Background: The role of endoplasmic reticulum (ER) stress in influenza A viral infection is unknown.
Results: Influenza A virus induces the IRE1 pathway of the ER stress response. Inhibition of IRE1 activity leads to decreased viral replication.
Conclusion: IRE1 is a potential therapeutic target for influenza A virus.
Significance: Targeting a host molecular mechanism is a novel therapeutic strategy that is less likely to be invalidated by viral mutagenesis.
Keywords: Endoplasmic Reticulum Stress, Epithelial Cell, Influenza Virus, Molecular Chaperone, Unfolded Protein Response, Tauroursodeoxycholic Acid
Abstract
Known therapies for influenza A virus infection are complicated by the frequent emergence of resistance. A therapeutic strategy that may escape viral resistance is targeting host cellular mechanisms involved in viral replication and pathogenesis. The endoplasmic reticulum (ER) stress response, also known as the unfolded protein response (UPR), is a primitive, evolutionary conserved molecular signaling cascade that has been implicated in multiple biological phenomena including innate immunity and the pathogenesis of certain viral infections. We investigated the effect of influenza A viral infection on ER stress pathways in lung epithelial cells. Influenza A virus induced ER stress in a pathway-specific manner. We showed that the virus activates the IRE1 pathway with little or no concomitant activation of the PERK and the ATF6 pathways. When we examined the effects of modulating the ER stress response on the virus, we found that the molecular chaperone tauroursodeoxycholic acid (TUDCA) significantly inhibits influenza A viral replication. In addition, a specific inhibitor of the IRE1 pathway also blocked viral replication. Our findings constitute the first evidence that ER stress plays a role in the pathogenesis of influenza A viral infection. Decreasing viral replication by modulating the host ER stress response is a novel strategy that has important therapeutic implications.
Introduction
Influenza A virus has been causing recurrent pandemics for centuries and continues to be a global health threat with a major economic burden (1). In the United States, seasonal influenza is estimated to cause 36,000 deaths and 226,000 hospitalizations annually. The annual Influenza virus epidemics are estimated to cost 10.4 billion in direct medical expenses and 16.4 billion in lost potential earnings (2, 3). Furthermore antigenic shift continues to cause recurrent pandemics with a devastating death toll. More than 40 million people died during the 1918 pandemic outnumbering the death toll of World War 1 (4, 5). With emergence of strains resistant to pharmacologic therapies, yearly vaccination remains the only available strategy for influenza A virus. The limited ability to produce large amounts of vaccines in a short time period represents a problem at times of pandemics. Therefore further understanding of host cellular mechanisms involved in the pathogenesis of influenza A viral infection and identification of therapeutic targets in the host are needed for the development of new treatments less susceptible to resistance.
The endoplasmic reticulum stress response, also known as the unfolded protein response (UPR),2 was originally discovered as an evolutionary conserved molecular signaling cascade the main function of which is to restore ER homeostasis and protein folding capacity during ER stress. Over the last decade, the growing body of knowledge about the UPR revealed a broader range of effects implicating it in multiple other cellular and disease processes including apoptosis, inflammation, and metabolism (6–12). While ER stress has been shown to be involved in the pathogenesis of other viruses (13), its role in influenza A viral infection is unknown.
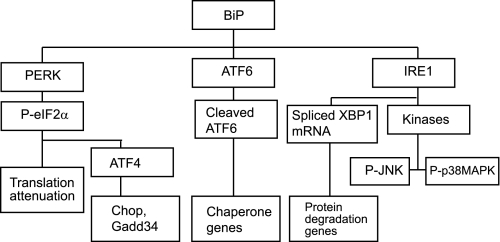
The adaptive effects of the UPR can be classified into three categories: (i) increasing protein folding capacity of the ER by transcriptional up-regulation of chaperone proteins, (ii) decreasing ER protein load by attenuation of global protein translation, (iii) increasing proteasome-mediated degradation of misfolded proteins. In addition to its adaptive effects, if ER stress is severe or prolonged, the UPR is also known to mediate apoptotic signals to protect the organism from necrotic cell death and inflammation. The factors that determine the balance between the adaptive versus apoptotic effects are still not well understood (14). The upstream mediators of the UPR are three ER resident transmembrane proteins, activating transcription factor 6 (ATF6), PKR-like ER kinase (PERK), and inositol-requiring enzyme 1 (IRE1), usually held inactive by binding immunoglobulin protein (BiP) at their luminal N-terminal side. BiP is a major chaperone protein and is considered the master regulator of the UPR (7). During ER stress BiP is released from ATF6, PERK, and IRE1 because of competitive binding to the increasing levels of mis-folded proteins and thus allowing the activation of the UPR (Fig. 1). When released from BIP, ATF6 translocates to the Golgi where it gets cleaved by resident proteases. Cleaved ATF6 functions as a transcription factor for chaperone genes. PERK and IRE1 homodimerize, when released from BIP, which induces their auto-phosphorylation and activation. PERK is a serine/threonine kinase that phosphorylates and inactivates eIF2α (eukaryotic translation initiation factor 2α). Phosphorylation of eIF2α induces global shut down of protein translation. Certain mRNAs, for example activating transcription factor 4 (ATF4) and BiP, escape that inhibition and gain a translational advantage (7). The third ER stress regulator, IRE1, has an endoribonuclease domain as well as a kinase domain. The endonuclease activity induces splicing of a 26-base intron in the XBP1 mRNA leading to a reading frameshift and translation into an active transcription factor for genes involved in ER-associated degradation (ERAD). The downstream effects of the IRE1 kinase function include phosphorylation of JNK and p38 MAP kinases, both of which are implicated in mediating some of the apoptotic effects of the UPR (7, 14, 15) (Fig. 1).
FIGURE 1.
Simplified diagram of the unfolded protein response.
ER stress is induced in the setting of certain viral infections such as hepatitis B virus, Japanese encephalitis virus, Enterovirus 71, and Moloney murine leukemia virus (MoMuLV)-ts1 (16–19). In some instances it plays a role in their pathogenesis. For instance, Japanese encephalitis virus, bovine diarrhea virus, tula virus, severe acute respiratory syndrome coronavirus (SARS-CoV), and West Nile virus have all been shown to induce their apoptotic effects through the UPR (13, 17, 20–22). Oxidative stress in the setting of hepatitis C infection has been shown to be mediated by ER stress (23). Certain viruses have been shown to modulate the ER stress response or preferentially activate the different pathways of the UPR. Hepatitic C virus induces the ATF6 pathway while blocking the IRE1 pathway, while hepatitis B virus induces ATF6 and IRE1 but not PERK (13, 16, 24). Herpes simplex virus 1 is thought to have evolutionary developed a virulence factor enabling dephosphorylation of eIF2α, one of the downstream effectors of the PERK pathway (25, 26).
