Background: UDP-galactopyranose mutase (UGM) requires the reduced FAD coenzyme to interconvert UDP-galactopyranose and UDP-galactofuranose.
Results: Structural perturbations of the coenzyme inhibit bond cleavage in the substrate.
Conclusion: Concerted bond breaking and formation between substrate and coenzyme occur during UGM catalysis.
Significance: Mechanistic understanding of UGM offers new insight for clinically relevant inhibitor design.
Keywords: Carbohydrate Metabolism, Enzyme Mechanisms, FAD, Galactose Metabolism, Tuberculosis, UDP-galactopyranose Mutase
Abstract
UDP-galactopyranose mutase (UGM) requires reduced FAD (FADred) to catalyze the reversible interconversion of UDP-galactopyranose (UDP-Galp) and UDP-galactofuranose (UDP-Galf). Recent structural and mechanistic studies of UGM have provided evidence for the existence of an FAD-Galf/p adduct as an intermediate in the catalytic cycle. These findings are consistent with Lewis acid/base chemistry involving nucleophilic attack by N5 of FADred at C1 of UDP-Galf/p. In this study, we employed a variety of FAD analogues to characterize the role of FADred in the UGM catalytic cycle using positional isotope exchange (PIX) and linear free energy relationship studies. PIX studies indicated that UGM reconstituted with 5-deaza-FADred is unable to catalyze PIX of the bridging C1–OPβ oxygen of UDP-Galp, suggesting a direct role for the FADred N5 atom in this process. In addition, analysis of kinetic linear free energy relationships of kcat versus the nucleophilicity of N5 of FADred gave a slope of ρ = −2.4 ± 0.4. Together, these findings are most consistent with a chemical mechanism for UGM involving an SN2-type displacement of UDP from UDP-Galf/p by N5 of FADred.
Introduction
UDP-galactopyranose mutase (UGM)2 is a flavoprotein that catalyzes the isomerization of UDP-galactopyranose (UDP-Galp; 1 in Fig. 1) and UDP-galactofuranose (UDP-Galf; 2). This reaction is essential for many pathogenic species of bacteria, protozoa, and fungi because UDP-Galf serves as the activated Galf donor during cell wall biosynthesis in these organisms (1, 2). Of particular clinical importance is the causative agent of tuberculosis, Mycobacterium tuberculosis, the cell wall of which possesses a galactan chain of ∼35 Galf residues that is essential for viability (3). Given the global prevalence of tuberculosis (World Health Organization Media Center) (4) and the increasing incidence of multidrug-resistant strains (6), UGM has become an attractive drug target because mammalian glycans do not contain Galf residues. UGM has also attracted much attention because it requires a reduced FAD coenzyme (FADred; 3) to catalyze a reversible ring contraction/expansion reaction that is redox-neutral (7). This has raised questions about the catalytic function of the coenzyme during turnover.
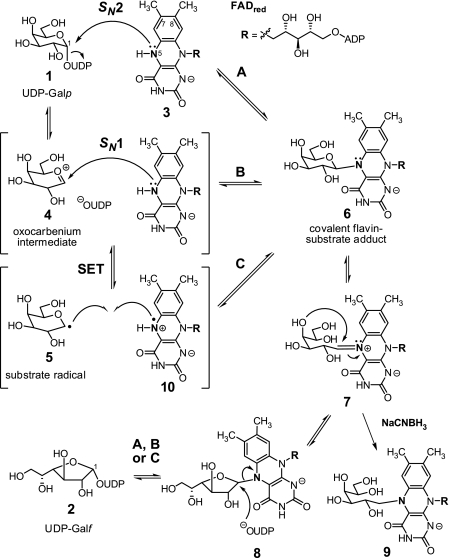
FIGURE 1.
Putative chemical mechanisms for UGM catalysis.
Several previous studies have provided important insights into the chemical mechanism of the UGM-catalyzed reaction. Using positional isotope exchange (PIX) studies, Blanchard and co-workers (8) demonstrated that the anomeric C1–OPβ bond is broken and reformed during turnover. Using NaCNBH3 as a chemical quenching agent to trap species 9 (Fig. 1), Kiessling and co-workers (9, 10) provided evidence for the intermediacy of the iminium ion (7) in the UGM catalytic cycle. Species 7 is likely derived from the FAD-substrate adducts 6 and 8, where N5 of the flavin is covalently linked to the anomeric carbon of the substrate (9, 10). Structures of reduced UGM determined in the presence of UDP-Galp by saturation transfer difference NMR spectroscopy (11) and x-ray crystallography (10) revealed that N5 of FADred is in close proximity to the anomeric carbon of the substrate, providing compelling evidence for the participation of N5 in nucleophilic attack at C1 of the substrate to form 6 and 8 during turnover.
