Background: Pathway choice for the repair of double strand breaks is not fully understood.
Results: Human Ku blocks exonuclease 1-mediated DNA processing in the presence of Mre11 or Mre11/Rad50.
Conclusion: Unlike in yeast, the displacement of Ku from DNA ends is not mediated by Mre11 or the Mre11/Rad50 complex.
Significance: Pathway choice between nonhomologous end-joining and homologous recombination is likely more complex than simple competition between the two pathways.
Keywords: DNA Damage, DNA Damage Response, DNA Repair, Enzymes, Homologous Recombination, DNA End Resection, Exonuclease 1, Ku70/80, Mre11/Rad50
Abstract
DNA double strand breaks (DSB) are repaired by nonhomologous end-joining (NHEJ) or homologous recombination (HR). Recent genetic data in yeast shows that the choice between these two pathways for the repair of DSBs is via competition between the NHEJ protein, Ku, and the HR protein, Mre11/Rad50/Xrs2 (MRX) complex. To study the interrelationship between human Ku and Mre11 or Mre11/Rad50 (MR), we established an in vitro DNA end resection system using a forked model dsDNA substrate and purified human Ku70/80, Mre11, Mre11/Rad50, and exonuclease 1 (Exo1). Our study shows that the addition of Ku70/80 blocks Exo1-mediated DNA end resection of the forked dsDNA substrate. Although human Mre11 and MR bind to the forked double strand DNA, they could not compete with Ku for DNA ends or actively mediate the displacement of Ku from the DNA end either physically or via its exonuclease or endonuclease activity. Our in vitro studies show that Ku can block DNA resection and suggest that Ku must be actively displaced for DNA end processing to occur and is more complicated than the competition model established in yeast.
Introduction
DNA double-strand breaks (DSB)3 can be generated spontaneously by endogenous sources, such as DNA replication-associated errors and by products of cellular metabolism, or exogenous agents including ionizing irradiation and radiomimetic chemicals (1). Upon exposure to DSBs, prompt and precise DSB repair is critical because unrepaired DSBs can result in genomic instability, cell death, and tumorigenesis (2). There are two major DSB repair pathways, homologous recombination (HR) and nonhomologous end joining (NHEJ) (3). HR involves a series of steps including damage sensing, DNA resection, strand invasion, DNA synthesis, and ligation. HR is likely initiated when the Mre11/Rad50/Nbs1 (MRN) complex recognizes the DSB and recruits CtIP and Exo1 to mediate DNA resection. Resection of the DSB ends generates 3′-single strand DNA overhangs that are bound with RPA and then replaced by Rad51 for homologous template invasion and HR completion (4, 5). In NHEJ, Ku quickly recognizes the broken DNA ends where it then functions as a platform to assemble other NHEJ factors including DNA-PKcs, Artemis, XLF, and DNA ligase IV/XRCC4. After minimal processing, the DNA ends are ligated via the DNA ligase IV/XRCC4 dimer (2).
Although much work has been performed to identify and characterize factors that are required for repair by both DSB repair pathways, important questions are still unresolved, including what is the mechanism that modulates the pathway choice/switching between NHEJ and HR. Two factors are believed to play major roles in the choice of HR over NHEJ. First, the cell cycle phase is of importance as HR requires a homologous template; therefore, HR is believed to only be active during S and G2 phases of the cell cycle when a sister chromatid is available. NHEJ does not require a homologous template and is thus not restricted to a certain phase of the cell cycle but it is believed to be the dominant repair pathway in the G0 and G1 phases. The second factor is DNA end resection. The prevalent model for DNA end resection comes from genetic data generated from studies in Sacchromyces cerevisiae. The studies in yeast suggest a competition for DSB ends between the HR factors, Mre11 or Mre11/Rad50/Xrs2 (MRX), and the NHEJ factor, yeast Ku (yKu) (6, 7). As both pathways require initial DNA damage sensing and processing, the choice between the two pathways may reside in which repair protein complex is initially assembled at DSB sites and how DNA ends are processed before ligation. Both MRX and yKu are recruited to DSB sites independently and simultaneously (8) and lack of either complex results in an increase in the binding of the other complex and thus an increase in the other repair pathway (7–8). For example, in the absence of the MRX complex, yKu is accumulated at DSB sites and blocks Exo1-mediated DNA end resection (7). This data implicates that the MRX complex can mediate the dissociation of yKu from DSBs to allow DNA end resection to occur. The data in yeast suggests that competition and physical displacement of Ku from DNA ends via the MRX complex pushes the pathway choice to HR-mediated DSB repair instead of NHEJ.
