Abstract
The production of mitochondrial reactive oxygen species occurs as a consequence of aerobic metabolism. Mitochondrial oxidants are increasingly viewed less as byproducts of metabolism and more as important signaling molecules. Here, I review several notable examples, including the cellular response to hypoxia, aspects of innate immunity, the regulation of autophagy, and stem cell self-renewal capacity, where evidence suggests an important regulatory role for mitochondrial oxidants.
Introduction
Mitochondria are believed to be the major source of intracellular reactive oxygen species (ROS)2 generation. Working with isolated mitochondria, some estimates have suggested that as much as 3–5% of the oxygen consumed is ultimately diverted toward ROS production (1). Although these estimates are potential benchmarks, the experimental conditions, including substrate concentration and ATP levels, in which many of these measurements were made, as well as the functional alterations that occur with mitochondrial isolation, argue for caution with regard to such in vitro determinations. As such, the precise in vivo amount of mitochondrial superoxide or hydrogen peroxide production remains elusive. Although the magnitude of ROS production remains in doubt, the location of the one-electron reduction of molecular oxygen is less uncertain. Both Complexes I and III of the electron transport chain are thought to be the major sites of ROS production (2, 3), although clearly other electron complexes, as well as other mitochondrial enzymes, can generate ROS. Once generated, superoxide rapidly and spontaneously dismutates to hydrogen peroxide. This conversion is accelerated in the presence of the enzyme superoxide dismutase. To avoid the potential damaging effects of ROS, mitochondria express a number of protein antioxidants, including SOD2, as well as other scavenging enzymes such as peroxiredoxins 3 and 5. Although the activities of intracellular antioxidants and peroxidases determine the magnitude of ROS, the level of ROS generated is also believed to be dependent on certain intrinsic properties of mitochondrial energetics. For instance, a high proton motive force (e.g. high ψm) is believed to favor the production of ROS, whereas mitochondrial uncoupling agents result in a lower ψm and decreased ROS formation. Similarly, mutations in either mitochondrial DNA or nuclear DNA that lead to disruption in any of the components of the electron transport chain also predispose to ROS formation presumably by impeding the flow of electrons down the cytochrome chain.
Although the regulation of mitochondrial ROS release was experimentally appreciated, for many years, the prevailing view was that mitochondrial oxidants were autonomously produced solely as a byproduct of metabolism. That notion has slowly given way to a more nuanced view of mitochondrial oxidants as potential regulators of a number of intracellular pathways. Initial reports suggested that increased supply of metabolic substrates augmented mitochondrial oxidant production and that the release of oxidants could in turn activate JNK, a stress-responsive kinase (4). Subsequent studies have suggested that the link between excess metabolic supply and mitochondrial ROS production may have important implications for human disease (5, 6). Furthermore, the release of mitochondrial oxidants and the subsequent activation of kinases such as JNK were shown to be activated not only by alterations in substrate supply but also by exogenous ligands such as TNF-α that appear to increase mitochondrial ROS levels directly. These latter studies demonstrated that the target of the mitochondrial ROS was not JNK itself but rather a cysteine-dependent phosphatase, the redox-dependent inactivation of which led to sustained JNK activity (7). Thus, similar to other examples of redox-dependent signal transduction, much of the specific signaling revolves around the oxidation of reactive cysteine residues in specific target proteins. Although counterexamples exist, in most cases, the specific ROS molecule involved in signaling is believed to be hydrogen peroxide (8). Here, I will highlight a subset of these emerging trends that together suggest a complex signaling system that is initiated by the release of mitochondrial oxidants. Although not exhaustive, these examples highlight the growing importance of mitochondrial ROS as physiological and pathophysiological regulators of a diverse range of biological phenomena.
Hypoxia, Oxidants, and Regulation of Hypoxia-inducible Factor 1α (HIF-1α)
Accumulating evidence suggests that mitochondrial oxidants may be important regulators of the cellular response to low oxygen. In particular, there are a number of studies that suggest that, under hypoxic conditions (oxygen concentrations between 1 and 5%), mitochondria might actually increase their release of ROS. This release appears to come from Complex III; however, the precise molecular basis for the seemingly counterintuitive relationship between ambient oxygen levels and ROS production remains obscure. Nonetheless, evidence suggests that the release of ROS under these conditions functions as an important physiological regulator of HIF-1α. HIF-1α consists of a labile α-subunit and a constitutively expressed β-subunit. The stability of the α-subunit is regulated by oxygen levels such that, under normoxic conditions, this subunit undergoes proteasomal degradation, whereas under hypoxic conditions, it is stabilized.