Three mechanistic hypotheses have been proposed to explain the formation of 6 and 8 (Fig. 1). Generation of these intermediates may occur via nucleophilic attack by N5 of FADred at the anomeric carbon of 1 (or 2) concerted with cleavage of the C1–OPβ bond (Fig. 1, path A), which is reminiscent of typical SN2-type substitutions. Alternatively, formation of 6 and 8 may take place in a stepwise fashion similar to SN1-type substitutions (Fig. 1, path B), where elimination of UDP to produce an oxocarbenium intermediate (such as 4) precedes the nucleophilic attack by N5. It is also possible that the electron-deficient nature of 4 could facilitate single-electron transfer (SET) from FADred to form a radical pair (such as 5 and 10), followed by covalent bond formation to afford 6 and 8 (Fig. 1, path C). Indirect evidence for an oxocarbenium intermediate has come in the form of a significant rate reduction observed with UDP-[2-F]Galf (12) and the inability of UGM to displace UDP from the linear substrate analog UDP-galactitol (13).
In this study, we employed PIX and linear free energy relationships (LFERs) to further investigate the role of N5 of FADred during UGM catalysis. To determine whether N5 is indeed necessary for cleavage of the anomeric bond of UDP-Galp, PIX was monitored for the double-labeled UDP-Galp substrate in the presence of UGM reconstituted with reduced 5-deaza-FAD. The results were then corroborated by considering the perturbations to the rate of isomerization imposed upon changing the nucleophilicity of N5. Together, the results of these experiments are most consistent with an SN2-type displacement of UDP from the substrate by N5 of FADred (Fig. 1, path A).
EXPERIMENTAL PROCEDURES
UGM Expression and Purification
The glf gene encoding UGM was introduced into a pET-24b+ vector to generate the recombinant pQZ-1 plasmid (7). The plasmid was then transformed into Escherichia coli BL21 StarTM (DE3) for overexpression of UGM as a C-terminal His6 fusion protein. Cells were cultured in LB medium with 50 mg/liter kanamycin at 37 °C until the absorbance at 600 nm reached 0.6. Protein overexpression was induced by the addition of isopropyl β-d-thiogalactopyranoside to 0.1 mm, and the culture was allowed to incubate for an additional 18 h at 18 °C. The cells were then harvested by centrifugation, disrupted by sonication, and purified using nickel-nitrilotriacetic acid resin (Qiagen). UGM was purified as the holoenzyme with a bright yellow color and a distinct UV-visible absorption profile characteristic of tightly bound FAD. The purified enzyme was then flash-frozen in storage buffer (100 mm potassium Pi (pH 7.5) containing 15% glycerol) and stored at −80 °C.
Preparation of Double-labeled UDP-Galp
Double-labeled UDP-Galp was prepared according to published methods (8), where the double labels were introduced by incubating 100 mg of d-[1-13C]galactose (99%; Cambridge Isotope Laboratories) in 200 μl of H218O (95%, normalized; Fluka) at 55 °C for 2 days. This led to ∼80% incorporation of 18O at C1 as determined by mass spectrometry. A solution of 80 mm double-labeled galactose was then treated with pyruvate kinase and galactose kinase in the presence of 125 mm phosphoenolpyruvate and 6 mm ATP in 50 mm Tris buffer (pH 7.5) to generate [1-13C,1-18O]galactose 1-phosphate. The enzymes were removed by filtration (YM-10 membrane) following complete consumption of the labeled galactose as monitored by TLC. The filtrate was adjusted to pH 8.5 and treated with galactose-1-phosphate uridyltransferase, UDP-glucose pyrophosphorylase, and inorganic pyrophosphatase in the presence of 0.6 mm UDP-glucose and 100 mm UTP to form UDP-[1-13C,1-18O]Galp. The reaction was monitored by HPLC (see below for conditions). Enzymes were then removed by filtration (YM-10 membrane), and the product was purified using a DEAE-cellulose column eluted with a 0–0.1 m gradient of NH4HCO3 in water. The isolated UDP-Galp was determined to be 95% pure based on HPLC analysis. The extent of isotopic double-label incorporation into the UDP-Galp product was determined by 1H NMR and 13C NMR spectrometries and confirmed by high resolution electrospray ionization mass spectrometry (negative-ion mode), demonstrating peaks at m/z 566.0518 and 568.0530, corresponding to the 16O (calculated m/z 566.0511) and 18O (calculated m/z 568.0553) isotopologues, respectively.