Although the studies in yeast produce a very compelling model for DSB repair pathway choice, the functional relationship between the HR and NHEJ pathways and the choice between the two may not directly apply to the human system. First, in yeast, the MRX complex is essential for classical NHEJ but it is not required for NHEJ in human cells and appears to only be involved in an alternative end-joining pathway. Second, Ku is very abundant (∼5 × 105 copies in HeLa cells) and NHEJ is more dominant in human cells, whereas yeast uses HR as its major DSB repair pathway (9). For example, Ku deficiency causes severe radiosensitization in humans but Ku deficiency does not result in radiosensitivity in S. cerevisiae unless the HR protein Rad52 is also deleted (10). Finally, there are structural differences between yKu and human Ku (11–12). As there are clear marked differences between the human and yeast Ku proteins, in this report we wanted to examine in vitro with purified human proteins whether Ku and Mre11 or Mre11/Rad50 compete with each other similarly as yKu and MRX for DNA ends and modulate DNA end resection. An in vitro assay system was established to biochemically study the interplay between human proteins in HR and NHEJ by using Ku, Mre11, Mre11/Rad50, and Exo1 and a specific DNA substrate with forked structure at one end. It was found that human Ku blocks Exo1-mediated resection and that neither Mre11 nor Mre11/Rad50 could actively mediate the displacement of Ku from the DNA end either physically or via its exonucleases or endonucleases activities to free the end for Exo1-mediated resection. Our data is in contrast with previous in vitro data generated with purified yeast proteins and suggest that the displacement of human Ku from DNA ends is more complicated than the direct competition with the MRX complex, which was established in yeast.
EXPERIMENTAL PROCEDURES
Oligonucleotide Substrate and DNA End Labeling
The forked dsDNA was synthesized with a long single strand of 5′-CGCGCCCAGCTTTCCCAGCTAATAAACTAAAAACTCCTAAGG-3′ (42 nt) and a short single strand of 5′-CCTTAGGAGTTTTTAGTTTATTGGGCGCG-3′ (29 nt) (Invitrogen). The underlined nucleotides represented the hairpin sequence. 5′-End labeling of the long strand of the forked DNA was performed by incubating 2 μl of long strand of the forked DNA (10 μm), 1 μl of T4 polynucleotide kinase (10,000 units/ml), 5 μl of [γ-32P]ATP (3000 Ci/mmol, 10 mCi/ml), and 1 μl of cold ATP (200 μm) in a 20-μl reaction system at 37 °C for 30 min and stopped by heat inactivation at 65 °C for 20 min. 3′-End labeling of the short strand of the forked DNA was performed in a 50-μl reaction system containing 5 μl of short strand of the forked DNA (10 μm), 1 μl of terminal transferase (400 units/reaction), 5 μl of 3′-labeled [α-32P]Cordycepin 5′-triphosphate (5000 Ci/mmol, 10 mCi/ml) (Roche Applied Science), and 5 μl of CoCl2 (25 mm) at 37 °C for 15 min and stopped by addition of 5 μl of 0.2 m EDTA (pH 8.0). Both 5′- and 3′-labeled DNA were applied to a MicroSpin G-25 column to remove free 32P-nucleotide (GE Healthcare). The radiolabeled strand was annealed with the same molar amount of its complementary strand by incubating them together at 95 °C and cooled down to room temperature slowly. Labeling efficiency and DNA quality were examined using a 16% TBE native gel electrophoresis.
Gel Mobility Shifting Assay
DNA binding of Ku was performed using 10 nm forked DNA in a reaction system containing 25 mm Tris-HCl (pH 7.9), 50 mm KCl, 2 mm MgCl2, 1 mm EDTA, and 5% glycerol. Binding of Mre11 or Mre11/Rad50 to DNA in the absence or presence of Ku was performed using forked DNA in a binding buffer containing 25 mm MOPS (pH 7.0), 50 mm KCl, 1 mm DTT, and 1 mm MnCl2. All reactions were performed in a 10-μl system at room temperature for 15 min and 2 μl of loading dye (0.25% bromphenol blue, 0.25% xylene cyanol FF, and 30% glycerol) was added. The samples were separated via a 4% Tris glycine native gel, dried, and analyzed by Typhoon 9410 (GE Healthcare).
Nuclease Assay
All nuclease assays were performed in a 10-μl reaction. Exo1-mediated DNA resection was performed using 3′-end labeled forked DNA. To study the effect of Ku on Exo1-mediated DNA resection, 10 nm Exo1 and increasing concentrations of Ku (as indicated in Fig. 1B) were incubated with 3′-end labeled forked DNA in a reaction buffer containing 20 mm Hepes (pH 8.0), 50 mm KCl, 5 mm MgCl2, and 1 mm DTT at 37 °C for 1 h.
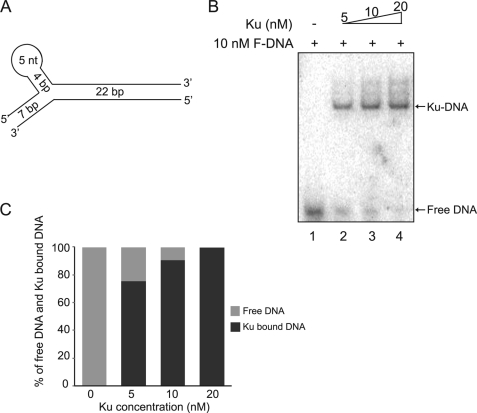
FIGURE 1.
