Background: Plant nitrate reductase activity is regulated by phosphorylation and subsequent 14-3-3 protein binding.
Results: Steady-state and pre-steady kinetics revealed the electron transfer rates between all three redox-active cofactors and identified the heme-to-molybdenum transfer as key regulatory point.
Conclusion: 14-3-3 proteins inhibit domain movement in nitrate reductase.
Significance: This is the first description of 14-3-3-regulated electron transfer in a multidomain metallo-enzyme.
Keywords: Electron Transfer, Metabolic Regulation, Nitrogen Metabolism, Plant Biochemistry, Protein Phosphorylation, 14-3-3 Proteins, Domain Movement, Nitrate Reductase, Post-translational Regulation, Steady-state and Pre-steady-state Kinetics
Abstract
14-3-3 proteins regulate key processes in eukaryotic cells including nitrogen assimilation in plants by tuning the activity of nitrate reductase (NR), the first and rate-limiting enzyme in this pathway. The homodimeric NR harbors three cofactors, each of which is bound to separate domains, thus forming an electron transfer chain. 14-3-3 proteins inhibit NR by binding to a conserved phosphorylation site localized in the linker between the heme and molybdenum cofactor-containing domains. Here, we have investigated the molecular mechanism of 14-3-3-mediated NR inhibition using a fragment of the enzyme lacking the third domain, allowing us to analyze electron transfer from the heme cofactor via the molybdenum center to nitrate. The kinetic behavior of the inhibited Mo-heme fragment indicates that the principal point at which 14-3-3 acts is the electron transfer from the heme to the molybdenum cofactor. We demonstrate that this is not due to a perturbation of the reduction potentials of either the heme or the molybdenum center and conclude that 14-3-3 most likely inhibits nitrate reductase by inducing a conformational change that significantly increases the distance between the two redox-active sites.
Introduction
Protein phosphorylation is a ubiquitous mechanism of post-translational control of protein function in both prokaryotes and eukaryotes. Phosphorylation frequently triggers binding of specific effector proteins that in turn alter the function of the target protein. In plants, two groups of proteins are known to function in this way: those with forkhead-associated domains and 14-3-3 proteins (1). Although forkhead-associated domains are small domains of proteins with otherwise diverse functions, 14-3-3 proteins, named based on their chromatographic and electrophoretic properties (2), function solely as regulatory elements. 14-3-3 proteins are abundant in eukaryotes and recognize conserved binding motifs that contain a phosphorylated serine or threonine residue (3), although recognition of non-canonical and non-phosphorylated motifs has also been reported (4–7). Crystal structures of 14-3-3 (8–10) reveal an elongated dimeric structure with a basket-like shape and highly conserved α-helices. Each monomer forms a large groove suitable for target binding. Thus a 14-3-3 dimer can (i) function as a scaffolding protein by bringing together different proteins, (ii) modify the structure of a given protein by binding to distinct areas of the same protein, or (iii) bind and modify simultaneously two sites of a protein complex (11–14).
Arabidopsis encodes 13 distinct 14-3-3 proteins (14), the most in one organism, reflecting the functional importance as well as specificity of this protein family in Arabidopsis. Based on exon numbers, 14-3-3 proteins are divided into an ϵ group (6–7 exons, five members) and a plant-specific non-ϵ group (four exons, seven members in three subgroups) (15). Thus Arabidopsis 14-3-3 proteins form a functionally heterogeneous group (with four subgroups) on the basis of both their diverse gene and expression pattern as well as cellular localization (16, 17).
In plants, nitrate reductase (NR)4 was one of the first proteins recognized to be regulated by 14-3-3 proteins (18–20). NR catalyzes the first and rate-limiting step in primary nitrogen metabolism, the cytosolic reduction of nitrate to nitrite (21), using reducing equivalents provided by NADH. NR has a modular structure with an N-terminal molybdenum-containing domain, a b-type cytochrome domain, and a C-terminal FAD domain (see Fig. 1) (22), each connected by a hinge region. Product nitrite is transported into the plastids and further reduced in a 6-electron process to ammonia, which is subsequently incorporated into glutamine by the eponymous synthetase.
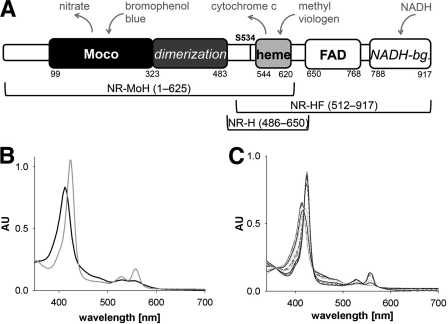
FIGURE 1.
Domain structure and spectral properties of NR and NR fragments. A, domain structure of NR. Numbers indicate first/last residues of a domain, and cofactors are written in bold. The dimerization domain and NADH-binding (NADH-bg.) lobe are italicized, and the regulatory crucial serine residue Ser-534 is labeled (as S534). Above, electron donors and acceptors are shown in gray. Below, NR fragments and their respective lengths are depicted (NR-MoH, Mo-heme fragment; NR-HF, heme-FAD fragment; NR-H, heme fragment). B and C, UV-visible spectra of Mo-heme (6.9 μm) following the reduction with dithionite (gray trace) and reoxidation with nitrate (black trace) (B) and oxidized heme-FAD fragment (5.4 μm) (black trace) (C) in the course of titration with a stock solution of 200 μm NADH (gray traces). AU, absorbance units.
NR is tightly regulated to avoid deleterious accumulation of nitrite in the cell (23, 24), being down-regulated at night when photosynthetically generated reducing equivalents are no longer available. This diurnal regulation is post-translationally controlled and involves phosphorylation of a conserved serine residue (Ser-534 in Arabidopsis) in the Hinge 1 region intervening between the molybdenum- and heme-binding domains and subsequent binding of an inhibitory 14-3-3 protein (25–28). The precise molecular mechanism by which this inhibition occurs, however, is not known.
There are four steps in the overall catalytic cycle of NR: 1) the initial 2-electron reduction of the FAD cofactor by NADH; 2) sequential single-electron electron transfer from FAD to heme; 3) two single-electron transfers from the heme onto the molybdenum cofactor, eventually reducing the metal from the MoVI to the MoIV state; and 4) the reduction of nitrate to nitrite. Using a fully defined in vitro expression system for NR from Arabidopsis thaliana, which enabled us to obtain the enzyme in adequate amounts and purity to examine its kinetic properties, we have recently shown that steps 3 or 4, individually or in combination, are the points of 14-3-3-mediated inhibition (18). Of the several 14-3-3 proteins examined, inhibition was found to be most effective using 14-3-3ω (18).