Preparation of 7/8-Substituted FAD Analogues
The 7/8-substituted FAD analogues used in this study were prepared according to published methods (14). The identity of each compound was verified by 1H NMR and 31P NMR spectroscopies as well as by high resolution electrospray ionization mass spectrometry.
Preparation of Apo-UGM and Reconstitution with Other FAD Analogues
FAD was removed from the purified enzyme by the addition of a 2.6-fold volume excess of 3 m KBr in 20% glycerol to a 10 mg/ml enzyme stock in storage buffer (15). The resulting mixture was incubated on ice for 2 min before precipitating the enzyme by the addition of a 1.8-fold volume excess of saturated ammonium sulfate (pH 2.5). The enzyme precipitate was pelleted by centrifugation at 18,000 × g for 10 min. The clear yellow supernatant containing released FAD was decanted, and the protein pellet was redissolved in 4 ml of storage buffer. The high salt treatment was repeated a second time, and the resulting apo-UGM was dialyzed against storage buffer. Reconstitution of UGM with FAD analogues was carried out by incubating a 2.5 mg/ml solution of apo-UGM with an excess of flavin cofactor for 15 min at room temperature. The reconstituted enzyme/cofactor solution was then diluted with storage buffer to a concentration compatible with the enzyme assays. In the LFER studies, the molar ratio of cofactor to apo-UGM was ∼100:1. Enzyme concentrations were determined by the Bradford assay using bovine serum albumin as the protein standard.
PIX Time Course Experiments
PIX experiments were carried out at 27 °C in NMR tubes containing 50 mm potassium Pi (pH 7.5), 10% D2O (for locking the 13C NMR signal), 7 mm Na2S2O4, and ∼14 mm double-labeled UDP-Galp in a total volume of ∼630 μl. Apo-UGM was preincubated with the respective cofactor for 15 min before dilution into the reaction solution. In the 5-deaza-FAD/UGM and apo-UGM samples, the final enzyme concentration was 15 μm, and the 5-deaza-FAD concentration was ∼820 μm. In the FAD/UGM positive control, the final enzyme and FAD concentrations were 10 and 220 μm, respectively. All 13C NMR spectra were recorded using a 600-MHz Varian NMR spectrometer except for the reaction with apo-UGM, which was recorded using a 500-MHz Varian NMR spectrometer, at the Nuclear Magnetic Resonance Facility of the Department of Chemistry and Biochemistry of the University of Texas at Austin.
UGM Kinetic Assay
Initial rates for the conversion of UDP-Galf to UDP-Galp by reconstituted UGM were determined according to a previously described discontinuous assay (12). Reactions were run in 50 mm potassium Pi (pH 7.5) containing 7 mm Na2S2O4 and 10–500 μm UDP-Galf in a total volume of 30 μl. The temperature was maintained at 37 °C using a water bath. Enzyme concentrations varied from 0.015 to 0.2 μm depending on the analogue tested. Reactions were initiated upon the addition of UDP-Galf, quenched with a 3-fold excess of methanol, and centrifuged to remove precipitated protein. The resulting supernatant was dried in a speed vacuum concentrator prior to HPLC analysis. The fraction of reaction was determined based on the relative integrations of the UDP-Galf and UDP-Galp peaks and used to calculate the initial rate of reaction.
HPLC Analysis
HPLC analysis was performed at room temperature using a Varian Microsorb-MV 100-5 C18 column (250 × 4.6 mm) with UV detection at 262 nm. Solvent A contained 50 mm potassium Pi and 2.5 mm tetrabutylammonium hydrogen sulfate (pH 6.9) in H2O, and solvent B contained 50 mm potassium Pi and 2.5 mm tetrabutylammonium hydrogen sulfate (pH 6.9) in 50% H2O/acetonitrile. Isocratic elution was performed at 96% solvent A at a flow rate of 1 ml/min.
RESULTS
PIX Equilibration by Apoenzyme Is Not Accelerated in the Presence of 5-Deazaflavin
As described previously (8), the C1 13C NMR signal of UDP-Galp is split into a doublet by the adjacent β-phosphate and demonstrates an upfield shift of ∼0.03 ppm when the anomeric oxygen is replaced with 18O. The resulting two doublets were deconvoluted and integrated using Varian VnmrJ software and a multicomponent fit with Lorentzian line shapes. A 13C NMR example spectrum is provided in supplemental Fig. S2 along with the deconvolution. 13C NMR and mass spectrometry results indicated that the double-labeled UDP-Galp (11) was prepared with >99% incorporation of 13C at C1 and 77 ± 4% incorporation of 18O at the bridging C1–OPβ position. All remaining atoms in the substrate were at natural isotopic abundance.
