Background: The ARID2 protein is specific for a certain subset of SWI/SNF chromatin-remodeling complexes known as PBAF.
Results: ARID2 is essential for gene activation along the osteoblast phenotype.
Conclusion: ARID2-containing complexes play a major role in lineage commitment and differentiation.
Significance: Manipulating levels of ARID family subunits (of which there are three mutually exclusive alternatives in SWI/SNF) may influence precursor state and lineage determination without compromising full SWI/SNF function.
Keywords: Cell Differentiation, Chromatin Remodeling, DNA-binding Protein, Epigenetics, Gene Expression, Osteoblasts, Transcription Regulation, ARID2, PBAF, SWI/SNF
Abstract
Unfolding of the gene expression program that converts precursor cells to their terminally differentiated counterparts is critically dependent on the nucleosome-remodeling activity of the mammalian SWI/SNF complex. The complex can be powered by either of two alternative ATPases, BRM or BRG1. BRG1 is critical for development and the activation of tissue specific genes and is found in two major stable configurations. The complex of BRG1-associated factors termed BAF is the originally characterized form of mammalian SWI/SNF. A more recently recognized configuration shares many of the same subunits but is termed PBAF in recognition of a unique subunit, the polybromo protein (PBRM1). Two other unique subunits, BRD7 and ARID2, are also diagnostic of PBAF. PBAF plays an essential role in development, apparent from the embryonic lethality of Pbmr1-null mice, but very little is known about the role of PBAF, or its signature subunits, in tissue-specific gene expression in individual differentiation programs. Osteoblast differentiation is an attractive model for tissue-specific gene expression because the process is highly regulated and remains tightly synchronized over a period of several weeks. This model was used here, with a stable shRNA-mediated depletion approach, to examine the role of the signature PBAF subunit, ARID2, during differentiation. This analysis identifies a critical role for ARID2-containing complexes in promoting osteoblast differentiation and supports a view that the PBAF subset of SWI/SNF contributes importantly to maintaining cellular identity and activating tissue-specific gene expression.
Introduction
Osteoblast differentiation has several advantages as a model of tissue-specific gene expression. Osteoblasts, the cells responsible for bone formation, are derived from multipotent mesenchymal stem cells. The commitment of mesenchymal stem cells to the osteoblastic lineage and further differentiation to osteoblasts and ultimately to mature osteocytes is tightly regulated and synchronized over a period of several weeks (see Fig. 1A). Successive changes in osteogenic gene expression patterns are controlled by specific transcription factors acting in concert with chromatin modifiers that determine acetylation and methylation marks and with chromatin-remodeling complexes that alter nucleosome positioning in an ATPase-dependent manner (1–7).
FIGURE 1.
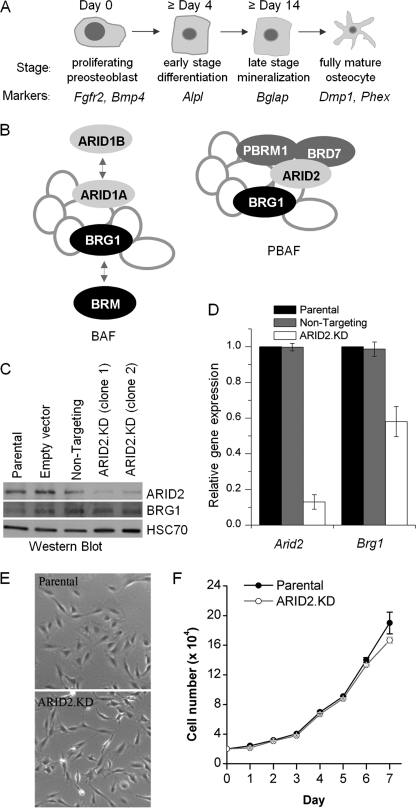
Generation of stable ARID2-depleted MC3T3-E1 cell lines. A, the program of osteoblast differentiation can be divided into four phases: (a) proliferative pre-osteoblasts expressing Fgfr2 and Bmp4; (b) early differentiation characterized by Alpl expression; (c) late differentiation distinguished by highly induced expression of Bglap (encoding osteocalcin); and (d) terminal differentiation to the fully mature osteocyte phenotype marked by expression of Dmp1 and Phex. B, the multisubunit SWI/SNF complex contains a core ATPase plus seven or more non-catalytic subunits. The BAF complex can contain either the BRG1 or the BRM ATPase; its signature subunits are the mutually exclusive ARID1A and ARID1B proteins. The PBAF complex is only known to contain BGR1; it is distinguished by the presence of three unique subunits: ARID2 (alias BAF200), polybromo/PBRM1 (alias BAF180), and BRD7. C, total cell lysates from parental cells, cells transfected with an empty vector, vectors encoding a non-targeting control sequence, or either of two lines expressing ARID2-targeting sequences (ARID2.KD) were probed in a Western blot with antibodies specific to either ARID2 or BRG1. The constitutively expressed HSC70 protein was used as a loading control. D, expression of Arid2 and Brg1 in ARID2-depleted cells (ARID2.KD, clone 1) or the cell line transfected with the non-targeting control sequence, relative to the parental cells, was determined by qRT-PCR, normalized to Gapdh expression. Shown are mean values relative to the parental line with the S.E. (n = 3). E, representative pictures of the general morphology of parental cells and ARID2-depleted cells. F, the growth rate of parental and ARID2.KD cells. Cells were seeded in 6-well plates at 2 × 104 cells/well, and the medium was changed on day 4 after seeding. Each point on the growth curve shows the mean value with the S.E. from three wells.
Unfolding of the gene expression program that converts precursor cells to osteoblasts is critically dependent on the nucleosome-remodeling activity of the mammalian SWI/SNF complex (reviewed in Refs. 8–10). Understanding how SWI/SNF acts to maintain or advance specific stages of differentiation is basic to understanding molecular mechanisms of tissue development. The complex can be powered by either of two ATPases: BRM or the BRM-related gene product BRG1. BRG1-containing SWI/SNF generally acts to induce osteogenic genes, whereas BRM-specific complexes can play auxiliary, complementary, or opposing roles. In osteoblast precursors, BRM-specific complexes, sometimes acting in concert with p130 and repressor E2Fs, primarily restrain differentiation, helping to maintain the committed precursor state until the pre-osteoblasts receive appropriate signals for differentiation (2, 3).
BRG1 participates in two related SWI/SNF complexes, designated BAF2 or PBAF (11). BAF and PBAF share most subunits but can be distinguished by the presence of the polybromo (PBRM1) and BRD7 subunits in PBAF (see Fig. 1B) and by the choice of another component that occurs as related alternatives: the ARID family subunit. ARID family proteins (reviewed in Ref. 12) are characterized by a consensus AT-rich interaction domain, which comprises a DNA binding motif of loose specificity (13). Mammalian cells encode 15 ARID family proteins, three of which are identified as SWI/SNF subunits. The family also includes a set of histone demethylases (14), so it is closely linked with chromatin regulation. The SWI/SNF-associated ARID subunits are large proteins containing multiple protein interactive domains. BAF complexes contain ARID1A or ARID1B (15, 16), whereas PBAF contains ARID2 (alias BAF200) (17). In addition to the N-terminal ARID motif, ARID2 contains two classic C2H2 zinc fingers at the carboxyl terminus (18).