In light of the growing evidence of diverse interactions between viral infections and ER stress we investigated the effects of influenza A viral infection on the different pathways of the UPR and the potential role of ER stress in viral replication.
EXPERIMENTAL PROCEDURES
Reagents
Tunicamycin (#654380) and tauroursodeoxycholic acid (#580549) were purchased from Calbiochem. 3,5-Dibromosalicylaldehyde was purchased from Sigma Aldrich (#122130). Abs used in this study were obtained from a variety of sources. BIP Ab (cs-3183), PERK Ab (cs-13073), ph-p38MAP kinase Ab (cs-9211), and ph-eIF2α Ab (cs-9721) were obtained from Cell Signaling Biotechnology (Beverly, MA). p38MAP kinase Ab (sc-7149), JNK Ab (sc-474), HRP-conjugated Abs anti-rabbit (sc-2004), and isotype control IgG Abs mouse (sc-2025) were obtained from Santa Cruz Biotechnology (Santa Cruz, CA). ph-JNK Ab (#559309) was obtained from Calbiochem. Influenza NP mouse Ab (MCA400) was obtained from AbD Serotec. Influenza NS1 mouse Ab was obtained from Dr. Jonathan Yewdell at the National Institute of Allergy and Infectious Diseases (Bethesda, MD). Influenza M1 mouse Ab (MA1–34775) was obtained from Thermo Scientific (Rockford, IL).
Human Tracheobronchial Epithelial Cells
Human tracheobronchial epithelial (HTBE) cells were obtained from Dr. Joseph Zabner and the Cell Culture Core under a protocol approved by the University of Iowa Institutional Review Board. Epithelial cells were isolated from tracheal and bronchial mucosa by enzymatic dissociation and cultured in Laboratory of Human Carcinogenesis (LHC)-8e medium on plates coated with collagen/albumin for study up to passage 10 (27, 28). All of the experiments were conducted using cells from at least three different donors. For infection, cells at 50–80% confluence were treated with Influenza A strain PR/8/34 (H1,N1) (MOI 1). The initial stock (105 cfu/μl) was aliquoted and kept frozen at −80 °C, and a fresh aliquot was thawed for each experiment.
Cellular Protein
Whole-cell protein extracts were prepared by lysis of cell monolayers in 200 ml of lysis buffer (50 mm Tris, pH 7.4, 150 mm NaCl, and 1% Nonidet P-40), a protease inhibitor mixture (Roche Applied Science, Indianapolis, IN), and a phosphatase inhibitor mixture (no. 524625; Calbiochem). The lysates were sonicated for 20 s and kept at 4 °C for 30 min. After 5 min of centrifugation (14,000 rpm at 4 °C), the supernatant was saved as a whole-cell lysate. Protein determinations were made using a protein measurement kit (Bradford protein assay, no. 500-0006) from Bio-Rad. Extracts were stored at −80 °C.
Immunoblot Analysis
Protein (25–30 mg) was mixed 1:1 with 23 sample buffer (20% glycerol, 4% SDS, 10% 2-ME, 0.05% bromphenol blue, and 1.25 m Tris, pH 6.8), loaded onto a 12 or 15% (ISG15) SDS-PAGE gel, and run at 150 V for 90 min. Cell proteins were transferred to an Immuno-Blot polyvinylidene difluoride membrane (Bio-Rad) with a Bio-Rad semidry transfer system according to the manufacturer's instructions. The polyvinylidene difluoride membrane was then incubated with primary Ab (dilutions 1:500–1:1000) in 5% milk in Tris-buffered saline with 0.1% Tween 20 overnight. The blots were washed three times with Tris-buffered saline with 0.1% Tween 20 and incubated for 1 h with HRP-conjugated secondary anti-IgG Ab (dilution 1:2000–1:20,000). The blots were washed again three times with Tris-buffered saline with 0.1% Tween 20, and immunoreactive bands were developed using the chemiluminescent substrate ECL Plus (Amersham Biosciences, Piscataway, NJ). An autoradiograph was obtained with exposure times of 10 s to 2 min. Equal loading of proteins was confirmed with HRP-conjugated GAPDH (ab9482; Abcam, Cambridge, MA) (Ab dilution 1:20,000).
RNA Isolation
RNA was isolated from HTBEs using a kit, MirVana, (Applied Biosystems, Austin, TX) reagents according to manufacturer's instructions. After preparation, RNA samples were stored in a −80 °C freezer until use. RNA quality and quantity were assessed with an Experion Automated Electrophoresis System (Bio-Rad) using the Experion RNA StdSens Analysis Kit according to the manufacturer's protocol. RNA quality was considered adequate for use if the 28 S/18 S ratio was >1.2, and the RNA quality indicator was >7.
Real Time RT-PCR for mRNA Quantitation
Total RNA (1 μg) was reverse-transcribed to cDNA using the iScript cDNA Synthesis Kit (Bio-Rad) following the manufacturer's instructions. In the case of viral cDNA synthesis we used the SuperScript III First-Strand Synthesis System (#18080-051) from Invitrogen and the Uni-12 primer AGCAAAAGCAGG (29). PCR reactions were performed using 2 μl of cDNA and 48 μl of master mix, containing iQ SYBR Green Supermix (Bio-Rad), 15 pmol of forward primer, and 15 pmol of reverse primer, in a CFX96 Real-Time PCR Detection System (Bio-Rad) as follows: 3 min at 95 °C followed by 40 cycles of 10 s at 95 °C and 30 s at 55 °C and plate read. The fluorescence signal generated with SYBR Green I DNA dye was measured during annealing steps. Specificity of the amplification was confirmed using a melting curve analysis. Data were collected and recorded by CFX Manager Software (Bio-Rad) and expressed as a function of threshold cycle (CT). The relative quantity of the gene of interest was then normalized to the relative quantity of hypoxanthine phosphoribosyltransferase (ΔΔCT). The sample mRNA abundance was calculated by the equation 2−( ΔΔCT). Gene-specific primers were custom-synthesized and purchased from Integrated DNA Technologies (Iowa City, IA) based on design using gene- specific nucleotide sequences from the National Center for Biotechnology Information sequence databases and the PrimerQuest Web interface (Integrated DNA Technologies).