In the present work, we have used two different NR fragments to first characterize the individual steps within the catalytic cycle and to identify the specific process that is sensitive to 14-3-3ω binding. One fragment consists of the N-terminal residues 1–625, including the molybdenum- and heme-containing domains as well as Hinge 1 (Mo-heme fragment). The second fragment represents the C-terminal portion of NR comprising residues 512–917, starting from Hinge 1 all the way to the C terminus harboring the heme- and FAD-containing domains (heme-FAD fragment, see Fig. 1). Using steady-state, pre-steady-state, and rapid quench kinetics, we found that electron transfer from the heme to the molybdenum is the specific step that is slowed upon phosphorylation and binding of 14-3-3ω. Inhibition appears to trap the heme and molybdenum domains in a conformation unfavorable for electron transfer, most likely due to an increase in the distance between the two redox-active centers. To our knowledge, this is the first example of 14-3-3-mediated regulation of enzyme activity by modulating the rate of intramolecular electron transfer.
EXPERIMENTAL PROCEDURES
Cloning, Expression, and Purification of NR Fragments, CPK-17, and 14-3-3ω
Using plasmid pKK001 (kindly provided by N. Crawford, University of California San Diego) comprising the cDNA sequence of A. thaliana Nia2, a fragment encoding residues 1–625 was PCR-cloned into KpnI and HindIII restriction sites of pQE80L (Qiagen) and transformed into Escherichia coli TP1000. To express the Mo-heme fragment, pQE80-At-Mo-heme plasmid was transformed into E. coli TP1000 strain. Cells were grown in LB medium containing ampicillin (100 μg/ml), kanamycin (25 μg/ml), and sodium molybdate (500 μm) at room temperature until the A600 reached ∼0.8. Isopropyl-β-d-thiogalactoside was added to a final concentration of 100 μm, and the cells were further incubated for 48 h before harvesting. All steps during purification were performed on ice using buffers enriched with CompleteTM EDTA-free protease inhibitor mixture (Roche Applied Science) to prevent degradation. The pellet was resuspended in lysis buffer (100 mm potassium phosphate, pH 7.5, 300 mm sodium chloride, 15 mm imidazole) and mechanically disrupted using a cell disruptor (Constant Systems Ltd.) and sonication. After centrifugation, the supernatant was incubated in-batch with nickel-nitrilotriacetic acid (Ni-NTA) resin (Qiagen) for 30 min, washed, and eluted with elution buffer (100 mm potassium phosphate, pH 7.5, 300 mm sodium chloride, 500 mm imidazole). Afterward size exclusion chromatography was performed using assay buffer (50 mm Bis-Tris, pH 7.0, 50 mm potassium chloride, 5 mm magnesium acetate, 1 mm calcium chloride). Protein concentrations were determined spectrophotometrically based on the heme content using ϵ413 nm = 120 mm−1 cm−1. The molybdenum cofactor content was determined using HPLC Form A analysis (29). Protein concentration was corrected to the content of the cofactor, which was less abundant. SDS-PAGE was performed to confirm protein identity and purity, respectively.
The H600A variant of the NR-Mo-heme fragment was produced by PCR using the QuikChange® site-directed mutagenesis kit (Stratagene), transformed into E. coli DH5α strain, and subsequently sequenced to confirm the successful mutagenesis. Expression and purification were performed as for the NR-Mo-heme wild-type fragment.
Using the same template as above, the coding sequence of the heme-FAD fragment (residues 512–917) was PCR-cloned into KpnI and HindIII sites of pQE80L and expressed in E. coli DL41 cells at 18 °C overnight (100 μm isopropyl-β-d-thiogalactoside). Initial purification of the heme-FAD fragment was performed as for the Mo-heme fragment using Ni-NTA resin. Following buffer-exchange to 50 mm Tris acetate, pH 7.5, using PD10 columns (GE Healthcare), anion exchange chromatography (Source Q15, GE Healthcare) was performed. Protein containing fractions were pooled and further enriched via size exclusion chromatography (Superdex 200 16/60, GE Healthcare) using 20 mm Tris acetate, 200 mm NaCl, pH 7.0.
The coding sequence of the heme fragment (residues 486–650) was PCR-cloned into BamHI and HindIII sites of pET21a+ (Novagen) and expressed in E. coli strain ER2566 at 37 °C for 16 h (250 μm isopropyl-β-d-thiogalactoside). Purification of the heme fragment was performed as for the Mo-heme fragment except for the use of Tris/HCl instead of phosphate buffer throughout Ni-NTA purification. Protein-containing fractions were concentrated and applied onto a Superdex 200 16/60 column (GE Healthcare) equilibrated in 10 mm Tris/HCl, pH 8.0, 250 mm NaCl. Wild-type Arabidopsis NR was recombinantly expressed in Pichia pastoris KM71, and the calcium-dependent protein kinase CPK-17 as well as the inhibitor protein 14-3-3ω were expressed in E. coli and purified as described earlier (18).
Steady-state Kinetics and Viscosity Dependence
To examine the viscosity effect on the intramolecular electron transfer, kinetic parameters were determined using 35% glycerol and nitrate concentrations from 0 to 1 mm. NADH-dependent oxidation of cytochrome c (NADH:cytochrome c assay) by the heme-FAD fragment was performed and analyzed as described earlier (18) (supplemental Table S1).
Phosphorylation of the Mo-Heme Fragment
NR, Mo-heme, as well as the heme fragment (2–8 μm) were phosphorylated with 10-fold lower concentrations of CPK-17 kinase. The phosphorylation reaction was performed in assay buffer (50 mm Bis-Tris, pH 7.0, 50 mm potassium chloride, 5 mm magnesium acetate, 1 mm calcium chloride) supplemented with 400 μm ATP for 30–60 min at room temperature. Successful phosphorylation of all fragments was demonstrated by immunoblots using either an antibody specifically raised against the NR hinge phospho-peptide (residues 529–541 encompassing phospho-Ser-534) or a His tag antibody to monitor equal loading of the respective proteins (see supplemental Fig. S1).
Steady-state Kinetics and Inhibition Assays
Nitrate-dependent oxidation of reduced methyl viologen by the Mo-heme fragment or holo-NR was analyzed in assay buffer at room temperature under strictly anaerobic conditions in a glove box (Toepffer Lab Systems) using a Sunrise plate reader (Tecan). For each reaction, assay buffer and 40 nm enzyme were loaded into the reaction well and mixed with 0–5 mm KNO3. For inhibition assays, assay buffer, 40 nm enzyme, and 1 μm 14-3-3ω were mixed and incubated for 5 min at room temperature prior to nitrate addition. In a separate well, oxidized methyl viologen was reduced to an A595 of 1 by adding sodium dithionite. The reaction was started by the addition of reduced methyl viologen (0.4 mm). The decrease in A595 indicating oxidation of methyl viologen was recorded, and the reaction velocities were calculated based on the initial slopes. Kinetic data were analyzed using SigmaPlot (Systat) to determine the apparent Km and kcat values.
