Background: MTDH is overexpressed in solid tumors and is involved in metastasis and chemoresistance.
Results: Cytoplasmic MTDH associates with RNA and RNA-associated proteins, blocks Rad51 nuclear accumulation, and increases survival and drug resistance.
Conclusion: Cytoplasmic MTDH promotes cancer cell proliferation and resistance to treatment by acting as an RNA-binding protein.
Significance: Targeting MTDH may increase sensitivity to anti-cancer treatments.
Keywords: Cell Proliferation, Chemoresistance, Metastasis, RNA-binding Protein, Stress Granule, AEG-1/Lyric, Rad51, SND1, Metadherin
Abstract
Overexpression of metadherin (MTDH) has been documented in many solid tumors and is implicated in metastasis and chemoresistance. MTDH has been detected at the plasma membrane as well as in the cytoplasm and nucleus, and the function of MTDH in these locales remains under investigation. In the nucleus, MTDH acts as a transcription co-factor to induce expression of chemoresistance-associated genes. However, MTDH is predominantly cytoplasmic in prostate tumors, and this localization correlates with poor prognosis. Herein, we used endometrial cancer cells as a model system to define a new role for MTDH in the cytoplasm. First, MTDH was primarily localized to the cytoplasm in endometrial cancer cells, and the N-terminal region of MTDH was required to maintain cytoplasmic localization. Next, we identified novel binding partners for cytoplasmic MTDH, including RNA-binding proteins and components of the RNA-induced silencing complex. Nucleic acids were required for the association of MTDH with these cytoplasmic proteins. Furthermore, MTDH interacted with and regulated protein expression of multiple mRNAs, such as PDCD10 and KDM6A. Depletion of cytoplasmic MTDH was associated with increased stress granule formation, reduced survival in response to chemotherapy and the tyrosine kinase inhibitor BIBF1120, Rad51 nuclear accumulation, and cell cycle arrest at G2/M. Finally, in vivo tumor formation was abrogated with knockdown of cytoplasmic MTDH. Taken together, our data identify a novel function for cytoplasmic MTDH as an RNA-binding protein. Our findings implicate cytoplasmic MTDH in cell survival and broad drug resistance via association with RNA and RNA-binding proteins.
Introduction
The oncogene metadherin (MTDH,2 also known as AEG-1 and LYRIC) influences several oncogenic signaling pathways and transcription factors, such as Ras, Myc, PI3K/AKT, nuclear factor-κB (NF-κB), mitogen-activated protein kinase (MAPK), and Wnt pathways (1, 2). Overexpression of MTDH has been documented in all solid tumors examined to date, including breast, prostate, gastric, renal, colorectal, ovarian, and endometrial cancers (2–6). Moreover, MTDH overexpression has been implicated in metastasis (7) and resistance to the chemotherapeutic agents 5-fluorouracil, doxorubicin, paclitaxel, and cisplatin (8, 9). Although the underlying mechanism by which MTDH confers chemoresistance has not been established, one possibility is that MTDH functions as a transcription co-factor to promote expression of genes involved in resistance to chemotherapeutic agents (3, 10). Accordingly, MTDH has been shown to interact with NFκB, promyelocytic leukemia zinc finger (PLZF), and BRCA2 CDKN1A-interacting protein (BCCIP) (9, 11–13). However, a complete understanding of the biological functions and biochemical characteristics of MTDH remains elusive to date, particularly the significance of differential MTDH subcellular localization.
EXPERIMENTAL PROCEDURES
Immunostaining
Subcellular distribution of endogenous and exogenous MTDH and exogenous MTDH fragments in Hec50 cells was detected by microscopy and electron microscope as described in the supplemental Methods. Nuclear Rad51 accumulation was assessed by staining with anti-Rad51 (Ab-1, Calbiochem). For stress granule formation, cells were subjected to no treatment or 45 °C heat shock for 10 or 15 min.
MTDH Truncations
FLAG-tagged full-length or fragments of MTDH were generated by PCR, cloned in pCMV6 (OriGene), and expressed in Hec50 cells transiently (Rad51 experiments) or stably by selection with G418. A stable cell line expressing FLAG-tagged MTDH that is resistant to MTDH shRNA was also established (see also the supplemental Methods).
