Background: SHMT1 limits rates of dTMP biosynthesis in mammalian cells.
Results: Ubiquitin and SUMO modifications of SHMT1 occur on the same consensus site and determine nuclear import, export, and stability.
Conclusion: Competition between SHMT1 SUMOylation and ubiquitination mediates SHMT1 nuclear localization and stability.
Significance: SUMOylation and ubiquitination of SHMT1 affect nuclear de novo thymidylate synthesis capacity.
Keywords: One-carbon Pool, SUMO, SUMOylation, Ubiquitin-conjugating Enzyme (Ubc), Ubiquitination, SHMT1, dTMP, Folate, Thymidylate
Abstract
Serine hydroxymethyltransferase 1 (SHMT1) expression limits rates of de novo dTMP synthesis in the nucleus. Here we report that SHMT1 is ubiquitinated at the small ubiquitin-like modifier (SUMO) consensus motif and that ubiquitination at that site is required for SHMT1 degradation. SHMT1 protein levels are cell cycle-regulated, and Ub-SHMT1 levels are lowest at S phase when SHMT1 undergoes SUMO modification and nuclear transport. Mutation of the SUMO consensus motif increases SHMT1 stability. SHMT1 interacts with components of the proteasome in both the nucleus and cytoplasm, indicating that degradation occurs in both compartments. Ubc13-mediated ubiquitination is required for SHMT1 nuclear export and increases stability of SHMT1 within the nucleus, whereas Ubc9-mediated modification with Sumo2/3 is involved in nuclear degradation. These data demonstrate that SUMO and ubiquitin modification of SHMT1 occurs on the same lysine residue and determine the localization and accumulation of SHMT1 in the nucleus.
Introduction
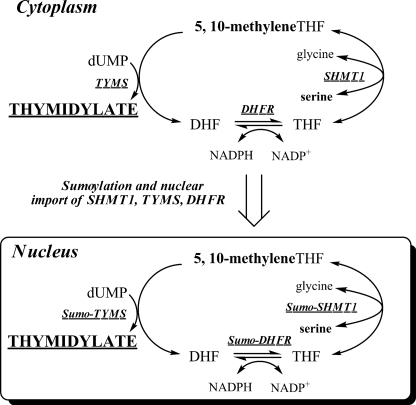
Folate-dependent de novo thymidylate biosynthesis in the nucleus requires three enzymes: thymidylate synthase, dihydrofolate reductase (DHFR)2, and serine hydroxymethyltransferase (SHMT1 and SHMT2α) (Fig. 1). Methylenetetrahydrofolate (methyleneTHF) is the one-carbon and two-electron donor for the thymidylate synthase-catalyzed conversion of dUMP to dTMP and is generated from serine and THF by SHMT. The NADPH-dependent reduction of DHF to regenerate THF is catalyzed by DHFR and permits subsequent rounds of de novo thymidylate synthesis. The enzymes that constitute the de novo dTMP synthesis pathway are posttranslationally modified by Ubc9 with the small ubiquitin-like modifier 1 (SUMO-1) and undergo nuclear translocation during S and G2/M phases (2–4) and in response to UV (5) for nuclear dTMP synthesis (4). Studies in cell cultures have shown that SHMT1 expression determined capacity for de novo dTMP synthesis (6). Regulation of cellular TTP pools is essential for maintaining both nuclear and mitochondrial DNA integrity by preventing uracil accumulation in DNA, and previous studies have demonstrated the importance of maintaining dNTP pool size throughout the cell cycle to avoid genome instability (7, 8).
FIGURE 1.
Folate-dependent de novo thymidylate biosynthesis. The de novo thymidylate biosynthesis pathway is comprised of SHMT1, thymidylate synthase (TYMS), and DHFR. During S phase, these enzymes are SUMOylated by Ubc9, which serves as a signal for nuclear import.
We have observed previously that SHMT1 interacts with Ubc13, an E2 conjugase for ubiquitination (2), although the functional significance of this interaction has yet to be determined. Previous studies have indicated that SUMO and ubiquitin (Ub) can modify the same consensus motif, ΨKXE/D, on target proteins and, in some cases, allow pathway choice (9). Although SUMO-1 modification is essential for nuclear translocation of the de novo dTMP pathway, the role of ubiquitination in the regulation of SHMT1 and de novo thymidylate biosynthesis is unknown. In this study, we demonstrate that Ubc13-mediated modification of SHMT1 leads to nuclear export and acts as a mediator of SHMT1 stability within the nucleus. Furthermore, we demonstrate that SHMT1 is degraded in the cytoplasm via the proteasome through Lys-48 linkage-specific polyubiquitination. This study indicates that competition between SHMT1 SUMOylation and ubiquitination determines SHMT1 nuclear localization by competing for the same consensus motif on SHMT1, and these posttranslational modifications also affect the stability and accumulation of SHMT1 in the nucleus and cytoplasm.
EXPERIMENTAL PROCEDURES
Cell Lines, Media, and Transfection Conditions
HeLa cells were obtained and maintained as reported previously (4). Cells were grown in α-minimal essential medium (Hyclone) supplemented with 10% fetal bovine serum (Hyclone) and penicillin/streptomycin (Mediatech) at 37 °C and 5% CO2. All transfections were performed using the Nucleofector II and kit R for HeLa cells according to the manufacturer's instructions (Lonza).
Vectors and Vector Construction
A construct encoding a V5-SHMT1 fusion protein, pcDNA3.1-SHMT1-V5-HisA, was generated to differentiate between endogenous SHMT1 and SUMO site mutants. The vector pcDNA3.1/V5-HisA was purchased from Invitrogen. The human SHMT1 cDNA from phiYFP-SHMT1 (4) was amplified by PCR using the forward primer 5′-ATATAAGCTTATGACGATGCCAGTCAAC-3′, where the boldface text indicates a HindIII restriction site (New England Biolabs, Inc.). The reverse primer was 5′-ATATCTCGAGGAAGTCAGGCAGGCCAGG-3′, where the boldface text indicates a XhoI restriction site. PCR reactions were conducted as follows: 95 °C for 45 s, 55 °C for 45 s, and 72 °C for 2 min. PCR products were gel-purified using the QIAquick gel extraction kit (Qiagen). Vectors and PCR products were restriction-digested with their respective restriction enzymes according to the manufacturer's protocol. Ligation was completed using T4 DNA ligase (Invitrogen) as indicated in the manufacturer's protocol. Ligation mixtures were transformed into Top10 cells (Invitrogen) and selected for ampicillin (Fisher) resistance. Following plasmid purification and isolation using plasmid mini-preps (Qiagen), constructs were sequenced at the Cornell University Life Sciences Core Laboratories Center. Mutations within the conserved SUMO motif were performed as reported previously (2). The pCMV-HA-Ub vector was generously provided by Dr. Shu-Bing Qian, Cornell University, Ithaca, NY. The K48R and K63R Ub mutants were generated using the primers 5′-ATCTTTGCAGGCAGGCAGCTGGAAGA-3′ and 5′-CTACAATATTCAAAGGGAGTCTACTC-3′, respectively, and reverse complements (the mutation is boldface ) using the QuikChange II site-directed mutagenesis kit (Stratagene) according to the manufacturer's instructions. All siRNA was purchased from Qiagen.