UV-visible Spectroscopy
Reductive titrations of the Mo-heme and heme-FAD fragments were performed using an HP-8452 diode array spectrophotometer. All non-protein solutions (buffer/substrates) were made anaerobic by bubbling with argon for at least 30 min at room temperature prior to use. Enzyme solutions were placed in glass tonometers on ice and gas-exchanged by evacuation and flushing with argon for at least 1 h. Reduction of 6–13 μm anaerobic oxidized heme-FAD fragment was performed by titration with a 100 μm stock solution of NADH, recording the absorption spectrum. For reduction of the Mo-heme fragment, a stock solution of ∼200 μm sodium dithionite was used.
Pre-steady-state Kinetics
Rapid-reaction kinetic experiments were performed with an Applied Photophysics Inc. SX-18MV stopped-flow instrument equipped with either a diode array detector or a photomultiplier at 10 °C in assay buffer. For the reaction of the heme-FAD fragment with NADH, ∼6 μm solutions made anaerobic as described above were mounted on the stopped-flow apparatus and mixed with varying concentrations of NADH, monitoring the reaction at 460 nm (flavin) or 425 nm (heme). For the Mo-heme fragment (untreated or phosphorylated and/or mixed with 14-3-3ω), 3 μm enzyme was made anaerobic as described above and titrated to full reduction with sodium dithionite. It was then mounted and mixed with varying concentrations of anaerobic nitrate. Heme reoxidation was followed by the absorbance decrease at 425 nm. The resulting time courses were fitted to sums of exponentials using an iterative non-linear least-squares Levenberg-Marquardt algorithm to obtain the concentration-dependent observed rate constants. Hyperbolic plots of observed rate constant versus substrate concentration were fitted using the equation kobs = klim [S]/(Kd + [S]) to obtain the limiting rate constant, klim, and the dissociation constant, Kd.
EPR Spectroscopy
Samples were prepared by reacting 7 μm of the phosphorylated Mo-heme fragment in the presence or absence of 14-3-3ω, made anaerobic as described above and pre-reduced with excess sodium dithionite (>3 mm), with 5 mm anaerobic KNO3. Small aliquots were removed and simultaneously quenched with 0.5 mm zinc acetate as the remaining sample was frozen for EPR analysis. After initial analysis, sample tubes were made anaerobic, and samples were thawed and allowed to react for 10–30 s. The sample tubes were promptly refrozen with small aliquots concurrently removed and quenched as before. This procedure was repeated a number of times. The quenched samples were clarified and analyzed via a modified nitrite assay (30). EPR spectra were recorded using a Bruker Instruments ER 300 spectrometer equipped with an ER 035M gaussmeter and a HP 5352B microwave frequency counter. Temperature was controlled at 150 K using a Bruker ER 4111VT liquid N2 cryostat. Signal intensity was quantified by double integration of the baseline-corrected EPR spectra using EWWIN version 6.1 and subsequently normalized.
Rapid Quench Experiments
Rapid quench-flow experiments were performed with an Applied Photophysics Inc. SX-20MV stopped-flow instrument equipped with a photomultiplier at 10 °C in assay buffer, but otherwise according to the procedure of Skipper et al. (31). Phosphorylated Mo-heme fragment (2.5 μm) in the presence or absence of 14-3-3ω was made anaerobic as described above, pre-reduced with sodium dithionite, and mixed with an anaerobic solution of 10 mm KNO3 containing 1.0 m zinc acetate as quenching agent. Quenching of protein activity was monitored at 557 nm, where the absorbance change reflected extrusion of the heme from the polypeptide; the quenched solution was removed from the stop syringe and subjected to nitrite quantification (30). Clarified samples (100 μl) were mixed with ice-cold dapsone (4,4′-diamino-diphenylsulfone; 14 mm in 2 n HCl), and then 50 μl of N-(1-naphthyl)-ethylenediamine (4 mm in H2O) was added, and the mixture was incubated at room temperature for 5–10 min. The resulting azo compound was measured at 550 nm and quantified. Additional Experimental Procedures can be found in Supplemental Information.
RESULTS
Characterization of the Mo-Heme and Heme-FAD Fragments of A. thaliana NR
Given our previous finding that 14-3-3-mediated inhibition of NR did not affect electron transfer from NADH to heme (18), we developed and investigated a simplified system that allowed a more detailed kinetic characterization of the underlying mechanism following 14-3-3 binding. We expressed and purified a Mo-heme fragment of A. thaliana NR that lacked the C-terminal FAD/NADH-binding domain (Figs. 1A and 2, A and E). This fragment was found to exhibit the expected spectral characteristics of a heme b5-containing protein (Fig. 1B) and showed activity in a methyl viologen:nitrate assay comparable with that seen with holo-NR (Fig. 3, A and B, supplemental Table S1). It is important to note that methyl viologen preferentially reduces the heme site, as confirmed here by creating a heme-deficient variant (see supplemental Fig. S2) of the Mo-heme fragment that was devoid of activity in the methyl viologen:nitrate assay (see below). The Mo-heme fragment also exhibited the same sensitivity to increased solution viscosity as holo-NR, confirming a similar motion of the isolated heme domain in the truncated enzyme as in the holo-enzyme (18) (Fig. 3, A and B).
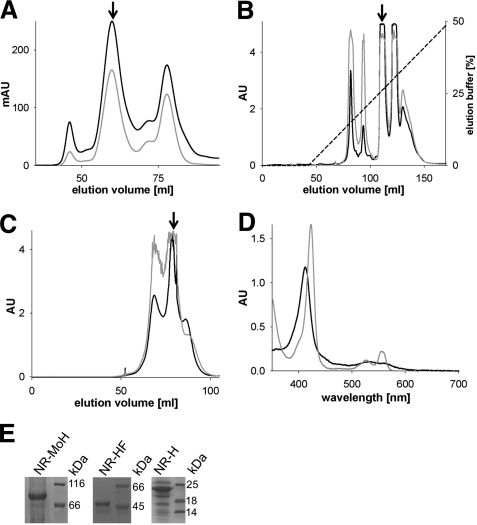
FIGURE 2.
Purification and characterization of A. thaliana nitrate reductase fragments. A, size exclusion chromatography of the Mo-heme fragment, with the fragment peak labeled with an arrow; absorption was recorded at 280 (black trace) and 413 nm (gray trace). mAU, milliabsorbance units. B and C, anion exchange chromatography (B) and size exclusion chromatography (C) of the heme-FAD fragment. Arrows mark the heme-FAD-containing peaks; the recorded absorptions are depicted as above. D, UV-visible spectroscopy of the heme fragment (9.3 μm) in its oxidized state (black trace) and after reduction with sodium dithionite (gray trace). E, SDS-PAGE analysis of purified and concentrated Mo-heme (NR-MoH), heme-FAD (NR-HF), and heme fragments (NR-H). The proteins of the molecular mass standard are labeled in kDa. All proteins were initially purified by affinity chromatography.