The choice of ARID1A over ARID1B favors cell cycle arrest over continued proliferation (19, 20), and the proteins have distinct expression profiles during embryogenesis and the cell cycle (21). ARID1A is recognized as a tumor suppressor (22–25), and ARID2 may have such a role as well (26). Induction of the osteogenic marker, alkaline phosphatase, is impaired in either ARID1A-depleted or ARID1B-depleted pre-osteoblasts (19) in a milder version of the BRG1 depletion phenotype, implying a specific requirement for BAF in osteoblast differentiation. However, these studies did not consider the role of the PBAF configuration, a significant question because identifying specific gene expression patterns determined by the alternative subunits offers the potential to influence parameters such as the rate of bone formation, the size of progenitor pools, and the balance of lineage determination.
PBAF plays a required role in development (27, 28) but has not been studied in specific differentiation systems. In this study, to determine whether PBAF is required during osteoblast differentiation, we used an shRNA approach to target ARID2 in MC3T3-E1 pre-osteoblasts. The specific analysis described here identifies a critical role for ARID2-containing complexes in promoting osteoblast differentiation and supports a view that the PBAF subset of SWI/SNF contributes importantly to maintaining cellular identity and tissue-specific gene expression.
EXPERIMENTAL PROCEDURES
Materials
Ascorbic acid, β-glycerol phosphate, sodium phosphate mono- and dibasic, Alizarin Red S, puromycin, and protease inhibitors were obtained from Sigma. Penicillin-streptomycin solution was purchased from Mediatech (Herndon, VA). Fetal bovine serum (FBS) was purchased from Atlanta Biologicals, and α-modified essential medium was purchased from Irvine Scientific (Santa Ana, CA).
Cell Culture
MC3T3-E1 cells, obtained from the ATCC (catalogue number: CRL-2593), were maintained in α-modified essential medium supplemented with 10% FBS and 1% penicillin-streptomycin solution. Differentiation of low passage MC3T3-E1 cells was induced by the addition of 50 μg/ml ascorbic acid and 10 mm β-glycerol phosphate to the standard growth medium. The medium was changed every 3–4 days, and the inducing agents were replaced with each medium change.
shRNA and Isolation of Stable ARID2 Knockdown Cell Lines
To generate stable ARID2-depleted cell lines, MC3T3-E1 cells were transfected with pRS vectors containing ARID2 shRNA-encoding sequences or a scrambled sequence purchased from OriGene (Rockville, MD) and selected with puromycin (4 μg/ml). The specific knockdown sequences were 5′-TGA GAC GAG CCA GTG CTC ACT AAT CAG CA-3′ for clone 1 (S4C1) and 5′-CAG CCA CAA GAC ACT GTT ATC ATA GCA CC-3′ for clone 2 (S3.AA4), respectively. Puromycin-resistant clones were expanded and then screened by Western blot for ARID2 expression. The decreased expression of ARID2 was confirmed by quantitative reverse transcription PCR (qRT-PCR).
Growth Analysis of ARID2 Stable Knockdown (ARID2.KD) Cell Line
For growth analysis, parental and ARID2.KD (clone 1) cells were seeded into 6-well plates (in triplicate) at 20,000 cells/well and cultured in α-modified essential medium supplemented with 10% FBS and 1% penicillin-streptomycin medium. To determine the cell number, cells were trypsinized and counted using a Vi-CELL cell viability analyzer (Beckman Coulter) every 24 h for 7 days. Data are presented as mean cell number per well with the S.E. of triplicate determinations in each line. The cells were photographed with an EVOS AME i2111 phase contrast microscope (Advanced Microscopy Group, Bothell, WA).
Staining for Alkaline Phosphatase Activity and Mineralization
These assays were carried out as described previously (3).
Immunoblotting
The antibodies used were: ARID2 (H-182, sc-98299), BRG1 (H-88, sc-10768), and HSC70 (B-6, sc-7298) from Santa Cruz Biotechnology, Inc., (Santa Cruz, CA). The procedure was previously described (29).
qRT-PCR
Total RNA was isolated using TRIzol (Invitrogen) according to the manufacturer's instructions. Quality and concentration of RNA were determined using a NanoDrop ND-1000 system. Total RNA (5 μg) was primed with oligo(dT) and reverse-transcribed using the SuperScript III first strand synthesis supermix kit (Invitrogen). SYBR Green-based quantitative PCRs were carried out using an ABI PRISM 7500 real time PCR system (Applied Biosystems, Foster City, CA). The gene-specific primers used for PCR are: Arid2 (forward, 5′-GCA GCC AAT TTC CAC TCC TGT TG-3′; reverse, 5′-GAT TGG TGA CAG GAG TCC TCT G-3′), Brg1 (forward, 5′-GAA AGT GGC TCT GAA GAG GAG G-3′; reverse, 5′-TCC ACC TCA GAG ACA TCA TCG C-3′), Alpl (forward, 5′-CCA GAA AGA CAC CTT GAC TGT GG-3′; reverse, 5′-TCT TGT CCG TGT CGC TCA CCA T-3′), Bglap (forward, 5′-GCA ATA AGG TAG TGA ACA GAC TCC-3′; reverse, 5′-GCG TTT GTA GGC GGT CTT CAA G-3′), Fgfr2 (forward, 5′-GTC TCC GAG TAT GAG TTG CCA G-3′; reverse, 5′-CCA CTG CTT CAG CCA TGA CTA C-3′), Bmp4 (forward, 5′-GCC GAG CCA ACA CTG TGA GGA-3′; reverse, 5′-GAT GCT GCT GAG GTT GAA GAG G-3′), Dmp1 (forward, 5′-TTG TGG GAA AAA GAC CTT GG-3′; reverse, 5′-AAT CAC CCG TCC TCT CTT CA-3′), Phex (forward, 5′-CTG GCT GTA AGG GAA GAC TTC C-3′; reverse, 5′-GCT CCT AAA AGC ACA GCA GTG TC-3′), and Gapdh (forward, 5′-GTC TCC TCT GAC TTC AAC AGC G-3′; reverse, 5′-ACC ACC CTG TTG CTG TAG CCA A-3′). Expression of Gapdh was used as an internal control. Data are presented as means ± S.E.