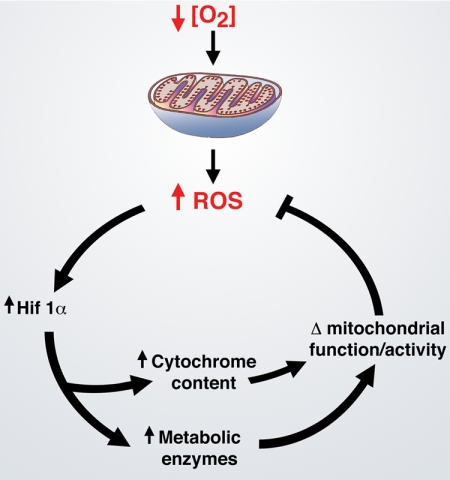
The first indication that there might be a connection between hypoxia, mitochondrial ROS production, and HIF-1α activation came from the analysis of ρ0 cells. These cells are generated by culturing in the presence of ethidium bromide for several weeks to inhibit replication of mitochondrial DNA selectively. Such efforts lead to cells that lack the capacity for electron transport because they cannot produce a sufficient number of key cytochrome components and other molecules encoded exclusively by mitochondrial DNA. In these initial reports, the parental cell line increased ROS levels during hypoxia, whereas ρ0 cells did not. In addition, ρ0 cells also appeared impaired in their ability to activate HIF-1α under hypoxic conditions (9). In contrast, some follow-up reports working with a similar but not identical paradigm disputed how important the hypoxic release of mitochondrial ROS was for HIF-1α stabilization (10, 11). The studies were complicated by the difficulty of measuring ROS levels in living cells under hypoxic conditions. In addition, although useful, ρ0 cells are an imperfect model of mitochondrial deficiencies. Many of these objections were resolved in subsequent studies. Indeed, using a FRET-based approach, more sensitive measurements were possible, allowing for a clearer demonstration that hypoxia triggers ROS release (12). Similarly, three studies either using RNAi-based approaches to inhibit components of Complex III or using murine embryonic stem cells lacking cytochrome c showed that intact mitochondrial function is required for the hypoxic induction of HIF-1α (12–14). Furthermore, using a variety of genetic or pharmacological approaches, these studies demonstrated that it was the mitochondrial release of hydrogen peroxide that serves as the signaling intermediary for this induction. Interestingly, these results suggest that, under low oxygen conditions, HIF-1α may be uniquely poised to respond to a rise in ROS and then to feedback and inhibit the production of ROS levels (Fig. 1). This latter feedback activity is suggested by a number of other studies in which HIF-1α was shown to have an important role in mitochondrial function and ROS generation. Included among these relevant HIF-1α activities is the transcriptional regulation of metabolic enzymes such as lactate dehydrogenase A and PDK1 (pyruvate dehydrogenase kinase 1) that control the flow of carbon substrates into the mitochondria (15, 16). Similarly, HIF-1α appears to regulate the expression of certain cytochrome components directly and to regulate specific microRNAs that can in turn regulate the expression of components of the electron transport chain (17, 18). Taken together, these studies suggest that HIF-1α functions in a homeostatic network both to respond to mitochondrial oxidant production and to regulate mitochondrial oxidant generation.
FIGURE 1.
HIF-1α and mitochondrial oxidants. Shown is the potential role of HIF-1α in regulating ROS levels under low oxygen conditions. Evidence suggests that low oxygen levels actually increase the levels of mitochondrial ROS. Once released, these oxidants appear to stabilize HIF-1α protein and thereby increase HIF-1α activity. In turn, through a variety of mechanisms, HIF-1α alters mitochondrial carbon metabolism by regulating such enzymes as lactate dehydrogenase A and PDK1 and potentially alters mitochondrial activity by augmenting cytochrome content. The end result of these HIF-1α-regulated activities is a potential reduction of the increased ROS levels induced by hypoxic stress.
Another interesting area in which there might be a convergence between mitochondrial oxidants and the activation of HIF-1α comes from the analysis of a series of rare tumors caused by germ line mutations in components of Complex II of the electron transport chain. There are four subunits of mammalian Complex II (SdhA, SdhB, SdhC, and SdhD), and unlike Complex I, III, IV, or V, all of the components of Complex II are nuclear encoded. The activity of Complex II is also unique in that it has a role both in electron transport and in the Krebs cycle, where it oxidizes succinate to fumarate. Genetic evidence suggests that mutations in SdhB, SdhC, and SdhD can lead to an autosomal dominant tumor syndrome characterized by the development of exceedingly rare tumors, including paragangliomas and pheochromocytomas (19). Significant evidence suggests that tumors in this syndrome develop, at least in part, through the chronic activation of HIF-1α. This activation may be secondary to a Complex II-dependent defect in succinate metabolism. A rise in succinate could act to inhibit the 2-oxoglutarate-dependent dioxygenase that regulates the proline hydroxylation and hence the stability of HIF-1α (20, 21). Nonetheless, there are also some examples in which specific Sdh mutations appear to activate HIF-1α through an increase in mitochondrial oxidants (22). Such mutations alter the conformation of Complex II presumably to become an important site of ROS generation. The relative contribution of changes in succinate levels versus changes in mitochondrial ROS remains unclear, and it remains possible that, in this condition, different mutations lead to activation of HIF-1α through different pathways.
Mitochondrial Oxidants and the Inflammasome
Several recent observations have suggested that mitochondrial oxidants can also act as important signaling molecules to regulate the inflammatory response. These reports have centered mostly on the activation of the inflammasome, a molecular platform required for a fully functional innate immune system. In particular, several groups have looked at activation of NLRP3 (NOD-like receptor, pyrin domain-containing 3), an intracellular receptor that senses a wide range of damage signals, including microorganisms, endogenous danger signals such as ATP and uric acid, and environmental irritants such as silica and asbestos. Interestingly, many of the known activators of the NLRP3 inflammasome are also known to generate intracellular ROS (23). Once activated, NLRP3 associates with a number of other protein partners to form a high molecular weight intracellular complex that functions to regulate the maturation and secretion of proinflammatory cytokines such as IL-1β. Prior to these most recent works, there was already some indication that ROS play a key role in the activation of the inflammasome. For instance, it had been observed that known activators of NLRP3 caused the redox-dependent dissociation of thioredoxin-interacting protein from thioredoxin. Once free from thioredoxin, thioredoxin-interacting protein bound to NLRP3, and this binding appeared to be required for the subsequent robust activation of the inflammasome (24).