Preparation of cDNA and Real-time PCR
HeLa cells were cell cycle-arrested as described below and total RNA was isolated using a RNeasy kit (Qiagen) according to the manufacturer's protocol after 1-h treatment of samples with DNase I (Qiagen) at 37 °C to remove residual DNA. Total RNA was converted to cDNA using Superscript III first-strand cDNA synthesis kit (Invitrogen) using oligo(dT) primers according to the manufacturer's protocol. Real-time PCR was completed using Quantifast SYBR Green PCR kit (Qiagen) and SHMT1 primers (Qiagen). PCR products were quantified using Applied Biosystems 7500 real-time PCR system.
Immunoblotting
Cellular proteins were quantified, separated, transferred, and detected as described previously (2). GAPDH, Lamin-A, and Ubc13 immunoblots were generated as described previously (2, 4). All antibodies were diluted in 5% nonfat dry milk (Carnation) containing 1% Nonidet P-40 (US Biologicals). For Ub detection, mouse anti-Ub antibody (BIOMOL) was diluted 1:1000. For V5 tag detection, mouse anti-V5 antibody (Invitrogen) was diluted 1:5000. Mouse anti-β-actin antibody (Abcam) was used at a 1:5000 dilution. For detection of HA-tagged Ub, mouse anti-HA antibody (Santa Cruz Biotechnology, Inc.) was used. For detection of Ub, V5, HA, and β-actin, goat anti-mouse IgG-HRP (Thermo Scientific) was diluted 1:10,000. For detection of SUMO-2/3 conjugates, rabbit anti-SUMO-2/3 (Cell Signaling) was used at a dilution of 1:1000. Goat anti-rabbit-HRP secondary antibody was used at a dilution of 1:20,000 (Thermo Scientific). Bands were quantified by densitometry using the gel analyzer tool in ImageJ. All values for the SHMT1 protein were normalized to values obtained for GAPDH, Lamin-A, or actin.
Immunoprecipitation
Immunoprecipitations were conducted using the Dynabeads protein G immunoprecipitation kit (Invitrogen). For whole cell immunoprecipitation, HeLa cells were lysed using mammalian protein extraction reagent (Pierce) supplemented with 1 mm N-ethylmaleimide (Sigma), 100 μm calpain inhibitor 1 (N-acetyl-L-leucyl-L-leucyl-L-norleucinal, Sigma), 2 mm β-mercaptoethanol (Calbiochem), 0.1 mm EDTA (Fisher Scientific), 1 mm PMSF (Alexis Biochemicals), and 1:1000 protease inhibitor mixture (Sigma). For immunoprecipitations of nuclear and cytosolic extracts, nuclei were purified using the Active Motif nuclear extract kit according to the manufacturer's protocol. 1 mg of total protein per sample was incubated with 5 μg of either anti-Lys-63-linkage Ub (Millipore), anti-Ub (BIOMOL), or anti-V5 tag (Invitrogen) antibody overnight at 4 °C. For the nuclear immunoprecipitations, anti-SHMT1 antibody was used following the same protocol. The beads were collected and washed four times with 1 ml PBS. 40 μl of SDS-PAGE sample buffer was added to the beads to elute with heating at 100 °C.
Cell Cycle Synchronization and Analysis
HeLa cells at 60% confluence were arrested at various cell cycle stages using 30 μm lovastatin for G1 (Sigma), 2 mm hydroxyurea for S phase (Sigma), or 100 ng/ml nocodazole for G2/M phase (Sigma). Preparation for FACS was performed as described previously (2). FACS analysis was performed at the Biomedical Sciences Flow Cytometry Core Laboratory at Cornell University.
Proteasomal Inhibition and Half-life Analysis
HeLa cells were treated with 50 μg/ml cycloheximide (Sigma) to stabilize cellular protein levels and 100 μm ALLN (Sigma) to stabilize Ub linkages. Proteasome inhibition was achieved using 20 μm MG132 (Sigma). Cells were isolated at 0, 1, 2, 4, and 8 h or 0, 2, 4, and 8 h as described in the figure legends. For determination of SHMT1 nuclear versus cytoplasmic half-life, nuclei and cytosol were isolated from cells using a nuclear extraction kit (Active Motif) at the appropriate time points according to the manufacturer's protocol.
Immunofluorescence
The SHMT1 antibody was labeled with Cy3 dye using the Amersham Biosciences Cy3 bis-reactive labeling kit (GE Life Sciences) according to the manufacturer's protocol. HeLa cells were grown on coverslips in 6-well plates. Cells were treated with DMSO (Fisher) or 50 μm MG132 (Sigma) for 3 h. Cells were washed, medium was added back, and cells were incubated for 12 h for recovery. Cells were then fixed using 4% formaldehyde solution at room temperature for 10 min. Fixation was quenched in 100 mm glycine in PBS for 5 min. Cells were permeabilized in 0.1% Triton X-100 (Fisher) in PBS for 5 min at room temperature. Immunohistochemistry was performed using antibodies against SHMT1, 20S α + β subunits (Abcam), and the 19 S S7 subunit (Abcam) diluted in 1% BSA (Sigma) in PBS at 1 μg/ml, 1:500 and 1:1000, respectively. Primary antibodies were incubated on the fixed and permeabilized cells for 1 h at room temperature. Following four washes of each coverslip with 3 ml of PBS, secondary antibodies were incubated on coverslips for 1 h at room temperature. Alexa Fluor 555 donkey anti-rabbit secondary antibody was used for detection of 20 S α and β subunits and the 19 S S7 subunit diluted in 1% BSA in PBS at 1:1000. Coverslips were washed as stated previously stated. DRAQ5 (Alexis Biochemicals) was used for nuclear staining according to the manufacturer's protocol. Cells were mounted to slides using Mowiol mounting solution (Calbiochem) and allowed to harden overnight at 4 °C and then visualized using confocal microscopy.
Leptomycin B Treatment
HeLa cells were treated with or without 20 μm MG132 and either 0, 2.5, or 5 ng/ml of the nuclear export inhibitor leptomycin B (LMB) (Sigma) for 24 h. Nuclei were isolated from cells as stated above and lysed using SDS-PAGE loading buffer for immunoblotting.