XBP1 mRNA Splicing Assay
To determine the degree of XBP1 mRNA splicing, total cellular RNA was isolated as described above. Then total XBP1 cDNA was synthesized and amplified using the Superscript III One Step RT PCR with platinum TaqDNA polymerase kit (#12574-018) (Invitrogen) following the manufacturer's instructions. To amplify both the spliced and the unspliced variants of XBP1 cDNA, we used forward and reverse primers that circumvent the spliced segment. The PCR product was then separated by gel electrophoresis using a 12% acrylamide gel. Staining with a SYBR green fluorescent dye was then performed for visualization of the separated DNA bands.
Influenza Measurements
Viral titers were determined as previously described (30, 31) by end point dilution assay and expressed as 50% tissue culture infectious dose (TCID50). Briefly, 10-fold serial dilutions of the supernatant from influenza virus-infected cells were mixed with 5 × 105 Madin-Darby canine kidney (MDCK) cells in DMEM and incubated at 37 °C for 24 h. Culture supernatants were removed, and DMEM containing 0.0002% l-1-(tosylamido-2phenyl) ethyl chloromethyl ketone- treated trypsin (Worthington Diagnostics) and penicillin (100 units/ml)/Streptomycin (100 mg/ml) was added to each well. Following 4 days of incubation at 37 °C, supernatants were mixed with an equal volume of 0.5% chicken RBC, the agglutination pattern was read, and the TCID50 values were calculated using Reed-Muench accumulative method.
Scanning Electron Microscopy
Cells were fixed with 2.5% glutaraldehyde in 0.1 m sodium cacodylate buffer (pH 7.2) for 30 min. Samples were kept in paraformaldehyde at 4 °C until used for microscopy. Samples were mounted and processed according to previously described techniques (32, 33) and viewed on a Hitachi S-4800 scanning electron microscope operating at 2 kV accelerating voltage.
Statistical Analysis
Two-tailed Student's t test was used for group comparisons. Statistical significance was determined based on p value of less than 0.05.
RESULTS
Influenza A/PR/8/34 Infects HTBEs
To detect differences in ER stress markers, we set up our infection model to infect the majority of the cells in the system. We infected confluent HTBE monolayer cell cultures with influenza A virus at increasing MOI. We determined the percent infectivity 12 h postinfection (h.p.i) by performing immunofluorescent microscopy using an anti-influenza NS1 antibody. NS1 is a nonstructural protein only expressed during an active influenza infection. At an MOI of 1, more than 80% of the cells stained positive for NS1 protein (data not shown). An MOI of 1 was chosen for all subsequent experiments. Efficient infection was confirmed by quantitative RT-PCR (q-RT-PCR) demonstrating a time dependent increase in viral M1 and NP RNA levels (data not shown).
Influenza A/PR/8/34 Infection Induces ER Stress in HTBE Cells
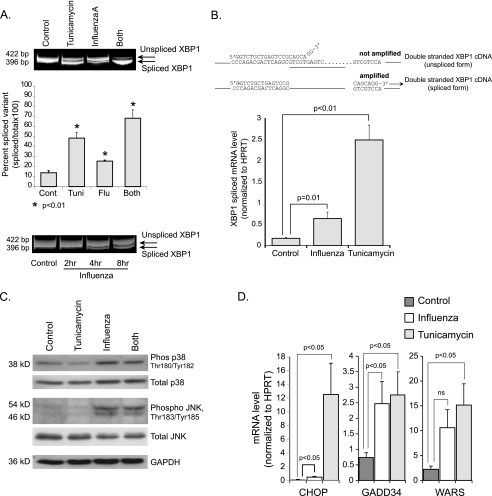
To investigate the effects of influenza A viral infection on the unfolded protein response, we tested activation of the three pathways activated by ER stress. We first examined the IRE1 pathway by using a number of different assays. As a positive control, we treated cells with the N-linked glycosylation inhibitor, tunicamycin, at 1 μg/ml. The endonuclease activity of IRE1 was assessed by detecting the degree of splicing of XBP1 mRNA, 12 h post-treatment. XBP1 splicing occurs quickly with IRE1 activation. This was performed by PCR amplification followed by gel electrophoresis to separate the amplified unspliced and spliced variants of XBP1 cDNA. Detailed description of this assay is outlined under “Experimental Procedures.” Influenza A virus infection induced significant splicing of XBP1 mRNA as compared with the noninfected HTBE controls (Fig. 2A). The degree of XBP1 mRNA splicing in response to influenza A virus infection was found to increase in a time-dependent manner (Fig. 2A). Notably, IRE1 is not known to be activated by any stimulus other than ER stress. We next determined the degree of splicing of XBP1 mRNA by q-RT-PCR using a primer previously validated to amplify only the spliced variant (34). The results also showed an increase in XBP1 mRNA splicing with influenza A virus infection (Fig. 2B).
FIGURE 2.
Influenza A viral infection induces the ER stress response. A, DNA gel showing spliced and unspliced XBP1 separated by electrophoresis following conventional PCR amplification. RNA was isolated from indicated groups at 12 h.p.i. Corresponding densitometry analysis from three independent experiments is shown. The same assay was performed on control and influenza A-infected HTBE cells at different time points (bottom). B, quantitative RT PCR from the indicated groups at a 12 h time point using a primer that amplifies only the spliced variant of XBP1 mRNA. C, Western blot analysis of ph-p38 and ph-JNK. Total cellular proteins were isolated from the indicated groups at 12 h.p.i. D, quantitative RT-PCR data showing induction of a number of ER stress genes by influenza A viral infection. RNA was isolated from the indicated groups of HTBE cells at 12 h post-treatment.
In addition to its endonuclease domain, IRE1 has a protein kinase domain that is one of many activators of JNK and p38 MAP kinase (Fig. 1). Immunoblot analysis of total cellular proteins, harvested 12 h.p.i, showed increased levels of both phosphorylated (activated) proteins (Fig. 2C). Taken together these results demonstrate activation of IRE1 by the influenza A viral infection.