Given the wide range of known activators of the inflammasome, it seemed likely that there might be a common mediator for NLRP3 activation. As mentioned above, one candidate appeared to be ROS. Preliminary evidence initially hinted that cytoplasmic members of the NADPH oxidase family (e.g. NOX1, NOX2, etc.) might be the enzymatic source. However, recent definitive genetic experimentation argues against this possibility (25, 26). In contrast, there appears to be a robust correlation between mitochondrial oxidants and inflammasome activation. For instance, pharmacological agents that block Complex I or III and result in the increased release of mitochondrial ROS also produce an increase in secreted IL-1β, a hallmark of NLRP3 activation (26). Indeed, following activation, the NLRP3 complex was observed to co-localize to the mitochondria (26). Furthermore, antioxidants specifically targeted to the mitochondria appeared to block inflammasome activation and inflammation in general (25–27). These and other data suggest a model wherein a wide range of danger signals converge to cause the increased generation of mitochondrial ROS. A rise in mitochondrial oxidant production could therefore represent the common currency of all of these divergent stress signals with subsequent activation of the NLRP3 inflammasome. Interestingly, mitochondrial oxidants may play a similar role in the execution of the apoptotic response and in the formation of the apoptosome. At present, it is unclear if there are quantitative or qualitative differences in the ROS generated during inflammatory or apoptotic conditions. Unfortunately, the mechanisms through which inflammatory signals can regulate mitochondrial function and the precise mechanism by which the release of ROS can trigger NLRP3 activation are undefined. Finally, very recently, another link between mitochondrial function and innate immunity was established. In particular, it has been recently demonstrated that, in macrophages, stimulation of Toll-like receptors directly recruits mitochondria to the macrophage phagosome (28). The mitochondria that are recruited produce ROS in a fashion that is similar to the more widely characterized phagocytic NADPH oxidase. Indeed, this recent study suggests an interesting functional overlap between these two ROS-generating systems in host defense and innate immunity.
Autophagy, Mitophagy, and ROS
Another area in which mitochondrial oxidants have been recently implicated is in the regulation of autophagy (29). The process of autophagy was first characterized in yeast. Although there are numerous types of autophagy, the majority of work has involved the analysis of macroautophagy. In this process, both proteins and organelles are engulfed by a double-membrane vesicle known as the autophagosome. The contents of the autophagosome are then delivered to the lysosome, where they undergo degradation. The metabolic breakdown products of this process are in turn used as substrates for new biosynthesis. In the absence of an effective autophagy network, there is an accumulation of damaged organelles, including dysfunctional mitochondria. Indeed, mitochondria isolated from autophagy-deficient tissues have impaired respiratory function (30). Similarly, cells with impaired autophagy have high basal levels of ROS, which appears to emanate from the mitochondria (31). Autophagy occurs under all conditions and is essential for housekeeping functions, including the removal of large protein complexes and the recycling of damaged organelles. In addition, a number of cellular stresses can induce a higher rate of autophagic flux. In this context, the classical stimulus for autophagy is starvation. Under starved conditions, the increase in autophagy is presumably a mechanism to increase biosynthetic intermediates in a situation in which there are few external nutrients.
Genetic analysis initially in simple organisms such as yeast has defined a set of ∼30 specific genes that are essential for autophagic flux. These genes (named ATG1, ATG2, etc.) are involved in various processes, including formation and maturation of the double-membrane autophagosome and coordinating the eventual fusion of this structure with the lysosome. Functional analysis has demonstrated that autophagic flux is negatively regulated by the mTOR (mammalian target of rapamycin) pathway and positively regulated by Class III PI3K activity. As mentioned above, one classical stimulus for autophagy is starvation. Recent evidence suggests that starvation of cells triggers the induction of autophagy through an increase in mitochondrial ROS levels. This rise in ROS levels was evident within 15 min of starvation (32). Inhibition of the rise in ROS through pharmacological means resulted in impaired activation of starvation-induced autophagy. Furthermore, a specific cysteine residue on the essential autophagy gene ATG4 was shown to be redox-sensitive. This led to a model wherein starvation increased mitochondrial oxidants and, in particular, increased hydrogen peroxide levels. This mitochondrion-derived hydrogen peroxide could in turn oxidize a critical cysteine residue of the ATG4 gene product. Once oxidized, ATG4 was shown to regulate positively the activity of another essential autophagy gene, ATG8, which in turn increased overall autophagosome formation. Although these results are instructive, it is likely that ATG4 is not the only redox-sensitive target in the autophagic machinery, nor is this result likely to be the only redox connection to the process of autophagy. For instance, prolonged autophagy appears to be linked to cell death. In this context, autophagy and apoptosis may both be viewed as alternative forms of programmed cell death. Interestingly, similar to cell death by apoptosis, autophagic cell death is accompanied by high levels of ROS. The accumulation of ROS under these conditions appears to be at least partially the result of a selective autophagic degradation of the peroxide-scavenging enzyme catalase (33). These results, coupled with the redox-dependent activation of ATG4, suggest that ROS levels are important both in the induction of stress-induced autophagy and in the execution of autophagic cell death.