Colony Formation
Stable MTDH or control shRNA Hec50 cells were established as reported previously (14). Cells were treated with mitomycin C (MMC) for 24 h and then cultured in normal medium for 2 weeks. Resulting colonies were stained with crystal violet and counted.
Cell Viability
Stable MTDH or control shRNA Hec50 cells were treated with BIBF1120 (LC Laboratories), and then viability was assessed with the WST-1 assay.
MTDH Protein Co-immunoprecipitation Assays
Cell lysates from Hec50-FLAG-MTDH (with mutated shRNA binding site) transfected with MTDH shRNA or parental Hec50 cells were used to identify and validate MTDH-interacting proteins as described in the supplemental Methods. Where indicated, lysates were treated with Benzonase nuclease (50 units/ml, EMD Chemicals) prior to and during immunoprecipitation (IP).
Pulldown of MTDH-associated RNAs
Magna RIPTM (RNA-binding protein immunoprecipitation) kit and microarray were used to pull down MTDH-associated RNAs and to identify mRNAs that associate with MTDH in the absence or presence of 100 nm BEZ235 (LC Laboratories). The full list of RNAs associated with MTDH is provided in supplemental Tables 2–5. Microarray data have been deposited at GEO (www.ncbi.nlm.nih.gov/geo) under accession number GSE30588.
Cell Cycle Analysis
The cell cycle profile in control and MTDH-depleted Hec50 cells cultured in medium containing 10% FBS or 0.1% FBS for 2 days was analyzed by Click-iT EdU flow cytometry assay kits (Invitrogen). Total DNA was stained with FxCycleTM violet.
In Vivo Tumor Formation
For xenograft studies (15), 6–8-week-old athymic NCr-NU/NU female mice (NCI-Frederick, National Institutes of Health) were injected subcutaneously (s.c.) with 10 × 106 Hec50 cells in 100 μl of DMEM. Tumors were measured weekly, and the cross-sectional area (mm2) was calculated as length (mm) × width (mm) × π/4. The University of Iowa Institutional Animal Care and Use Committee approved experimental protocols.
RESULTS AND DISCUSSION
MTDH Is Primarily Cytoplasmic in Endometrial and Ovarian Cancer Cell Lines
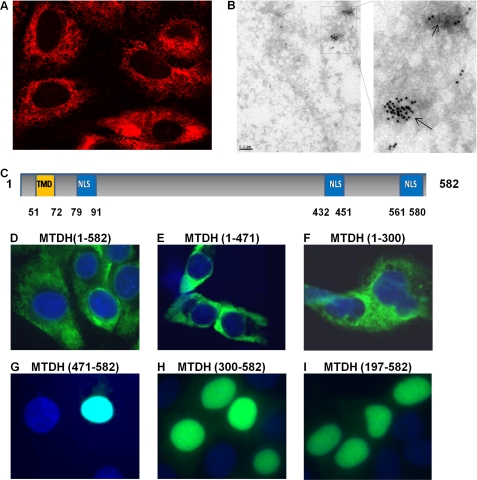
Expression of MTDH has been observed in the ER/perinuclear region, nucleus, and nucleolus and at the plasma membrane (3, 16–18). In these studies, distribution depended on the cell types examined and the experimental approach. We therefore first investigated the relative subcellular localization of MTDH in multiple endometrial and ovarian cancer cell lines. MTDH was detected predominantly in the cytoplasm, although a low level of punctate staining was also detected in the nucleus of all tested endometrial and ovarian cancer cell lines, similar to early reports that MTDH is an ER/nuclear envelope-associated protein (16) (Fig. 1, A and B, and supplemental Fig. 1). Other anti-MTDH antibodies that target the N and C terminus of MTDH were used to confirm subcellular distribution of MTDH (supplemental Fig. 2). Exogenously expressed FLAG-tagged MTDH was also localized in the cytoplasm as evidenced by staining with FLAG antibody (Fig. 1D).
FIGURE 1.
MTDH is primarily cytoplasmic in cancer cell lines. A and B, subcellular distribution of MTDH and MTDH fragments was detected in Hec50 cells by confocal microscopy (A) and electron microscope (B) using a rabbit antibody against MTDH (amino acids 315–461). C, schematic representation of MTDH highlighting the NLS peptides. Orange box, transmembrane domain (TMD). Blue boxes, three NLS regions. D–I, distribution of exogenous FLAG-tagged full-length MTDH (D), MTDH C-terminal truncation mutants (E and F), and MTDH N-terminal truncation mutants (G–I) was detected with FLAG antibody. Nuclei were stained with DAPI.