We then examined the differential mRNA expression of genes downstream of the UPR in response to the viral infection and found that influenza A virus caused a significant induction of growth arrest and DNA-damage inducible protein 153 (GADD 153 or CHOP), GADD34, and tryptophanyl t-RNA synthetase (WARS) (Fig. 2D). Although not specific for any particular one of the three ER stress pathways, induction of these genes is suggestive of activation of the ER stress response by influenza A virus.
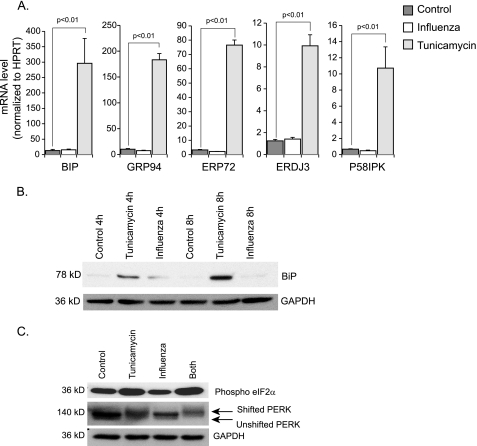
Having found strong evidence of IRE1 activation, we next asked if influenza A virus also activates the other two main ER stress pathways, PERK and ATF6. When we examined the mRNA expression of ER stress genes, chaperone genes including BIP, GRP94, ERDJ3, ERP72, and P58IPK were not induced by the infection (Fig. 3A). These genes are at least partially dependent on ATF6 for their full up-regulation (35, 36). This suggested that influenza A virus infection did not activate the ATF6 pathway. Consistent with the lack of mRNA induction, BIP protein expression, as detected by Western blot analysis, was not significantly enhanced by the infection at 4 and 8 h.p.i (Fig. 3B). The same was true at 12 h.p.i. (data not shown).
FIGURE 3.
Effect of influenza A viral infection on ATF6 and PERK pathways. A, quantitative RT-PCR data showing mRNA levels of different ER stress genes. RNA was isolated from control, influenza A infected, and tunicamycin-treated HTBE cells at a 12 h time point. Results are shown as averages of independent experiments with S.E. B, Western blot analysis for BiP. Total cellular proteins were isolated from similar groups as in A at 4 and 8 h time points. C, Western blot analysis of PERK and ph-eIF2α. Total cellular proteins were isolated from the indicated groups at 12 h.p.i.
To examine the PERK pathway, we looked for phosphorylation-induced changes in PERK molecular weight (a marker of activation). We performed Western blot analysis for PERK on control cells and cells treated with influenza A virus, tunicamycin, or both. A shift in migration of the band is an indication of PERK phosphorylation. The shift in migration was seen in the tunicamycin-treated cells but not with influenza A virus. Furthermore, the virus did not enhance the tunicamycin effect when cells were treated with both. Phosphorylation of eIF2α, a substrate of PERK, was not induced by the virus either (Fig. 3C). These data suggest that influenza A viral infection does not induce activation of PERK. However, and in light of CHOP and GADD34 induction, which we have shown earlier, there is the possibility that influenza A virus induces activation of PERK to a slight extent, detectable only in very sensitive downstream genes like CHOP and GADD34. Note that both these genes can be downstream of multiple pathways other than PERK. Taken together, our results suggest that influenza A viral infection leads to activation of a non-canonical UPR, with the IRE1/XBP1 axis strongly activated, but with little or no concomitant activation of the PERK and ATF6 pathways.
Influenza A/PR/8/34 Infection Modulates the UPR in HTBES
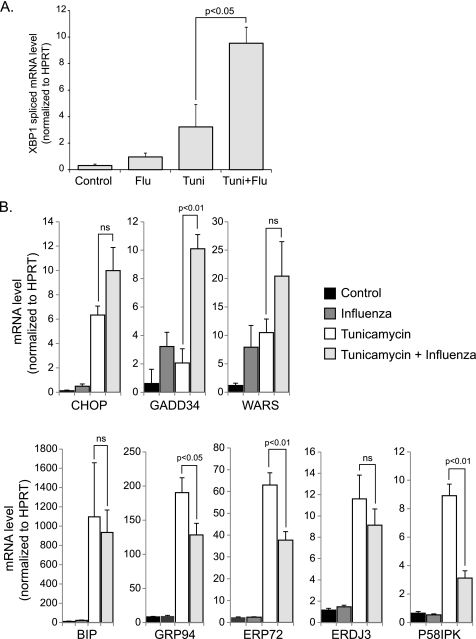
The differential activation of UPR pathways led us to investigate whether influenza A virus could modulate a pre-existing ER stress response. To test this hypothesis, we co-treated HTBE cells with both tunicamycin and influenza A virus then compared the ER stress response to tunicamycin treatment alone using q-RT-PCR of ER stress genes as well as spliced XBP1 mRNA. We found that influenza A virus infection potentiated some of the effects of tunicamycin, in particular splicing of XBP1 mRNA and induction of GADD34 (Fig. 4, A and B). Interestingly however, the virus inhibited the tunicamycin induction of the ATF6-driven genes Erdj3, Erp72, P58ipk, and GRP94 (Fig. 4B). These results suggest that influenza A virus modulates the ER stress response by decreasing the activation of the ATF6 pathway.
FIGURE 4.
Effects of influenza A viral infection on the ER stress response to tunicamycin. A, quantitative RT-PCR of spliced variant XBP1 mRNA at 12 h.p.i using the same primer set as in Fig. 2B. B, quantitative RT-PCR data of ER stress genes. RNA was isolated from the indicated groups at 12 h post-treatment. Results are shown as averages of independent experiments with S.E.