The selective removal of mitochondria by the autophagic machinery, a process termed mitophagy, has been recently linked to a host of degenerative and metabolic disorders. One area in which this link is particularly intriguing comes from the study of two gene products linked to hereditary forms of Parkinson disease. Work from several laboratories has described a pathway wherein the PINK1 kinase (PTEN-induced kinase 1) is involved in recruiting the usually cytosolic protein parkin to the mitochondria (34, 35). Mutations in both PINK1 and parkin have been previously linked to early-onset forms of Parkinson disease. Parkin is an E3 ubiquitin ligase, and its recruitment to the mitochondria implies that it might ubiquinate specific mitochondrial proteins that in turn act to stimulate the selective removal of the damaged organelle. One such protein target may be the outer mitochondrial membrane protein VDAC1, which appears to undergo parkin-dependent polyubiquitination (35). It is currently unclear what signals damaged mitochondria produce that allow for the selective recruitment of molecules such as parkin. One hypothesis is that a reduction in mitochondrial membrane potential, perhaps caused by increased ROS levels, might be a precipitating factor. How such a signal is perceived or transduced remains unknown at this point. The intersection of disease-associated genes, mitochondrial function, autophagy, and oxidative stress appears to extend to other genes implicated in Parkinson disease. For instance, another gene product linked to hereditary Parkinson disease is DJ-1. Cellular and animal models have demonstrated that loss of this gene results in an increase in ROS levels, as well as aberrant appearing mitochondria (36–38). Furthermore, DJ-1 has a highly conserved and reactive cysteine residue (Cys-106 in human DJ-1) that appears to be required for the protein to function as a regulator of redox homeostasis (39, 40). Together with the emerging data on PINK1 and parkin, these observations suggest that many of the protein products of Parkinson disease susceptibility genes share a common function of regulating mitochondrial dynamics and oxidative stress.
Energy Metabolism, Mitochondria, and Stem Cells
A final area in which the release of mitochondrial oxidants appears to play an increasingly important role is in the biology of stem cells. Much of this work grew out of the broader connection between mitochondrial oxidants and aging. Since the late 1950s when Denham Harman first articulated his “free radical theory of aging,” there has been significant interest in the connection between mitochondrial oxidants and organismal aging (41, 42). Many believe there is an inherent connection between a decline in stem cell function and the aging process, and as such, understanding the role of metabolism and oxidative stress within stem cells seems a particularly fruitful area of investigation. Progress in this area has been spurred by the analysis of various instructive mouse genetic models. Perhaps the first indication that a rise in ROS might be particularly deleterious for stem cells came from the study of mice deficient for the ATM (ataxia telangiectasia mutated) kinase. The ATM kinase is involved in the response to DNA damage, and patients carrying defective ATM alleles suffer from a syndrome characterized by neurological and immunological defects, cancer predisposition, and an accelerated aging phenotype. Evidence from mouse models and human patient samples suggests that ATM deficiency results in an increase in basal ROS levels (43). The precise source of these oxidants is unclear; however, this is a growing connection between ATM and metabolic and mitochondrial function (44–46). Observations with atm−/− mice demonstrated that hematopoietic stem cells (HSCs) lacking ATM develop a profound impairment in stem cell self-renewal (47). The process of self-renewal involves the capacity of HSCs to divide and give rise to new daughter HSCs rather than to mature progeny (i.e. red cells, white cells, platelets, etc.). In the absence of a robust self-renewal capacity, stem cell numbers rapidly deplete. This rapid depletion of stem cell number was observed in the atm−/− mice, yet surprisingly, these animals could be rescued by the simple administration of the antioxidant N-acetylcysteine (47). Indeed, HSCs from atm−/− mice exhibited high levels of basal ROS, and a follow-up study suggested that the redox-dependent activation of p38 MAPK was at least partially responsible for the decline in HSC self-renewal capacity (48). Subsequent to these observations, other groups working with the FoxO family of transcription factors furthered strengthen the association between a rise in oxidant levels and a decline in HSC self-renewal capacity (49). Again, as in the case of HSCs deficient for ATM, HSCs deficient for FoxO family members exhibited a redox-dependent impairment of stem cell self-renewal.
The connection between mitochondrial oxidants and stem cell function has been further expanded by several recent observations. For instance, the Polycomb repressor Bmi1 has been shown to be essential for stem cell renewal, as Bmi1−/− mice also show a rapid postnatal depletion of both hematopoietic and neural stem cells. More recently, the absence of Bmi1 was shown to result in a rise in mitochondrial oxidants (50). This rise in mitochondrial oxidants was of sufficient magnitude to result in oxidative damage to nuclear DNA and the activation of the DNA damage response pathway. Genetic inhibition of the DNA damage response could partially rescue the Bmi1 phenotype without altering the levels of ROS. These results, as well as the above results with p38 MAPK activation, argue that stem and progenitor dysfunction may result from the activation of specific redox-sensitive pathways rather than the random destructive effects of ROS.
This general theme has also been expanded by three recent studies in which the tumor suppressor and metabolic regulator LKB1 was shown to regulate HSC quiescence (51–53). In the absence of LKB1, stem cells appeared to proliferate initially, giving rise to increased progenitor cell number. Over time, this led to exhaustion of the HSC compartment, with eventual pancytopenia in the LKB1-deficient mice. Although LKB1 can regulate a number of intracellular pathways, including the FoxO family of transcription factors, the effects of conditional LKB1 ablation appeared to be independent of FoxO activity or ROS levels. Several of the groups suggested, however, that a decrease in the transcriptional coactivator peroxisome proliferator-activated receptor-γ coactivator 1α (PGC-1α) may play a role in the observed Lkb1 phenotype. PGC-1α is a master regulator of mitochondrial biogenesis and indeed, similar to the Bmi1-deficient animals, LKB1-deficient HSCs had alterations in mitochondrial number and function. As such, stem cells might be particularly sensitive to decreased mitochondrial function or increased mitochondrial oxidants. These results might have important implications connecting the age-dependent decline in mitochondrial and stem cell function. This connection was furthered underscored by observations that telomere dysfunction, a hallmark of aging, can also regulate mitochondrial function (54). Again, as with the data regarding LKB1, the connection appears to be through PGC-1α. In this example, however, telomere shortening causes activation of the DNA damage response pathway, leading to p53 activation. Interestingly, p53 appears to be a direct repressor of PGC-1α expression. The absence of PGC-1α appears to cause a decline in mitochondrial number and function and a rise in ROS levels. Finally, although the role of inappropriate mitochondrial ROS production appears to contribute to a pathological decline in stem cell number and function, there is also evidence that oxidants might play a physiological role as well. The best evidence comes from analysis in Drosophila, where a rise in ROS appears to be required for proper differentiation of the myeloid progenitor cells (55). In this situation, oxidants appear to regulate the activity of many of the factors discussed above, including Polycomb repressors, as well as FoxO transcription factors. It remains unclear whether a similar physiological role for the regulated release of mitochondrial oxidants also exists in the differentiation of mammalian myeloid cells.