Several lines of evidence substantiate a critical function for MTDH in the cytoplasm of cancer cells. First, a previous study demonstrated that cytoplasmic MTDH correlates with poor prognosis or aggressive disease in prostate cancer patients (12). Second, MTDH has been reported to promote chemoresistance by facilitating translation of the multidrug resistance gene (MDR1), which is involved in drug efflux (19). Third, MTDH was shown to co-localize with the RNA-induced silencing complex (RISC) component staphylococcal nuclease domain-containing-1 (SND1) (20, 21). Finally, we recently reported that depletion of MTDH alters expression of several MTDH downstream genes, and MTDH is mainly cytoplasmic in the endometrial cancer cell lines used in this study (14).
As shown in Fig. 1C, MTDH contains three functional nuclear localization signals (NLSs) including residues 79–91, 432–451, and 561–580 (22). We investigated distribution of FLAG-tagged MTDH fragments with the different NLSs in Hec50 endometrial cancer cells. Full-length MTDH (MTDH 1–582) or MTDH truncated at the C terminus (MTDH 1–300 and 1–471) were all detected in the cytoplasm (Fig. 1, D–F). However, MTDH fragments with an N-terminal deletion (MTDH 197–582, 300–582, and 471–582) were nuclear (Fig. 1, G–I). Our data indicate that the N terminus of MTDH (residues 1–197) may play a dominant role in MTDH localization in the cytoplasm.
We hypothesized that deletion of the N-terminal region or regulation of this region by post-translation modification will release MTDH from the cytoplasmic membrane system and promote translocation to the nucleus. For example, cytoplasmic but not nuclear MTDH is monoubiquitinated (22), indicating that MTDH subcellular distribution may be regulated by post-translation modification. MTDH phosphorylation at multiple sites, including Thr-143 (23), Ser-298 (24), Ser-84, Ser-415, Ser-426, Ser-308 (25), and Ser-496 (26), has been reported in mammalian cells (supplemental Fig. 3A), but no studies have examined how MTDH phosphorylation affects its distribution. We made several site-specific mutations at MTDH phosphorylation sites, but all of these mutants retained cytoplasmic localization (supplemental Fig. 3, B–D).
MTDH Interacts with RNA-binding Proteins
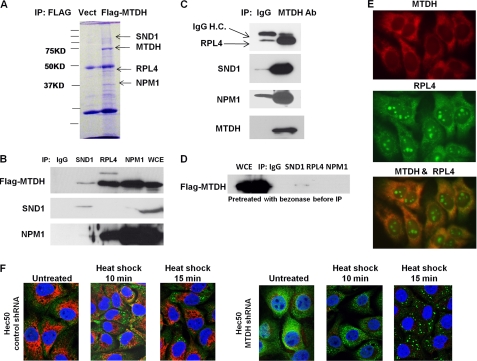
To explore the function of cytoplasmic MTDH, we employed the strategy of FLAG-tagged MTDH co-IP coupled with mass spectrometry to identify novel MTDH-interacting proteins in the cytoplasm. A total of 64 potential MTDH-interacting proteins were thus identified (supplemental Table 1). The most represented proteins were ribosomal protein L4 (RPL4), nucleophosmin 1 (NPM1), SND1, and MTDH (Fig. 2A). The interactions between SND1, NPM1, and RPL4 with MTDH were confirmed by co-IP (Fig. 2, B and C). The finding that SND1, which is part of RNA-induced silencing complex, interacts with MTDH is consistent with two recent studies that identified a metastatic role for the association of SND1 with MTDH (21) and a novel role for MTDH in RNAi-mediated gene silencing (20). Interestingly, pretreatment of protein lysates with Benzonase nuclease, which degrades nucleic acids, disrupted the interaction of MTDH with SND1 and RPL4 (Fig. 2D), demonstrating that nucleic acids are required for MTDH association with these proteins. Mutation of phosphorylation sites in MTDH did not prevent association with SND1 and RPL4 (supplemental Fig. 4). Co-localization of MTDH and RPL4, as determined by immunofluorescence, was observed only in the cytoplasm and not in the nucleus (Fig. 2E). Our data indicate that MTDH associates with multiple cytoplasmic proteins, and this interaction is dependent on the presence of nucleic acids.