ER Stress Affects Influenza A Viral Replication
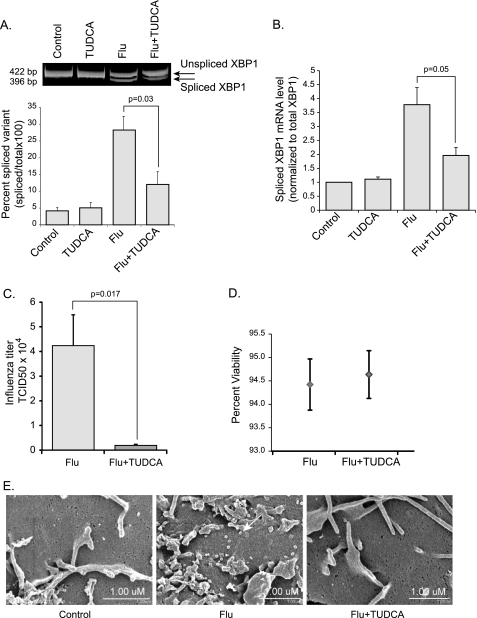
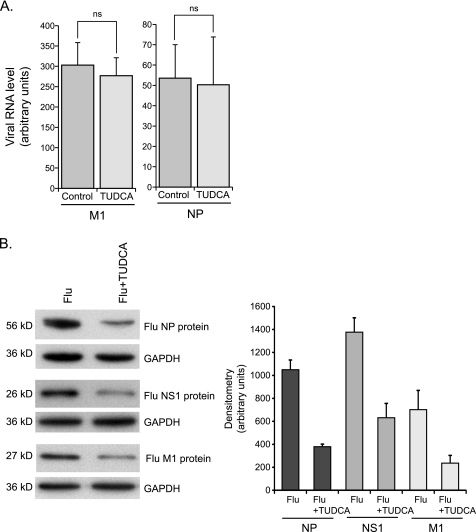
To further understand the role of the UPR in the pathogenesis of influenza A viral infection, we examined the effect of modulating the ER stress response on viral replication. Molecular and chemical chaperones are compounds known to alleviate ER stress by stabilizing proteins in their native conformation and therefore enhancing the ER folding capacity (37). A compound known to have these properties is the naturally occurring bile salt derivative, Tauroursodeoxycholic acid (TUDCA). TUDCA has been shown, in vitro and in vivo, to reduce ER stress in response to tunicamycin (11). We infected control HTBE cells and cells pretreated with TUDCA (500 μg/ml) for 16 h with influenza A virus. To verify that TUDCA is reducing the ER stress response induced by the virus we harvested total RNA 5 h.p.i. and evaluated the degree of XBP1 mRNA splicing as described earlier. We saw a significant decrease in XBP1 mRNA splicing in the TUDCA pretreated cells as compared with the control cells in response to the infection. Data were obtained from at least three independent experiments with cells from three different donors (Fig. 5, A and B). We infected TUDCA pretreated and control HTBE cells and measured viral titers in the supernatant 12 h.p.i using a modified MDCK infection assay with a hemagglutination readout. Surprisingly, viral titers were significantly lower in the TUDCA pretreated group indicating that TUDCA inhibited viral replication (Fig. 5C). To ensure the difference in viral titers was not due to differences in cell death between the groups, we performed a trypan blue cell viability assay. TUDCA pretreated and control HTBE cells were stained with trypan blue 12 h.p.i with influenza A virus. Percent cell viability was calculated and was unchanged between the groups (Fig. 5D). We next performed scanning electron microscopy to examine the influenza A virions on the surface of infected cells. At a magnification of ×35,000, the budding virions (∼80 nm) were visualized. There were significantly less virions on the surface of the TUDCA pretreated cells as compared with the naïve cells (Fig. 5E). To pinpoint the mechanism by which TUDCA altered viral replication, we examined levels of influenza genomic M1 and NP RNA as well as levels of three influenza proteins, M1, NP, and NS1. We found that while viral RNA replication did not change, the expression levels of viral proteins were markedly decreased when cells were pretreated with TUDCA (Fig. 6, A and B). This suggests that the inhibitory effect of TUDCA occurs at the viral protein synthesis or protein stability level. These results indicate that influenza A virus requires either impaired ER function, or IRE1 activation, or both, for efficient replication.
FIGURE 5.
Effects of modulating the ER stress response on influenza A viral replication. Naive HTBE cells and cells pretreated for 16 h with TUDCA (500 μg/ml) were infected with influenza A virus. A, degree of XBP1mRNA splicing was determined as described previously at 5 h.p.i. Corresponding densitometry analysis from three independent experiments is shown at the bottom. B, XBP1 mRNA splicing in the same groups was also measured using qRT-PCR. C, influenza A viral titers were measured in the supernatant of infected naïve and TUDCA-pretreated HTBE cells 12 h.p.i. D, trypan blue cell viability assay was performed on the same groups described in C. Results are shown as averages from independent experiments with S.E. E, scanning electron micrographs showing the surface of a control HTBE cell versus cells infected with influenza A virus (MOI of 1) with or without TUDCA pretreatment. Cells were fixed 12 h.p.i. The white arrow in the flu alone panel points at one of many influenza A virions on the surface of the naïve HTBE cell.
FIGURE 6.
TUDCA inhibits viral protein expression. A, quantitative RT-PCR of M1 and NP genomic viral RNA 12 h.p.i of control and TUDCA-pretreated cells. Results are shown as averages with S.E. B, Western blot analysis of influenza NP, NS1, and M1 proteins with corresponding densitometry displayed as averages with S.E. Proteins were isolated from independent experiments with similar groups as in A at 6 h.p.i.
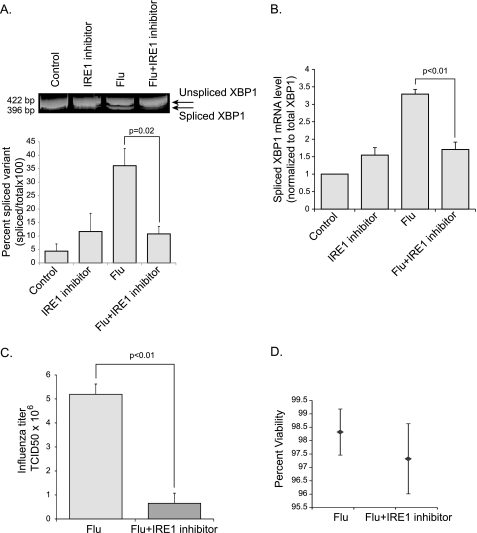
To test this hypothesis further, we examined the effects of a recently described selective inhibitor of the IRE1 endoribonuclease, 3,5-dibromosalicylaldehyde (IRE1 inhibitor) (38). Like TUDCA, the IRE1 inhibitor decreased the influenza A virus induced splicing of XBP1 mRNA (Fig. 7, A and B). Importantly, viral titers were inhibited when HTBE cells were co-treated with 3,5-dibromosalicylaldehyde (Fig. 7C). Cell viability was not affected by the IRE1 inhibitor treatment (Fig. 7D). Taken together these results indicate that activation of the IRE1 branch of the UPR is important for efficient influenza A viral replication.