Conclusion
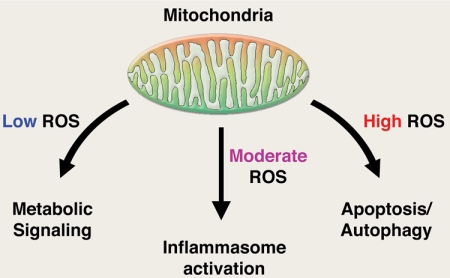
The preceding examples implicate mitochondrial oxidants as regulators of a diverse range of biological activities. In most of these examples, many unanswered questions remain. For instance, very little is known regarding the regulation of mitochondrial oxidant production. For now, it would appear that mitochondrial ROS increase both in the setting of increased substrate supply and during starvation. Similarly, mitochondrial ROS have been shown to increase during both hyperoxia and hypoxia. Clearer molecular insight regarding how such seemingly paradoxical regulation is achieved would appear to be crucial to further our understanding of such complex systems. In addition, the target(s) for mitochondrial oxidants are often poorly characterized. For instance, although there is compelling evidence that mitochondrial oxidants regulate the inflammasome, there is little insight regarding what precisely oxidants do in this or similar circumstances. Indeed, in many of the described examples, although genetic or pharmacological evidence strongly implicates mitochondrial oxidants as playing an important biological role, the precise molecular target for mitochondrial ROS is often mysterious or poorly understood. Finally, although perhaps too simplistic, it may be convenient to view the release of mitochondrial oxidants as a cellular early warning system. It is not presumably strictly by chance that many of the pathways in which mitochondrial ROS have been implicated can be broadly viewed as lethal or sublethal stresses. So, too, the specific cytosolic pathways implicated to date that respond to a rise in mitochondrial ROS are often activated by endogenous or exogenous stress. Given the ancient origin of the present day eukaryotic mitochondria, it is tempting to speculate that the persistently observed release of mitochondrial oxidants is not some evolutionary oversight. Rather, perhaps these data are telling us that mitochondria should be viewed as more than just autonomous factories that produce ATP. Much like the canary in the mine, these organelles may also serve as a constant sentry to warn us of impending trouble. In such a scenario, multiple diverse stresses would converge on the mitochondria, culminating in the release of ROS (Fig. 2). The release of oxidants would be interpreted as a signal that a stress had been encountered, with the intensity or duration of ROS release a potential determining factor in the ultimate biological outcome. Low intensity ROS production may be important in metabolic adaptation such as seen with nutrient excess or under conditions of low oxygen as discussed. Moderate ROS production stimulated by endogenous or exogenous danger signals might be involved in regulating inflammatory mediators. Finally, high level ROS production might signal the induction of pathways such as apoptosis or autophagy capable of inducing cell death. In each case, different redox-sensitive cytosolic pathways would be mobilized. Although many details remain to be elucidated, the preceding examples suggest that our initial view of mitochondrial ROS as being produced in an unregulated and unintentional fashion needs to reevaluated. The speed of recent discoveries suggests that such reevaluation is already under way.
FIGURE 2.
Mitochondrial oxidants as signaling molecules. Shown is a potential model in which the intensity of mitochondrial oxidant production generates a gradient of biological responses. See text for details.
Acknowledgment
I am grateful to I. Rovira for help in the preparation of this manuscript.
This work was supported, in whole or in part, by National Institutes of Health intramural funds. This work was also supported by the Ellison Medical Foundation. This is the sixth article in the Thematic Minireview Series on Redox Sensing and Regulation.
- ROS
- reactive oxygen species
- HIF-1α
- hypoxia-inducible factor 1α
- HSC
- hematopoietic stem cell
- PGC-1α
- peroxisome proliferator-activated receptor-γ coactivator 1α.
