANCM-FAAP24 in the FA core complex has not been identified thus far, although FANCM-FAAP24 was originally identified through protein association in a FANCA-specific immunoprecipitation assay (5, 43, 55). FANCM−/− cells are sensitive to camptothecin, a topoisomerase inhibitor. Susceptibility to camptothecin is a unique feature identified only for FANCD1/BRCA2 and FANCN/PALB2 but not for components of the FA core complex (52). These observations suggest that FANCM may act downstream of FANCD2, and therefore, the upstream FA core complex may be recruited to DNA through other mechanisms, such as the DNA-binding activity of FANCA. Based on our observations, FANCA seems to be capable of recruiting the FA core complex to stalled replication forks through its ssDNA-binding activity and its preferential recognition of 5′-flap and splayed arm structures.
Another interesting insight to emerge from these studies is that FANCA has a higher affinity for ssRNA than for ssDNA. There is currently limited information to explain how the RNA-binding activity of FANCA could be linked to its functions. However, we think this activity may be physiologically relevant to RNA-related processes, such as transcription, translation, and RNA stability. First, the FA core complex has been reported to be involved in regulating gene expression through transcriptional (25) and post-transcriptional (56) mechanisms. Second, besides the nucleus, FANCA does localize to the cytoplasm (57–59), which supports a possible function in RNA metabolism. Third, FANCA has been shown to functionally interact with PKR (protein kinase regulated by RNA), a critical factor in translational control as well as regulation of cell proliferation and apoptosis (60). We speculate that, through its RNA-binding activity, FANCA may actively participate in these important biological processes. Further investigation into this issue should help us understand the unusually disproportional contribution of FANCA to FA.
The nucleic acid-binding domain of FANCA is located at its C terminus, where an imperfect leucine zipper and an ATR phosphorylation site are found (Fig. 1A) (61). It would be interesting to test whether the partial leucine zipper and the phosphorylation site have any effect on nucleic acid binding. It is very intriguing that, by analyzing the FANCA variants (1380 public entries as of March 23, 2011) available in the Fanconi Anemia Mutation Database, we found that ∼90% of the reported disease-causing point mutations of FANCA are located at the C terminus (from amino acids 772 to 1455), where the nucleic acid-binding domain is identified, further supporting the idea that FANCA is likely to exert its functions through its affinity for nucleic acids.
Acknowledgments
We thank Drs. Murray Deutscher and Mary Lou King (University of Miami) for reagents. We are grateful to Dr. Wei Yang (NIDDK, National Institutes of Health) and Dr. Murray Deutscher for critical comments and discussion.
This work was supported, in whole or in part, by National Institutes of Health Grant R01 HL105631 (to Y. Z.). This work was also supported by a new investigator research grant from the Florida Biomedical Research Program (to Y. Z.).
- FA
- Fanconi anemia
- ICL
- interstrand cross-link
- RPA
- replication protein A
- ssDNA
- single-stranded DNA
- dsDNA
- double-stranded DNA
- ssRNA
- single-stranded RNA.