FIGURE 7.
Effect of IRE1 inhibitor on influenza A viral replication. Control HTBE cells and cells co-treated with 3,5-dibromosalicylaldehyde (IRE1 inhibitor) (40 μm) were infected with influenza A virus. A, degree of XBP1mRNA splicing was determined as described previously at 5 h.p.i. Corresponding densitometry analysis from three independent experiments is shown at the bottom. B, XBP1 mRNA splicing in the same groups was also measured using qRT-PCR. C, influenza A viral titers were measured in the supernatant of control HTBE cells and cells co-treated with the IRE1 inhibitor by hemagglutination assay 8 h.p.i. D, trypan blue cell viability assay was performed on the same groups described in C. Results are shown as averages from independent experiments with S.E.
DISCUSSION
Over the last decade, several laboratories have reported the induction of ER stress in the setting of some viral infections. However, very little is known about the mechanisms by which ER stress is induced or about the role of the UPR in the pathogenesis of these viruses. Until our study, the role of ER stress in influenza A viral infection has been unknown. The only studies linking influenza infection to ER stress are ones showing that hemagglutinin A induces ER stress. In 1988 investigators showed that ER stress can be induced by overexpressing a mutated misfolded form of the influenza A hemagglutinin protein in simian cells (39). In a more recent study, overexpression of wild type influenza A hemagglutinin protein was shown to induce NF-κB activation. The mechanism proposed by the investigators was the induction of ER stress but it was not directly proven (40).
Our data constitute the first evidence that infection with wild type influenza A virus induces ER stress. Furthermore we showed that influenza A viral infection induces the ER stress response, in primary human airway epithelial cells, in a pathway-specific manner. The virus specifically induces the IRE1 branch of the UPR with little or no concomitant activation of the PERK and ATF6 pathways. We also showed that the virus modulates the stress response in the setting of a preexisting stress by decreasing the activation of the ATF6 pathway as indicated by its inhibitory effect on ATF6-driven genes.
A few studies have shown differential activation of the ER stress pathways by certain viruses. However, unlike influenza A, hepatitis C virus induces only the ATF6 pathway whereas hepatitis B virus induces both ATF6 and IRE1 but not PERK. The different effects on the ER stress pathways might be explained by the differences between the host cells these viruses infect, airway epithelial cells in the case of influenza and hepatocytes in the case of hepatitis B and C. Hepatocytes are specialized secretory cells that are actively synthesizing and secreting a significantly larger amount of proteins compared with airway epithelial cells and therefore are more sensitive to ER stress (41). In this case it is conceivable that up-regulation of chaperones is crucial for hepatitis B and C viral replication by preventing severe ER stress and eventual cell death. This could explain why these viruses have evolved mechanisms to preferentially induce the ATF6 pathway known to activate chaperone gene expression. In airway epithelial cells, the effects of a virus on ER stress are different.
Although the mechanism by which influenza A virus induces ER stress could be the accumulation of viral proteins in the ER lumen, the differential effects on the different pathways of the UPR are indicative of additional levels of interaction between the virus and the ER stress signaling cascade. To date the only known mechanism of activation of PERK, ATF6, and IRE1 is their release from BiP. The mechanism behind differential activation of the different arms of the UPR is unknown (14).
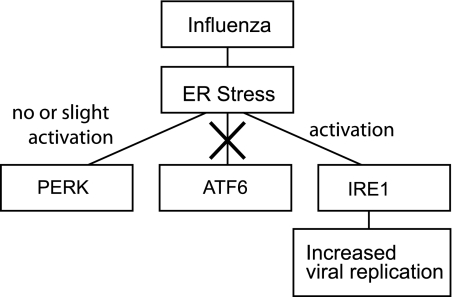
We showed that by modulating the ER stress response with the chemical chaperone TUDCA, viral replication was inhibited. This suggests that the UPR plays a facilitating role in the replication cycle of the virus. The combined effects of influenza A viral infection on the ER stress pathways seem to be aiming at optimal activation of IRE1 pathway. By inhibiting the ATF6 pathway, the activation of which is known to increase the ER folding capacity and to alleviate the ER stress response, the virus may be insuring optimal and sustained activation of IRE1. The inhibition of influenza A viral replication with TUDCA, which alleviates the ER stress response, suggests that one or more of the downstream effects of IRE1, the only ER stress pathway activated by the virus, is required for viral replication (Fig. 8). This was confirmed in our studies by the inhibition of viral replication with the use of the IRE1 inhibitor, 3,5-dibromosalicylaldehyde.
FIGURE 8.
Diagram summarizing the interactions between the ER stress pathways and influenza A viral infection.
The implication of the UPR in influenza A viral replication is a novel finding that could lead to new therapies for influenza A virus respiratory infection. By targeting a host cellular mechanism, such therapies are less susceptible to viral resistance mechanisms. We are interested in studying the effects of chemical chaperones, in particular TUDCA, on in vivo models of influenza A viral infection. We anticipate the effects to be more complex than merely inhibiting viral replication. The disease manifestations of influenza A viral infection are in part due to the host immune response and inflammatory cytokines (42). The UPR is known to intersect with cytokine release mechanisms through NF-κB activation (12). Therefore modulation of the ER stress response by chemical chaperones may cause a change in the cytokine milieu created by the infection and therefore a change in the disease phenotype. Furthermore, MHC class I expression on the cell surface, which is important in antigen presentation and in mediating T cell immunity, is dependent on the ER function (43–45). Improving the ER folding capacity with chemical chaperones may render dendritic cells more efficient in presenting viral antigens and therefore accelerating viral clearance. The combined effect of all these potential mechanisms is difficult to predict and an area of future study.
This work was supported, in whole or in part, by National Institutes of Health Grants R01 HL079901, R01 HL 096625, and R21 HL109589 (to M. M. M.) and NIDDK Grants R01 DK 084058 (to D. T. R.) and NCRR1 UL1 RR024979 from the National Center for Research Resources (NCRR), part of the National Institutes of Health.
- UPR
- unfolded protein response
- ER
- endoplasmic reticulum
- ERAD
- ER-associated degradation
- h.p.i.
- h postinfection
- TUDCA
- tauroursodeoxycholic acid
- HTBE
- human tracheobronchial epithelial.