Chromatin Immunoprecipitation (ChIP) Assays
ChIP assays were performed with the EZ-ChIPTM system (Millipore, Lake Placid, NY), according to the manufacturer's directions, modified to include preclearing of lysates with 60 μl of a 50% slurry of protein G/salmon sperm DNA for 1 h at 4 °C and again overnight. The antibodies used were: ARID2 (H-182x, sc-98299x), BRG1 (H-88, sc-10768), BRM (N-19, sc-6450), Pol II (H-224, sc-9001), RUNX2 (H-300, sc-33170), and osterix (OSX) (A-13x, sc-22536x) from Santa Cruz Biotechnology. Monoclonal antibodies specific for ARID1A (PSG3) and ARID1B (KMN1) were described previously (16, 20) and are available from Millipore (04-080) and Santa Cruz Biotechnology (sc-32761 and sc-32762). PCR conditions were 40 cycles at 30 s at 95 °C, 30 s at 72 °C, and 30 s at 60 °C. The primers used for ChIP are: Alpl (forward, 5′-AAG GGG TCT CCT TCT GCT TC-3′; reverse, 5′-CTT TGT CCC TCG ATG GTT GT-3′), Bglap (forward, 5′-GAG TGT CGT CCC CAA TCC-3′; reverse, 5′-CTG CTA CCA CCG AGG CTG-3′), Fgfr2 (forward, 5′-GAG GCT TTG GAT GAC TCT GC-3′; reverse, 5′-CCA TGG GAA AAG GCA ATC TA-3′), Bmp4 (forward, 5′-TCC CAT GGG TAT TTT TGG AA-3′; reverse, 5′-CTT CCC TCC TCC CTT AAT CG-3′), Dmp1 (forward, 5′-GAA GGT CTC TAC CCG CCT CT-3′; reverse, 5′-CCC ATA CAC CCA CAC TTC CT-3′), Phex (forward, 5′-CCT GAG TTT GGG GTG AAA TG-3′; reverse, 5′-TGA CAC CAG ACC TCA GCA AG-3′), and 3′-untranslated region (3′-UTR) of Alpl (forward, 5′-TTG TTC CTC TTG CCT CAG GT-3′; reverse, 5′-TGA CAA TCA CAT GGC CTC TC-3′).
RESULTS
Generation of Stable ARID2-depleted Cell Lines
Osteoblast-specific gene expression can be studied in MC3T3-E1 cells, which are cell culture-adapted, non-transformed precursor cells committed to the osteoblast lineage but not yet differentiated. The cells proliferate indefinitely as precursors, but when stimulated by bone anabolic agents, they undergo terminal differentiation in a tightly synchronized program lasting more than 2 weeks (30, 31). Induction of alkaline phosphatase is an early stage event, coordinated closely with differentiation-associated cell cycle arrest. Within a week, the arrested cells are laying down a collagenous extracellular matrix, which undergoes mineralization apparent by day 21.
Depletion of BRG1 severely impairs the ability of these cells to mineralize or to induce the key markers of osteoblast differentiation, i.e. alkaline phosphatase activity and osteocalcin (3). In addition to markers of osteoblast differentiation, gene array analysis identified two other classes of genes dependent on BRG1-mediated regulation. Expression of the genes encoding bone morphogenic protein, BMP4, and the fibroblast growth factor receptor, FGFR2, is characteristic of osteoblast precursors. Both genes are implicated in lineage commitment in mesenchymal stem cells, with FGFR2 particularly implicated in osteogenesis (32–35). Expression of these genes drops sharply in BRG1-depleted cells. The third class of genes regulated by BRG1 in pre-osteoblasts is negatively regulated and comprises genes considered diagnostic of the fully mature osteocyte stage. Expression of these genes rises sharply in BRG1-depleted precursor cells. The appropriate peak expression time for these genes is shown in Fig. 1A.
Because the SWI/SNF complexes coordinately control broad patterns of gene expression, they afford a means of understanding the temporal dynamics of gene expression during differentiation. Thus, it is important to distinguish the specific roles of different SWI/SNF subsets. To this end, we investigated here whether ARID2-containing SWI/SNF plays a specific role in any of the osteogenic gene regulatory patterns now linked with BRG1. To address this question, MC3T3-E1-derived cell lines were generated that stably express shRNA sequences targeting ARID2. Two independent lines were isolated using two different shRNA sequences. Western blot analysis (Fig. 1C) shows strong depletion of ARID2 levels in these lines when compared with control lines isolated after transfection with an empty vector or a vector carrying a non-targeting shRNA sequence. The effect on BRG1 levels is relatively mild. The ARID2-depleted lines both showed the same phenotype in a standard assay of osteoblast differentiation. Results with clone 1 are shown here, with quantification of ARID2 mRNA levels for this line shown in Fig. 1D. Quantification shows severe depletion of ARID2 accompanied by partial depletion of BRG1; this is not unexpected as ARID2 depletion may make a portion of BRG1 extraneous (17). Depletion of ARID2 does not impair the overall health of the cells as the general morphology remains indistinguishable from the parental cells (Fig. 1E), and the doubling time remains essentially the same (Fig. 1F).
ARID2 Is Required for Normal Osteoblast Differentiation
Normally differentiating osteoblasts reach a mineralization phenotype that can be detected as Alizarin Red S reactive deposits by day 21 (Fig. 2A). By day 28, it is clear that ARID2-depleted cells are severely impaired for this phenotype, indicating that ARID2-containing SWI/SNF plays a specific crucial role in osteogenic differentiation. This phenotype was examined further at the level of gene expression.
FIGURE 2.
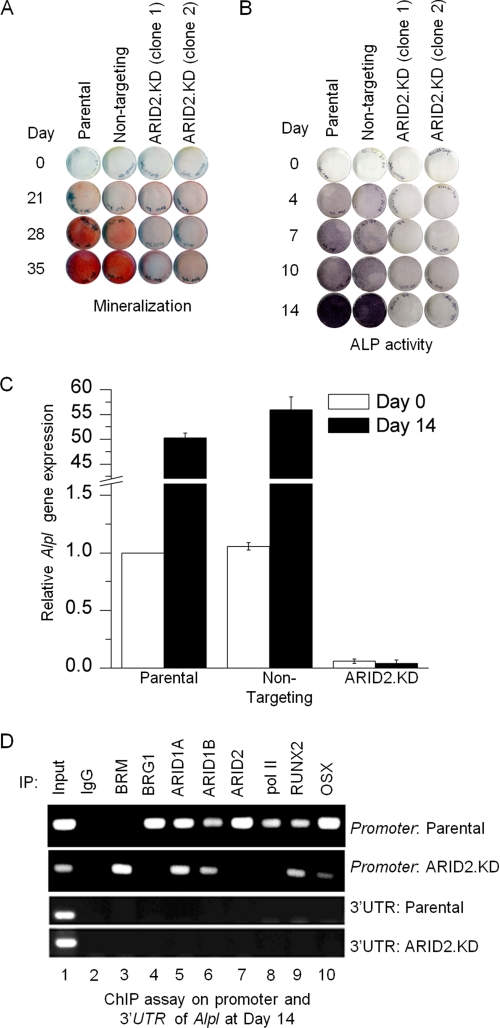
Stable depletion of ARID2 represses osteoblast differentiation. A, parental cells, a cell line isolated after transfection with a vector encoding a non-targeting control sequence, or ARID2-depleted cells were treated with differentiation medium for the time intervals indicated and then stained with Alizarin Red S to reveal the presence of calcium-containing compounds in the cell matrix (mineralization). B, the same panel of cell lines was induced for the time intervals indicated and stained with 5-bromo-4-chloro-3-indolyl phosphate/nitro blue tetrazolium to reveal alkaline phosphatase (ALP) activity as the production of a purple-black deposit following substrate hydrolysis. C, parental cells, the cell line transfected with the non-targeting control sequence, or ARID2-depleted cells were induced for the time intervals indicated, at which point total RNA was isolated and qRT-PCR was performed to determine Alpl expression levels. Shown here are mean values relative to the parental line with S.E. (n = 3). D, ChIP analysis was performed on the Alpl promoter from parental cells and ARID2-depleted cells at day 14 after induction to determine the association pattern of the ATPase and ARID family subunits as well as selected transcriptional activators, as indicated in the figure. The same immunoprecipitated DNA was also amplified with primers corresponding to the 3′-UTR of Alpl as a negative control. IP, immunoprecipitation.