FIGURE 2.
MTDH associates with multiple cytoplasmic proteins, and depletion of MTDH increases formation of stress granules induced by heat shock. A, Coomassie Brilliant Blue-stained gel of ell lysates from parental Hec50 or Hec50-FLAG-MTDH (with mutated shRNA binding site) + MTDH shRNA cells following IP with anti-FLAG and separation by SDS-PAGE. Indicated bands were analyzed by LC-MS/MS. Vect, vector. B, control IgG or MTDH antibody was used to co-IP RPL4, SND1, and NPM1 followed by blotting with the indicated antibodies. WCE, whole cell extract. C, the reciprocal of B was performed. Ab, antibody; IgG H.C., immunoglobulin G heavy chain. D, lysates from C were treated with Benzonase nuclease prior to and during IP and then blotted with anti-MTDH. E, co-localization of MTDH with RPL4 was detected by immunostaining. F, Hec50 cells expressing either control shRNA or MTDH shRNA were subjected to no treatment or 45 °C heat shock for 10 or 15 min. Red, MTDH; green, stress granule marker G3BP; blue, DAPI.
MTDH Knockdown Increases Formation of Stress Granules in Response to Stress
SND1 was recently found to physiologically interact with G3BP, a component of stress granules (SGs) (27). SGs are dynamic dense structures that rapidly form in the cytosol in response to stress stimuli, such as heat shock and arsenite, and serve as a reservoir for nontranslated mRNAs (28). Although cytosolic mRNAs such as actin aggregate into SGs in response to stress, there is a complete lack of localization of ER-associated transcripts such as MDR1 with SGs in arsenite-treated cells (29, 30). Importantly, mRNAs that escape from SGs resume translation faster than mRNAs aggregated in SGs, which may increase cellular resistance to stress (29, 30). Knockdown of endogenous SND1 retards aggregation of small SGs into large SGs, although it has no effect on SG formation (27). Based on the observations that MTDH interacts with both SND1 and mRNAs, we tested whether MTDH regulates SG formation or aggregation. As shown in Fig. 2F, formation of SGs induced by 45 °C heat shock was significantly increased in MTDH knockdown Hec50 cells as compared with control shRNA-transfected cells, suggesting that MTDH prevents SG formation. Heat shock had no effect on MTDH levels in control shRNA-transfected cells (supplemental Fig. 5). Our data reveal a previously unrecognized cytoplasmic function of MTDH in stress granule formation.
Cytoplasmic MTDH Associates with mRNAs
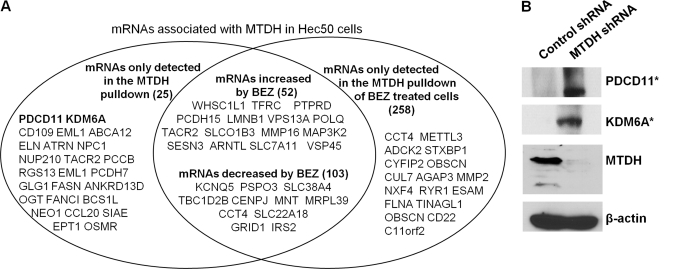
Because the interaction of MTDH with SND1, RPL4, or NPM1 depended on the presence of nucleic acid, we conducted a bioinformatics analysis to identify novel functional domains in MTDH. We found that MTDH has homology with the RNA-binding protein leucine tRNA synthase and contains several putative RNA binding domains (supplemental Figs. 6 and 7). Given the involvement of MTDH in RNA and protein complex formation, we therefore examined whether MTDH is a novel RNA-binding protein and whether it regulates translation of specific mRNAs. We conducted RNA-binding protein immunoprecipitation (31) followed by microarray analysis (RIP-chip) to identify MTDH-associated mRNA targets. As shown in Fig. 3A and supplemental Tables 2–5, various mRNAs interacted with MTDH, and some of these encode proteins that are linked to chemoresistance, such as ABCA 12 (32). Others have shown that MTDH increases MDR1 protein levels without affecting MDR1 transcription in a mechanism that involves increased association of MDR1 mRNA with polysomes (19). This study also reported that the association of MDR1 mRNA with polysomes is profoundly inhibited by PI3K inhibitor LY294002. Thus, we tested the effect of PI3K/mTOR inhibition using BEZ235 (dual PI3K/mTOR inhibitor) on the association of MTDH with target mRNAs. BEZ235 treatment reduced the association of some mRNAs with MTDH, increased the association of some mRNAs with MTDH, and also induced the association of novel mRNAs with MTDH (Fig. 3A and supplemental Tables 2–5). These data demonstrate that MTDH can associate with various mRNAs, and inhibition of PI3K/mTOR alters the pattern of mRNAs associated with MTDH.