REFERENCES
- 1. de Winter J. P., Joenje H. (2009) The genetic and molecular basis of Fanconi anemia. Mutat. Res. 668, 11–19 [DOI] [PubMed] [Google Scholar]
- 2. Joenje H., Patel K. J. (2001) The emerging genetic and molecular basis of Fanconi anaemia. Nat. Rev. Genet. 2, 446–457 [DOI] [PubMed] [Google Scholar]
- 3. Kutler D. I., Singh B., Satagopan J., Batish S. D., Berwick M., Giampietro P. F., Hanenberg H., Auerbach A. D. (2003) A 20-year perspective on the International Fanconi Anemia Registry (IFAR). Blood 101, 1249–1256 [DOI] [PubMed] [Google Scholar]
- 4. Niedernhofer L. J., Lalai A. S., Hoeijmakers J. H. (2005) Fanconi anemia cross-linked to DNA repair. Cell 123, 1191–1198 [DOI] [PubMed] [Google Scholar]
- 5. Thompson L. H., Hinz J. M. (2009) Cellular and molecular consequences of defective Fanconi anemia proteins in replication-coupled DNA repair: mechanistic insights. Mutat. Res. 668, 54–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wang W. (2007) Emergence of a DNA-damage response network consisting of Fanconi anaemia and BRCA proteins. Nat. Rev. Genet. 8, 735–748 [DOI] [PubMed] [Google Scholar]
- 7. D'Andrea A. D. (2010) Susceptibility pathways in Fanconi anemia and breast cancer. N. Engl. J. Med. 362, 1909–1919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Moldovan G. L., D'Andrea A. D. (2009) How the Fanconi anemia pathway guards the genome. Annu. Rev. Genet. 43, 223–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Yuan F., Song L., Qian L., Hu J. J., Zhang Y. (2010) Assembling an orchestra: Fanconi anemia pathway of DNA repair. Front. Biosci. 15, 1131–1149 [DOI] [PubMed] [Google Scholar]
- 10. Alter B. P. (2003) Cancer in Fanconi anemia, 1927–2001. Cancer 97, 425–440 [DOI] [PubMed] [Google Scholar]
- 11. Kee Y., D'Andrea A. D. (2010) Expanded roles of the Fanconi anemia pathway in preserving genomic stability. Genes Dev. 24, 1680–1694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Vaz F., Hanenberg H., Schuster B., Barker K., Wiek C., Erven V., Neveling K., Endt D., Kesterton I., Autore F., Fraternali F., Freund M., Hartmann L., Grimwade D., Roberts R. G., Schaal H., Mohammed S., Rahman N., Schindler D., Mathew C. G. (2010) Mutation of the RAD51C gene in a Fanconi anemia-like disorder. Nat. Genet. 42, 406–409 [DOI] [PubMed] [Google Scholar]
- 13. Kim Y., Lach F. P., Desetty R., Hanenberg H., Auerbach A. D., Smogorzewska A. (2011) Mutations of the SLX4 gene in Fanconi anemia. Nat. Genet. 43, 142–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Cybulski K. E., Howlett N. G. (2011) FANCP/SLX4: a Swiss army knife of DNA interstrand cross-link repair. Cell Cycle 10, 1757–1763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Smogorzewska A., Matsuoka S., Vinciguerra P., McDonald E. R., 3rd, Hurov K. E., Luo J., Ballif B. A., Gygi S. P., Hofmann K., D'Andrea A. D., Elledge S. J. (2007) Identification of the FANCI protein, a monoubiquitinated FANCD2 paralog required for DNA repair. Cell 129, 289–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Crossan G. P., van der Weyden L., Rosado I. V., Langevin F., Gaillard P. H., McIntyre R. E., Sanger Mouse Genetics Project, Gallagher F., Kettunen M. I., Lewis D. Y., Brindle K., Arends M. J., Adams D. J., Patel K. J. (2011) Disruption of mouse Slx4, a regulator of structure-specific nucleases, phenocopies Fanconi anemia. Nat. Genet. 43, 147–152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Stoepker C., Hain K., Schuster B., Hilhorst-Hofstee Y., Rooimans M. A., Steltenpool J., Oostra A. B., Eirich K., Korthof E. T., Nieuwint A. W., Jaspers N. G., Bettecken T., Joenje H., Schindler D., Rouse J., de Winter J. P. (2011) SLX4, a coordinator of structure-specific endonucleases, is mutated in a new Fanconi anemia subtype. Nat. Genet. 43, 138–141 [DOI] [PubMed] [Google Scholar]