The key genetic markers of differentiation are induction of the alkaline phosphatase gene, Alpl, which occurs early, and induction of the osteocalcin gene, Bglap, which correlates with late-stage osteoblast differentiation. The level of alkaline phosphatase activity can be detected in an in situ enzymatic assay. Normal cells show a positive colorimetric response within 4 days of transfer to osteogenic medium, but ARID2-depleted cells are severely impaired for induction of this marker, showing very low levels even up to day 14 (Fig. 2B). A previous analysis showed a similar pattern in ARID1A/B-depleted lines (19).
The effect of ARID2 depletion on induction of Alpl was determined quantitatively by qRT-PCR analysis of RNA collected at day 14 of exposure to differentiation medium (Fig. 2C). ARID2-depleted cells are severely impaired for induction of Alpl, and in fact, do not even sustain the low level of expression seen in the parental pre-osteoblasts at day 0, establishing the finding that ARID2-containing complexes play an important role in osteogenic gene activation. BRG1 is already known to target Alpl directly during activation (1), and ChIP assays here confirm that the ARID2 subunits, including ARID2, occupy the promoter as well (Fig. 2D). A parallel ChIP assay in the ARID2-depleted cells shows that ARID2 depletion does not impair promoter association by the other ARID family subunits, and occupation by the canonical osteogenic transcription factors RUNX2 and OSX remains detectable. Other association patterns are radically altered, however. In the ARID2-depleted cells, the promoter does not switch from occupation by BRM-containing SWI/SNF, which characterizes repression, to BRG1-SWI/SNF, which characterizes activation. In addition, RNA polymerase II (pol II) is not detected at the promoter, consistent with a previous finding that pol II recruitment to Alpl is BRG1-dependent (1). Thus, ARID2-containing complexes act directly at the Alpl promoter and play a vital role in pol II recruitment. Probing the same immune complexes with primers complementary to the Alpl 3′-untranslated region serves as a negative control.
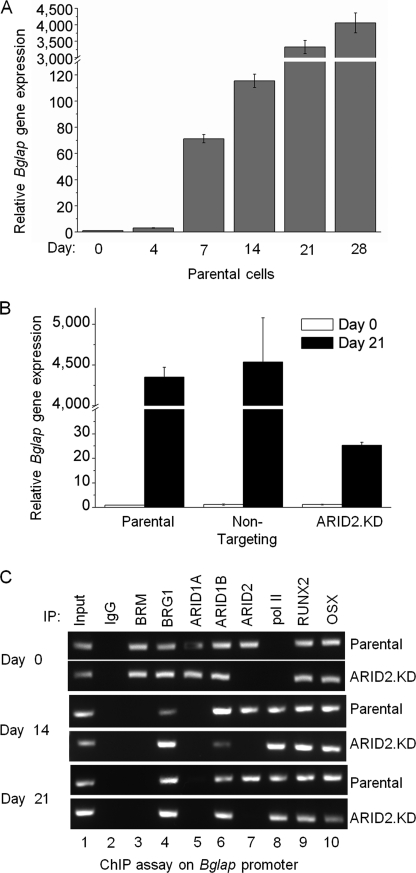
Progression to the mineralization phenotype requires expression of late-stage genes as well. The best-studied late-stage marker is Bglap, the gene encoding osteocalcin. Bglap expression increases dramatically at 2–3 weeks after induction (Fig. 3A). Comparison with the ARID2-depleted line (Fig. 3B) shows that expression increases somewhat despite ARID2 deficiency, but not on the same scale as in normal cells. At day 21, expression is almost 200-fold less than in the parental line. We have shown previously that the ARID1A/B proteins target Bglap directly with ARID1A dissociating upon induction (3). A ChIP assay here shows that ARID2 also targets this promoter directly and that ARID2 occupation continues through peak activation (Fig. 3C). On Bglap, BRG1-SWI/SNF occupies the promoter constitutively in parental cells, but pol II is not recruited until BRM-SWI/SNF dissociates (3). Probing the promoter in ARID2-depleted cells shows that depletion does not affect SWI/SNF and pol II occupation patterns on Bglap as was seen on Alpl. In this case, BRM-SWI/SNF successfully dissociates, and pol II is successfully recruited; this correlates with modest induction of Bglap (Fig. 3B) when compared with profoundly impaired expression of Alpl (Fig. 2C). The poor expression of Bglap despite recruitment of pol II suggests that ARID2-containing complexes may have a role in transcriptional progression as well as initiation. The dissociation of BRM and recruitment of pol II indicate that transcription signals are still operable in ARID2-depleted cells, but not fully able to reach fruition.
FIGURE 3.
ARID2 is essential for activation of expression of Bglap (encoding osteocalcin). A, expression of Bglap during differentiation in MC3T3-E1 cells was determined by qRT-PCR with Bglap-specific primers. Shown are mean values relative to uninduced (Day 0) with S.E. (n = 3). B, parental cells, the cell line transfected with the non-targeting control sequence, or ARID2-depleted cells were cultured in differentiation medium, and total RNA was isolated at days 0 and 21 as indicated; qRT-PCR was performed with Bglap-specific primers. Shown here are mean values relative to the parental line with S.E. (n = 3). C, ChIP analysis was performed on parental and ARID2-depleted cells at days 0, 14, and 21 after induction to determine the association pattern of the ATPase and ARID family subunits and selected transcriptional activators on the Bglap promoter. IP, immunoprecipitation.