FIGURE 3.
MTDH associates with mRNAs and alters protein expression. A, the association of MTDH with mRNAs was identified by RNA immunoprecipitation and subsequent microarray. Listed are representative mRNAs associated with MTDH and the effect of 100 nm BEZ235 (BEZ) on the association of MTDH with mRNAs (see also supplemental Tables 2–5). B, the effect of MTDH depletion on expression of proteins encoded by MTDH-associated mRNAs was detected by Western blotting with antibodies against PDCD10 and KDM6A. * denotes proteins encoded by mRNAs found to associate with MTDH.
To determine whether MTDH association with mRNAs alters their translation, we examined how MTDH knockdown affects protein levels of select MTDH-associated mRNAs. As shown in Fig. 3B, an increase in protein levels of PDCD11 and KDM6A was observed in MTDH-depleted cells, suggesting that cytoplasmic MTDH negatively regulates translation of these mRNAs.
Depletion of Cytoplasmic MTDH Provides Survival Advantage to Cancer Cells
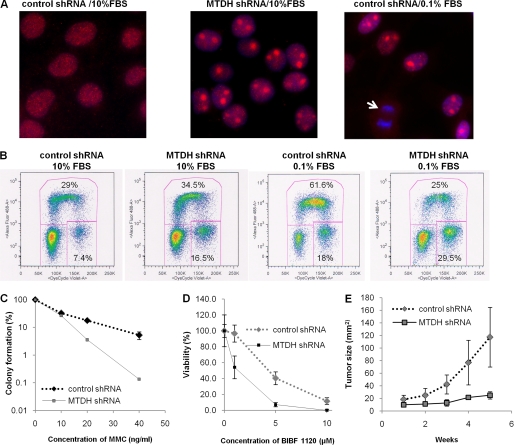
We next performed a series of experiments to understand the functional consequences of cytoplasmic MTDH expression. First, to test whether MTDH affects the response to DNA damage, we examined distribution of the homologous repair protein Rad51 by immunostaining in Hec50 cells with control or MTDH shRNA. A significant increase in nuclear accumulation of Rad51 was observed with MTDH depletion in full serum (Fig. 4A). It should be noted that the nuclear Rad51 is much larger than the typically reported size of Rad51 foci that form in response to DNA damage. However, a similar Rad51 accumulation was observed in serum-starved Hec50 cells without MTDH knockdown (Fig. 4A). This specific Rad51 staining was not detected in mitotic cells, which are indicated in Fig. 4A by the arrow. Restoring expression of full-length MTDH or MTDH (1–471) but not MTDH (1–300) or MTDH (471–582) inhibited Rad51 nuclear accumulation (supplemental Fig. 8).
FIGURE 4.
Depletion of cytoplasmic MTDH induces nuclear Rad51 accumulation, alters cell cycle profile, increases sensitivity to MMC and BIBF1120, and inhibits tumor proliferation in vivo. A, immunostaining of Rad51 in the nuclei was examined in control (left) or MTDH-depleted (center) Hec50 cells cultured in 10% FBS or control Hec50 cells cultured in 0.1% FBS (right). The arrow denotes mitotic cells. B, cell cycle profile was analyzed by flow cytometry after EdU pulse incorporation for 1 h in control or MTDH-depleted Hec50 cells cultured for 2 days in either 10% FBS or 0.1% FBS. C, colony formation of control or MTDH knockdown Hec50 cells was assessed after treatment with the indicated doses of MMC (ng/ml) for 1 day followed by culture in normal medium for 2 weeks. Results represent the average of three experiments. D, viability of control or MTDH-depleted cells in the presence of BIBF1120 for 3 days was detected by WST-1 assay. E, tumor volume was examined weekly in athymic mice injected with control or MTDH-depleted Hec50 cells (n = 4 animals per group). Error bars in C–E indicate S.D.