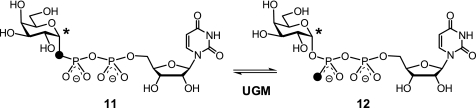
PIX experiments were conducted under reducing conditions (7 mm sodium dithionite) in NMR tubes. The change in the fraction (f) of 18O versus 16O bound to 13C1 was monitored versus time (t) by 13C NMR spectroscopy. (13C NMR stack plots are shown in supplemental Fig. S2.) Complete scrambling of the Pβ oxygens (Fig. 2) predicts the fraction of 18O at the bridging C1–OPβ position to ultimately reach an equilibrium value of feq = 0.26 (the product of the initial 18O enrichment in our double-labeled UDP-Galp substrate and the statistical factor of one-third). In the presence of 10 μm UGM reconstituted with FADred, PIX scrambling (11 ⇆ 12) was >90% complete within 20 min of incubation. In contrast, PIX was considerably slower in the presence of either 15 μm apo-UGM or UGM reconstituted with 5-deaza-FADred (Fig. 3), occurring over a time scale of >7 h. This permitted f to be measured every 60 min and subsequently fit using Equation 1 to extract the first-order rate constant for PIX (16),
where feq is the final value of f after complete equilibration of the exchangeable oxygens, and Δf is the difference between the initial value of f and feq. The first-order rate constant (kPIX) describes the approach to the PIX equilibrium under the experimental conditions and is equivalent to the positional exchange rate (17) normalized for the total initial substrate concentration, which was held constant and saturating in the experiments. The parameters kPIX and Δf were both allowed to float during nonlinear fitting, whereas feq was fixed at 0.26.
FIGURE 2.
Rationale for PIX experiments.
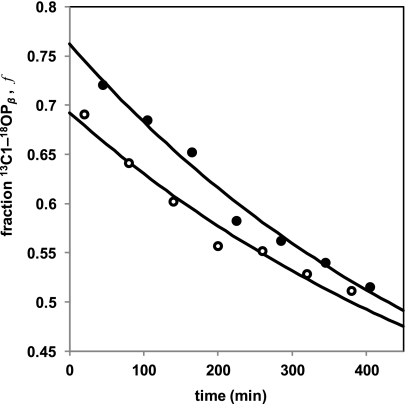
FIGURE 3.
Comparison of PIX rates for apo-UGM (○) versus UGM reconstituted with 5-deaza-FAD (●). Each time course is a plot of the fraction (f) of UDP-Galp possessing 18O at the bridging C1–OPβ position as determined by 13C NMR versus time. As a reference, the PIX rate was ∼100 times faster when UGM was reconstituted with FADred. Time courses were determined in separate experiments leading to the difference in the ordinate intercepts (see supplemental data for details). Reactions were run at 27 °C in potassium Pi (pH 7.5), 10% D2O, and 7 mm Na2S2O4 with 15 μm enzyme. Progress curves were obtained by fitting Equation 1 to the observed time courses using nonlinear regression.
The observed values of kPIX for apo-UGM and UGM reconstituted with 5-deaza-FADred were 0.0015 ± 0.0001 and 0.0017 ± 0.0001 min−1, respectively, ∼100-fold smaller than the kPIX for UGM reconstituted with FADred. These values of kPIX are significantly different from zero (p < 0.0001); however, they are not significantly different from one another (p > 0.3). No PIX scrambling was observed over a 24-h period when UGM was omitted from the reaction solution. Additional details regarding data fitting and statistical analysis are provided in the supplemental data.