ARID2 Is Required to Maintain Expression of Genes Characteristic of Committed Pre-osteoblasts
Another pattern of BRG1-dependent gene regulation revealed by earlier gene array analysis (3) is maintenance of expression of certain osteogenic genes already elevated in committed, but undifferentiated, osteoblasts. This includes several forms of collagen, as well as two genes encoding products that play important roles in signal transduction: the anabolic growth factor BMP4 and the growth factor receptor FGFR2.
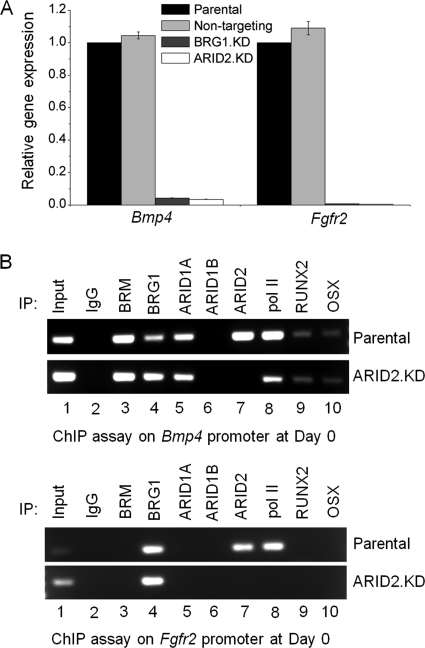
Expression of Bmp4 and Fgfr2 is sharply reduced in BRG1-depleted cells, and the same effect is seen in ARID2-depleted cells (Fig. 4A), indicating that ARID2-containing SWI/SNF plays a major role in maintaining active expression of these genes. ChIP assays (Fig. 4B) show direct occupation by ARID2 and BRG1 of both promoters in the pre-osteoblasts. ARID1B was not detected on either promoter, and ARID1A was detected only on Bmp4. Depletion of ARID2 results in a diminished signal for pol II on Bmp4 and no detected signal on Fgfr2, concordant with the respective decline in expression of each gene in the depleted cells.
FIGURE 4.
ARID2 is required to maintain expression of genes characteristic of committed pre-osteoblasts. A, total RNA was isolated from parental cells, the non-targeting control line, and BRG1-depleted and ARID2-depleted pre-osteoblasts, and qRT-PCR was carried out to assess the relative expression of Bmp4 and Fgfr2. Shown are mean values relative to parental cells with S.E. (n = 3). B, parental and ARID2-depleted pre-osteoblasts were analyzed by ChIP assay for association of the ATPase and ARID family subunits as well as selected transcriptional activators with the Bmp4 and Fgfr2 promoters. IP, immunoprecipitation.
ARID2 Is Not Required for BRG1-dependent Repression of Markers of Fully Mature Osteocytes
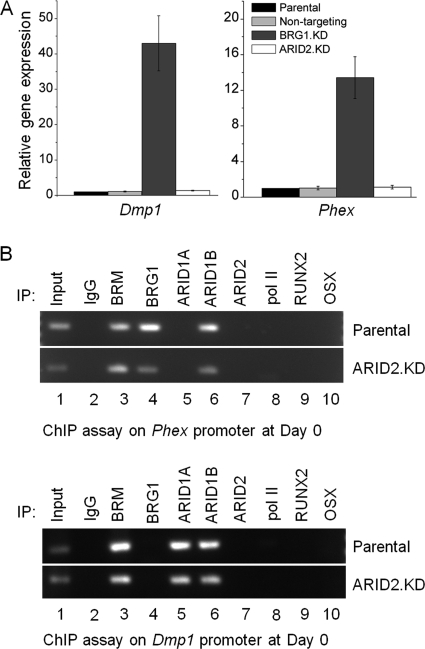
SWI/SNF specifically containing the ATPase BRG1 is essential for activated expression of osteoblast markers during differentiation. However, BRG1-dependent effects are not limited to gene activation. Analysis of BRG1-depleted MC3T3-E1 cells on an osteogenic gene array showed that repression of markers associated with full maturation to the terminal osteocyte phenotype requires the combined action of BRM and BRG1 (3). Dentin matrix protein 1, encoded by the Dmp1 gene, plays a key role in controlling osteocyte formation and phosphate homeostasis in bone and is expressed predominantly in osteocytes (36–38). Expression of the Phex gene, which encodes a phosphate-regulating enzyme, is also characteristic of osteocytes (39). BRG1-depleted pre-osteoblasts show premature expression of both these classic osteocyte marker genes, but parallel analysis shows that their expression is essentially unaffected in ARID2-depleted cells (Fig. 5A). Thus, this repression function is independent of ARID2-containing configurations, in strong contrast to the required role of ARID2-containing SWI/SNF in activation.
FIGURE 5.
ARID2 is not required for BRG1-dependent repression of markers of fully mature osteocytes in pre-osteoblasts. A, total RNA was isolated from parental cells, the non-targeting control line, and BRG1-depleted and ARID2-depleted pre-osteoblasts, and qRT-PCR was carried out to assess the relative expression of Dmp1 and Phex. Shown are mean values relative to the parental cells with S.E. (n = 3). B, parental and ARID2-depleted pre-osteoblasts were analyzed by ChIP assay for association of the ATPase and ARID family subunits as well as selected transcriptional activators with the Dmp1 and Phex promoters. IP, immunoprecipitation.
ChIP analysis (Fig. 5B) shows that Phex is targeted directly by both BRM and BRG1 in undifferentiated pre-osteoblasts, consistent with the requirement for both SWI/SNF ATPases in Phex repression. ARID2 was not detected on the Phex promoter, as expected from the lack of effect of ARID2 depletion on Phex expression. Dmp1 likewise is not targeted by ARID2. These primer probes were performed on the same set of chromatin immunoprecipitations probed for Fgfr2 in Fig. 4, which thus serves as a positive control for ARID2. Somewhat surprisingly, ChIP analysis does not show occupation of Dmp1 by BRG1 either. Repressor BAF complexes can act at sites very distal to the transcriptional start (40), so they may be outside the detection range of the primers used here, although BRM complexes were readily detected. Alternatively, BRG1-dependent repression of Dmp1 may be exerted indirectly. ChIP analysis shows that Dmp1 and Phex are each targeted by at least one of the ARID1 subunits, consistent with a conclusion that repression of premature expression of these genes is linked predominantly with other SWI/SNF compositions and not with ARID2-containing complexes.
Overall, this analysis defines the ARID2-containing SWI/SNF configuration (PBAF) as crucial to the activation of osteogenic genes and to osteoblast differentiation. As yet, there is no indication that ARID2 plays a role in osteogenic gene repression in these cells.
DISCUSSION
This study identifies a critical role for ARID2-containing complexes in promoting osteoblast commitment and differentiation. Key osteogenic markers are targeted directly by the ARID2-conaining complexes, which in some cases contribute substantially to recruitment of RNA polymerase II. These findings represent an important advance over the very few studies to date that have considered the role of PBAF in development or differentiation and support a view that the PBAF subset of SWI/SNF plays a major role in maintaining cellular identity and activating tissue-specific gene expression.