Ku and forked DNA form a 1:1 complex. A, schematic structure of the forked DNA substrate. B, Ku bound to the forked DNA substrate. The binding reaction was performed using 10 nm radiolabeled forked DNA and increasing concentrations of Ku as indicated in the figure. Free DNA and Ku-bound DNA were separated via a 4% Tris glycine native gel and analyzed using a PhosphorImager. Free DNA and Ku-DNA complex are indicated by arrows. C, quantitation of free and Ku-bound DNA. The band intensities of both free DNA and Ku-bound DNA were quantitated by PhosphorImager analysis. The relative ratios of the band intensities of free DNA and Ku-bound DNA in each binding reaction are expressed as a percentage.
Mre11-mediated DNA digestion was performed using 5′-end labeled forked DNA in a reaction buffer containing 25 mm MOPS (pH 7.0), 50 mm KCl, 1 mm DTT, and 1 mm MnCl2 at 37 °C for 1 h. To study the effect of Ku, increasing concentrations of Ku were added to the above reaction buffer. To differentiate nuclease activities of Mre11, 12 nm Mre11 was used for exonuclease activity and 60 nm Mre11 was used for endonuclease activity. To study whether the Mre11 helped Exo1 to process DNA in the presence of Ku, increasing concentrations of Ku were incubated with 10 nm Exo1, 120 nm Mre11, and 10 nm 5′-end labeled forked DNA in a reaction buffer containing 20 mm Hepes (pH 8.0), 50 mm KCl, 5 mm MgCl2, 0.5 mm MnCl2, and 1 mm DTT at 37 °C for 1 h.
Mre11/Rad50-mediated exonuclease activity was performed in the same reaction buffer as for Mre11. For endonuclease activity of Mre11/Rad50, ATP (1 mm) was added to the same reaction buffer. To study the effect of Ku, increasing concentrations of Ku were added to the above reaction buffer. The effect of Mre11/Rad50 on Exo1-mediated DNA resection in the presence of Ku was examined in a two-step procedure. Increasing concentrations of Ku and 50 nm Mre11/Rad50 were incubated together with 10 nm 5′-end labeled forked DNA with or without ATP (1 mm) in a 10-μl reaction buffer containing 25 mm MOPS (pH 7.0), 40 mm KCl, 1 mm DTT, and 0.5 mm MnCl2 at 37 °C for 1 h. In the second step, 10 nm Exo1 and 5 mm MgCl2 were added to the reaction to a final volume of 12 μl and incubated for another 1 h at 37 °C. All reactions were stopped by adding an equal volume of formamide dye (95% formamide, 0.05% bromphenol blue, 0.05% xylene cyanol FF, 18 mm EDTA, 0.025% SDS), boiled at 95 °C for 5 min, and cooled down on ice for 5 min. The samples were separated by 16% urea-PAGE, dried, and analyzed in Typhoon 9410.
siRNAs and Transfection
Mre11 siRNAs were purchased from Thermo Fisher Scientific. The sequence for siRNA oligonucleotides against Mre11 was ACAGGAGAAGAGAUCAACUdTdT. Transfection of siRNA oligonucleotides was performed using Lipofectamine RNAi MAX (Invitrogen) according to the manufacturer's procedures.
Live Cell Imaging and Laser Microirradiation
Live cell imaging combined with laser microirradiation was carried out as described previously with modifications (13, 14). Fluorescence was monitored by using an Axiovert 200M microscope (Carl Zeiss, Inc.), with a Plan Apochromat ×63/NA 1.40 oil immersion objective (Carl Zeiss, Inc.). A 365-nm pulsed nitrogen laser (Spectra Physics) was directly coupled to the epifluorescence path of the microscope and used to generate DSBs in a defined area of the nucleus. For quantitative analyses, standardized microirradiation conditions (minimal laser output of 75% for 5 pulses) were used to generate the same amount of DNA damage in each experiment. Time-lapse images were taken by an AxioCamHRm camera and the fluorescence intensities of microirradiated and nonirradiated areas within the cell nucleus were determined using the Axiovison Software, version 4.8 (Carl Zeiss, Inc.). To eliminate the influence of nuclear background fluorescence, the fluorescence intensity of an undamaged site in the same nuclei was subtracted from the fluorescence intensity of the accumulation spot for every cell at each time point. Nonspecific photobleaching and UV lamp output fluctuation were compensated for by correcting the accumulation site fluorescence intensity (IN) of each time point based on pre-laser background intensity using the formula: IN(t) = Idt/Ibt × IbpreIR, where Idt represents the difference between the accumulation spot intensity and the undamaged site background intensity of each time point, Ibt represents the background intensity of each time point, and IbpreIR represents the background intensity before irradiation. Relative fluorescence intensity (RF) was calculated using the formula: RF(t) = (INt − INpreIR)/(INmax − INpreIR), where INpreIR means IN of the microirradiated area before laser damage and INmax is the maximum IN in the microirradiated area of all time points. Each data point is the average of at least 10 independent measurements.