- 18. Auerbach A. D. (2009) Fanconi anemia and its diagnosis. Mutat. Res. 668, 4–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Park S. J., Ciccone S. L., Beck B. D., Hwang B., Freie B., Clapp D. W., Lee S. H. (2004) Oxidative stress/damage induces multimerization and interaction of Fanconi anemia proteins. J. Biol. Chem. 279, 30053–30059 [DOI] [PubMed] [Google Scholar]
- 20. Christianson T. A., Bagby G. C. (2000) FANCA protein binds FANCG proteins in an intracellular complex. Blood 95, 725–726 [PubMed] [Google Scholar]
- 21. Garcia-Higuera I., Kuang Y., Denham J., D'Andrea A. D. (2000) The Fanconi anemia proteins FANCA and FANCG stabilize each other and promote the nuclear accumulation of the Fanconi anemia complex. Blood 96, 3224–3230 [PubMed] [Google Scholar]
- 22. Garcia-Higuera I., Kuang Y., Näf D., Wasik J., D'Andrea A. D. (1999) Fanconi anemia proteins FANCA, FANCC, and FANCG/XRCC9 interact in a functional nuclear complex. Mol. Cell. Biol. 19, 4866–4873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Reuter T., Herterich S., Bernhard O., Hoehn H., Gross H. J. (2000) Strong FANCA/FANCG but weak FANCA/FANCC interaction in the yeast two-hybrid system. Blood 95, 719–720 [PubMed] [Google Scholar]
- 24. Tremblay C. S., Huang F. F., Habi O., Huard C. C., Godin C., Lévesque G., Carreau M. (2008) HES1 is a novel interactor of the Fanconi anemia core complex. Blood 112, 2062–2070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Tremblay C. S., Huard C. C., Huang F. F., Habi O., Bourdages V., Lévesque G., Carreau M. (2009) The Fanconi anemia core complex acts as a transcriptional co-regulator in Hairy Enhancer of Split 1 signaling. J. Biol. Chem. 284, 13384–13395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Otsuki T., Furukawa Y., Ikeda K., Endo H., Yamashita T., Shinohara A., Iwamatsu A., Ozawa K., Liu J. M. (2001) Fanconi anemia protein FANCA associates with BRG1, a component of the human SWI/SNF complex. Hum. Mol. Genet. 10, 2651–2660 [DOI] [PubMed] [Google Scholar]
- 27. Qiao F., Moss A., Kupfer G. M. (2001) Fanconi anemia proteins localize to chromatin and the nuclear matrix in a DNA damage- and cell cycle-regulated manner. J. Biol. Chem. 276, 23391–23396 [DOI] [PubMed] [Google Scholar]
- 28. Papadopoulo D., Porfirio B., Moustacchi E. (1990) Mutagenic response of Fanconi anemia cells from a defined complementation group after treatment with photoactivated bifunctional psoralens. Cancer Res. 50, 3289–3294 [PubMed] [Google Scholar]
- 29. Mirchandani K. D., McCaffrey R. M., D'Andrea A. D. (2008) The Fanconi anemia core complex is required for efficient point mutagenesis and Rev1 foci assembly. DNA Repair 7, 902–911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Yang Y. G., Herceg Z., Nakanishi K., Demuth I., Piccoli C., Michelon J., Hildebrand G., Jasin M., Digweed M., Wang Z. Q. (2005) The Fanconi anemia group A protein modulates homologous repair of DNA double-strand breaks in mammalian cells. Carcinogenesis 26, 1731–1740 [DOI] [PubMed] [Google Scholar]
- 31. San Filippo J., Sung P., Klein H. (2008) Mechanism of eukaryotic homologous recombination. Annu. Rev. Biochem. 77, 229–257 [DOI] [PubMed] [Google Scholar]
- 32. Mao G., Pan X., Zhu B. B., Zhang Y., Yuan F., Huang J., Lovell M. A., Lee M. P., Markesbery W. R., Li G. M., Gu L. (2007) Identification and characterization of OGG1 mutations in patients with Alzheimer disease. Nucleic Acids Res. 35, 2759–2766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zhang Y., Yuan F., Presnell S. R., Tian K., Gao Y., Tomkinson A. E., Gu L., Li G. M. (2005) Reconstitution of 5′-directed human mismatch repair in a purified system. Cell 122, 693–705 [DOI] [PubMed] [Google Scholar]