The BAF complex is critical from the earliest stages of development, with knock-out of BRG1 or ARID1A fatal by the peri-implantation stage or embryonic day 6.5, respectively (41, 42). A modified BAF complex is critical to embryonic stem cell pluripotency (43). Probing the role of PBAF in embryonic stem cells via depletion of BRD7 suggested that the majority of BRG1-dependent transcriptional regulation in these cells requires BAF specifically, although activation and repression targets of PBAF were identifiable (44). The role of PBAF appears to become increasingly important as development proceeds. Depletion of BRD7 identified an essential role for PBAF in maintaining expression of critical neural crest specifiers in eight-cell Xenopus embryos (45). Knock-out of the polybromo (PBRM1/BAF180) subunit becomes fatal in mice by about embryonic days 12.5–15. At this point, the embryos succumb to severe heart defects with impaired expression of certain retinoic acid-response genes (27, 28, 45). BAF and PBAF make distinguishable contributions to activation of hormone-responsive promoters (reviewed in Refs. 10 and 46).
In earlier studies, exploring the role of PBAF through the ARID2 subunit showed that ARID2 depletion impairs the ability of interferon to induce the interferon-response gene IFITM1 in HeLa cells, whereas a panel of other interferon-response genes is unaffected (17). Overexpression of ARID2 can activate transient expression of a set of cardiac reporter genes in NIH3T3 cells (18), and depletion of ARID2 in human lymphoid CEM cells impairs Tat-activated transcription of the HIV-LTR (47). No previous studies have followed the role of PBAF through a tissue-specific transcription program.
The current study focuses on an individual model of lineage commitment and differentiation. The effect of BRG1 depletion on endogenous osteogenic gene expression was reported previously and revealed distinct BRG1-dependent transcriptional programs (2, 3). This information was used to examine the specific contribution of ARID2-containing complexes. Biologically, maturation of osteoblasts to the mature mineralization phenotype is severely impaired in ARID2-depleted cells. The requirement for ARID2 in maintaining expression of Bmp4 and Fgfr2 in pre-osteoblasts implies a significant role for PBAF in establishing osteoblast commitment. The requirement for ARID2 in transcriptional activation of Alpl and Bglap shows that PBAF plays an important role in activating major markers of tissue-specific gene expression. Alpl activation is an example of cooperativity between BAF and PBAF (19) (Fig. 2). Common gene targets have been noted before, both cooperative and antagonistic (44), with a suggestion that hybrid complexes are capable of forming on individual promoters (48). Nevertheless, the lack of requirement for ARID2 in maintaining early repression of the late-stage markers Dmp1 and Phex clearly distinguishes the role of ARID2-containing complexes from the totality of BRG1 functions. BRD7 and polybromo are also implicated in proliferative controls and tumor suppression (49–53), and the ARID2-depleted osteoblast precursor lines derived here will be useful in future studies to help elucidate the contribution of PBAF to those processes.
This work was supported, in whole or in part, by National Institutes of Health Grants GM073257 (to E. M.) and T32-CA134268 (to S. F.) through the United States Public Health Service.
- BAF
- BRG1-associated factor
- PBAF
- polybromo BAF
- qRT-PCR
- quantitative reverse transcription PCR
- KD
- knockdown
- pol II
- RNA polymerase II.
REFERENCES
- 1. Flowers S., Beck G. R., Jr., Moran E. (2010) Transcriptional activation by pRB and its coordination with SWI/SNF recruitment. Cancer Res. 70, 8282–8287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Flowers S., Beck G. R., Jr., Moran E. (2011) Tissue-specific gene targeting by the multiprotein mammalian DREAM complex. J. Biol. Chem. 286, 27867–27871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Flowers S., Nagl N. G., Jr., Beck G. R., Jr., Moran E. (2009) Antagonistic roles for BRM and BRG1 SWI/SNF complexes in differentiation. J. Biol. Chem. 284, 10067–10075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Schroeder T. M., Kahler R. A., Li X., Westendorf J. J. (2004) Histone deacetylase 3 interacts with Runx2 to repress the osteocalcin promoter and regulate osteoblast differentiation. J. Biol. Chem. 279, 41998–42007 [DOI] [PubMed] [Google Scholar]
- 5. Sinha K. M., Yasuda H., Coombes M. M., Dent S. Y., de Crombrugghe B. (2010) Regulation of the osteoblast-specific transcription factor Osterix by NO66, a Jumonji family histone demethylase. EMBO J. 29, 68–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Westendorf J. J. (2007) Histone deacetylases in control of skeletogenesis. J. Cell. Biochem. 102, 332–340 [DOI] [PubMed] [Google Scholar]
- 7. Young D. W., Pratap J., Javed A., Weiner B., Ohkawa Y., van Wijnen A., Montecino M., Stein G. S., Stein J. L., Imbalzano A. N., Lian J. B. (2005) SWI/SNF chromatin-remodeling complex is obligatory for BMP2-induced, Runx2-dependent skeletal gene expression that controls osteoblast differentiation. J. Cell. Biochem. 94, 720–730 [DOI] [PubMed] [Google Scholar]
- 8. de la Serna I. L., Ohkawa Y., Imbalzano A. N. (2006) Chromatin remodelling in mammalian differentiation: lessons from ATP-dependent remodellers. Nat. Rev. Genet. 7, 461–473 [DOI] [PubMed] [Google Scholar]
- 9. Smith C. L., Peterson C. L. (2005) ATP-dependent chromatin-remodeling. Curr. Top. Dev. Biol. 65, 115–148 [DOI] [PubMed] [Google Scholar]
- 10. Wilson B. G., Roberts C. W. (2011) SWI/SNF nucleosome remodelers and cancer. Nat. Rev. Cancer 11, 481–492 [DOI] [PubMed] [Google Scholar]
- 11. Wang W., Côté J., Xue Y., Zhou S., Khavari P. A., Biggar S. R., Muchardt C., Kalpana G. V., Goff S. P., Yaniv M., Workman J. L., Crabtree G. R. (1996) Purification and biochemical heterogeneity of the mammalian SWI/SNF complex. EMBO J. 15, 5370–5382 [PMC free article] [PubMed] [Google Scholar]