FAD Analogues Yield Negative ρ Value in LFER Studies
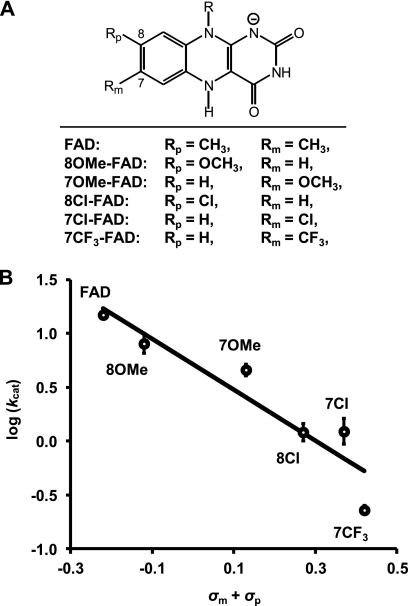
Following reconstitution of apo-UGM with each FAD analog (Fig. 4A), the turnover number (kcat) for conversion of UDP-Galf to UDP-Galp (Fig. 1, 2 → 1) was determined from plots of the initial rate (vi) (12, 15) versus substrate concentration (si) according to the structural relation (Equation 2),
where e0 is the total enzyme concentration, and Km is the Michaelis constant. Controls were also performed with ratios of 1000:1 for the 8-methoxy-FAD and 7-chloro-FAD analogues versus apo-UGM. No significant differences were observed in the fitted kinetic parameters, implying that the variations in kcat were not a result of subsaturating flavin concentrations. The resulting plot of log10(kcat) versus the sum of the substituent constants at positions 7 and 8 of the FAD analogues, denoted σm and σp, respectively, is shown in Fig. 4B. The values of σm and σp are based on the ionization of phenylacetic acid in water (18); however, substituent constants based on the ionization of benzoic acid in water gave similar results. On the basis of an analysis of variance, we found no evidence for either unequal expression of para versus meta effects in the LFER, i.e. ρm ≠ ρp(19), or non-additivity, i.e. higher order terms of σ. The value of the susceptibility factor (ρ) estimated from the linear correlation was −2.4 ± 0.4 and is significantly different from zero (p < 0.01). Additional details regarding data fitting and statistical analysis are provided in the supplemental data.
FIGURE 4.
LFER between kcat for UGM-catalyzed isomerization of UDP-Galf to UDP-Galp and electron-withdrawing effects due to para/meta substitution versus N5 of FADred. A, list of FAD analogues prepared for measuring the LFER. B, Hammett plot for the correlation of log10(kcat) versus the sum of σp and σm. The fit was obtained from unweighted least-squares linear regression, and error bars denote 1 S.E. of log10(kcat) above and below the observed value. OMe, methoxy; Cl, chloro; CF3, trifluoromethyl.
DISCUSSION
Evidence for the cleavage of the anomeric UDP-Galp bond during turnover by UGM was first provided by Blanchard and co-workers (8) using PIX. In this study, incubation of UGM with UDP-[1-13C]Galp enriched with 18O at the bridging oxygen between C1 and the β-phosphate (11) was monitored by 13C NMR spectroscopy. When 18O is replaced with 16O at the bridging position in the double-labeled substrate during turnover (see 12), a well resolved downfield shift of the 13C1 resonance occurs, with the change in the ratio of the 13C–18O and 13C–16O signals indicative of bond cleavage and reformation during catalysis (Fig. 2). It has also been shown that UGM reconstituted with 5-deaza-FAD does not catalyze interconversion of UDP-Galf and UDP-Galp (15). This observation was initially interpreted as support for a mechanism involving SET (Fig. 1, path C) because 5-deaza-FAD is restricted to 2-electron processes (20). Nevertheless, this indirect evidence does not exclude the possibility of a nucleophilic role for N5 of FADred and, in particular, an SN2-type process if it is required for expulsion of the UDP moiety.
As an initial test of the hypothesis that N5 is directly involved in the cleavage of the anomeric bond of UDP-Galp to form the flavin-substrate adduct (6 and 8), we considered the ability of UGM to utilize 5-deaza-FAD to catalyze PIX of the Pβ oxygens in UDP-Galp. Reconstitution of apo-UGM with 5-deaza-FAD showed no effect on an observable low background rate of PIX despite the comparable binding affinities of 5-deaza-FAD and FADred (15). Therefore, the observation that 5-deaza-FAD is unable to substitute for FADred in catalyzing PIX of the Pβ oxygens implies that N5 of FADred is necessary for cleavage of the anomeric C1–OPβ bond. The background PIX is most likely due to residual (<1%) holo-UGM in the apo-UGM preparation, which is consistent with the tight binding of FADred to UGM (KD < 10 nm) (15) and the observed formation of UDP-Galf in all reactions (see supplemental data). It should also be noted that PIX would be independent of SET if the reaction were to proceed via the oxocarbenium ion intermediate 4 (Fig. 1, paths B and C). Thus, the inability of reduced 5-deaza-FAD to promote anomeric bond cleavage/reformation of 1 and 2 cannot be ascribed to an inability to facilitate SET and is more likely due to the absence of N5 serving as the nucleophile in C1–OPβ bond cleavage during the concerted attack at C1 of the substrate (Fig. 1, path A).