RESULTS
Establishment of in Vitro DNA End Resection System with 1:1 Stoichiometry of Ku and DNA
Previous data generated in yeast showed that Ku may block DNA ends and that the MRX complex may actively displace Ku for DNA ends resection to occur. To study the mechanism of human Ku dissociation from the DNA end and determine whether it is similar to the yeast system, we first established in vitro binding conditions of human Ku70/80 to a model dsDNA substrate. The model dsDNA substrate used in the study is a forked DNA (F-DNA) substrate, which contains a 22-bp linear region connected to a branched structure that contains a 7-bp linear DNA on one end and a stem loop on the other (Fig. 1A). This forked DNA substrate was chosen because it allows Ku binding at the 22-bp linear region in only one direction and prevents its translocation off the DNA via the branched structure. The forked DNA structure with a 22-bp linear region also mimics the average 20-bp linker DNA found in nucleosomes, which likely only allows one directional loading of protein onto DNA in vivo. In addition, as 14 nt are required for Ku70/80 binding to DNA, the length of the linear region (22 bp) limits the binding of only one Ku to the model DNA substrate and allows the generation of a homogenous protein-DNA complex (15). Finally, a similar structured DNA was used previously to solve the x-ray crystal structure of Ku70/80 as it assured uniform complex formation of Ku and DNA (16).
First, quantitative binding of Ku to the model DNA substrate was performed by incubating increasing concentrations of Ku with 10 nm 32P-labeled forked DNA. One Ku (5–20 nm) and DNA band was observed in the 4% native Tris glycine gel representing 1:1 stoichiometry (Fig. 1B, lanes 2–4). The band intensities of both free DNA and Ku-bound DNA from the same binding reaction were measured to calculate their relative ratio. The values of ratios were expressed as percentage of total amount of DNA, which included free DNA and Ku-bound DNA and plotted as a histogram (Fig. 1C). It was found that 5 nm Ku was able to bind ∼80% of the model forked DNA, whereas 10 to 20 nm Ku occupied nearly 100% of the model DNA (Fig. 1C). It should be noted that at higher concentrations of Ku70/80 a small amount of higher order Ku-DNA species were observed (Fig. 1B and data not shown). These are likely due to the ability of Ku to unstably bind to shorter stretches of DNA, but the vast majority of the Ku-DNA species observed were 1 Ku with 1 DNA (15). In summary, a stable 1:1 stoichiometric binding was formed between 5 and 20 nm Ku and 10 nm forked DNA and the open end of the forked DNA substrate is clearly bound by Ku.
Ku Blocks Exo1-mediated DNA End Resection in Vitro
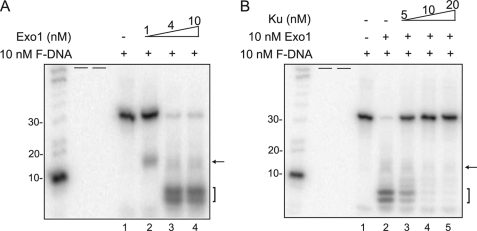
As yKu has been implicated in blocking Exo1-mediated end resection in the absence of the MRX complex, we wanted to establish an in vitro DNA end resection system with human proteins to test if human Ku could block end resection by human Exo1 using the defined DNA substrate (7). Recombinant human Exo1 and Ku70/80 were purified from Sf9 insect cells following expression via recombinant baculoviruses (supplemental Fig. S1A and data not shown). First, the exonuclease activity of Exo1 was studied by incubating various concentrations of Exo1 (1–10 nm) with the forked DNA substrate, which was radiolabeled at the 3′-end at 37 °C for 1 h. After the end of the reaction, DNA was denatured and separated by a 16% urea denaturing gel. The efficiency of DNA resection was determined by observing reduction of the original DNA substrate and appearance of resected DNA fragments. Low concentration of Exo1 (1 nm) resected DNA modestly as it produced an intermediate DNA fragment of ∼15 nt as marked by an arrow (Fig. 2A, lane 2). More thorough DNA resection was observed with increased concentrations of Exo1 as the amount of the original DNA was nearly lost and two newly resected DNA fragments were observed as marked by a bracket (Fig. 2A, lanes 3 and 4). To examine whether human Ku blocks human Exo1-mediated DNA resection, DNA was incubated with 10 nm Exo1 and increasing concentrations of Ku. Only very limited Exo1 resection was observed in the reaction when 5 nm Ku was added to the reaction (Fig. 2B, lane 3). As shown in Fig. 1, B and C, addition of 5 nm Ku resulted in ∼80% forked DNA bound with Ku and ∼20% unbound. Thus the limited resection observed in the presence of 5 nm Ku70/80 is likely due to Exo1-mediated resection of the 20% unbound DNA. When the Ku:forked DNA ratio was at a 1:1 molar ratio (as measured in Fig. 1, B and C) Exo1-mediated resection of the model DNA substrate was completely abrogated. This data shows that similar to the data generated with yeast; human Ku70/80 can bind to DNA substrates and block Exo1-mediated DNA end resection.
FIGURE 2.