REFERENCES
- 1. Chance B., Sies H., Boveris A. (1979) Hydroperoxide metabolism in mammalian organs. Physiol. Rev. 59, 527–605 [DOI] [PubMed] [Google Scholar]
- 2. Murphy M. P. (2009) How mitochondria produce reactive oxygen species. Biochem. J. 417, 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Brand M. D. (2010) The sites and topology of mitochondrial superoxide production. Exp. Gerontol. 45, 466–472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Nemoto S., Takeda K., Yu Z. X., Ferrans V. J., Finkel T. (2000) Role for mitochondrial oxidants as regulators of cellular metabolism. Mol. Cell. Biol. 20, 7311–7318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Pérez-Matute P., Zulet M. A., Martínez J. A. (2009) Reactive species and diabetes: counteracting oxidative stress to improve health. Curr. Opin. Pharmacol. 9, 771–779 [DOI] [PubMed] [Google Scholar]
- 6. Tiganis T. (2011) Reactive oxygen species and insulin resistance: the good, the bad and the ugly. Trends Pharmacol. Sci. 32, 82–89 [DOI] [PubMed] [Google Scholar]
- 7. Kamata H., Honda S., Maeda S., Chang L., Hirata H., Karin M. (2005) Reactive oxygen species promote TNF-α-induced death and sustained JNK activation by inhibiting MAP kinase phosphatases. Cell 120, 649–661 [DOI] [PubMed] [Google Scholar]
- 8. Murphy M. P., Holmgren A., Larsson N. G., Halliwell B., Chang C. J., Kalyanaraman B., Rhee S. G., Thornalley P. J., Partridge L., Gems D., Nyström T., Belousov V., Schumacker P. T., Winterbourn C. C. (2011) Unraveling the biological roles of reactive oxygen species. Cell Metab. 13, 361–366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chandel N. S., Maltepe E., Goldwasser E., Mathieu C. E., Simon M. C., Schumacker P. T. (1998) Mitochondrial reactive oxygen species trigger hypoxia-induced transcription. Proc. Natl. Acad. Sci. U.S.A. 95, 11715–11720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Vaux E. C., Metzen E., Yeates K. M., Ratcliffe P. J. (2001) Regulation of hypoxia-inducible factor is preserved in the absence of a functioning mitochondrial respiratory chain. Blood 98, 296–302 [DOI] [PubMed] [Google Scholar]
- 11. Srinivas V., Leshchinsky I., Sang N., King M. P., Minchenko A., Caro J. (2001) Oxygen sensing and HIF-1 activation do not require an active mitochondrial respiratory chain electron transfer pathway. J. Biol. Chem. 276, 21995–21998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Guzy R. D., Hoyos B., Robin E., Chen H., Liu L., Mansfield K. D., Simon M. C., Hammerling U., Schumacker P. T. (2005) Mitochondrial complex III is required for hypoxia-induced ROS production and cellular oxygen sensing. Cell Metab. 1, 401–408 [DOI] [PubMed] [Google Scholar]
- 13. Brunelle J. K., Bell E. L., Quesada N. M., Vercauteren K., Tiranti V., Zeviani M., Scarpulla R. C., Chandel N. S. (2005) Oxygen sensing requires mitochondrial ROS but not oxidative phosphorylation. Cell Metab. 1, 409–414 [DOI] [PubMed] [Google Scholar]
- 14. Mansfield K. D., Guzy R. D., Pan Y., Young R. M., Cash T. P., Schumacker P. T., Simon M. C. (2005) Mitochondrial dysfunction resulting from loss of cytochrome c impairs cellular oxygen sensing and hypoxic HIF-α activation. Cell Metab. 1, 393–399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kim J. W., Tchernyshyov I., Semenza G. L., Dang C. V. (2006) HIF-1-mediated expression of pyruvate dehydrogenase kinase: a metabolic switch required for cellular adaptation to hypoxia. Cell Metab. 3, 177–185 [DOI] [PubMed] [Google Scholar]
- 16. Semenza G. L., Jiang B. H., Leung S. W., Passantino R., Concordet J. P., Maire P., Giallongo A. (1996) Hypoxia response elements in the aldolase A, enolase 1, and lactate dehydrogenase A gene promoters contain essential binding sites for hypoxia-inducible factor 1. J. Biol. Chem. 271, 32529–32537 [DOI] [PubMed] [Google Scholar]
- 17. Fukuda R., Zhang H., Kim J. W., Shimoda L., Dang C. V., Semenza G. L. (2007) HIF-1 regulates cytochrome oxidase subunits to optimize efficiency of respiration in hypoxic cells. Cell 129, 111–122 [DOI] [PubMed] [Google Scholar]
- 18. Chan S. Y., Zhang Y. Y., Hemann C., Mahoney C. E., Zweier J. L., Loscalzo J. (2009) MicroRNA-210 controls mitochondrial metabolism during hypoxia by repressing the iron-sulfur cluster assembly proteins ISCU1/2. Cell Metab. 10, 273–284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Baysal B. E., Ferrell R. E., Willett-Brozick J. E., Lawrence E. C., Myssiorek D., Bosch A., van der Mey A., Taschner P. E., Rubinstein W. S., Myers E. N., Richard C. W., 3rd, Cornelisse C. J., Devilee P., Devlin B. (2000) Mutations in SDHD, a mitochondrial complex II gene, in hereditary paraganglioma. Science 287, 848–851 [DOI] [PubMed] [Google Scholar]
- 20. Pollard P. J., Brière J. J., Alam N. A., Barwell J., Barclay E., Wortham N. C., Hunt T., Mitchell M., Olpin S., Moat S. J., Hargreaves I. P., Heales S. J., Chung Y. L., Griffiths J. R., Dalgleish A., McGrath J. A., Gleeson M. J., Hodgson S. V., Poulsom R., Rustin P., Tomlinson I. P. (2005) Accumulation of Krebs cycle intermediates and overexpression of HIF-1α in tumors which result from germ line FH and SDH mutations. Hum. Mol. Genet. 14, 2231–2239 [DOI] [PubMed] [Google Scholar]