Nuclear Localization of SHMT1 as a Function of Ubc13 Expression
cDNAs encoding YFP-SHMT1 (4), Ubc13 (Open Biosystems), or siRNA against Ubc13 (Qiagen) were transfected into HeLa cells. Cells were visualized using confocal microscopy with DRAQ5 as the nuclear control. Cells with nuclear SHMT1 versus cytosolic SHMT1 were counted (n = 100) for each of the following treatments: YFP-SHMT1 with endogenous Ubc13 levels, YFP-SHMT1 and Ubc13 siRNA, YFP-SHMT1 and Ubc13 overexpression, and YFP-SHMT1 with ubc13 overexpression and 2.5 ng/ml LMB treatment.
RESULTS
SHMT1 Protein Levels and Ubiquitination Change with the Cell Cycle
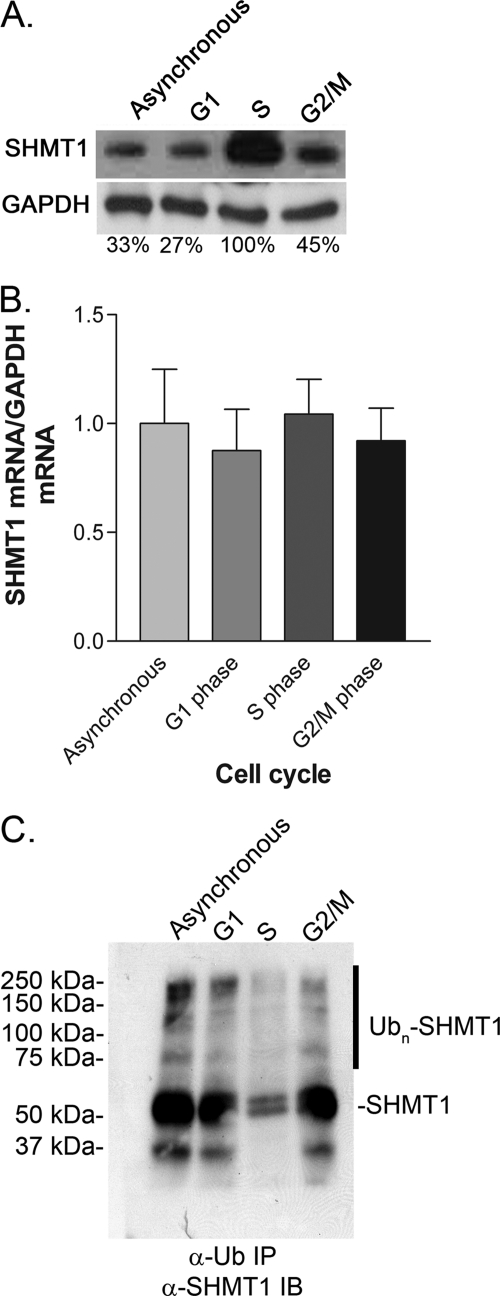
The levels of cellular SHMT1 protein and Ub-modified SHMT1 (Ub-SHMT1) were quantified as a function of the cell cycle in HeLa cells (Fig. 2). Protein levels of SHMT1 were elevated during S phase (Fig. 2A), whereas mRNA levels of SHMT1 did not change as a function of the cell cycle (B). Ub-SHMT1 was detected in whole-cell extracts at all phases of the cell cycle but was diminished at S phase. These data demonstrate that SHMT1 is a cell cycle-regulated protein and that Ub-SHMT1 levels are lowest at S phase when SHMT1 undergoes SUMO modification and nuclear localization.
FIGURE 2.
SHMT1 protein levels change with the cell cycle. HeLa cells were cell cycle-blocked by 24-h treatment with lovastatin (30 μm) for G1 phase, hydroxyurea (1 mm) for S phase, and nocodazole (100 ng/ml) for G2/M phase. A, SHMT1 protein levels were determined by immunoblotting of 20 μg of protein extract. B, SHMT1 mRNA levels were quantified from whole cell extract. C, Ub-SHMT1 levels were quantified by immunoprecipitation (IP) of ubiquitinated substrates from whole cell extracts (20 μg) followed by SHMT1 immunoblotting (IB). All experiments were repeated at least three times with similar results.
Proteasome Inhibition Stabilizes SHMT1 and Increases Ubiquitinated SHMT1 Levels
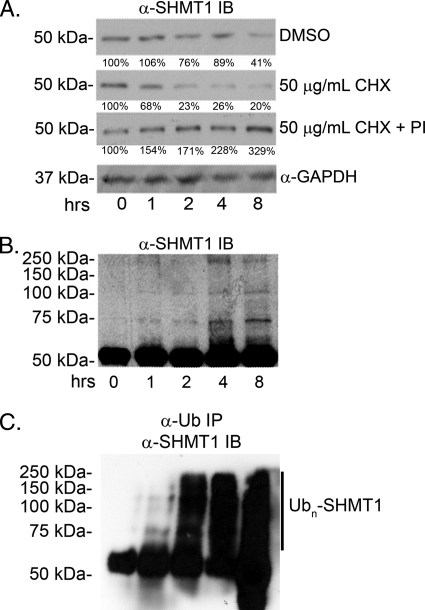
HeLa cells were treated with the translation inhibitor cycloheximide (CHX) and proteasome inhibitors MG132 and ALLN to determine whether Ub modification mediates SHMT1 half-life. SHMT1 exhibited a half-life of ∼2 h in HeLa cells treated with CHX, whereas SHMT1 levels increased over an 8-h interval in HeLa cells treated with CHX and proteasome inhibitors (Fig. 3A), indicating that SHMT1 is subject to Ub-dependent proteasomal processing. Cells treated with CHX and proteasome inhibitors accumulated higher molecular weight SHMT1 immunoreactive bands over time (Fig. 3B), indicating that Ub-SHMT1 is an intermediate in SHMT1 degradation by the proteasome. These higher molecular weight bands were identified as SHMT1 ubiquitilated high-molecular weight species through Ub immunoprecipitation and SHMT1 immunoblotting (Fig. 3C).
FIGURE 3.