REFERENCES
- 1. Rajagopal S., Treanor J. (2007) Pandemic (avian) influenza. Semin. Respir. Crit. Care Med. 28, 159–170 [DOI] [PubMed] [Google Scholar]
- 2. Fiore A. E., Shay D. K., Broder K., Iskander J. K., Uyeki T. M., Mootrey G., Bresee J. S., Cox N. S. (2008) Prevention and control of influenza: recommendations of the Advisory Committee on Immunization Practices (ACIP). MMWR Recomm. Rep. 57, 1–60 [PubMed] [Google Scholar]
- 3. Molinari N. A., Ortega-Sanchez I. R., Messonnier M. L., Thompson W. W., Wortley P. M., Weintraub E., Bridges C. B. (2007) The annual impact of seasonal influenza in the US: measuring disease burden and costs. Vaccine 25, 5086–5096 [DOI] [PubMed] [Google Scholar]
- 4. Johnson N. P., Mueller J. (2002) Updating the accounts: global mortality of the 1918–1920 “Spanish” influenza pandemic. Bull. Hist. Med. 76, 105–115 [DOI] [PubMed] [Google Scholar]
- 5. Taubenberger J. K., Morens D. M. (2006) 1918 Influenza: the mother of all pandemics. Emerg. Infect. Dis. 12, 15–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hotamisligil G. S. (2010) Endoplasmic reticulum stress and the inflammatory basis of metabolic disease. Cell 140, 900–917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Xu C., Bailly-Maitre B., Reed J. C. (2005) Endoplasmic reticulum stress: cell life and death decisions. J. Clin. Investig. 115, 2656–2664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Zhang K., Shen X., Wu J., Sakaki K., Saunders T., Rutkowski D. T., Back S. H., Kaufman R. J. (2006) Endoplasmic reticulum stress activates cleavage of CREBH to induce a systemic inflammatory response. Cell 124, 587–599 [DOI] [PubMed] [Google Scholar]
- 9. Gupta S., Cuffe L., Szegezdi E., Logue S. E., Neary C., Healy S., Samali A. (2010) Mechanisms of ER stress-mediated mitochondrial membrane permeabilization. Int. J. Cell Biol. 2010, 170215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Nakajima S., Hiramatsu N., Hayakawa K., Saito Y., Kato H., Huang T., Yao J., Paton A. W., Paton J. C., Kitamura M. (2011) Selective abrogation of BiP/GRP78 blunts activation of NF-κB through the ATF6 branch of the UPR: involvement of C/EBPβ and mTOR-dependent dephosphorylation of Akt. Mol. Cell. Biol. 31, 1710–1718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ozcan U., Yilmaz E., Ozcan L., Furuhashi M., Vaillancourt E., Smith R. O., Görgün C. Z., Hotamisligil G. S. (2006) Chemical chaperones reduce ER stress and restore glucose homeostasis in a mouse model of type 2 diabetes. Science 313, 1137–1140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Pahl H. L., Baeuerle P. A. (1996) Activation of NF-κB by ER stress requires both Ca2+ and reactive oxygen intermediates as messengers. FEBS Lett. 392, 129–136 [DOI] [PubMed] [Google Scholar]
- 13. He B. (2006) Viruses, endoplasmic reticulum stress, and interferon responses. Cell Death Diff. 13, 393–403 [DOI] [PubMed] [Google Scholar]
- 14. Rutkowski D. T., Kaufman R. J. (2004) A trip to the ER: coping with stress. Trends Cell Biol. 14, 20–28 [DOI] [PubMed] [Google Scholar]
- 15. Naidoo N. (2009) ER and aging-protein folding and the ER stress response. Ageing Res. Reviews 8, 150–159 [DOI] [PubMed] [Google Scholar]
- 16. Li B., Gao B., Ye L., Han X., Wang W., Kong L., Fang X., Zeng Y., Zheng H., Li S., Wu Z., Ye L. (2007) Hepatitis B virus X protein (HBx) activates ATF6 and IRE1-XBP1 pathways of unfolded protein response. Virus Res. 124, 44–49 [DOI] [PubMed] [Google Scholar]
- 17. Su H. L., Liao C. L., Lin Y. L. (2002) Japanese encephalitis virus infection initiates endoplasmic reticulum stress and an unfolded protein response. J. Virol. 76, 4162–4171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Jheng J. R., Lau K. S., Tang W. F., Wu M. S., Horng J. T. (2010) Endoplasmic reticulum stress is induced and modulated by enterovirus 71. Cellular Microbiol. 12, 796–813 [DOI] [PubMed] [Google Scholar]
- 19. Kim H. T., Waters K., Stoica G., Qiang W., Liu N., Scofield V. L., Wong P. K. (2004) Activation of endoplasmic reticulum stress signaling pathway is associated with neuronal degeneration in MoMuLV-ts-1 induced spongiform encephalomyelopathy. Lab. Invest. 84, 816–827 [DOI] [PubMed] [Google Scholar]
- 20. Li X. D., Lankinen H., Putkuri N., Vapalahti O., Vaheri A. (2005) Tula hantavirus triggers pro-apoptotic signals of ER stress in Vero E6 cells. Virology 333, 180–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ye Z., Wong C. K., Li P., Xie Y. (2008) A SARS-CoV protein, ORF-6, induces caspase-3-mediated, ER stress and JNK-dependent apoptosis. Biochim Biophys Acta 1780, 1383–1387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Medigeshi G. R., Lancaster A. M., Hirsch A. J., Briese T., Lipkin W. I., Defilippis V., Früh K., Mason P. W., Nikolich-Zugich J., Nelson J. A. (2007) West Nile virus infection activates the unfolded protein response, leading to CHOP induction and apoptosis. J. Virol. 81, 10849–10860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Tardif K. D., Waris G., Siddiqui A. (2005) Hepatitis C virus, ER stress, and oxidative stress. Trends Microbiol. 13, 159–163 [DOI] [PubMed] [Google Scholar]
- 24. Tardif K. D., Mori K., Kaufman R. J., Siddiqui A. (2004) Hepatitis C virus suppresses the IRE1-XBP1 pathway of the unfolded protein response. J. Biol. Chem. 279, 17158–17164 [DOI] [PubMed] [Google Scholar]