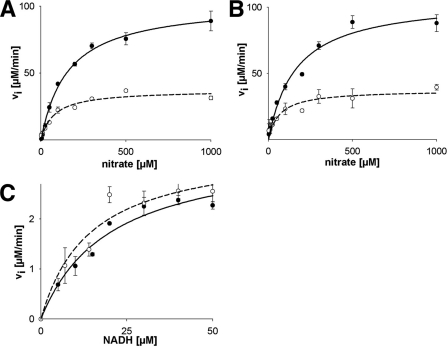
FIGURE 3.
Steady-state kinetics of holo-NR, Mo-heme fragment, and heme-FAD fragment under normal or viscous conditions. A, methyl viologen:nitrate assay of holo-NR using 0.4 mm methyl viologen in assay buffer (solid trace) or in assay buffer including 35% glycerol (dashed trace). B, methyl viologen:nitrate assay (details as above) of Mo-heme fragment in assay buffer (solid trace) or in assay buffer with 35% glycerol (dashed trace). C, NADH:cytochrome c activity assay of the heme-FAD fragment using 0.1 mm cytochrome c in assay buffer (solid trace) or in assay buffer including 35% glycerol (dashed trace). Error bars in all panels indicate S.D.
We also prepared an enzyme fragment comprising the C-terminal portion of NR that included the Hinge 1 region and both the heme-binding and the FAD-binding domains (Figs. 1A and 2, B, C, and E). This heme-FAD fragment exhibited an absorption spectrum in the UV-visible region expected for a heme-flavo-protein (Fig. 1C) and showed similar activity in an NADH:cytochrome c assay as holo-NR. The behavior of this fragment was neither viscosity-dependent (Fig. 3C and supplemental Table S1) nor affected by phosphorylation of Ser-534 and complexation with 14-3-3ω, indicating that neither the initial reduction of the FAD site by NADH nor the subsequent transfer of reducing equivalents from the FAD to the heme site were affected by phosphorylation and binding of 14-3-3ω, consistent with previous findings using the holo-enzyme (18). Nevertheless, to more fully characterize the heme-FAD fragment, we examined its reaction with saturating NADH (200 μm) by stopped-flow. The rate constant for the initial reduction of FAD, as reflected in the reaction observed at 460 nm, was determined to be 638 s−1 at 10 °C, and the subsequent reduction of the heme, as followed at 557 nm, was 583 s−1 (Fig. 4, A and B, and Table 1). These rate constants were found to be unchanged upon phosphorylation and binding of 14-3-3ω.
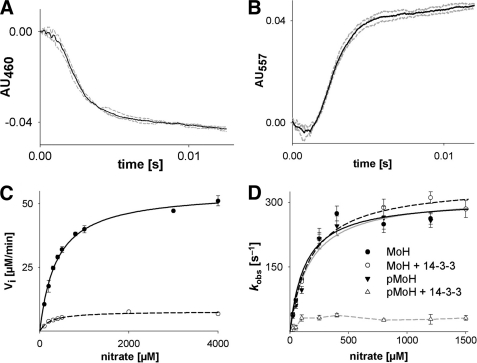
FIGURE 4.
Steady-state and pre-steady-state kinetics of NR fragments. A and B, kinetics of the reduction of the FAD (A) and heme cofactor (B) in 8 μm heme-FAD fragment using 50 μm NADH recorded at 460 (AU460) and 557 nm (AU557), respectively. The average (black trace) of three individual measurements (gray traces) is highlighted. AU, absorbance units. C, steady-state kinetics of the Mo-heme fragment recorded by the methyl viologen:nitrate assay. Mo-heme (black line) or phosphorylated Mo-heme with excess 14-3-3ω was recorded (dashed line). D, pre-steady-state nitrate reoxidation kinetics of the Mo-heme fragment (MoH) (3 μm). Where indicated, the fragment was phosphorylated (pMoH) and incubated with 10 μm 14-3-3ω (+ 14-3-3). Relaxation rates were plotted against the nitrate concentration and fitted using SigmaPlot (see Table 1 for kobs and Kd). Error bars in panels C and D indicate S.D.
TABLE 1.
Rapid reaction kinetic parameters for NR-fragments
| Sample | kobs | Kd |
|---|---|---|
| s−1 | μm | |
| Mo-heme fragmenta | 310 ± 25 | 131 ± 37 |
| Mo-heme fragment + 14–3-3ωa | 345 ± 14 | 176 ± 27 |
| Phosphorylated Mo-heme fragmenta | 319 ± 25 | 168 ± 43 |
| Phosphorylated Mo-heme fragment + 14–3-3ωa | ∼35 | NDb |
| Heme-FAD fragmentc | 638 ± 22 | |
| Heme-FAD fragmentd | 583 ± 32 |
a Heme re-oxidation after pre-reduction with sodium dithionite.
b ND = not detectable.
c FAD reduction after reduction with NADH.
d Heme reduction after reduction with NADH.
Kinetic Studies of 14-3-3-inhibited Mo-Heme Fragment
In contrast to the activity of the heme-FAD fragment, that of the Mo-heme fragment was found to be inhibited upon phosphorylation of Ser-534 and binding of 14-3-3ω and to a degree comparable with that seen previously with holo-enzyme (18); kcat was reduced from 23 s−1 for the uninhibited fragment (after adjustment for the level of cofactor saturation) to 3.3 s−1 upon inhibition (Fig. 4C). These results indicate that 14-3-3ω inhibits NR at one or both of the two reaction steps involved in the methyl viologen:nitrate assay, electron transfer from heme to molybdenum and nitrate reduction at the molybdenum center.
To more specifically examine the effect of phosphorylation and 14-3-3ω binding on the Mo-heme fragment, pre-steady-state kinetics were performed under standard and inhibiting conditions. The protein was first reduced by titration with sodium dithionite (following the spectral change associated with heme reduction (Fig. 1B)) under strictly anaerobic conditions and then reacted with varying concentrations of nitrate in a stopped-flow apparatus following the reoxidation of the heme at 425 nm. In this experiment, the fully reduced molybdenum center first reacts with nitrate in a reaction that results in no detectable absorption change followed by electron transfer from the heme to the now-oxidized molybdenum center.