Proteasome inhibition stabilizes SHMT1 and increases ubiquitinated SHMT1 levels. HeLa cells were treated with DMSO (control), CHX, or CHX and 20 μm MG132 + 100 μm ALLN proteasome inhibitors (PI) for 8 h. SHMT1 immunoblot analyses were performed as described under “Experimental Procedures.” A, total cellular protein extracts (20 μg) were separated by electrophoresis, and SHMT1 protein levels were determined by immunoblotting (IB). The half-life of endogenous SHMT1 when CHX treated is ∼2 h. DMSO control and CHX + PI treatments were stable over 8 h. B, SHMT1 was visualized in cell extracts by immunoblotting as described above. Accumulation of higher molecular weight bands was observed in CHX + PI treatment over 8 h. C, Ub-SHMT1 was visualized in total cell protein extracts (20 μg) by immunoprecipitation (IP) of Ub-conjugated proteins followed by SHMT1 immunoblotting. All experiments were repeated at least three times with similar results.
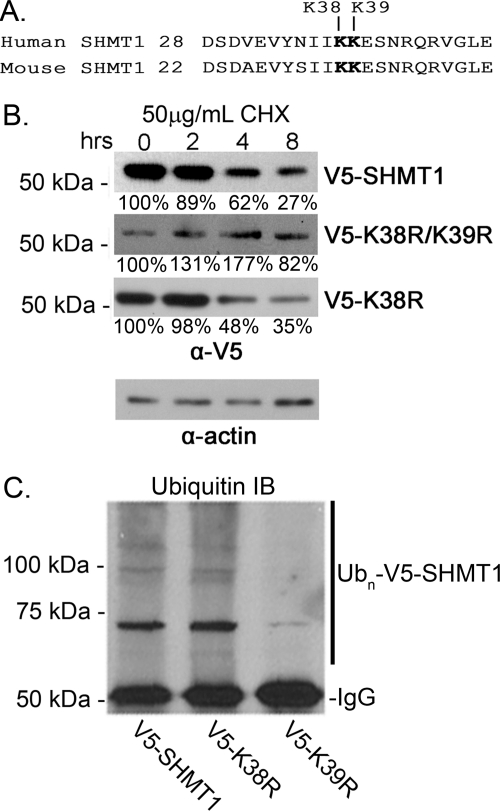
SHMT1 K39 Is the Ubiquitin Modification Site
We observed previously that SHMT1 SUMO-1 modification occurs at either Lys-38 or Lys-39 (Fig. 4A) and that mutation of either lysine to arginine resulted in the ablation of SUMOylation (2, 4). To determine whether ubiquitination occurs at this site, expression vectors encoding V5-SHMT1, V5-K38R SHMT1, and V5-K38R/K39R were transfected into HeLa cells. The cells were treated with CHX to determine the role of Lys-38 and Lys-39 in SHMT1 turnover and half-life. V5-SHMT1 and V5-K38R SHMT1 exhibited similar half-lives, whereas V5-K38R/K39R SHMT1 was stable over time (Fig. 4B). Immunoprecipitation of the V5 tag followed by immunoblotting with anti-Ub antibody indicated that mutation of Lys-39 results in a marked reduction of Ub-SHMT (Fig. 4C). These data demonstrate that Lys-39 is a modification site for ubiquitination and that Lys-39 ubiquitination leads to proteasomal processing.
FIGURE 4.
SHMT1 Lys-39 is the Ub modification site. HeLa cells were transfected with expression vectors encoding V5-tagged wild-type SHMT1, K38R SHMT1, K38R/K39R SHMT1, or K39R SHMT1. A, the conserved SUMO motif (IKKE) is also a site for ubiquitination. B, total cellular protein extracts (20 μg) were separated by electrophoresis, and V5-SHMT1 protein levels were determined by immunoblot analyses against V5. V5 wild-type SHMT1 and V5-K38R SHMT1 both exhibited a half-life of ∼4 h. Mutation of both K38R and K39R resulted in SHMT1 stability. Actin was used as a loading control. C, Ub-V5-SHMT1 was visualized in total cell protein extracts (20 μg) by V5 immunoprecipitation followed by Ub immunoblot (IB) analyses. UB-V5-SHMT1 is reduced in K39R mutants compared with wild-type SHMT1 and K38R mutants. All experiments were repeated at least three times with similar results.
Colocalization of SHMT1 with the 19 S Cap and 20 S Core of the Proteasome in the Nucleus and the Cytosol
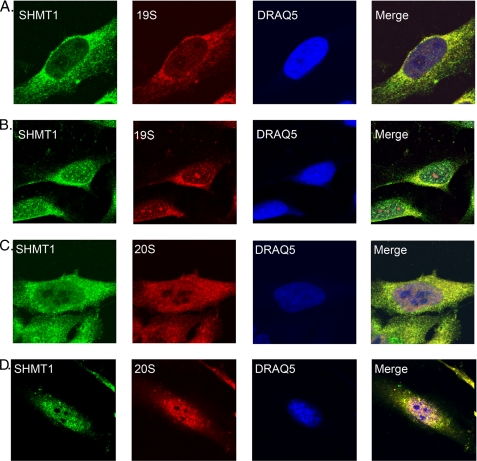
Immunolocalization and biochemical studies have shown that proteasomes are localized to the nuclei and cytosol (10, 11). To determine whether degradation of SHMT1 occurs within the nucleus and cytoplasm, immunochemistry was performed using HeLa cells treated with either DMSO or MG132. The proteasome 20 S catalytic core, the 19 S cap, and SHMT1 colocalized in both the cytoplasm and nucleus with discrete puncta formation following MG132 treatment as compared with control DMSO treatments, indicating that SHMT1 associates with the proteasome in both compartments (Fig. 5).
FIGURE 5.
SHMT1 colocalizes with the 19 S cap and 20 S core of the proteasome in the nucleus and the cytosol. Cells were treated with DMSO (A and C) or 50 μm MG132 (B and D) for 3 h. Cells were washed, medium was added back, and cells were incubated for a 12-h recovery. Immunohistochemistry was performed using antibodies against SHMT1, 20S α and β subunits, and the 19 S S7 subunit. DRAQ5 was used for nuclear staining. Cells were then visualized using confocal microscopy. Colocalization of SHMT1 with 19 S and 20 S are seen in controls, most prevalently within the cytosol. Upon treatment with MG132 and recovery, colocalization of 19 S and 20 S with SHMT1 is observed within puncta in the nucleus and cytoplasm. This experiment was repeated twice with the same result.