- 21. Selak M. A., Armour S. M., MacKenzie E. D., Boulahbel H., Watson D. G., Mansfield K. D., Pan Y., Simon M. C., Thompson C. B., Gottlieb E. (2005) Succinate links TCA cycle dysfunction to oncogenesis by inhibiting HIF-α prolyl hydroxylase. Cancer Cell 7, 77–85 [DOI] [PubMed] [Google Scholar]
- 22. Guzy R. D., Sharma B., Bell E., Chandel N. S., Schumacker P. T. (2008) Loss of the SdhB, but not the SdhA, subunit of complex II triggers reactive oxygen species-dependent hypoxia-inducible factor activation and tumorigenesis. Mol. Cell. Biol. 28, 718–731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Tschopp J., Schroder K. (2010) NLRP3 inflammasome activation: the convergence of multiple signaling pathways on ROS production? Nat. Rev. Immunol. 10, 210–215 [DOI] [PubMed] [Google Scholar]
- 24. Zhou R., Tardivel A., Thorens B., Choi I., Tschopp J. (2010) Thioredoxin-interacting protein links oxidative stress to inflammasome activation. Nat. Immunol. 11, 136–140 [DOI] [PubMed] [Google Scholar]
- 25. Bulua A. C., Simon A., Maddipati R., Pelletier M., Park H., Kim K. Y., Sack M. N., Kastner D. L., Siegel R. M. (2011) Mitochondrial reactive oxygen species promote production of proinflammatory cytokines and are elevated in TNFR1-associated periodic syndrome (TRAPS). J. Exp. Med. 208, 519–533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Zhou R., Yazdi A. S., Menu P., Tschopp J. (2011) A role for mitochondria in NLRP3 inflammasome activation. Nature 469, 221–225 [DOI] [PubMed] [Google Scholar]
- 27. Nakahira K., Haspel J. A., Rathinam V. A., Lee S. J., Dolinay T., Lam H. C., Englert J. A., Rabinovitch M., Cernadas M., Kim H. P., Fitzgerald K. A., Ryter S. W., Choi A. M. (2011) Autophagy proteins regulate innate immune responses by inhibiting the release of mitochondrial DNA mediated by the NALP3 inflammasome. Nat. Immunol. 12, 222–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. West A. P., Brodsky I. E., Rahner C., Woo D. K., Erdjument-Bromage H., Tempst P., Walsh M. C., Choi Y., Shadel G. S., Ghosh S. (2011) TLR signaling augments macrophage bactericidal activity through mitochondrial ROS. Nature 472, 476–480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Scherz-Shouval R., Elazar Z. (2011) Regulation of autophagy by ROS: physiology and pathology. Trends Biochem. Sci. 36, 30–38 [DOI] [PubMed] [Google Scholar]
- 30. Wu J. J., Quijano C., Chen E., Liu H., Cao L., Fergusson M. M., Rovira I. I., Gutkind S., Daniels M. P., Komatsu M., Finkel T. (2009) Mitochondrial dysfunction and oxidative stress mediate the physiological impairment induced by the disruption of autophagy. Aging 1, 425–437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Mathew R., Karp C. M., Beaudoin B., Vuong N., Chen G., Chen H. Y., Bray K., Reddy A., Bhanot G., Gelinas C., Dipaola R. S., Karantza-Wadsworth V., White E. (2009) Autophagy suppresses tumorigenesis through elimination of p62. Cell 137, 1062–1075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Scherz-Shouval R., Shvets E., Fass E., Shorer H., Gil L., Elazar Z. (2007) Reactive oxygen species are essential for autophagy and specifically regulate the activity of Atg4. EMBO J. 26, 1749–1760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Yu L., Wan F., Dutta S., Welsh S., Liu Z., Freundt E., Baehrecke E. H., Lenardo M. (2006) Autophagic programmed cell death by selective catalase degradation. Proc. Natl. Acad. Sci. U.S.A. 103, 4952–4957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Narendra D., Tanaka A., Suen D. F., Youle R. J. (2008) Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J. Cell Biol. 183, 795–803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Geisler S., Holmström K. M., Skujat D., Fiesel F. C., Rothfuss O. C., Kahle P. J., Springer W. (2010) PINK1/parkin-mediated mitophagy is dependent on VDAC1 and p62/SQSTM1. Nat. Cell Biol. 12, 119–131 [DOI] [PubMed] [Google Scholar]
- 36. Irrcher I., Aleyasin H., Seifert E. L., Hewitt S. J., Chhabra S., Phillips M., Lutz A. K., Rousseaux M. W., Bevilacqua L., Jahani-Asl A., Callaghan S., MacLaurin J. G., Winklhofer K. F., Rizzu P., Rippstein P., Kim R. H., Chen C. X., Fon E. A., Slack R. S., Harper M. E., McBride H. M., Mak T. W., Park D. S. (2010) Loss of the Parkinson disease-linked gene DJ-1 perturbs mitochondrial dynamics. Hum. Mol. Genet. 19, 3734–3746 [DOI] [PubMed] [Google Scholar]
- 37. Andres-Mateos E., Perier C., Zhang L., Blanchard-Fillion B., Greco T. M., Thomas B., Ko H. S., Sasaki M., Ischiropoulos H., Przedborski S., Dawson T. M., Dawson V. L. (2007) DJ-1 gene deletion reveals that DJ-1 is an atypical peroxiredoxin-like peroxidase. Proc. Natl. Acad. Sci. U.S.A. 104, 14807–14812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Yang Y., Gehrke S., Haque M. E., Imai Y., Kosek J., Yang L., Beal M. F., Nishimura I., Wakamatsu K., Ito S., Takahashi R., Lu B. (2005) Inactivation of Drosophila DJ-1 leads to impairments of oxidative stress response and phosphatidylinositol 3-kinase/Akt signaling. Proc. Natl. Acad. Sci. U.S.A. 102, 13670–13675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Canet-Avilés R. M., Wilson M. A., Miller D. W., Ahmad R., McLendon C., Bandyopadhyay S., Baptista M. J., Ringe D., Petsko G. A., Cookson M. R. (2004) The Parkinson disease protein DJ-1 is neuroprotective due to cysteine sulfinic acid-driven mitochondrial localization. Proc. Natl. Acad. Sci. U.S.A. 101, 9103–9108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Meulener M. C., Xu K., Thomson L., Ischiropoulos H., Bonini N. M. (2006) Mutational analysis of DJ-1 in Drosophila implicates functional inactivation by oxidative damage and aging. Proc. Natl. Acad. Sci. U.S.A. 103, 12517–12522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Harman D. (1956) Aging: a theory based on free radical and radiation chemistry. J Gerontol. 11, 298–300 [DOI] [PubMed] [Google Scholar]