We next analyzed how cytoplasmic MTDH affects cell cycle progression. When cells were cultured in full serum, flow cytometry analysis showed that 16.5% of MTDH-depleted cells were at the G2/M phase as compared with 7.4% of control shRNA-transfected cells (Fig. 4B). However, after culture in 0.1% FBS, 25% of MTDH-depleted cells were in S phase and 29.5% of cells were at G2/M as compared with 61.6% of control cells at S and 18% at G2/M (Fig. 4B). These data suggest that MTDH is involved in cell cycle regulation.
Expression of MTDH confers resistance to multiple chemotherapeutic agents (8, 9), but the specific contribution of cytoplasmic MTDH to drug resistance has not been evaluated. Therefore, we explored how depletion of cytoplasmic MTDH alters sensitivity to the DNA-damaging agent MMC. Decreased colony formation was observed with MTDH knockdown as compared with control shRNA-transfected cells after treatment with MMC (Fig. 4C). In addition to DNA-damaging agents, MTDH depletion also increased sensitivity to targeted agents such as the triple angiokinase inhibitor BIBF1120 (Fig. 4D). Finally, we examined how MTDH affects tumor formation and progression by injecting MTDH shRNA or control shRNA Hec50 cells into athymic mice. MTDH knockdown significantly reduced malignancy as determined by tumor volume (Fig. 4E). Collectively, our data indicate that cytoplasmic MTDH functions in cell cycle regulation, proliferation, sensitivity to anti-cancer treatments, and tumor progression in vivo.
In conclusion, we define a novel function for cytoplasmic MTDH that may contribute to drug resistance and metastasis. Our study indicates that cytoplasmic MTDH associates with multiple cytoplasmic proteins and mRNAs and regulates mRNA translation. Furthermore, we reveal that MTDH interacts with mRNAs encoding proteins involved in numerous oncogenic signaling pathways and inhibits translation of these mRNAs. Our functional data reveal that cytoplasmic MTDH provides a strong survival advantage to cancer cells and identify MTDH as an emerging therapeutic target for cancers in which it is overexpressed.
Supplementary Material
Acknowledgments
We thank the University of Iowa Central Microscopy Facility, DNA Core Facility, and Proteomic Facility for technical assistance and Kristina W. Thiel for assistance in manuscript preparation.
This work was supported, in whole or in part, by National Institutes of Health Grant R01CA99908-7 (to K. K. L.). This work was also supported by the Department of Obstetrics and Gynecology Research Development Fund and Institutional Research Grant IRG-77-004-31 from the American Cancer Society (to X. M.), administered through the Holden Comprehensive Cancer Center at the University of Iowa.

This article contains supplemental Methods, Tables 1–5, and Figs. 1–8.
- MTDH
- metadherin
- IP
- immunoprecipitation
- SG
- stress granule
- NLS
- nuclear localization signal
- MMC
- mitomycin C
- ER
- endoplasmic reticulum
- mTOR
- mammalian target of rapamycin.