Ku blocks Exo1-mediated DNA end resection. A, Exo1 has 5′ to 3′ exonuclease activity in vitro. Exo1-mediated DNA end resection assays were performed using 10 nm 3′-end labeled forked DNA and various concentrations of Exo1. Resected DNA was analyzed via a 16% urea PAGE and analyzed by PhosphorImager analysis (arrow indicates intermediate fragment and the bracket indicates completely digested fragments). Radiolabeled 10-nt DNA marker (Invitrogen) was used to measure the size of the resected DNA. B, Ku blocks Exo1-mediated DNA end resection. The effect of Ku on Exo1-mediated DNA end resection was performed by incubating 10 nm Exo1, 10 nm 3′-end labeled forked DNA, and increasing concentrations of Ku as indicated in the figure. Resected products were analyzed as stated in A. Short black bars represent empty lanes.
Mre11 or Mre11/Rad50 Could Not Compete with Ku for DNA Binding
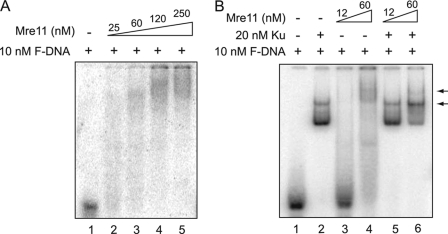
In S. cerevisiae, either Mre11 or Rad50 deficiency causes excess Ku accumulation at DSBs, which suggests that the MR and Ku complexes compete for the same DSB ends (7). In S. cerevisiae, Mre11 itself rather than its nuclease activity is required for DNA resection and repair of HO-induced DSBs suggesting that structurally Mre11 can regulate and compete for DNA ends (7, 17). Taking this into consideration, we next wanted to test the competition between human Ku and purified recombinant Mre11 or Mre11/Rad50 for DNA ends in vitro (supplemental Fig. S1, B and C). First, quantitative binding of Mre11 to the model DNA substrate was performed by incubating varying concentrations of Mre11 with the F-DNA. The gel shift data shows that at low concentrations, the Mre11-DNA complex produced smeared signals in the native gel (Fig. 3A, lanes 2 and 3). Increasing the concentration (120 and 250 nm) of Mre11 resulted in the formation of high order Mre11-DNA complexes (Fig. 3A). The data suggests that Mre11 has relatively low DNA binding affinity and likely forms heterogeneous complexes with the F-DNA substrate.
FIGURE 3.
Mre11 could not compete with Ku for DNA binding. A, Mre11 binds to the forked DNA substrate. DNA binding reactions were performed using 5′-end labeled forked DNA and increasing concentrations of Mre11 as indicated in the figure. Free DNA and Mre11-bound DNA were separated via a 4% Tris glycine gel and analyzed via PhosphorImager analysis. B, DNA binding of Ku and Mre11 together. DNA binding reactions were performed using 20 nm Ku and either 12 or 60 nm Mre11 with 10 nm 5′-end labeled forked DNA.
To study the competition between Mre11 and Ku70/80 for binding to F-DNA in vitro, F-DNA was incubated with 20 nm Ku and either 12 or 60 nm Mre11 simultaneously. Because the Mre11-DNA and Ku-DNA complexes migrated differently in the native gel (Figs. 1B and 3A, respectively), direct competition between Ku and Mre11 for DNA binding can be determined by the disappearance of one of the protein-DNA signals. Ku-DNA formed the only major protein-DNA complex when 12 nm Mre11 was added to the reaction (Fig. 3B, lane 5). The Ku-DNA complex was the dominant protein-DNA complex when 60 nm Mre11 was added, suggesting that Mre11 could not compete with Ku for DNA binding (Fig. 3B, lane 6). However, a new band with higher molecular weight than the Ku-DNA complex was observed when 60 nm Mre11 and 20 nm Ku were incubated together (Fig. 3B, lane 6). Furthermore, the addition of 60 nm Mre11 appears to result in a switch in the dominant Ku-DNA complex from one Ku binding to the F-DNA to two Ku. This suggests that Mre11 and Ku may be able to simultaneously bind the same DNA substrate and possibly change the binding of Ku to DNA. Furthermore, the data suggests that Mre11 may interact with Ku itself. To test whether Ku and Mre11 can interact independent of DNA, we tested protein-protein interaction with purified proteins. After incubation of purified Ku and Mre11 at 4 °C, each protein was immunoprecipitated via a specific antibody and tested whether they interact in the absence of DNA in vitro. The results show that Ku and Mre11 fail to interact in vitro (supplemental Fig. S2). We conclude that Ku and Mre11 do not physically interact and Mre11 is not able to block Ku from binding to DNA or mediate its dissociation from DNA.