The observed rate constant for heme reoxidation in this experiment exhibited hyperbolic dependence on [nitrate] (Fig. 4D), with a limiting kobs of 310 s−1 and a Kdnitrate of 131 μm (Table 1). The observed [nitrate] dependence suggests that the intrinsic rate constant for electron transfer is extremely rapid and is rate-limited by the chemistry of nitrate reduction, which is expected to be the sole [nitrate]-dependent step overall. Neither phosphorylation of the Mo-heme fragment nor the addition of 14-3-3ω to non-phosphorylated enzyme resulted in a significant change of this behavior. With phosphorylated Mo-heme fragment in the presence of 14-3-3ω, however, the kobs was reduced to a value of 35 s−1 and seemed to have lost its dependence on [nitrate], at least above ∼50 μm (Fig. 4D and Table 1). This result specifically eliminates the possibility that the residual steady-state activity exhibited by the inhibited fragment (Fig. 4C) was due to a small fraction of uninhibited enzyme present in the sample (that either had not been successfully phosphorylated or in equilibrium did not have bound 14-3-3ω). Were this the case, the observed kinetics would have reflected the same kobs as seen with uninhibited enzyme, but the extent of reaction (as reflected in the magnitude of the absorbance change) would have reflected the extent of uninhibited fragment present. The observed loss of [nitrate] dependence on heme reoxidation in the inhibited fragment suggests, but does not prove, that the intrinsic rate constant for electron transfer from heme to molybdenum has become rate-limiting upon inhibition by 14-3-3.
14-3-3 Does Not Impair Nitrate Reduction
There are two possible explanations for the observation that phosphorylation and 14-3-3ω binding significantly reduce the rate at which the heme center is reoxidized by electron transfer to the molybdenum center. (i) The chemistry of nitrate reduction at the molybdenum center is compromised; or (ii) the intrinsic rate constant associated with electron transfer from heme to the molybdenum center is slowed. To distinguish between these, two experiments were performed. First, a heme-deficient variant, H600A, of the Mo-heme fragment was prepared (see supplemental Fig. S2) to examine a system in which there is no electron transfer from heme. We found an ∼20-fold reduction of the methyl viologen:nitrate activity for the H600A variant, whereas it was as active as the wild-type protein in the bromphenol blue:nitrate assay. This shows that bromphenol blue directly reduces the molybdenum center, bypassing the heme domain. Next, we investigated the bromphenol blue:nitrate activity of both wild-type and the H600A variant at high viscosity and in the presence or absence of phosphorylation/14-3-3ω binding (Fig. 5A). The 14-3-3ω-treated wild-type enzyme exhibited full activity in both cases, demonstrating that catalytic turnover at the molybdenum site was unaffected.
FIGURE 5.
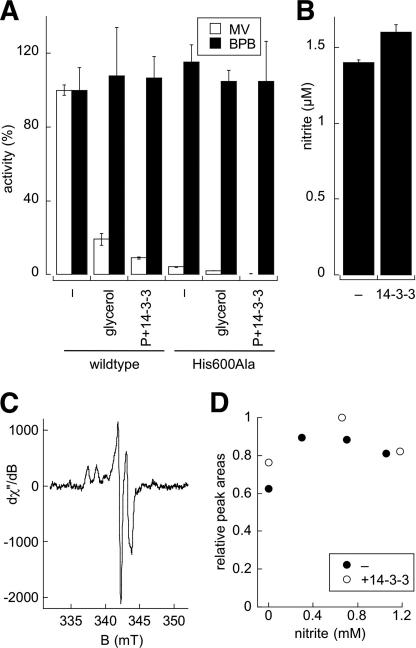
Rate-limiting step in 14-3-3ω inhibition. A, methyl viologen:nitrate (MV, white bars) or bromphenol blue:nitrate activities (BPB, black bars) of the wild-type Mo-heme fragment and a heme-deficient H600A variant under standard conditions (−), in the presence of 35% glycerol, or after phosphorylation and 14-3-3ω incubation (P+14-3-3). B, nitrite production by pre-reduced, phosphorylated Mo-heme fragment without (−) or with 14-3-3ω in a single-turnover experiment using a mix of nitrate and quenching reagent. Error bars in panels A and B indicate S.D. C, representative EPR spectrum of the phospho-Mo-heme fragment following the reaction with nitrate. D, steady-state EPR MoV analysis as a function of nitrite produced. Pre-reduced phospho-Mo-heme fragment (7 μm) ± 14-3-3ω was reacted with 5 mm nitrate under anaerobic conditions, and aliquots were removed and quenched with zinc acetate for nitrite quantification as the EPR tubes were frozen. After initial EPR analysis, samples were thawed and incubated for 10–30 s, and the procedure was repeated again. dχ/dB is the first derivative of the out-of-phase magnetic susceptibility of the sample as a function of the magnetic flux, B.
In a second experiment, a solution of nitrate and zinc acetate as quenching reagent was mixed with pre-reduced, phosphorylated Mo-heme fragment with or without 14-3-3ω in a stopped-flow apparatus, and the extent of nitrite accumulation after a very short reaction time (sufficient to allow only a single turnover) was examined. The extremely rapid reaction of nitrate at the molybdenum site precluded a traditional double-mixing experiment with a delay between the mixing of substrate with enzyme and subsequent quenching. With the quenching reagent present in the nitrate solution, the reaction is expected to proceed only approximately for the 1–2-ms dead time of the stopped-flow instrument. An absorption spectrum taken 10 ms after mixing confirmed that the enzyme was fully oxidized at this point and that reaction had occurred. Precipitated protein from the stopped-flow effluent was sedimented, and the nitrite in the supernatant was quantified colorimetrically. As shown in Fig. 5B, we found that comparable amounts of nitrite were produced by the phosphorylated Mo-heme fragment in the absence and presence of 14-3-3ω (1.4 ± 0.02 and 1.6 ± 0.05 μm, respectively, using 2.5 μm of the functional fragment), indicating that the rate of reduction of nitrate at the molybdenum center was not significantly affected by phosphorylation and binding of 14-3-3ω.
Rate-limiting Step in 14-3-3ω Inhibition
The above results demonstrate that the rate of nitrate reduction at the molybdenum center is not affected significantly by phosphorylation and binding of 14-3-3ω and indicate that 14-3-3ω inhibition is instead due to a lowering of the rate of electron transfer from heme to molybdenum. According to the prevailing theoretical model for electron transfer in (bio)chemical systems, there are two principal factors that influence the rate of electron transfer between a given donor:acceptor pair: (i) the overall driving force for the reaction (in this case the difference in reduction potentials for the heme and molybdenum center) and (ii) the distance between donor and acceptor.
To ascertain whether there is any change in driving force for electron transfer upon inhibition, the reduction potential for the heme in various constructs (Mo-heme, heme-FAD, and heme fragments, Figs. 1A and 2E) was determined (see supplemental Fig. S3). We found that the heme has essentially the same reduction potential in the Mo-heme, heme-FAD, and heme fragments and, importantly, it is not affected by phosphorylation and binding of 14-3-3ω (Table 2). Although it had been suggested that 14-3-3 binding might raise the reduction potential of the heme and thus impede electron transfer (32), we find this not to be the case (at least for the Arabidopsis enzyme).
TABLE 2.