REFERENCES
- 1. Emdad L., Sarkar D., Su Z. Z., Randolph A., Boukerche H., Valerie K., Fisher P. B. (2006) Activation of the nuclear factor κB pathway by astrocyte elevated gene-1: implications for tumor progression and metastasis. Cancer Res. 66, 1509–1516 [DOI] [PubMed] [Google Scholar]
- 2. Yoo B. K., Emdad L., Lee S. G., Su Z. Z., Santhekadur P., Chen D., Gredler R., Fisher P. B., Sarkar D. (2011) Astrocyte elevated gene-1 (AEG-1): a multifunctional regulator of normal and abnormal physiology. Pharmacol. Ther. 130, 1–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hu G., Chong R. A., Yang Q., Wei Y., Blanco M. A., Li F., Reiss M., Au J. L., Haffty B. G., Kang Y. (2009) MTDH activation by 8q22 genomic gain promotes chemoresistance and metastasis of poor prognosis breast cancer. Cancer Cell 15, 9–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Song H., Li C., Lu R., Zhang Y., Geng J. (2010) Expression of astrocyte elevated gene-1: a novel marker of the pathogenesis, progression, and poor prognosis for endometrial cancer. Int. J. Gynecol. Cancer 20, 1188–1196 [DOI] [PubMed] [Google Scholar]
- 5. Li C., Liu J., Lu R., Yu G., Wang X., Zhao Y., Song H., Lin P., Sun X., Yu X., Zhang Y., Ning X., Geng J. (2011) AEG-1 overexpression: a novel indicator for peritoneal dissemination and lymph node metastasis in epithelial ovarian cancers. Int. J. Gynecol. Cancer 21, 602–608 [DOI] [PubMed] [Google Scholar]
- 6. Meng F., Luo C., Ma L., Hu Y., Lou G. (2011) Clinical significance of astrocyte elevated gene-1 expression in human epithelial ovarian carcinoma. Int. J. Gynecol. Pathol. 30, 145–150 [DOI] [PubMed] [Google Scholar]
- 7. Hu G., Wei Y., Kang Y. (2009) The multifaceted role of MTDH/AEG-1 in cancer progression. Clin. Cancer Res. 15, 5615–5620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Yoo B. K., Gredler R., Vozhilla N., Su Z. Z., Chen D., Forcier T., Shah K., Saxena U., Hansen U., Fisher P. B., Sarkar D. (2009) Identification of genes conferring resistance to 5-fluorouracil. Proc. Natl. Acad. Sci. U.S.A. 106, 12938–12943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Liu H., Song X., Liu C., Xie L., Wei L., Sun R. (2009) Knockdown of astrocyte elevated gene-1 inhibits proliferation and enhancing chemo-sensitivity to cisplatin or doxorubicin in neuroblastoma cells. J. Exp. Clin. Cancer Res. 28, 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Xiang H., Shi X. L., Li Q. X., Zhang W., Wang J. (2011) [Myxoinflammatory fibroblastic sarcoma: a clinicopathologic study of six cases with review of literature]. Zhonghua Bing Li Xue Za Zhi 40, 94–98 [PubMed] [Google Scholar]
- 11. Ash S. C., Yang D. Q., Britt D. E. (2008) LYRIC/AEG-1 overexpression modulates BCCIPα protein levels in prostate tumor cells. Biochem. Biophys. Res. Commun. 371, 333–338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Thirkettle H. J., Mills I. G., Whitaker H. C., Neal D. E. (2009) Nuclear LYRIC/AEG-1 interacts with PLZF and relieves PLZF-mediated repression. Oncogene 28, 3663–3670 [DOI] [PubMed] [Google Scholar]
- 13. Sarkar D., Park E. S., Emdad L., Lee S. G., Su Z. Z., Fisher P. B. (2008) Molecular basis of nuclear factor-κB activation by astrocyte elevated gene-1. Cancer Res. 68, 1478–1484 [DOI] [PubMed] [Google Scholar]
- 14. Meng X., Brachova P., Yang S., Xiong Z., Zhang Y., Thiel K. W., Leslie K. K. (2011) Knockdown of MTDH sensitizes endometrial cancer cells to cell death induction by death receptor ligand TRAIL and HDAC inhibitor LBH589 co-treatment. PloS One 6, e20920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Dai D., Holmes A. M., Nguyen T., Davies S., Theele D. P., Verschraegen C., Leslie K. K. (2005) A potential synergistic anticancer effect of paclitaxel and amifostine on endometrial cancer. Cancer Res. 65, 9517–9524 [DOI] [PubMed] [Google Scholar]
- 16. Sutherland H. G., Lam Y. W., Briers S., Lamond A. I., Bickmore W. A. (2004) 3D3/lyric: a novel transmembrane protein of the endoplasmic reticulum and nuclear envelope, which is also present in the nucleolus. Exp. Cell Res. 294, 94–105 [DOI] [PubMed] [Google Scholar]