- 34. Gari K., Décaillet C., Stasiak A. Z., Stasiak A., Constantinou A. (2008) The Fanconi anemia protein FANCM can promote branch migration of Holliday junctions and replication forks. Mol. Cell 29, 141–148 [DOI] [PubMed] [Google Scholar]
- 35. Yuan F., El Hokayem J., Zhou W., Zhang Y. (2009) FANCI protein binds to DNA and interacts with FANCD2 to recognize branched structures. J. Biol. Chem. 284, 24443–24452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Tulpule A., Lensch M. W., Miller J. D., Austin K., D'Andrea A., Schlaeger T. M., Shimamura A., Daley G. Q. (2010) Knockdown of Fanconi anemia genes in human embryonic stem cells reveals early developmental defects in the hematopoietic lineage. Blood 115, 3453–3462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wold M. S. (1997) Replication protein A: a heterotrimeric, single-stranded DNA-binding protein required for eukaryotic DNA metabolism. Annu. Rev. Biochem. 66, 61–92 [DOI] [PubMed] [Google Scholar]
- 38. Fanning E., Klimovich V., Nager A. R. (2006) A dynamic model for replication protein A (RPA) function in DNA processing pathways. Nucleic Acids Res. 34, 4126–4137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Oakley G. G., Patrick S. M. (2010) Replication protein A: directing traffic at the intersection of replication and repair. Front. Biosci. 15, 883–900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sobeck A., Stone S., Costanzo V., de Graaf B., Reuter T., de Winter J., Wallisch M., Akkari Y., Olson S., Wang W., Joenje H., Christian J. L., Lupardus P. J., Cimprich K. A., Gautier J., Hoatlin M. E. (2006) Fanconi anemia proteins are required to prevent accumulation of replication-associated DNA double-strand breaks. Mol. Cell. Biol. 26, 425–437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wang L. C., Stone S., Hoatlin M. E., Gautier J. (2008) Fanconi anemia proteins stabilize replication forks. DNA Repair 7, 1973–1981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ciccia A., Ling C., Coulthard R., Yan Z., Xue Y., Meetei A. R., Laghmani el H., Joenje H., McDonald N., de Winter J. P., Wang W., West S. C. (2007) Identification of FAAP24, a Fanconi anemia core complex protein that interacts with FANCM. Mol. Cell 25, 331–343 [DOI] [PubMed] [Google Scholar]
- 43. Meetei A. R., Medhurst A. L., Ling C., Xue Y., Singh T. R., Bier P., Steltenpool J., Stone S., Dokal I., Mathew C. G., Hoatlin M., Joenje H., de Winter J. P., Wang W. (2005) A human ortholog of archaeal DNA repair protein Hef is defective in Fanconi anemia complementation group M. Nat. Genet. 37, 958–963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Gari K., Décaillet C., Delannoy M., Wu L., Constantinou A. (2008) Remodeling of DNA replication structures by the branch point translocase FANCM. Proc. Natl. Acad. Sci. U.S.A. 105, 16107–16112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kim J. M., Kee Y., Gurtan A., D'Andrea A. D. (2008) Cell cycle-dependent chromatin loading of the Fanconi anemia core complex by FANCM/FAAP24. Blood 111, 5215–5222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Ciccia A., McDonald N., West S. C. (2008) Structural and functional relationships of the XPF/MUS81 family of proteins. Annu. Rev. Biochem. 77, 259–287 [DOI] [PubMed] [Google Scholar]
- 47. Medhurst A. L., Laghmani el H., Steltenpool J., Ferrer M., Fontaine C., de Groot J., Rooimans M. A., Scheper R. J., Meetei A. R., Wang W., Joenje H., de Winter J. P. (2006) Evidence for subcomplexes in the Fanconi anemia pathway. Blood 108, 2072–2080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ling C., Ishiai M., Ali A. M., Medhurst A. L., Neveling K., Kalb R., Yan Z., Xue Y., Oostra A. B., Auerbach A. D., Hoatlin M. E., Schindler D., Joenje H., de Winter J. P., Takata M., Meetei A. R., Wang W. (2007) FAAP100 is essential for activation of the Fanconi anemia-associated DNA damage response pathway. EMBO J. 26, 2104–2114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Mosedale G., Niedzwiedz W., Alpi A., Perrina F., Pereira-Leal J. B., Johnson M., Langevin F., Pace P., Patel K. J. (2005) The vertebrate Hef ortholog is a component of the Fanconi anemia tumor suppressor pathway. Nat. Struct. Mol. Biol. 12, 763–771 [DOI] [PubMed] [Google Scholar]