Summary of redox potentials for various preparations of NR
| Protein | Reduction potential (mV vs. SHE)a |
|---|---|
| Mo-heme fragment | −187 ± 1 |
| Phosphorylated Mo-heme fragment + 14-3-3ω | −186 ± 1 |
| Heme fragment | −186 ± 3 |
| Heme-FAD fragment | −173 ± 2 |
a Determined using indigo carmine as standardization dye (−116 mV at pH 7.0, 22 °C); n = 2.
Having excluded inhibition of nitrate reduction at the molybdenum center and any change in the reduction potential in the heme as the basis for inhibition of the Mo-heme fragment, we finally examined whether binding of 14-3-3ω to the phosphorylated Mo-heme fragment alters the reduction potential of the molybdenum center. It is important to note that two half-potentials of the molybdenum center (i.e. the MoVI/MoV and MoV/MoIV couples) needed to be considered, as well as the midpoint potential of the center (i.e. the average of the two half-potentials). In the absence of a spectral signature associated with oxidation-reduction of the molybdenum center, we instead examined the extent of steady-state accumulation of the EPR-active MoV state in the absence and presence of phosphorylation/14-3-3ω binding, which is dependent on the separation between the two half-potentials for the molybdenum center. Any change in the relative values for the two half-potentials would alter the amount of MoV that accumulates during turnover; increasing the MoVI/MoV and/or decreasing the MoV/MoIV couple upon inhibition would increase or decrease the fractional amount of enzyme that accumulates in the steady state as MoV. The experiment involved turning the Mo-heme fragment over with nitrate, using sodium dithionite as reductant (which directly reduces the molybdenum center). At different times after the reaction was initiated, parallel aliquots were removed and either frozen for EPR analysis (Fig. 5C) or quenched for nitrite quantification. Samples at several different reaction times (having different levels of accumulated nitrite) were collected to avoid any complication due to possible nitrite inhibition of the enzyme. Similar results were obtained with the phosphorylated Mo-heme fragment in the presence or absence of 14-3-3ω (Fig. 5D), with essentially no difference in the amount of accumulated MoV as a function of nitrite concentration. This implies that 14-3-3ω binding does not affect the relative values of the MoVI/MoV and MoV/MoIV half-potentials. We note that the steady-state EPR spectrum seen here (Fig. 5C) closely resembles the pre-steady-state spectra published earlier (31) for the full-length enzyme.
We conclude that neither the heme nor the molybdenum reduction potentials are perturbed upon inhibition of the Mo-heme fragment, and consequently, that there is no alteration in the driving force for the reaction. The reduced rate of electron transfer upon binding of 14-3-3ω thus appears to be due to a significant change in the distance between the heme and molybdenum center as the result of a 14-3-3ω-induced conformational change that reorients the molybdenum- and heme-containing domains of the protein.
DISCUSSION
14-3-3 proteins are universal modulators of a diverse range of cellular functions, including hormone signaling in humans (i.e. serontonin N-acetyltransferase (33)), control of stomatal pore opening in plants (plasma membrane ATPase (34)), and regulation of neurotransmitter synthesis in brain (i.e. tyrosine hydroxylase (35)). In most cases, 14-3-3 proteins are believed to trigger conformational changes in their targets, leading to alterations in activity, stability, oligomerization, or cellular localization of binding partners (36). Although numerous crystal structures of 14-3-3 proteins have been determined in both the apo-peptide-bound and the phospho-peptide-bound state (37), to date, their specific mode of action has been established in only two cases: the increase in substrate affinity of serotonin N-acetyltransferase in the 14-3-3 complexed state (33) and the oligomerization of plant plasma membrane ATPase (34).
In plants, 14-3-3 proteins are important in regulating nitrogen and carbon metabolism (38), with NR being the first and best studied target (1). We have previously shown that phosphorylation and binding of 14-3-3ω reduces turnover of NR by a factor of 10 (18). In the present work, we have shown the C-terminal heme-FAD fragment of NR to be unaffected by 14-3-3 binding and determined the individual rate constants for reduction of FAD by NADH (638 s−1) and subsequent electron transfer from FADH2 to heme (583 s−1) (Fig. 6). By contrast, the N-terminal Mo-heme fragment closely mimics the steady-state behavior of holo-NR, including inhibition by 14-3-3ω, making it an extremely valuable tool in dissecting the effect of inhibition on specific steps of the catalytic cycle. The present work also demonstrates that the electron donor methyl viologen preferentially reduces the heme domain, whereas bromphenol blue targets the molybdenum center of NR. The bromphenol blue:nitrate assay has thus been particularly useful in demonstrating that 14-3-3ω binding to phosphorylated NR does not impair the chemistry of nitrate reduction directly at the molybdenum center.
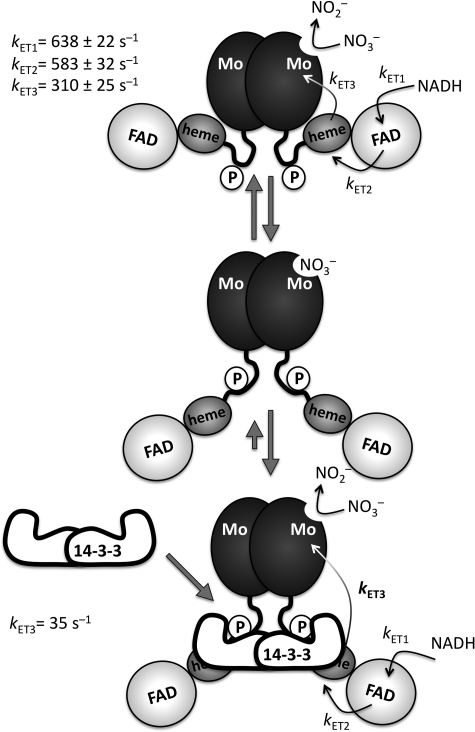
FIGURE 6.
Model of NR inhibition by 14-3-3. Domains are depicted according to their size. Given the known sensitivity of NR activity to solution viscosity and proteolytic degradation, an extended hinge, carrying the phosphorylated (P) Ser-534, is depicted. Note that electron transfer rates (kET1–kET3) shown in the upper panel were determined for the non-phosphorylated fragments. NR is depicted in a closed conformation illustrating effective electron transfer (top panel). The middle panel depicts the nitrate-bound open confirmation, which is proposed to bind 14-3-3 with higher affinity based on our steady-state kinetics (18). The lower panel illustrates the 14-3-3-inhibited enzyme in its stabilized open conformation with a reduced electron transfer rate (kET3) from heme to molybdenum.