- 17. Kang D. C., Su Z. Z., Sarkar D., Emdad L., Volsky D. J., Fisher P. B. (2005) Cloning and characterization of HIV-1-inducible astrocyte elevated gene-1, AEG-1. Gene 353, 8–15 [DOI] [PubMed] [Google Scholar]
- 18. Brown D. M., Ruoslahti E. (2004) Metadherin, a cell surface protein in breast tumors that mediates lung metastasis. Cancer Cell 5, 365–374 [DOI] [PubMed] [Google Scholar]
- 19. Yoo B. K., Chen D., Su Z. Z., Gredler R., Yoo J., Shah K., Fisher P. B., Sarkar D. (2010) Molecular mechanism of chemoresistance by astrocyte elevated gene-1. Cancer Res. 70, 3249–3258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Yoo B. K., Santhekadur P. K., Gredler R., Chen D., Emdad L., Bhutia S., Pannell L., Fisher P. B., Sarkar D. (2011) Increased RNA-induced silencing complex (RISC) activity contributes to hepatocellular carcinoma. Hepatology 53, 1538–1548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Blanco M. A., Aleckovi M., Hua Y., Li T., Wei Y., Xu Z., Cristea I. M., Kang Y. (2011) Identification of staphylococcal nuclease domain-containing 1 (SND1) as a metadherin-interacting protein with metastasis-promoting functions. J. Biol. Chem. 286, 19982–19992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Thirkettle H. J., Girling J., Warren A. Y., Mills I. G., Sahadevan K., Leung H., Hamdy F., Whitaker H. C., Neal D. E. (2009) LYRIC/AEG-1 is targeted to different subcellular compartments by ubiquitinylation and intrinsic nuclear localization signals. Clin Cancer Res. 15, 3003–3013 [DOI] [PubMed] [Google Scholar]
- 23. Dephoure N., Zhou C., Villén J., Beausoleil S. A., Bakalarski C. E., Elledge S. J., Gygi S. P. (2008) A quantitative atlas of mitotic phosphorylation. Proc. Natl. Acad. Sci. U.S.A. 105, 10762–10767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Villén J., Beausoleil S. A., Gerber S. A., Gygi S. P. (2007) Large-scale phosphorylation analysis of mouse liver. Proc. Natl. Acad. Sci. U.S.A. 104, 1488–1493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Olsen J. V., Blagoev B., Gnad F., Macek B., Kumar C., Mortensen P., Mann M. (2006) Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell 127, 635–648 [DOI] [PubMed] [Google Scholar]
- 26. Olsen J. V., Vermeulen M., Santamaria A., Kumar C., Miller M. L., Jensen L. J., Gnad F., Cox J., Jensen T. S., Nigg E. A., Brunak S., Mann M. (2010) Quantitative phosphoproteomics reveals widespread full phosphorylation site occupancy during mitosis. Sci. Signal. 3, ra3. [DOI] [PubMed] [Google Scholar]
- 27. Gao X., Ge L., Shao J., Su C., Zhao H., Saarikettu J., Yao X., Yao Z., Silvennoinen O., Yang J. (2010) Tudor-SN interacts with and co-localizes with G3BP in stress granules under stress conditions. FEBS Lett. 584, 3525–3532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Buchan J. R., Parker R. (2009) Eukaryotic stress granules: the ins and outs of translation. Mol. Cell 36, 932–941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Yagüe E., Raguz S. (2010) Escape from stress granule sequestration: another way to drug resistance? Biochem. Soc. Trans. 38, 1537–1542 [DOI] [PubMed] [Google Scholar]
- 30. Unsworth H., Raguz S., Edwards H. J., Higgins C. F., Yagüe E. (2010) mRNA escape from stress granule sequestration is dictated by localization to the endoplasmic reticulum. FASEB J. 24, 3370–3380 [DOI] [PubMed] [Google Scholar]
- 31. Zong X., Tripathi V., Prasanth K. V. (2011) RNA splicing control: yet another gene regulatory role for long nuclear noncoding RNAs. RNA Biol. 8, 968–977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Park S., Shimizu C., Shimoyama T., Takeda M., Ando M., Kohno T., Katsumata N., Kang Y. K., Nishio K., Fujiwara Y. (2006) Gene expression profiling of ATP binding cassette (ABC) transporters as a predictor of the pathologic response to neoadjuvant chemotherapy in breast cancer patients. Breast Cancer Res. Treat 99, 9–17 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.