- 25. He B., Gross M., Roizman B. (1997) The γ(1)34.5 protein of herpes simplex virus 1 complexes with protein phosphatase 1α to dephosphorylate the α subunit of the eukaryotic translation initiation factor 2 and preclude the shutoff of protein synthesis by double-stranded RNA-activated protein kinase. Proc. Natl. Acad. Sci. U.S.A. 94, 843–848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Cheng G., Feng Z., He B. (2005) Herpes simplex virus 1 infection activates the endoplasmic reticulum resident kinase PERK and mediates eIF-2α dephosphorylation by the γ(1)34.5 protein. J. Virol. 79, 1379–1388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Look D. C., Rapp S. R., Keller B. T., Holtzman M. J. (1992) Selective induction of intercellular adhesion molecule-1 by interferon-γ in human airway epithelial cells. Am. J. Physiol. 263, L79–L87 [DOI] [PubMed] [Google Scholar]
- 28. Hunter J. A., Finkbeiner W. E., Nadel J. A., Goetzl E. J., Holtzman M. J. (1985) Predominant generation of 15-lipoxygenase metabolites of arachidonic acid by epithelial cells from human trachea. Proc. Natl. Acad. Sci. U.S.A. 82, 4633–4637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hoffmann E., Stech J., Guan Y., Webster R. G., Perez D. R. (2001) Universal primer set for the full-length amplification of all influenza A viruses. Arch. Virol. 146, 2275–2289 [DOI] [PubMed] [Google Scholar]
- 30. Langlois R. A., Meyerholz D. K., Coleman R. A., Cook R. T., Waldschmidt T. J., Legge K. L. (2010) Oseltamivir treatment prevents the increased influenza virus disease severity and lethality occurring in chronic ethanol consuming mice. Alcohol Clin. Exp. Res. 34, 1425–1431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Meyerholz D. K., Edsen-Moore M., McGill J., Coleman R. A., Cook R. T., Legge K. L. (2008) Chronic alcohol consumption increases the severity of murine influenza virus infections. J. Immunol. 181, 641–648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Post D. M., Phillips N. J., Shao J. Q., Entz D. D., Gibson B. W., Apicella M. A. (2002) Intracellular survival of Neisseria gonorrhoeae in male urethral epithelial cells: importance of a hexaacyl lipid A. Inf. Immun. 70, 909–920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ketterer M. R., Shao J. Q., Hornick D. B., Buscher B., Bandi V. K., Apicella M. A. (1999) Infection of primary human bronchial epithelial cells by Haemophilus influenzae: macropinocytosis as a mechanism of airway epithelial cell entry. Inf. Immun. 67, 4161–4170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hirota M., Kitagaki M., Itagaki H., Aiba S. (2006) Quantitative measurement of spliced XBP1 mRNA as an indicator of endoplasmic reticulum stress. J. Toxicol. Sci. 31, 149–156 [DOI] [PubMed] [Google Scholar]
- 35. Wu J., Rutkowski D. T., Dubois M., Swathirajan J., Saunders T., Wang J., Song B., Yau G. D., Kaufman R. J. (2007) ATF6α optimizes long-term endoplasmic reticulum function to protect cells from chronic stress. Dev. Cell 13, 351–364 [DOI] [PubMed] [Google Scholar]
- 36. Adachi Y., Yamamoto K., Okada T., Yoshida H., Harada A., Mori K. (2008) ATF6 is a transcription factor specializing in the regulation of quality control proteins in the endoplasmic reticulum. Cell Struct. Funct. 33, 75–89 [DOI] [PubMed] [Google Scholar]
- 37. Welch W. J., Brown C. R. (1996) Influence of molecular and chemical chaperones on protein folding. Cell Stress Chaperones 1, 109–115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Volkmann K., Lucas J. L., Vuga D., Wang X., Brumm D., Stiles C., Kriebel D., Der-Sarkissian A., Krishnan K., Schweitzer C., Liu Z., Malyankar U. M., Chiovitti D., Canny M., Durocher D., Sicheri F., Patterson J. B. (2011) Potent and selective inhibitors of the inositol-requiring enzyme 1 endoribonuclease. J. Biol. Chem. 286, 12743–12755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kozutsumi Y., Segal M., Normington K., Gething M. J., Sambrook J. (1988) The presence of malfolded proteins in the endoplasmic reticulum signals the induction of glucose-regulated proteins. Nature 332, 462–464 [DOI] [PubMed] [Google Scholar]
- 40. Pahl H. L., Baeuerle P. A. (1995) Expression of influenza virus hemagglutinin activates transcription factor NF-κB. J. Virol. 69, 1480–1484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ji C., Kaplowitz N., Lau M. Y., Kao E., Petrovic L. M., Lee A. S. (2011) Liver-specific loss of glucose-regulated protein 78 perturbs the unfolded protein response and exacerbates a spectrum of liver diseases in mice. Hepatology 54, 229–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Bermejo-Martin J. F., Ortiz de Lejarazu R., Pumarola T., Rello J., Almansa R., Ramírez P., Martin-Loeches I., Varillas D., Gallegos M. C., Serón C., Micheloud D., Gomez J. M., Tenorio-Abreu A., Ramos M. J., Molina M. L., Huidobro S., Sanchez E., Gordón M., Fernández V., Del Castillo A., Marcos M. A., Villanueva B., López C. J., Rodríguez-Domínguez M., Galan J. C., Cantón R., Lietor A., Rojo S., Eiros J. M., Hinojosa C., Gonzalez I., Torner N., Banner D., Leon A., Cuesta P., Rowe T., Kelvin D. J. (2009) Th1 and Th17 hypercytokinemia as early host response signature in severe pandemic influenza. Crit. Care 13, R201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ulianich L., Terrazzano G., Annunziatella M., Ruggiero G., Beguinot F., Di Jeso B. (2011) ER stress impairs MHC Class I surface expression and increases susceptibility of thyroid cells to NK-mediated cytotoxicity. Biochim. Biophys. Acta 1812, 431–438 [DOI] [PubMed] [Google Scholar]
- 44. Dolan B. P., Bennink J. R., Yewdell J. W. (2011) Translating DRiPs: progress in understanding viral and cellular sources of MHC class I peptide ligands. Cell Mol Life Sci. 68, 1481–1489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Dolan B. P., Li L., Takeda K., Bennink J. R., Yewdell J. W. (2010) J. Immunol. 184, 1419–1424 [DOI] [PMC free article] [PubMed] [Google Scholar]