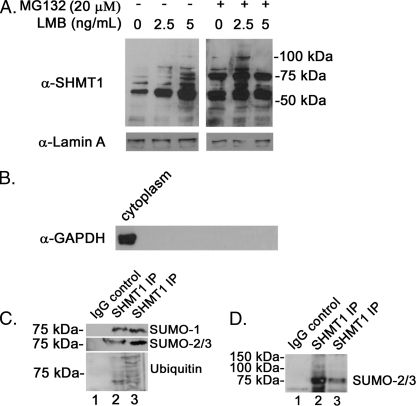
Inhibition of Nuclear Export Leads to Accumulation of SHMT1 in the Nucleus
To determine whether SHMT1 is degraded within the nucleus or if it is shuttled to the cytoplasm prior to degradation, HeLa cells were treated with the nuclear export inhibitor LMB for 24 h with or without MG132. Nuclei were isolated, and SHMT1 protein levels were quantified by Western blot analysis. Nuclear SHMT1 levels increased following LMB treatment, with an accumulation of higher molecular weight immunoreactive bands in both the MG132-treated and untreated cells (Fig. 6A). Nuclear extracts isolated from MG132-treated cells exhibited a prominent 75-kDa band that was identified to be a SUMO-2/3 conjugate of SHMT1 (Fig. 6, C and D). In the LMB-treated cells without MG132, nuclear extracts accumulated higher molecular weight SHMT1 immunoreactive bands consistent with canonical ubiquitination ladders (Fig. 6A). These were identified as Ub-SHMT1 through immunoprecipitation (Fig. 6C). Increases in nuclear SUMO-2/3-SHMT1 conjugates and Ub conjugates were observed in the samples treated with LMB and MG132 (Fig. 6, C and D). These data demonstrate that nuclear Ub-SHMT1 conjugates accumulate when SHMT1 nuclear export is inhibited and that mixed SUMO-2/3 and Ub SHMT1 conjugates accumulate when both SHMT1 nuclear export and degradation are impaired, indicating a role for Ub modification in nuclear export and SUMO-2/3 modification in SHMT1 degradation.
FIGURE 6.
Inhibition of nuclear export leads to accumulation of SHMT1 in the nucleus. A, HeLa cells were treated with or without 20 μm MG132 and varying levels of the nuclear export inhibitor LMB for 24 h. Nuclei were isolated from cells and lysed using SDS-PAGE loading buffer. SHMT1 immunoblot analyses were completed as described in Fig. 2. Lamin A immunoblots were performed to control for loading. B, nuclear purity was assessed by GAPDH immunoblotting. C, SHMT1 immunoprecipitations (IP) were performed using nuclear extracts from cells treated with 2.5 ng/ml LMB (lane 2) or 2.5 ng/ml LMB and 20 μm MG132 (lane 3) as described in Fig. 3. Non-immune IgG was used as a negative control for samples treated as in lane 3. D, SHMT1 immunoprecipitations were performed using nuclear extracts from cells treated with 2.5 ng/ml LMB (lane 3) or 2.5 ng/ml LMB and 20 μm MG132 (lane 2). Non-immune IgG was used as a negative control for samples treated as in lane 2. A darker exposure of this blot shows the presence of multiple higher molecular weight SHMT1-SUMO-2/3 conjugates in samples treated with MG132 (lane 3). All experiments were repeated at least three times with similar results.
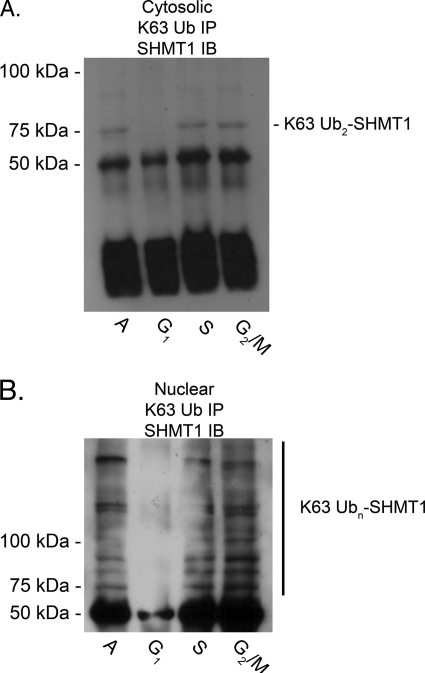
Lys-63 Polyubiquitination of SHMT1 Is Cell Cycle- and Compartment-specific
Ubc13 catalyzes the formation of Lys-63-linked polyUb chains, which is important for exporting p53 out of the nucleus (12). It has also been reported to interact with SHMT1 (2). The cell cycle dependence and compartment specificity of SHMT1 Lys-63 Ub linkages was determined in nuclear and cytosolic fractions of HeLa cells. Lys-63 ubiquitination was present in S and G2/M while absent from G1 in both nuclear and cytosolic fractions. SHMT1 Lys-63 polyubiquitination was extensive in the nuclear fraction (Fig. 7B), whereas only bands consistent with diubiquitination were observed in the cytosolic fraction (Fig. 7A). Lys-63 ubiquitination of SHMT1 exhibited a distinct cell cycle profile in both the cytosolic and nuclear fractions compared with ubiquitination in general (Figs. 7 and 2C).
FIGURE 7.
Lys-63 polyubiquitination of SHMT1 is cell cycle- and compartment-specific. HeLa cells were treated with cell cycle blocking agents as reported above. Cytosolic (A) and nuclear (B) fractions were isolated, and immunoprecipitation (IP) against Lys-63 linkage-specific Ub was performed. Antibody protein complexes were isolated from 20 μg of extract protein using protein G-conjugated Dynabeads. Immunoblot (IB) analyses were performed against SHMT1 as described in Fig. 2. In the cytosolic fraction (A), Lys-63-linked diUb was the most prevalent band and was diminished in G1 phase compared with S and G2/M phases. In the nuclear fraction (B), extensive Lys-63 polyubiquitination was observed in S and G2/M phases. Lys-63 ubiquitination was diminished in G1 phase in the nuclear fraction. All experiments were repeated at least three times with similar results. A, asynchronous.