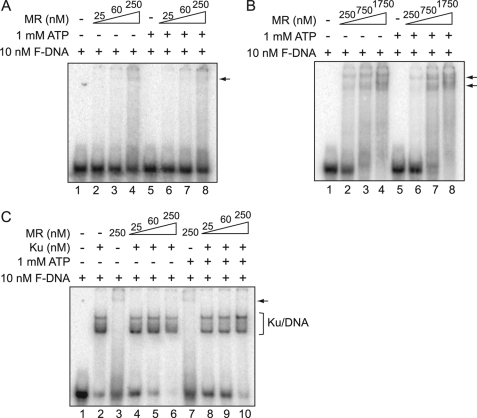
Rad50 makes a complex with Mre11 and confers allosteric regulation on Mre11 by coordinating the binding of Mre11 to DNA. In addition, ATP hydrolysis mediated by the ABC ATPase of Rad50 allows an open to close conformation of Mre11/Rad50 (18). Hence, we repeated the gel shift assays with Mre11/Rad50 to determine whether DNA binding was different from Mre11 and if Mre11/Rad50 could outcompete Ku for or mediate the dissociation of Ku from DNA ends. The condition of the Mre11/Rad50 binding reaction was similar to Mre11 except ATP was also included in a set of reactions to test a potential effect of ATP hydrolysis in binding to the F-DNA substrate. Like Mre11, Mre11/Rad50 binding also showed smeared signals regardless of the presence of ATP (Fig. 4A). However, its binding affinity was much lower than Mre11 because most DNA was left unbound and the smeared signals were weak even with 250 nm Mre11/Rad50 (Fig. 4A, lanes 4 and 8). When the concentration of MR was increased, a number of sharp bands were observed, possibly representing different stoichiometry between Mre11/Rad50 and DNA (Fig. 4B, lanes 3, 4 and 7, 8). In addition, unlike Mre11 alone, a number of sharp bands were generated by the Mre11/Rad50-DNA complex at higher Mre11/Rad50 concentrations, which suggests that Mre11/Rad50-DNA complexes are more uniform than the Mre11-DNA complex. Data also showed that ATP hydrolysis may affect the DNA binding affinity of the Mre11/Rad50 complex as a slight reduction in DNA binding was observed with the inclusion of ATP (Fig. 4B). The potential competition between Mre11/Rad50 and Ku for DNA binding was studied by incubating increasing concentrations of Mre11/Rad50 (25–250 nm) and Ku (20 nm) together. As shown in Fig. 4C, even high concentrations of Mre11/Rad50 (250 nm) with or without ATP cannot compete with Ku for DNA ends. Furthermore, unlike Mre11, it appears that Mre11/Rad50 cannot bind to the same DNA substrate as Ku. Taken together, the data shows Mre11 has higher binding affinity to the forked DNA substrate than Mre11/Rad50 and Mre11 partially changed Ku-DNA binding. However, neither Mre11 nor Mre11/Rad50 was able to suppress Ku binding or mediate its dissociation from DNA.
FIGURE 4.
Mre11/Rad50 (MR) complex cannot out compete with Ku for DNA binding. A and B, MR complex binds to forked DNA. DNA binding reactions were performed using 5′-end labeled fork DNA and increasing concentrations of MR complex with or without 1 mm ATP as indicated in the figures. The free DNA and MR-bound DNA were separated via a 4% Tris glycine gel and analyzed via PhosphorImager analysis. C, MR complex cannot compete with Ku for DNA binding. DNA binding reactions were performed using 10 nm 5′-end labeled forked DNA, 20 nm Ku, and increasing concentrations of MR complex as indicated in the figure with or without 1 mm ATP. Free DNA and DNA-bound complexes were visualized as stated in A and B.
Ku Blocks Mre11-mediated Forked DNA Digestion
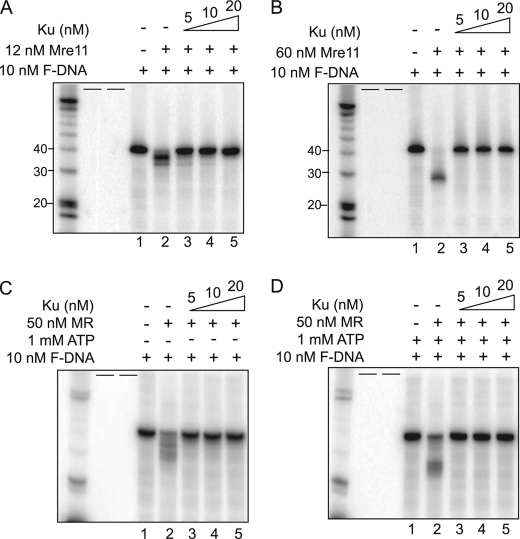
In Shizosaccharomyces pombe, cells with an Mre11 nuclease defect exhibited radiosensitivity that could be suppressed by Ku depletion suggesting a functional role of Mre11 in regulating Exo1-mediated DNA resection and HR (6, 19). Likewise, the importance of the nuclease activity of Mre11 in DNA resection and HR in mammalian cells was revealed using Mre11 nuclease dead mice (20). As the nuclease activity of Mre11 likely plays a role in DNA end resection in higher organisms, we wanted to test if the enzymatic activity of Mre11 or Mre11/Rad50 mediated the dissociation of Ku from DNA. First, using the forked DNA labeled at the 5′-end of the long strand as a template, conditions were optimized that activates the endonucleases