To characterize the role of N5 of FADred during the UGM-catalyzed isomerization more closely, we next examined the kinetic LFERs associated with changes in the nucleophilicity of N5. If adduct 6 (or 8) is formed via an SN2-type substitution (Fig. 1, path A), and if this step is at least partially rate-limiting to steady-state turnover, then changes in the nucleophilicity of N5 should be reflected in the rate of steady-state turnover. In contrast, the rate of steady-state turnover is expected to be much less sensitive to the nucleophilicity of N5 if adduct formation proceeds by an SN1 or a SET pathway (Fig. 1, paths B and C), where formation of the oxocarbenium species (4) would be substantially rate-limiting. Thus, as the electron density at N5 of FADred is decreased by substitution of the isoalloxazine moiety, a decrease in the steady-state reaction rate is expected for mechanism A, whereas little or no effect is expected for mechanisms B and C.
To test this hypothesis, several FAD analogues (Fig. 4A) containing electron-withdrawing or electron-donating groups at positions 7 and/or 8 of the isoalloxazine moiety (meta and para, respectively, to the N5 position) were chemoenzymatically synthesized according to published procedures (14). The nucleophilicity of N5 is represented as the sum of the σm and σp substituent constants for the substituent at the meta and para positions, respectively, of the FAD analogues. These substituent constants are based on the ionization of phenylacetic acid in water (18). Except for the para-methoxy substituent, these values are nearly identical to the Hammett substituent constants obtained for ionization of benzoic acid in water (21). In the case of p-methoxy, σp is significantly more negative with benzoic acid likely due to hydrogen bond stabilization of a trans-quinoidal resonance structure in H2O (22). Although a better correlation was obtained with the values of σm and σp based on ionization of phenylacetic acid, the same conclusions were drawn with those based on benzoic acid (see supplemental data). Previous studies of many flavoenzymes (14, 19, 23, 24) and model systems (25) reconstituted with 7- and 8-substituted flavin analogues have established a precedence for significant LFERs correlating Hammett substituent constants with a variety of parameters related to flavin structure and reactivity.
A linear correlation of log10(kcat) for the UGM-catalyzed reaction versus the sum of σm and σp for the FAD analogues was observed with a slope of −2.4 as shown in the Hammett plot of Fig. 4B. This implies that the rate of steady-state turnover by UGM is indeed sensitive to the electron density at N5 of FADred. The large negative ρ value suggests a substantial decrease in electron density on the flavin in the transition state of the step(s) that limits steady-state turnover. For the conversion of 2 to 1, formation of adduct 8 via an SN2-type reaction (2 → 8) and/or formation of the iminium species (8 → 7) would involve substantial loss of electron density from the flavin N5 center. If formation of 8 (Fig. 1) were to occur via an SN1 process, the expectation would be a ρ value approximating zero. Similarly, rate-determining product dissociation in either mechanism would also be expected to yield a ρ value near zero.
Alone, the results of our LFER studies cannot distinguish between rate-limiting SN2-type adduct formation (2 → 8) and iminium formation (8 → 7). However, our PIX studies strongly suggest that if an SN1-type mechanism were operative, then cleavage of the anomeric bond to form an oxocarbenium intermediate (4) would be energetically demanding and contribute significantly to limiting kcat. Because this step is significantly impaired when N5 of FADred is absent in the active site of UGM, the most consistent interpretation of our PIX and LFER studies is that adduct formation (2 → 8) occurs by an SN2-type reaction that is mediated by N5 of FADred.
In summary, 5-deaza-FAD is not able to support PIX of the β-phosphate oxygens of UDP-Galp, and kcat for the isomerization of UDP-Galf to UDP-Galp exhibits a negative LFER with respect to decreasing electron density at N5 of FADred. These observations represent a direct experimental evaluation of nucleophilic participation by FADred during UGM catalysis. Together, they support a mechanism in which the covalent FADred-Galp/f intermediates (6/8) are formed through a concerted SN2-type displacement involving N5 of FADred as the nucleophile. Thus, as has been characterized recently in several other flavoenzymes (14, 26–28), UGM appears to utilize reduced flavin in a covalent manner to mediate chemical transformations that do not involve redox chemistry, illustrating the catalytic versatility of the ubiquitous flavin coenzyme (29). Finally, these findings help explain the pharmacologically unsatisfactory results obtained with UGM inhibitors that mimic a transition state for generation of an oxocarbenium intermediate (5, 30–32). Therefore, analogues that specifically target the nucleophilic addition may offer more promising leads for developing inhibitors of UGM activity.
Supplementary Material
Acknowledgments
We thank Dr. Kenji Itoh for preparing UDP-Galf and Drs. Ben Shoulders, Yasushi Ogasawara, and Eita Sasaki for helpful suggestions and discussions. We also gratefully acknowledge Steve Sorey for assistance in collecting PIX data.
This work was supported, in whole or in part, by National Institutes of Health Grants GM035906 and GM054346 and Fellowship Award F32AI082906 from NIAID (to M. W. R.). This work was also supported by Welch Foundation Grant F-1511.