- 42. Balaban R. S., Nemoto S., Finkel T. (2005) Mitochondria, oxidants, and aging. Cell 120, 483–495 [DOI] [PubMed] [Google Scholar]
- 43. Perry J. J., Tainer J. A. (2011) All stressed out without ATM kinase. Sci. Signal. 4, pe18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Eaton J. S., Lin Z. P., Sartorelli A. C., Bonawitz N. D., Shadel G. S. (2007) Ataxia telangiectasia mutated kinase regulates ribonucleotide reductase and mitochondrial homeostasis. J. Clin. Invest. 117, 2723–2734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. GoDARTS and UKPDS Diabetes Pharmacogenetics Study Group, Wellcome Trust Case Control Consortium 2, Zhou K., Bellenguez C., Spencer C. C., Bennett A. J., Coleman R. L., Tavendale R., Hawley S. A., Donnelly L. A., Schofield C., Groves C. J., Burch L., Carr F., Strange A., Freeman C., Blackwell J. M., Bramon E., Brown M. A., Casas J. P., Corvin A., Craddock N., Deloukas P., Dronov S., Duncanson A., Edkins S., Gray E., Hunt S., Jankowski J., Langford C., Markus H. S., Mathew C. G., Plomin R., Rautanen A., Sawcer S. J., Samani N. J., Trembath R., Viswanathan A. C., Wood N. W., MAGIC investigators, Harries L. W., Hattersley A. T., Doney A. S., Colhoun H., Morris A. D., Sutherland C., Hardie D. G., Peltonen L., McCarthy M. I., Holman R. R., Palmer C. N., Donnelly P., Pearson E. R. (2011) Common variants near ATM are associated with glycemic response to metformin in type 2 diabetes. Nat. Genet. 43, 117–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Cosentino C., Grieco D., Costanzo V. (2011) ATM activates the pentose phosphate pathway promoting antioxidant defense and DNA repair. EMBO J. 30, 546–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Ito K., Hirao A., Arai F., Matsuoka S., Takubo K., Hamaguchi I., Nomiyama K., Hosokawa K., Sakurada K., Nakagata N., Ikeda Y., Mak T. W., Suda T. (2004) Regulation of oxidative stress by ATM is required for self-renewal of hematopoietic stem cells. Nature 431, 997–1002 [DOI] [PubMed] [Google Scholar]
- 48. Ito K., Hirao A., Arai F., Takubo K., Matsuoka S., Miyamoto K., Ohmura M., Naka K., Hosokawa K., Ikeda Y., Suda T. (2006) Reactive oxygen species act through p38 MAPK to limit the life span of hematopoietic stem cells. Nat. Med. 12, 446–451 [DOI] [PubMed] [Google Scholar]
- 49. Tothova Z., Kollipara R., Huntly B. J., Lee B. H., Castrillon D. H., Cullen D. E., McDowell E. P., Lazo-Kallanian S., Williams I. R., Sears C., Armstrong S. A., Passegué E., DePinho R. A., Gilliland D. G. (2007) FoxOs are critical mediators of hematopoietic stem cell resistance to physiologic oxidative stress. Cell 128, 325–339 [DOI] [PubMed] [Google Scholar]
- 50. Liu J., Cao L., Chen J., Song S., Lee I. H., Quijano C., Liu H., Keyvanfar K., Chen H., Cao L. Y., Ahn B. H., Kumar N. G., Rovira I. I., Xu X. L., van Lohuizen M., Motoyama N., Deng C. X., Finkel T. (2009) Bmi1 regulates mitochondrial function and the DNA damage response pathway. Nature 459, 387–392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Gan B., Hu J., Jiang S., Liu Y., Sahin E., Zhuang L., Fletcher-Sananikone E., Colla S., Wang Y. A., Chin L., DePinho R. A. (2010) Lkb1 regulates quiescence and metabolic homeostasis of hematopoietic stem cells. Nature 468, 701–704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Gurumurthy S., Xie S. Z., Alagesan B., Kim J., Yusuf R. Z., Saez B., Tzatsos A., Ozsolak F., Milos P., Ferrari F., Park P. J., Shirihai O. S., Scadden D. T., Bardeesy N. (2010) The Lkb1 metabolic sensor maintains hematopoietic stem cell survival. Nature 468, 659–663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Nakada D., Saunders T. L., Morrison S. J. (2010) Lkb1 regulates cell cycle and energy metabolism in hematopoietic stem cells. Nature 468, 653–658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Sahin E., Colla S., Liesa M., Moslehi J., Müller F. L., Guo M., Cooper M., Kotton D., Fabian A. J., Walkey C., Maser R. S., Tonon G., Foerster F., Xiong R., Wang Y. A., Shukla S. A., Jaskelioff M., Martin E. S., Heffernan T. P., Protopopov A., Ivanova E., Mahoney J. E., Kost-Alimova M., Perry S. R., Bronson R., Liao R., Mulligan R., Shirihai O. S., Chin L., DePinho R. A. (2011) Telomere dysfunction induces metabolic and mitochondrial compromise. Nature 470, 359–365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Owusu-Ansah E., Banerjee U. (2009) Reactive oxygen species prime Drosophila hematopoietic progenitors for differentiation. Nature 461, 537–541 [DOI] [PMC free article] [PubMed] [Google Scholar]