or exonuclease activity of Mre11 in vitro. Mre11 showed differential activities depending on its molar ratio to DNA (21). Mre11 (12 nm) induced DNA fragmentation, which is a sign of progressive digestion by the exonuclease activity of Mre11 (Fig. 5A, lane 2, and supplemental Fig. S3A), whereas at 60 nm, a single dominant fragment smaller than 30 nt was produced implying a cleavage in the middle of the linear region of forked DNA by the endonuclease activity of Mre11 (Fig. 5B, lane 2). To test if the exonuclease or endonucleases activity of Mre11 affected the binding of Ku to the F-DNA substrate, either 12 or 60 nm Mre11 was incubated with increasing concentrations of Ku and the ability of Mre11 to digest the F-DNA was determined. It was found that the exonucleolytic DNA resection induced by Mre11 was blocked by Ku (Fig. 5A, lanes 4 and 5). Similar to the Exo1-mediated digestion in the presence of Ku, at the lowest concentration of Ku (5 nm), there was a very small amount of Mre11 digestion of the F-DNA substrate, which is likely because the F-DNA is not fully occupied by 5 nm Ku (Fig. 5A, lane 3). Endonucleolytic DNA cleavage by Mre11 was completely suppressed by Ku at all concentrations (Fig. 5B, lanes 3–5). In summary, Mre11-mediated DNA processing was blocked by Ku and it is likely that neither the exonuclease or endonucleases activity of Mre11 can mediate the dissociation of Ku from DNA ends.
FIGURE 5.
Ku protects DNA ends from Mre11 or MR complex-mediated DNA processing. Mre11-mediated processing of DNA via its (A) exonuclease or (B) endonucleases activity is blocked by Ku. The reactions were performed using 12 (A) or 60 (B) nm Mre11, increasing concentrations of Ku as indicated in the figure, and 10 nm 5′-end labeled forked DNA. The reaction samples were separated via a 16% urea PAGE and analyzed via PhosphorImager. DNA processing was determined via the disappearance of the forked DNA substrate and appearance of processed DNA fragments. C and D, MR complex-mediated processing of DNA via its (C) exonuclease or (D) endonuclease activity is blocked by Ku. The reactions were performed using 50 nm MR complex without ATP (C) or with 1 mm ATP (D), increasing concentrations of Ku as indicated in the figure, and 10 nm 5′-labeled forked DNA. Processing of the forked DNA substrate was visualized as stated in A and B. Short black bars represent empty lanes.
Ku Blocks Mre11/Rad50-mediated Fork DNA Digestion
Next, we examined the nuclease activity of Mre11/Rad50 and its ability to mediate the dissociation of Ku from DNA by incubating Mre11/Rad50 and various concentrations of Ku in the in vitro DNA resection system. ATP hydrolysis confers allosteric conformation changes to Rad50 and switches the nuclease activity from exonuclease to endonuclease of Mre11 in Mre11/Rad50 (18, 22). Mre11/Rad50 induced DNA fragmentations by its exonuclease activity without ATP (Fig. 5C and supplemental Fig. S3B). The generated heterogenous bands in the denaturing gel were more disperse than those from the exonuclease activity of Mre11 (Fig. 5A, lane 2, and Fig. 5C, lane 2). In the presence of ATP, Mre11/Rad50 endonuclease activity generated a DNA fragment with a similar size to that from endonucleolytic cleavage by Mre11 and a number of other fragments (Fig. 5D, lane 2). DNA digestion by either exonuclease or endonuclease activity of Mre11/Rad50 was examined together with Ku. The data shows that neither the exonucleolytic (Fig. 5C, lanes 3–5) nor the endonucleolytic (Fig. 5D, lanes 3–5) of Mre11/Rad50 could mediate digestion of the F-DNA substrate in the presence of Ku70/80. Rad50-mediated allosteric conformational changes in Mre11/Rad50 did not improve the ability of Mre11 to abrogate the ability of Ku to block DNA ends.