This article contains supplemental data, Figs. S1–S3, Tables S1–S6, and additional equations and references.
- UGM
- UDP-galactopyranose mutase
- UDP-Galp
- UDP-galactopyranose
- UDP-Galf
- UDP-galactofuranose
- FADred
- reduced FAD
- PIX
- positional isotope exchange
- SET
- single-electron transfer
- LFER
- linear free energy relationship.
REFERENCES
- 1. Richards M. R., Lowary T. L. (2009) Chemistry and biology of galactofuranose-containing polysaccharides. ChemBioChem. 10, 1920–1938 [DOI] [PubMed] [Google Scholar]
- 2. Peltier P., Euzen R., Daniellou R., Nugier-Chauvin C., Ferrières V. (2008) Recent knowledge and innovations related to hexofuranosides: structure, synthesis, and applications. Carbohydr. Res. 343, 1897–1923 [DOI] [PubMed] [Google Scholar]
- 3. Pan F., Jackson M., Ma Y., McNeil M. (2001) Cell wall core galactofuran synthesis is essential for growth of mycobacteria. J. Bacteriol. 183, 3991–3998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Gandhi N. R., Nunn P., Dheda K., Schaaf H. S., Zignol M., van Soolingen D., Jensen P., Bayona J. (2010) Multidrug-resistant and extensively drug-resistant tuberculosis: a threat to global control of tuberculosis. Lancet 375, 1830–1843 [DOI] [PubMed] [Google Scholar]
- 5. Liautard V., Desvergnes V., Itoh K., Liu H. W., Martin O. R. (2008) Convergent and stereoselective synthesis of imino sugar-containing Galf and UDP-Galf mimics: evaluation as inhibitors of UDP-Gal mutase. J. Org. Chem. 73, 3103–3115 [DOI] [PubMed] [Google Scholar]
- 6. Peltier P., Beláňová M., Dianišková P., Zhou R., Zheng R. B., Pearcey J. A., Joe M., Brennan P. J., Nugier-Chauvin C., Ferrières V., Lowary T. L., Daniellou R., Mikušová K. (2010) Synthetic UDP-furanoses as potent inhibitors of mycobacterial galactan biogenesis. Chem. Biol. 17, 1356–1366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Zhang Q., Liu H. W. (2000) Galactopyranose mutase from Escherichia coli: an unusual role of reduced FAD in its Catalysis. J. Am. Chem. Soc. 122, 9065–9070 [Google Scholar]
- 8. Barlow J. N., Girvin M. E., Blanchard J. S. (1999) Positional isotope exchange catalyzed by UDP-galactopyranose mutase. J. Am. Chem. Soc. 121, 6968–6969 [Google Scholar]
- 9. Soltero-Higgin M., Carlson E. E., Gruber T. D., Kiessling L. L. (2004) A unique catalytic mechanism for UDP-galactopyranose mutase. Nat. Struct. Mol. Biol. 11, 539–543 [DOI] [PubMed] [Google Scholar]
- 10. Gruber T. D., Westler W. M., Kiessling L. L., Forest K. T. (2009) X-ray crystallography reveals a reduced substrate complex of UDP-galactopyranose mutase poised for covalent catalysis by flavin. Biochemistry 48, 9171–9173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Yuan Y., Bleile D. W., Wen X., Sanders D. A., Itoh K., Liu H. W., Pinto B. M. (2008) Investigation of binding of UDP-Galf and UDP-[3-F]Galf to UDP-galactopyranose mutase by STD-NMR spectroscopy, molecular dynamics, and CORCEMA-ST calculations. J. Am. Chem. Soc. 130, 3157–3168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zhang Q., Liu H. (2001) Mechanistic investigation of UDP-galactopyranose mutase from Escherichia coli using 2- and 3-fluorinated UDP-galactofuranose as probes. J. Am. Chem. Soc. 123, 6756–6766 [DOI] [PubMed] [Google Scholar]
- 13. Itoh K., Huang Z., Liu H. W. (2007) Synthesis and analysis of substrate analogues for UDP-galactopyranose mutase: implication for an oxocarbenium ion intermediate in the catalytic mechanism. Org. Lett. 9, 879–882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Thibodeaux C. J., Chang W. C., Liu H. W. (2010) Linear free energy relationships demonstrate a catalytic role for the flavin mononucleotide coenzyme of the type II isopentenyl diphosphate:dimethylallyl diphosphate isomerase. J. Am. Chem. Soc. 132, 9994–9996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Huang Z., Zhang Q., Liu H. W. (2003) Reconstitution of UDP-galactopyranose mutase with 1-deaza-FAD and 5-deaza-FAD: analysis and mechanistic implications. Bioorg. Chem. 31, 494–502 [DOI] [PubMed] [Google Scholar]