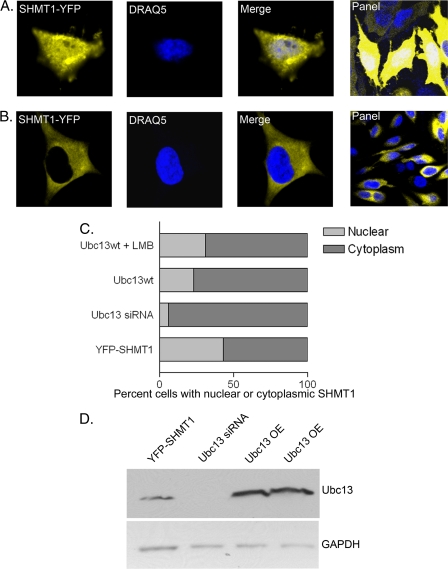
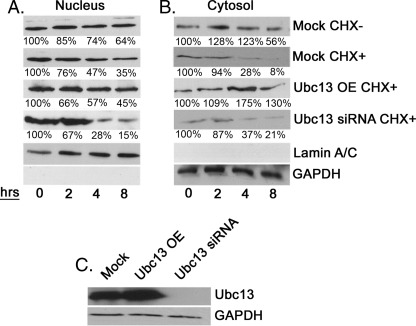
Ubc13 Affects SHMT1 Nuclear Accumulation
To determine whether Ubc13 mediates nuclear accumulation and/or SHMT1 half-life within the nucleus, a SHMT1-YFP expressing vector and either an Ubc13-expressing vector or Ubc13 siRNA were coexpressed in the presence and absence of LMB (Fig. 8). 43% of cells expressing YFP-SHMT1 with endogenous levels of Ubc13 accumulated SHMT1 in the nucleus (Fig. 8, A–C). Ubc13 overexpression led to fewer cells with nuclear SHMT1 (23%), indicating a role for Ubc13 in nuclear SHMT1 accumulation. Ubc13 overexpression with LMB treatment restored nuclear SHMT1 levels to 31%, which is statistically greater than levels observed for non-LMB-treated samples (Student's t test, p < 0.0001, n = 100), indicating a role for Ubc13 in facilitating SHMT1 nuclear export. Alternatively, overexpression of Ubc13 may have antagonistic effects on Ubc9 in the cytosol, although LMB would not be expected to restore nuclear SHMT1 if that were the case. Depletion of Ubc13 by siRNA treatment strongly inhibited SHMT1 accumulation in the nucleus with only 6% of cells exhibiting nuclear SHMT1. This may reflect competition between Ubc9 and Ubc13 in the nucleus leading to Sumo2/3-mediated SHMT1 degradation. The effects were not due to cell cycle differences as determined by cell sorting3. These data suggest that Ubc13 may be playing a role in both stabilization of SHMT1 within nuclei and export of SHMT1 out of nuclei.
FIGURE 8.
Ubc13 affects nuclear accumulation of SHMT1. Expression vectors encoding SHMT1-YFP, Ubc13, or siRNA against Ubc13 were transfected into HeLa cells. Cells were visualized using confocal microscopy with DRAQ5 as the nuclear control. Cells with nuclear SHMT1 (A) versus cytoplasm (B) were counted (n = 100) for each of the following treatments: YFP-SHMT1 with endogenous Ubc13 levels, YFP-SHMT1 and Ubc13 siRNA, YFP-SHMT1 and Ubc13 overexpression, and YFP-SHMT1 with Ubc13 overexpression and LMB treatment. C, YFP-SHMT1 samples with endogenous Ubc13 levels exhibited highest number of cells exhibiting nuclear SHMT1 (43% ± 4.9%). Overexpression of Ubc13 with LMB treatment had intermediate levels (31% ± 6.4%) between endogenous Ubc13 and Ubc13 overexpression alone (23% ± 3.5%). Ubc13 overexpression alone and Ubc13 overexpression and LMB treatment are statistically different with a p value of 0.0001 using Student's t test. Treatment with siRNA directed against Ubc13 exhibited the lowest number of cells with nuclear SHMT1 (6% ± 1.4%). Data are mean ± S.E. D, immunoblotting was performed on 20 μg of total cell extract to ensure Ubc13 knockdown and overexpression (OE) in cells with GAPDH as a loading control.
Ubc13 Affects Cytosolic but Not Nuclear Turnover of SHMT1
To determine whether Ubc13 stabilizes SHMT1 in the nucleus and the cytosol, the effect of Ubc13 expression on SHMT1 half-life was determined (Fig. 9). The stability of SHMT1 in nuclear extracts from cells treated with cycloheximide was diminished in cells treated with Ubc13 siRNA, supporting the hypothesis that Ubc9 and Ubc13 compete for SHMT1 modification, leading to either degradation in the nucleus or nuclear export, respectively. In cytoplasmic extracts, cells overexpressing Ubc13 exhibited elevated SHMT1 levels and increased SHMT1 stability, indicating that UBC9-mediated SHMT1 Lys-63 modification protects SHMT1 from degradation in the cytoplasm. Ubc13 knockdown led to a decrease in SHMT1 stability in the cytosolic fraction, supporting a role for Lys-63 ubiquitination in stabilizing SHMT1 in both the cytoplasm and nucleus.
FIGURE 9.
Ubc13 affects cytosolic turnover but not nuclear turnover of SHMT1. HeLa cells were subjected to mock, cDNA encoding Ubc13, or Ubc13 siRNA transfections and subjected to 50 μg/ml CHX for the times indicated. Nuclei and cytosol were isolated. A, SHMT1 was stable within the nucleus in all samples except for Ubc13 siRNA-treated samples. B, cytosolic SHMT1 was less stable within the cytosol. Ubc13 overexpression and knockdown increased half-life of SHMT1. C, to ensure that Ubc13 was overexpressed (OE) and knocked down, immunoblotting against Ubc13 on samples treated with cycloheximide for 8 h was performed. GAPDH was used as a loading control. All experiments were repeated at least three times with similar results.
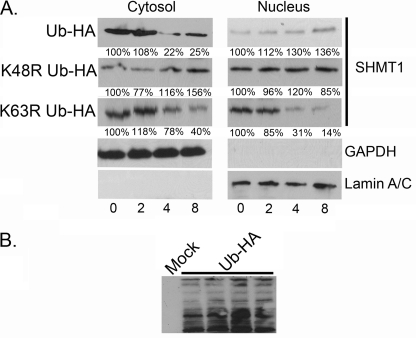
K48R and K63R Ubiquitin Mutants Affect Cytosolic and Nuclear SHMT1 Turnover
Ubc13 functions in Lys-63 Ub linkages, whereas the major Ub linkage implicated in degradation is Lys-48 modification. To determine the role of Lys-63 and Lys-48 Ub modifications on SHMT1 stability, Lys-48 and Lys-63 mutant Ub proteins were expressed in cells, and their effect on SHMT1 stability was determined in the cytoplasmic and nuclear compartments. SHMT1 levels were stable in nuclear extracts from cells transfected with either K48R or wild-type Ub, indicating that Lys-48 modifications do not influence nuclear SHMT1 stability (Fig. 10A). However, the stability of SHMT1 in cytosolic extracts from cells expressing K48R Ub was increased compared with extracts from cells expressing wild-type Ub, indicating a role for Lys-48 Ub linkages in SHMT1 turnover in the cytoplasm (Fig. 10A). In contrast, SHMT1 stability was unaffected in cytoplasmic extracts expressing K63R Ub relative to cytosolic extracts from cells expressing wild-type Ub, indicating that SHMT1 K63 Ub modification does not play a role in SHMT1 stability in the cytoplasm. However, SHMT1 stability was decreased in nuclear extracts from cells expressing K63R Ub relative to cytosolic extracts from cells expressing wild-type Ub, indicating a protective role for Lys-63 Ub modification of SHMT1 in the nucleus. These data indicate that SHMT1 is degraded via Lys-48 polyubiquitination only in the cytoplasm, whereas Lys-63 Ub modification is necessary for SHMT1 stability in the nucleus.