The results reported here demonstrate that the previously observed reduction of kcat upon 14-3-3ω binding to phosphorylated NR is due specifically to a reduction in the rate of electron transfer from heme to molybdenum, with the observed rate constant decreasing from 310 to 35 s−1, whereas nitrate reduction remains fast in the inhibited Mo-heme fragment. The observed rate constant for heme oxidation seen in the reaction of the reduced, inhibited fragment with nitrate must thus reflect the intrinsic electron transfer rate, whereas the rate constant seen with the uninhibited enzyme is a lower limit for the intrinsic electron transfer rate in the uninhibited fragment (because oxidation of the heme may well be rate-limited by the chemistry of nitrate reduction at the molybdenum center, which must occur prior to the electron transfer step). The observed nitrate concentration dependence of heme oxidation in the uninhibited enzyme suggests that electron transfer is indeed rate-limited by the chemistry of nitrate reduction. Our findings that the 14-3-3ω complexed enzyme (i) has full activity in steady-state kinetics monitoring the molybdenum-to-nitrate rate (in the bromphenol blue:nitrate assay), (ii) loses its [nitrate] dependence in rapid kinetics, and (iii) has an unchanged nitrite production rate in a single-turnover experiment support the conclusion that 14-3-3ω acts specifically by reducing the rate of electron transfer between heme and molybdenum and does not affect the rate of nitrate reduction at the molybdenum center.
To our knowledge, this is the first case in which a 14-3-3 protein is shown to act as a molecular switch in tuning electron transfer rates within a redox-active enzyme, and our results demonstrate that modulation of intramolecular electron transfer rates is an effective mechanism whereby enzyme activity can be tuned. The two principal factors that influence the rate of electron transfer between a given donor:acceptor pair are their relative reduction potentials (i.e. the thermodynamic driving force for the reaction) and the distance between them. We show here that phosphorylation and binding of 14-3-3ω do not significantly perturb the reduction potential of either the heme or the molybdenum center and conclude that inhibition of NR by 14-3-3ω is most likely due to an increase in the distance between the two centers, undoubtedly due to a significant reorientation of the molybdenum- and heme-containing domains, which are tethered together by the intervening Hinge 1 region (Fig. 6).
Based on the rapid kinetic results presented here and in conjunction with our earlier steady-state results (18), we propose two conformations for holo-NR: a “closed” form with a short heme-molybdenum distance favoring high electron transfer rates and an “open” form with a long heme-to-molybdenum distance that is trapped upon phosphorylation and binding of 14-3-3 (Fig. 6). This model is consistent with the observation that increasing solution viscosity significantly decreases steady-state turnover of both the holo-enzyme and the Mo-heme fragment (39). Once 14-3-3 has bound to phosphorylated NR and the open conformation is stabilized, electron transfer from the heme to the molybdenum center slows dramatically, resulting in the observed decrease in activity.
Supplementary Material
Acknowledgments
Technical assistance by Simona Jansen, Ulrika Beitnere (University of Cologne, Germany), and Judith Thevarajah (Technische Universität Braunschweig, Germany) is gratefully acknowledged.
This work was supported by the German Science Foundation (Grants Schw 759/6-1 and SFB 635 TP-A5 to G. S.), the National Institutes of Health (Grants GM 075036 and ES012658 to R. H.), and the Fonds der Chemischen Industrie (to G. S.).

This article contains supplemental Experimental Procedures, Table S1, and Figs. S1–S3.
- NR
- nitrate reductase
- CPK
- calcium-dependent protein kinase
- NTA
- nitrilotriacetic acid
- Bis-Tris
- 2-(bis(2-hydroxyethyl)amino)-2-(hydroxymethyl)propane-1,3-diol.
REFERENCES
- 1. Chevalier D., Morris E. R., Walker J. C. (2009) 14-3-3 and FHA domains mediate phospho-protein interactions. Annu. Rev. Plant Biol. 60, 67–91 [DOI] [PubMed] [Google Scholar]
- 2. Moore B. W., Perez V. J. (1967) in Physiological and Biochemical Aspects of Nervous Integration (Carlson F. ed) pp. 343–359, Prentice Hall, Englewood Cliffs, NJ [Google Scholar]
- 3. Yaffe M. B., Smerdon S. J. (2001) Phospho-serine/threonine-binding domains: you can't pSERious? Structure 9, R33–38 [DOI] [PubMed] [Google Scholar]
- 4. Fu H., Subramanian R. R., Masters S. C. (2000) 14-3-3 proteins: structure, function, and regulation. Annu. Rev. Pharmacol. Toxicol. 40, 617–647 [DOI] [PubMed] [Google Scholar]
- 5. Wang B., Yang H., Liu Y. C., Jelinek T., Zhang L., Ruoslahti E., Fu H. (1999) Isolation of high affinity peptide antagonists of 14-3-3 proteins by phage display. Biochemistry 38, 12499–12504 [DOI] [PubMed] [Google Scholar]
- 6. Aitken A. (2006) 14-3-3 proteins: a historic overview. Semin. Cancer Biol. 16, 162–172 [DOI] [PubMed] [Google Scholar]
- 7. Ottmann C., Yasmin L., Weyand M., Veesenmeyer J. L., Diaz M. H., Palmer R. H., Francis M. S., Hauser A. R., Wittinghofer A., Hallberg B. (2007) Phosphorylation-independent interaction between 14-3-3 and exoenzyme S: from structure to pathogenesis. EMBO J. 26, 902–913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Liu D., Bienkowska J., Petosa C., Collier R. J., Fu H., Liddington R. (1995) Crystal structure of the zeta isoform of the 14-3-3 protein. Nature 376, 191–194 [DOI] [PubMed] [Google Scholar]
- 9. Rittinger K., Budman J., Xu J., Volinia S., Cantley L. C., Smerdon S. J., Gamblin S. J., Yaffe M. B. (1999) Structural analysis of 14-3-3 phospho-peptide complexes identifies a dual role for the nuclear export signal of 14-3-3 in ligand binding. Mol. Cell 4, 153–166 [DOI] [PubMed] [Google Scholar]
- 10. Würtele M., Jelich-Ottmann C., Wittinghofer A., Oecking C. (2003) Structural view of a fungal toxin acting on a 14-3-3 regulatory complex. EMBO J. 22, 987–994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Sehnke P. C., DeLille J. M., Ferl R. J. (2002) Consummating signal transduction: the role of 14-3-3 proteins in the completion of signal-induced transitions in protein activity. Plant Cell 14, (suppl.) S339–S354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Shen W., Clark A. C., Huber S. C. (2003) The C-terminal tail of Arabidopsis 14-3-3ω functions as an autoinhibitor and may contain a tenth α-helix. Plant J. 34, 473–484 [DOI] [PubMed] [Google Scholar]