- 12. Wilsker D., Probst L., Wain H. M., Maltais L., Tucker P. W., Moran E. (2005) Nomenclature of the ARID family of DNA-binding proteins. Genomics 86, 242–251 [DOI] [PubMed] [Google Scholar]
- 13. Patsialou A., Wilsker D., Moran E. (2005) DNA-binding properties of ARID family proteins. Nucleic Acids Res. 33, 66–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Allis C. D., Berger S. L., Cote J., Dent S., Jenuwien T., Kouzarides T., Pillus L., Reinberg D., Shi Y., Shiekhattar R., Shilatifard A., Workman J., Zhang Y. (2007) New nomenclature for chromatin-modifying enzymes. Cell 131, 633–636 [DOI] [PubMed] [Google Scholar]
- 15. Inoue H., Furukawa T., Giannakopoulos S., Zhou S., King D. S., Tanese N. (2002) Largest subunits of the human SWI/SNF chromatin-remodeling complex promote transcriptional activation by steroid hormone receptors. J. Biol. Chem. 277, 41674–41685 [DOI] [PubMed] [Google Scholar]
- 16. Wang X., Nagl N. G., Wilsker D., Van Scoy M., Pacchione S., Yaciuk P., Dallas P. B., Moran E. (2004) Two related ARID family proteins are alternative subunits of human SWI/SNF complexes. Biochem. J. 383, 319–325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Yan Z., Cui K., Murray D. M., Ling C., Xue Y., Gerstein A., Parsons R., Zhao K., Wang W. (2005) PBAF chromatin-remodeling complex requires a novel specificity subunit, BAF200, to regulate expression of selective interferon-responsive genes. Genes Dev. 19, 1662–1667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zhang X., Azhar G., Zhong Y., Wei J. Y. (2006) Zipzap/p200 is a novel zinc finger protein contributing to cardiac gene regulation. Biochem. Biophys. Res. Commun. 346, 794–801 [DOI] [PubMed] [Google Scholar]
- 19. Nagl N. G., Jr., Patsialou A., Haines D. S., Dallas P. B., Beck G. R., Jr., Moran E. (2005) The p270 (ARID1A/SMARCF1) subunit of mammalian SWI/SNF-related complexes is essential for normal cell cycle arrest. Cancer Res. 65, 9236–9244 [DOI] [PubMed] [Google Scholar]
- 20. Nagl N. G., Jr., Wang X., Patsialou A., Van Scoy M., Moran E. (2007) Distinct mammalian SWI/SNF chromatin-remodeling complexes with opposing roles in cell cycle control. EMBO J. 26, 752–763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Flores-Alcantar A., Gonzalez-Sandoval A., Escalante-Alcalde D., Lomelí H. (2011) Dynamics of expression of ARID1A and ARID1B subunits in mouse embryos and in cells during the cell cycle. Cell Tissue Res. 345, 137–148 [DOI] [PubMed] [Google Scholar]
- 22. Guan B., Mao T. L., Panuganti P. K., Kuhn E., Kurman R. J., Maeda D., Chen E., Jeng Y. M., Wang T. L., Shih IeM. (2011) Mutation and loss of expression of ARID1A in uterine low-grade endometrioid carcinoma. Am. J. Surg. Pathol. 35, 625–632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Jones S., Wang T. L., Shih IeM., Mao T. L., Nakayama K., Roden R., Glas R., Slamon D., Diaz L. A., Jr., Vogelstein B., Kinzler K. W., Velculescu V. E., Papadopoulos N. (2010) Frequent mutations of chromatin-remodeling gene ARID1A in ovarian clear cell carcinoma. Science 330, 228–231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wang X., Nagl N. G., Jr., Flowers S., Zweitzig D., Dallas P. B., Moran E. (2004) Expression of p270 (ARID1A), a component of human SWI/SNF complexes, in human tumors. Int. J. Cancer 112, 636–642 [DOI] [PubMed] [Google Scholar]
- 25. Wiegand K. C., Shah S. P., Al-Agha O. M., Zhao Y., Tse K., Zeng T., Senz J., McConechy M. K., Anglesio M. S., Kalloger S. E., Yang W., Heravi-Moussavi A., Giuliany R., Chow C., Fee J., Zayed A., Prentice L., Melnyk N., Turashvili G., Delaney A. D., Madore J., Yip S., McPherson A. W., Ha G., Bell L., Fereday S., Tam A., Galletta L., Tonin P. N., Provencher D., Miller D., Jones S. J., Moore R. A., Morin G. B., Oloumi A., Boyd N., Aparicio S. A., Shih IeM., Mes-Masson A. M., Bowtell D. D., Hirst M., Gilks B., Marra M. A., Huntsman D. G. (2010) ARID1A mutations in endometriosis-associated ovarian carcinomas. N. Engl. J. Med. 363, 1532–1543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Li M., Zhao H., Zhang X., Wood L. D., Anders R. A., Choti M. A., Pawlik T. M., Daniel H. D., Kannangai R., Offerhaus G. J., Velculescu V. E., Wang L., Zhou S., Vogelstein B., Hruban R. H., Papadopoulos N., Cai J., Torbenson M. S., Kinzler K. W. (2011) Inactivating mutations of the chromatin-remodeling gene ARID2 in hepatocellular carcinoma. Nat. Genet. 43, 828–829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Huang X., Gao X., Diaz-Trelles R., Ruiz-Lozano P., Wang Z. (2008) Coronary development is regulated by ATP-dependent SWI/SNF chromatin-remodeling component BAF180. Dev. Biol. 319, 258–266 [DOI] [PubMed] [Google Scholar]
- 28. Wang Z., Zhai W., Richardson J. A., Olson E. N., Meneses J. J., Firpo M. T., Kang C., Skarnes W. C., Tjian R. (2004) Polybromo protein BAF180 functions in mammalian cardiac chamber maturation. Genes Dev. 18, 3106–3116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Nagl N. G., Jr., Zweitzig D. R., Thimmapaya B., Beck G. R., Jr., Moran E. (2006) The c-myc gene is a direct target of mammalian SWI/SNF-related complexes during differentiation-associated cell cycle arrest. Cancer Res. 66, 1289–1293 [DOI] [PubMed] [Google Scholar]
- 30. Beck G. R., Jr., Zerler B., Moran E. (2001) Gene array analysis of osteoblast differentiation. Cell Growth Differ. 12, 61–83 [PubMed] [Google Scholar]
- 31. Stein G. S., Lian J. B., Stein J. L., Van Wijnen A. J., Montecino M. (1996) Transcriptional control of osteoblast growth and differentiation. Physiol. Rev. 76, 593–629 [DOI] [PubMed] [Google Scholar]
- 32. Cordonnier T., Langonné A., Sohier J., Layrolle P., Rosset P., Sensébé L., Deschaseaux F. (2011) Consistent osteoblastic differentiation of human mesenchymal stem cells with bone morphogenetic protein 4 and low serum. Tissue Eng. Part C Methods 17, 249–259 [DOI] [PubMed] [Google Scholar]