- 50. Garcia-Higuera I., Taniguchi T., Ganesan S., Meyn M. S., Timmers C., Hejna J., Grompe M., D'Andrea A. D. (2001) Interaction of the Fanconi anemia proteins and BRCA1 in a common pathway. Mol. Cell 7, 249–262 [DOI] [PubMed] [Google Scholar]
- 51. Montes de Oca R., Andreassen P. R., Margossian S. P., Gregory R. C., Taniguchi T., Wang X., Houghtaling S., Grompe M., D'Andrea A. D. (2005) Regulated interaction of the Fanconi anemia protein FANCD2 with chromatin. Blood 105, 1003–1009 [DOI] [PubMed] [Google Scholar]
- 52. Singh T. R., Bakker S. T., Agarwal S., Jansen M., Grassman E., Godthelp B. C., Ali A. M., Du C. H., Rooimans M. A., Fan Q., Wahengbam K., Steltenpool J., Andreassen P. R., Williams D. A., Joenje H., de Winter J. P., Meetei A. R. (2009) Impaired FANCD2 monoubiquitination and hypersensitivity to camptothecin uniquely characterize Fanconi anemia complementation group M. Blood 114, 174–180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Rosado I. V., Niedzwiedz W., Alpi A. F., Patel K. J. (2009) The Walker B motif in avian FANCM is required to limit sister chromatid exchanges but is dispensable for DNA cross-link repair. Nucleic Acids Res. 37, 4360–4370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Bakker S. T., van de Vrugt H. J., Rooimans M. A., Oostra A. B., Steltenpool J., Delzenne-Goette E., van der Wal A., van der Valk M., Joenje H., te Riele H., de Winter J. P. (2009) Fancm-deficient mice reveal unique features of Fanconi anemia complementation group M. Hum. Mol. Genet. 18, 3484–3495 [DOI] [PubMed] [Google Scholar]
- 55. Meetei A. R., Sechi S., Wallisch M., Yang D., Young M. K., Joenje H., Hoatlin M. E., Wang W. (2003) A multiprotein nuclear complex connects Fanconi anemia and Bloom syndrome. Mol. Cell. Biol. 23, 3417–3426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Li Y., Youssoufian H. (1997) MxA overexpression reveals a common genetic link in four Fanconi anemia complementation groups. J. Clin. Invest. 100, 2873–2880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Näf D., Kupfer G. M., Suliman A., Lambert K., D'Andrea A. D. (1998) Functional activity of the Fanconi anemia protein FAA requires FAC binding and nuclear localization. Mol. Cell. Biol. 18, 5952–5960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Kruyt F. A., Waisfisz Q., Dijkmans L. M., Hermsen M. A., Youssoufian H., Arwert F., Joenje H. (1997) Cytoplasmic localization of a functionally active Fanconi anemia group A-green fluorescent protein chimera in human 293 cells. Blood 90, 3288–3295 [PubMed] [Google Scholar]
- 59. Du W., Li J., Sipple J., Chen J., Pang Q. (2010) Cytoplasmic FANCA-FANCC complex interacts and stabilizes the cytoplasm-dislocalized leukemic nucleophosmin protein (NPMc). J. Biol. Chem. 285, 37436–37444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Zhang X., Li J., Sejas D. P., Rathbun K. R., Bagby G. C., Pang Q. (2004) The Fanconi anemia proteins functionally interact with the protein kinase regulated by RNA (PKR). J. Biol. Chem. 279, 43910–43919 [DOI] [PubMed] [Google Scholar]
- 61. Collins N. B., Wilson J. B., Bush T., Thomashevski A., Roberts K. J., Jones N. J., Kupfer G. M. (2009) ATR-dependent phosphorylation of FANCA on serine 1449 after DNA damage is important for FA pathway function. Blood 113, 2181–2190 [DOI] [PMC free article] [PubMed] [Google Scholar]