FIGURE 10.
Ub Lys-63 modifications enhance SHMT1 stability in the nucleus, whereas Ub Lys-48 modifications decrease SHMT1 stability in the cytoplasm. HeLa cells were transfected with expression vectors encoding HA-Ub, HA-K48R Ub, or HA-K63R Ub. Following transfection, the cells were subjected to 50 μg/ml CHX for the times indicated, and SHMT1 immunoblotting performed on nuclear and cytosolic fractions as described in Fig. 2. A, SHMT1 was stable in nuclear extracts from cells expressing HA-WT Ub and HA-K48R Ub, whereas HA-K63R Ub expression decreased SHMT1 stability. In cytosolic fractions, SHMT1 stability was enhanced by HA-K48R Ub expression as compared with HA-WT Ub and HA-K63R Ub expression. Lamin A/C and GAPDH immunoblots were performed to control for nuclear and cytosolic purity and loading. B, a representative figure of HA immunoblotting is shown here to validate expression. All experiments were repeated at least three times with similar results.
DISCUSSION
Thymidine nucleotide synthesis and pool size is highly regulated, with expansion of the pool occurring during S-phase (13, 14). As the need for TTP diminishes toward mitotic entry, depletion of cellular pools of TTP is necessary to avoid growth inhibition and genetic instability (7). SHMT1 expression has been shown to be rate-limiting for de novo dTMP synthesis in both cell culture (15) and animal models (16). Previous studies have observed that DHFR is mono-ubiquitinated by MDM2 and that mono-ubiquitination lowers DHFR activity (17). However, ubiquitination of DHFR is not involved in DHFR degradation (17). Thymidylate synthase degradation has also been studied and has been found to be proteasomal but Ub-independent (18–20). Regulation of SHMT1 levels appears to be central to determining de novo dTMP synthesis capacity for DNA replication and repair. Here we report that SHMT1 levels are regulated in an Ub-dependent proteasomal manner in both the cytoplasm and in the nucleus. The ubiquitination observed is cell cycle-dependent, with greater SHMT1 ubiquitination occurring during the G1 and G2/M phases.
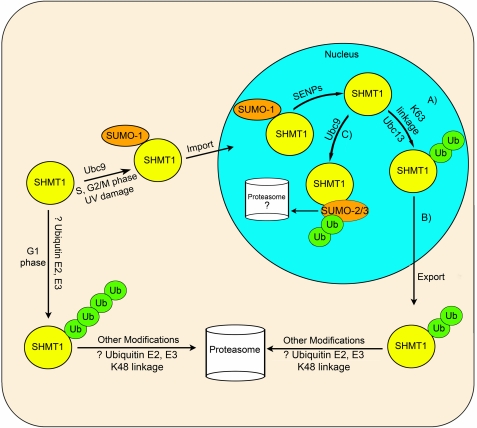
In this study, we demonstrate the interplay of SUMOylation and ubiquitination for the control of SHMT1 levels within the nucleus and cytoplasm. We have shown previously that mutation of the conserved SHMT1 SUMO motif prevents SHMT1 translocation to the nucleus (4). Here we show that mutation of the conserved SUMO site leads to the stabilization of SHMT1 in cycloheximide-treated samples and that there is a requirement for Lys-39 in SHMT1 degradation. Colocalization of SHMT1 with the 20 S catalytic core and 19 S cap of the proteasome within the cytoplasm and nucleus indicates that SHMT1 degradation occurs in both compartments. In the cytoplasm, SHMT1 degradation is facilitated by Lys-48 Ub linkages, whereas in the nucleus, Lys-63-linked ubiquitination prevents degradation, potentially by blocking Ubc9-mediated Sumo2/3 modification (Fig. 11). This finding suggests that Lys-63-linked ubiquitination may occur within the nucleus once SUMO1 modification that is required for nuclear import is removed and leads to the export of SHMT1 from the nucleus to the cytoplasm.
FIGURE 11.
Proposed interactions among SHMT1 ubiquitination and SUMOylation. In this model, SUMO-1 is conjugated to SHMT1 by Ubc9 during S, G2/M phases, and in response to UV damage, which leads to nuclear import. The Ub E2 conjugase Ubc13 acts to stabilize SHMT1 within the nucleus (A) and signals nuclear export of SHMT1 through Lys-63-specific Ub linkage (B). Following nuclear export, SHMT1 is degraded via the proteasome in a Lys-48 Ub linkage-specific manner by unknown Ub pathway E2s and E3s in the cytoplasm. C, Ubc9 catalyzes the formation of SUMO-2/3 conjugates, which may be involved in nuclear SHMT1 degradation as SHMT1 accumulates in the nucleus when nuclear export is blocked and proteasomal degradation is inhibited. Ubiquitination of SHMT1 also increases in the nucleus following proteasome inhibition. Following SUMO-2/3 addition to SHMT1, ubiquitination may occur in mixed SUMO/Ub chains mediating SHMT1 degradation. Degradation of SHMT1 may occur in both nuclear and cytosolic compartments.
Our proposed model for the interplay between ubiquitination and SUMOylation is as follows. Following the SUMO-1-dependent import of SHMT1 within the nucleus, the SUMO-1 moiety can be removed from the SUMO consensus motif of SHMT1 by the known activity of SUMO proteases (SENPs), but this has not been demonstrated directly in this study (Fig. 11). Following removal, ubiquitination can occur via Ubc13, creating Lys-63 Ub linkages. Although some Lys-63-linked Ub modifications have been observed to be signals for proteasomal degradation (21), our data does not support that hypothesis for SHMT1. Total cellular SHMT1 protein levels decline during G1 and G2/M phases with a concomitant increase in ubiquitination, which reflects predominately Lys-48 linkages in the cytoplasm (Fig. 2). Lys-63 ubiquitination occurs most during the S and G2/M phases but is absent in G1 phase. Lys-63-linked ubiquitination stabilizes SHMT1 in the nucleus and may also play a role as a mediator of SHMT1 export, as is the case for the p53 protein (22).
SHMT1 is also a substrate for UBC-9-mediated SUMOylation with SUMO-2/3. Here we propose that mixed SUMO-2/3-Ub chains may be acting to mediate the proteasomal degradation of SHMT1 within the nucleus, which has been observed previously for other proteins (22). The role of mixed SUMO-2/3-Ub chains seem to be compartment-specific to the nucleus. Future work must be done to understand which other Ub pathway enzymes are required for the degradation and stability of SHMT1 within the nucleus and cytosol.