- 16. Midelfort C. F., Rose I. A. (1976) A stereochemical method for detection of ATP terminal phosphate transfer in enzymatic reactions. Glutamine synthetase. J. Biol. Chem. 251, 5881–5887 [PubMed] [Google Scholar]
- 17. Mullins L. S., Raushel F. M. (1995) Positional isotope exchange as probe of enzyme action. Method Enzymol. 249, 398–425 [DOI] [PubMed] [Google Scholar]
- 18. Taft R. W., Jr., Ehrenson S., Lewis I. C., Glick R. E. (1959) Evaluation of resonance effects on reactivity by application of the linear inductive energy relationship. VI. Concerning the effects of polarization and conjugation on the mesomeric Order. J. Am. Chem. Soc. 81, 5352–5361 [Google Scholar]
- 19. Edmondson D. E., Ghisla S. (1999) Flavins and Flavoproteins, pp. 71–76, Agency for Scientific Publications, Berlin [Google Scholar]
- 20. Walsh C. (1986) Naturally occurring 5-deazaflavin coenzymes: biological redox roles. Acc. Chem. Res. 19, 216–221 [Google Scholar]
- 21. Hansch C., Leo A., Taft R. W. (1991) A survey of Hammett substituent constants and resonance and field parameters. Chem. Rev. 91, 165–195 [Google Scholar]
- 22. Taft R. W., Jr. (1960) Sigma values from reactivities. J. Phys. Chem. 64, 1805–1815 [Google Scholar]
- 23. Yorita K., Misaki H., Palfey B. A., Massey V. (2000) On the interpretation of quantitative structure-function activity relationship data for lactate oxidase. Proc. Natl. Acad. Sci. U.S.A. 97, 2480–2485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Roth J. P., Wincek R., Nodet G., Edmondson D. E., McIntire W. S., Klinman J. P. (2004) Oxygen isotope effects on electron transfer to O2 probed using chemically modified flavins bound to glucose oxidase. J. Am. Chem. Soc. 126, 15120–15131 [DOI] [PubMed] [Google Scholar]
- 25. Legrand Y. M., Gray M., Cooke G., Rotello V. M. (2003) Model systems for flavoenzyme activity: relationships between cofactor structure, binding, and redox properties. J. Am. Chem. Soc. 125, 15789–15795 [DOI] [PubMed] [Google Scholar]
- 26. Yu Q., Schaub P., Ghisla S., Al-Babili S., Krieger-Liszkay A., Beyer P. (2010) The lycopene cyclase CrtY from Pantoea ananatis (formerly Erwinia uredovora) catalyzes an FADred-dependent non-redox reaction. J. Biol. Chem. 285, 12109–12120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kommoju P. R., Bruckner R. C., Ferreira P., Jorns M. S. (2009) Probing the role of active site residues in NikD, an unusual amino acid oxidase that catalyzes an aromatization reaction important in nikkomycin biosynthesis. Biochemistry 48, 6951–6962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Rauch G., Ehammer H., Bornemann S., Macheroux P. (2007) Mutagenic analysis of an invariant aspartate residue in chorismate synthase supports its role as an active site base. Biochemistry 46, 3768–3774 [DOI] [PubMed] [Google Scholar]
- 29. Mansoorabadi S. O., Thibodeaux C. J., Liu H. W. (2007) The diverse roles of flavin coenzymes: nature's most versatile thespians. J. Org. Chem. 72, 6329–6342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Caravano A., Vincent S. P., Sinaÿ P. (2004) Efficient synthesis of a nucleoside-diphospho-exo-glycal displaying time-dependent inactivation of UDP-galactopyranose mutase. Chem. Commun. 1216–1217 [DOI] [PubMed] [Google Scholar]
- 31. Caravano A., Dohi H., Sinaÿ P., Vincent S. P. (2006) A new methodology for the synthesis of fluorinated exo-glycals and their time-dependent inhibition of UDP-galactopyranose mutase. Chem. Eur. J. 12, 3114–3123 [DOI] [PubMed] [Google Scholar]
- 32. Desvergnes S., Desvergnes V., Martin O. R., Itoh K., Liu H. W., Py S. (2007) Stereoselective synthesis of β-1-C-substituted 1,4-dideoxy-1,4-imino-d-galactitols and evaluation as UDP-galactopyranose mutase inhibitors. Bioorg. Med. Chem. 15, 6443–6449 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.