- 13. Lu G., Sehnke P. C., Ferl R. J. (1994) Phosphorylation and calcium binding properties of an Arabidopsis GF14 brain protein homolog. Plant Cell 6, 501–510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Mackintosh C. (2004) Dynamic interactions between 14-3-3 proteins and phospho-proteins regulate diverse cellular processes. Biochem. J. 381, 329–342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. DeLille J. M., Sehnke P. C., Ferl R. J. (2001) The Arabidopsis 14-3-3 family of signaling regulators. Plant Physiol. 126, 35–38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Yao Y., Du Y., Jiang L., Liu J. Y. (2007) Molecular analysis and expression patterns of the 14-3-3 gene family from Oryza sativa. J. Biochem. Mol. Biol. 40, 349–357 [DOI] [PubMed] [Google Scholar]
- 17. Paul A. L., Sehnke P. C., Ferl R. J. (2005) Isoform-specific subcellular localization among 14-3-3 proteins in Arabidopsis seems to be driven by client interactions. Mol. Biol. Cell 16, 1735–1743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lambeck I., Chi J. C., Krizowski S., Mueller S., Mehlmer N., Teige M., Fischer K., Schwarz G. (2010) Kinetic analysis of 14-3-3-inhibited Arabidopsis thaliana nitrate reductase. Biochemistry 49, 8177–8186 [DOI] [PubMed] [Google Scholar]
- 19. Kanamaru K., Wang R., Su W., Crawford N. M. (1999) Ser-534 in the Hinge 1 region of Arabidopsis nitrate reductase is conditionally required for binding of 14-3-3 proteins and in vitro inhibition. J. Biol. Chem. 274, 4160–4165 [DOI] [PubMed] [Google Scholar]
- 20. Bachmann M., Huber J. L., Athwal G. S., Wu K., Ferl R. J., Huber S. C. (1996) 14-3-3 proteins associate with the regulatory phosphorylation site of spinach leaf nitrate reductase in an isoform-specific manner and reduce dephosphorylation of Ser-543 by endogenous protein phosphatases. FEBS Lett. 398, 26–30 [DOI] [PubMed] [Google Scholar]
- 21. Lillo C. (2008) Signaling cascades integrating light-enhanced nitrate metabolism. Biochem. J. 415, 11–19 [DOI] [PubMed] [Google Scholar]
- 22. Campbell W. H. (1999) Nitrate reductase structure, function, and regulation: bridging the gap between biochemistry and physiology. Annu. Rev. Plant Physiol. Plant Mol. Biol. 50, 277–303 [DOI] [PubMed] [Google Scholar]
- 23. Deng H. (2006) Nitrite anions induce nitrosative deamination of peptides and proteins. Rapid Commun. Mass Spectrom. 20, 3634–3638 [DOI] [PubMed] [Google Scholar]
- 24. Spencer J. P., Whiteman M., Jenner A., Halliwell B. (2000) Nitrite-induced deamination and hypochlorite-induced oxidation of DNA in intact human respiratory tract epithelial cells. Free Radic. Biol. Med. 28, 1039–1050 [DOI] [PubMed] [Google Scholar]
- 25. Kaiser W. M., Brendle-Behnisch E. (1991) Rapid modulation of spinach leaf nitrate reductase activity by photosynthesis : I. modulation in Vivo by CO2 availability. Plant Physiol. 96, 363–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Huber J. L., Huber S. C., Campbell W. H., Redinbaugh M. G. (1992) Reversible light/dark modulation of spinach leaf nitrate reductase activity involves protein phosphorylation. Arch. Biochem. Biophys. 296, 58–65 [DOI] [PubMed] [Google Scholar]
- 27. MacKintosh C. (1992) Regulation of spinach leaf nitrate reductase by reversible phosphorylation. Biochim. Biophys. Acta 1137, 121–126 [DOI] [PubMed] [Google Scholar]
- 28. Mackintosh C., Douglas P., Lillo C. (1995) Identification of a protein that inhibits the phosphorylated form of nitrate reductase from spinach (Spinacia oleracea) leaves. Plant Physiol 107, 451–457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kuper J., Llamas A., Hecht H. J., Mendel R. R., Schwarz G. (2004) Structure of the molybdopterin-bound Cnx1G domain links molybdenum and copper metabolism. Nature 430, 803–806 [DOI] [PubMed] [Google Scholar]
- 30. Marzinzig M., Nussler A. K., Stadler J., Marzinzig E., Barthlen W., Nussler N. C., Beger H. G., Morris S. M., Jr., Brückner U. B. (1997) Improved methods to measure end products of nitric oxide in biological fluids: nitrite, nitrate, and S-nitrosothiols. Nitric Oxide 1, 177–189 [DOI] [PubMed] [Google Scholar]
- 31. Skipper L., Campbell W. H., Mertens J. A., Lowe D. J. (2001) Pre-steady-state kinetic analysis of recombinant Arabidopsis NADH:nitrate reductase: rate-limiting processes in catalysis. J. Biol. Chem. 276, 26995–27002 [DOI] [PubMed] [Google Scholar]
- 32. Cannons A. C., Barber M. J., Solomonson L. P. (1993) Expression and characterization of the heme-binding domain of Chlorella nitrate reductase. J. Biol. Chem. 268, 3268–3271 [PubMed] [Google Scholar]
- 33. Obsil T., Ghirlando R., Klein D. C., Ganguly S., Dyda F. (2001) Crystal structure of the 14-3-3ζ:serotonin N-acetyltransferase complex: a role for scaffolding in enzyme regulation. Cell 105, 257–267 [DOI] [PubMed] [Google Scholar]
- 34. Ottmann C., Marco S., Jaspert N., Marcon C., Schauer N., Weyand M., Vandermeeren C., Duby G., Boutry M., Wittinghofer A., Rigaud J. L., Oecking C. (2007) Structure of a 14-3-3 coordinated hexamer of the plant plasma membrane H+-ATPase by combining x-ray crystallography and electron cryomicroscopy. Mol. Cell 25, 427–440 [DOI] [PubMed] [Google Scholar]
- 35. Obsilova V., Nedbalkova E., Silhan J., Boura E., Herman P., Vecer J., Sulc M., Teisinger J., Dyda F., Obsil T. (2008) The 14-3-3 protein affects the conformation of the regulatory domain of human tyrosine hydroxylase. Biochemistry 47, 1768–1777 [DOI] [PubMed] [Google Scholar]
- 36. Gökirmak T., Paul A. L., Ferl R. J. (2010) Plant phospho-peptide-binding proteins as signaling mediators. Curr. Opin. Plant Biol. 13, 527–532 [DOI] [PubMed] [Google Scholar]
- 37. Obsil T., Obsilova V. (2011) Structural basis of 14-3-3 protein functions. Semin. Cell Dev. Biol. 22, 663–672 [DOI] [PubMed] [Google Scholar]
- 38. Huber S. C., MacKintosh C., Kaiser W. M. (2002) Metabolic enzymes as targets for 14-3-3 proteins. Plant Mol. Biol. 50, 1053–1063 [DOI] [PubMed] [Google Scholar]
- 39. Barbier G. G., Campbell W. H. (2005) Viscosity effects on eukaryotic nitrate reductase activity. J. Biol. Chem. 280, 26049–26054 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.