Acknowledgments
We thank Martha Field and Shu-Bing Qian for help in interpreting results.
This work was supported by National Institutes of Health Grant DK58144 (to P. J. S.).
D. D. Anderson, J. Y. Eom, and P. J. Stover, unpublished data.
- DHFR
- dihydrofolate reductase
- SHMT1
- cytoplasmic serine hydroxymethyltransferase
- TYMS
- thymidylate synthase
- DHF
- dihydrofolate
- CHX
- cycloheximide
- LMB
- leptomycin B
- Ub
- ubiquitin
- SUMO
- small ubiquitin-like modifier
- ALLN
- calpain inhibitor 1 N-acetyl-l-leucyl-l-leucyl-l-norleucinal
- DMSO
- dimethyl sulfoxide.
REFERENCES
- 1.Deleted in proof
- 2. Woeller C. F., Anderson D. D., Szebenyi D. M., Stover P. J. (2007) Evidence for small ubiquitin-like modifier-dependent nuclear import of the thymidylate biosynthesis pathway. J. Biol. Chem. 282, 17623–17631 [DOI] [PubMed] [Google Scholar]
- 3. Anderson D. D., Woeller C. F., Stover P. J. (2007) Small ubiquitin-like modifier-1 (SUMO-1) modification of thymidylate synthase and dihydrofolate reductase. Clin. Chem. Lab. Med. 45, 1760–1763 [DOI] [PubMed] [Google Scholar]
- 4. Anderson D. D., Stover P. J. (2009) SHMT1 and SHMT2 are functionally redundant in nuclear de novo thymidylate biosynthesis. PLoS ONE 4, e5839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Fox J. T., Shin W. K., Caudill M. A., Stover P. J. (2009) A UV-responsive internal ribosome entry site enhances serine hydroxymethyltransferase 1 expression for DNA damage repair. J. Biol. Chem. 284, 31097–31108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Herbig K., Chiang E. P., Lee L. R., Hills J., Shane B., Stover P. J. (2002) Cytoplasmic serine hydroxymethyltransferase mediates competition between folate-dependent deoxyribonucleotide and S-adenosylmethionine biosyntheses. J. Biol. Chem. 277, 38381–38389 [DOI] [PubMed] [Google Scholar]
- 7. Ke P. Y., Kuo Y. Y., Hu C. M., Chang Z. F. (2005) Control of dTTP pool size by anaphase-promoting complex/cyclosome is essential for the maintenance of genetic stability. Genes Dev. 19, 1920–1933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Samsonoff W. A., Reston J., McKee M., O'Connor B., Galivan J., Maley G., Maley F. (1997) Intracellular location of thymidylate synthase and its state of phosphorylation. J. Biol. Chem. 272, 13281–13285 [DOI] [PubMed] [Google Scholar]
- 9. Ulrich H. D. (2009) Regulating post-translational modifications of the eukaryotic replication clamp PCNA. DNA Repair 8, 461–469 [DOI] [PubMed] [Google Scholar]
- 10. Lafarga M., Berciano M. T., Pena E., Mayo I., Castaño J. G., Bohmann D., Rodrigues J. P., Tavanez J. P., Carmo-Fonseca M. (2002) Clastosome. A subtype of nuclear body enriched in 19 S and 20 S proteasomes, ubiquitin, and protein substrates of proteasome. Mol. Biol. Cell 13, 2771–2782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Brooks P., Fuertes G., Murray R. Z., Bose S., Knecht E., Rechsteiner M. C., Hendil K. B., Tanaka K., Dyson J., Rivett J. (2000) Subcellular localization of proteasomes and their regulatory complexes in mammalian cells. Biochem. J. 346, 155–161 [PMC free article] [PubMed] [Google Scholar]
- 12. Laine A., Topisirovic I., Zhai D., Reed J. C., Borden K. L., Ronai Z. (2006) Regulation of p53 localization and activity by Ubc13. Mol. Cell. Biol. 26, 8901–8913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Reichard P. (1988) Interactions between deoxyribonucleotide and DNA synthesis. Annu. Rev. Biochem. 57, 349–374 [DOI] [PubMed] [Google Scholar]
- 14. Spyrou G., Reichard P. (1988) Dynamics of the thymidine triphosphate pool during the cell cycle of synchronized 3T3 mouse fibroblasts. Mutat. Res. 200, 37–43 [DOI] [PubMed] [Google Scholar]
- 15. Oppenheim E. W., Adelman C., Liu X., Stover P. J. (2001) Heavy chain ferritin enhances serine hydroxymethyltransferase expression and de novo thymidine biosynthesis. J. Biol. Chem. 276, 19855–19861 [DOI] [PubMed] [Google Scholar]
- 16. MacFarlane A. J., Liu X., Perry C. A., Flodby P., Allen R. H., Stabler S. P., Stover P. J. (2008) Cytoplasmic serine hydroxymethyltransferase regulates the metabolic partitioning of methylenetetrahydrofolate but is not essential in mice. J. Biol. Chem. 283, 25846–25853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Maguire M., Nield P. C., Devling T., Jenkins R. E., Park B. K., Polański R., Vlatkovi N., Boyd M. T. (2008) MDM2 regulates dihydrofolate reductase activity through monoubiquitination. Cancer Res. 68, 3232–3242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Melo S. P., Yoshida A., Berger F. G. (2010) Functional dissection of the N-terminal degron of human thymidylate synthase. Biochem. J. 432, 217–226 [DOI] [PubMed] [Google Scholar]
- 19. Peña M. M., Melo S. P., Xing Y. Y., White K., Barbour K. W., Berger F. G. (2009) The intrinsically disordered N-terminal domain of thymidylate synthase targets the enzyme to the ubiquitin-independent proteasomal degradation pathway. J. Biol. Chem. 284, 31597–31607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Peña M. M., Xing Y. Y., Koli S., Berger F. G. (2006) Role of N-terminal residues in the ubiquitin-independent degradation of human thymidylate synthase. Biochem. J. 394, 355–363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Saeki Y., Kudo T., Sone T., Kikuchi Y., Yokosawa H., Toh-e A., Tanaka K. (2009) Lysine 63-linked polyubiquitin chain may serve as a targeting signal for the 26 S proteasome. EMBO J. 28, 359–371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Schimmel J., Larsen K. M., Matic I., van Hagen M., Cox J., Mann M., Andersen J. S., Vertegaal A. C. (2008) The ubiquitin-proteasome system is a key component of the SUMO-2/3 cycle. Mol. Cell. Proteomics 7, 2107–2122 [DOI] [PubMed] [Google Scholar]