- 33. Gluhak-Heinrich J., Guo D., Yang W., Harris M. A., Lichtler A., Kream B., Zhang J., Feng J. Q., Smith L. C., Dechow P., Harris S. E. (2010) New roles and mechanism of action of BMP4 in postnatal tooth cytodifferentiation. Bone 46, 1533–1545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Goldman D. C., Bailey A. S., Pfaffle D. L., Al Masri A., Christian J. L., Fleming W. H. (2009) BMP4 regulates the hematopoietic stem cell niche. Blood 114, 4393–4401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Huang H., Song T. J., Li X., Hu L., He Q., Liu M., Lane M. D., Tang Q. Q. (2009) BMP signaling pathway is required for commitment of C3H10T1/2 pluripotent stem cells to the adipocyte lineage. Proc. Natl. Acad. Sci. U.S.A. 106, 12670–12675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Fen J. Q., Zhang J., Dallas S. L., Lu Y., Chen S., Tan X., Owen M., Harris S. E., MacDougall M. (2002) Dentin matrix protein 1, a target molecule for Cbfa1 in bone, is a unique bone marker gene. J Bone Miner Res. 17, 1822–1831 [DOI] [PubMed] [Google Scholar]
- 37. George A., Sabsay B., Simonian P. A., Veis A. (1993) Characterization of a novel dentin matrix acidic phosphoprotein: implications for induction of biomineralization. J. Biol. Chem. 268, 12624–12630 [PubMed] [Google Scholar]
- 38. Toyosawa S., Shintani S., Fujiwara T., Ooshima T., Sato A., Ijuhin N., Komori T. (2001) Dentin matrix protein 1 is predominantly expressed in chicken and rat osteocytes but not in osteoblasts. J Bone Miner Res. 16, 2017–2026 [DOI] [PubMed] [Google Scholar]
- 39. Ruchon A. F., Tenenhouse H. S., Marcinkiewicz M., Siegfried G., Aubin J. E., DesGroseillers L., Crine P., Boileau G. (2000) Developmental expression and tissue distribution of Phex protein: effect of the Hyp mutation and relationship to bone markers. J Bone Miner Res. 15, 1440–1450 [DOI] [PubMed] [Google Scholar]
- 40. Chi T. H., Wan M., Zhao K., Taniuchi I., Chen L., Littman D. R., Crabtree G. R. (2002) Reciprocal regulation of CD4/CD8 expression by SWI/SNF-like BAF complexes. Nature 418, 195–199 [DOI] [PubMed] [Google Scholar]
- 41. Bultman S., Gebuhr T., Yee D., La Mantia C., Nicholson J., Gilliam A., Randazzo F., Metzger D., Chambon P., Crabtree G., Magnuson T. (2000) A Brg1-null mutation in the mouse reveals functional differences among mammalian SWI/SNF complexes. Mol. Cell 6, 1287–1295 [DOI] [PubMed] [Google Scholar]
- 42. Gao X., Tate P., Hu P., Tjian R., Skarnes W. C., Wang Z. (2008) ES cell pluripotency and germ layer formation require the SWI/SNF chromatin-remodeling component BAF250a. Proc. Natl. Acad. Sci. U.S.A. 105, 6656–6661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ho L., Ronan J. L., Wu J., Staahl B. T., Chen L., Kuo A., Lessard J., Nesvizhskii A. I., Ranish J., Crabtree G. R. (2009) An embryonic stem cell chromatin-remodeling complex, esBAF, is essential for embryonic stem cell self-renewal and pluripotency. Proc. Natl. Acad. Sci. U.S.A. 106, 5181–5186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kaeser M. D., Aslanian A., Dong M. Q., Yates J. R., 3rd, Emerson B. M. (2008) BRD7, a novel PBAF-specific SWI/SNF subunit, is required for target gene activation and repression in embryonic stem cells. J. Biol. Chem. 283, 32254–32263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Bajpai R., Chen D. A., Rada-Iglesias A., Zhang J., Xiong Y., Helms J., Chang C. P., Zhao Y., Swigut T., Wysocka J. (2010) CHD7 cooperates with PBAF to control multipotent neural crest formation. Nature 463, 958–962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Vicent G. P., Nacht A. S., Zaurín R., Ballaré C., Clausell J., Beato M. (2010) Minireview: role of kinases and chromatin remodeling in progesterone signaling to chromatin. Mol. Endocrinol. 24, 2088–2098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Easley R., Carpio L., Dannenberg L., Choi S., Alani D., Van Duyne R., Guendel I., Klase Z., Agbottah E., Kehn-Hall K., Kashanchi F. (2010) Transcription through the HIV-1 nucleosomes: effects of the PBAF complex in Tat-activated transcription. Virology 405, 322–333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ryme J., Asp P., Böhm S., Cavellán E., Farrants A. K. (2009) Variations in the composition of mammalian SWI/SNF chromatin-remodeling complexes. J. Cell. Biochem. 108, 565–576 [DOI] [PubMed] [Google Scholar]
- 49. Burrows A. E., Smogorzewska A., Elledge S. J. (2010) Polybromo-associated BRG1-associated factor components BRD7 and BAF180 are critical regulators of p53 required for induction of replicative senescence. Proc. Natl. Acad. Sci. U.S.A. 107, 14280–14285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Drost J., Mantovani F., Tocco F., Elkon R., Comel A., Holstege H., Kerkhoven R., Jonkers J., Voorhoeve P. M., Agami R., Del Sal G. (2010) BRD7 is a candidate tumor suppressor gene required for p53 function. Nat. Cell Biol. 12, 380–389 [DOI] [PubMed] [Google Scholar]
- 51. Varela I., Tarpey P., Raine K., Huang D., Ong C. K., Stephens P., Davies H., Jones D., Lin M. L., Teague J., Bignell G., Butler A., Cho J., Dalgliesh G. L., Galappaththige D., Greenman C., Hardy C., Jia M., Latimer C., Lau K. W., Marshall J., McLaren S., Menzies A., Mudie L., Stebbings L., Largaespada D. A., Wessels L. F., Richard S., Kahnoski R. J., Anema J., Tuveson D. A., Perez-Mancera P. A., Mustonen V., Fischer A., Adams D. J., Rust A., Chan-on W., Subimerb C., Dykema K., Furge K., Campbell P. J., Teh B. T., Stratton M. R., Futreal P. A. (2011) Exome sequencing identifies frequent mutation of the SWI/SNF complex gene PBRM1 in renal carcinoma. Nature 469, 539–542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Xia W., Nagase S., Montia A. G., Kalachikov S. M., Keniry M., Su T., Memeo L., Hibshoosh H., Parsons R. (2008) BAF180 is a critical regulator of p21 induction and a tumor suppressor mutated in breast cancer. Cancer Res. 68, 1667–1674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Zhou M., Liu H., Xu X., Zhou H., Li X., Peng C., Shen S., Xiong W., Ma J., Zeng Z., Fang S., Nie X., Yang Y., Zhou J., Xiang J., Cao L., Peng S., Li S., Li G. (2006) Identification of nuclear localization signal that governs nuclear import of BRD7 and its essential roles in inhibiting cell cycle progression. J. Cell. Biochem. 98, 920–930 [DOI] [PubMed] [Google Scholar]