Ku Blocks Digestion of DNA by Exo1-Mre11 and Exo1-Mre11/Rad50
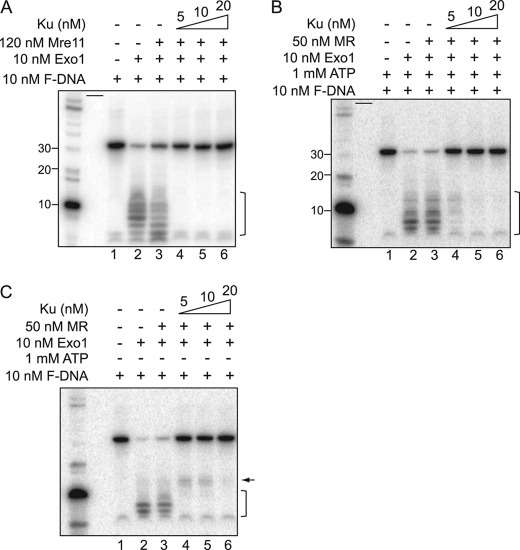
Mre11 or Mre11/Rad50 may directly or indirectly be involved in mediating the displacement of Ku from DNA ends to free them to facilitate Exo1-mediated DNA end resection (7). Next, it was tested if the presence of Mre11 or Mre11/Rad50 displaced Ku from DNA ends to allow Exo1-mediated DNA digestion. Previously, Mre11 and Exo1 were active with different metal ions, which were 1 mm MnCl2 and 5 mm MgCl2, respectively. However, we found that the presence of 1 mm MnCl2 inhibited 50% of the exonuclease activity of Exo1 (supplemental Fig. S4A, lane 4). To gain full activation of both nucleases, we used a reaction condition containing 5 mm MgCl2 and reduced concentration of MnCl2 (0.5 mm). In the new reaction condition, the exonuclease activity of Exo1 and Mre11 were fully active but the endonuclease activity of Mre11 was partially inhibited. To match the equivalent Mre11 endonuclease activity previously observed, the molar concentration of Mre11 was increased 2-fold (supplemental Fig. S4B). However, unlike Mre11, Mre11/Rad50 showed distinctive characteristics in a reaction buffer containing 0.5 mm MnCl2 and 5 mm MgCl2. In the presence of half of the amount of MnCl2 (0.5 mm), the endonucleases activity of Mre11/Rad50 was active without ATP, which is in contrast to previous conditions, which used 1 mm MnCl2 and 1 mm ATP (supplemental Fig. S4C). In addition, MgCl2 appeared to inhibit the endonuclease activity of Mre11/Rad50 and only allows its exonuclease activity (supplemental Fig. S4D, lanes 4 and 5). Hence, we performed the in vitro DNA resection assay with Mre11/Rad50 and Exo1 sequentially. First, F-DNA was incubated with Mre11/Rad50 and Ku in a buffer containing 0.5 mm MnCl2 for 1 h and then Exo1 and 5 mm MgCl2 were added for another 1 h. Exo1-mediated DNA resection was promoted by the endonuclease activity of Mre11, as shown by an increase in smaller resected DNA fragments (Fig. 6A, lane 3). Although the Exo1-mediated digestion of the F-DNA substrate was increased in the presence of Mre11, this addition did not result in any increase in digestion of the F-DNA substrate in the presence of Ku (Fig. 6A, lane 4-6). Next, Mre11/Rad50 and Exo1 together in the DNA resection system were determined. Data showed that neither the exonuclease nor the endonuclease activity of Mre11/Rad50 enhanced Exo1-mediated DNA resection (Fig. 6, B and C, lane 3). However, a small DNA fragment, although very weak, was observed when the active endonuclease of Mre11/Rad50 was preincubated with 5 to 10 nm Ku and Exo1 was then added (Fig. 6C, lanes 4 and 5). The size was ∼15 nt and is similar to the intermediate product of DNA resection by Exo1 alone (Fig. 2A, lane 2). Therefore, we presume that the endonuclease activity of Mre11/Rad50 might modestly promote Exo1-mediated DNA resection when Ku was at an equal or lower molar ratio than DNA; but was not able to completely dissociate Ku from DNA ends to allow further Exo1-mediated DNA processing (Fig. 6C, lane 6).
FIGURE 6.
Ku blocks digestion of DNA by Exo1-Mre11 and Exo1-Mre11/Rad50. A, the reactions were performed using 120 nm Mre11, 10 nm Exo1, increasing concentrations of Ku as indicated in the figure, and 10 nm 3′-end labeled forked DNA. The samples were separated via a 16% urea PAGE and analyzed via PhosphorImager. B and C, Exo1-mediated DNA end resection was not recovered by the MR complex via its (B) exonuclease ATP or (C) endonucleases activity in the presence of Ku. Processing of the forked DNA substrate was visualized as stated in A. Short black bars represent empty lanes.
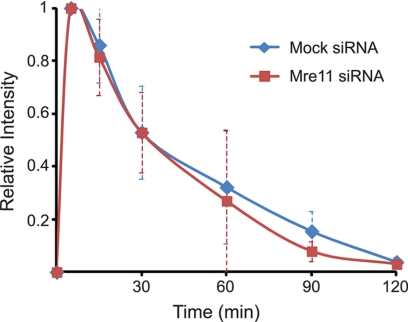
Finally, it was determined if Mre11 mediated the dissociation and/or affected the dynamics of Ku70/80 at DSBs in vivo. Localization and dynamics of YFP-tagged Ku80 were examined in the human cell line U2OS after knockdown of the Mre11 protein using specific siRNAs (supplemental Fig. S5) (14). The initial localization and dynamics/dissociation of Ku80 from laser-generated DSBs was not affected in the Mre11 siRNA-treated cells suggesting that Mre11 does not mediate the dissociation of Ku from DSBs in vivo (Fig. 7). Together, the data shows that human Mre11 and Mre11/Rad50 are unlikely responsible for mediating the active displacement of human Ku from DNA ends to allow Exo1-mediated DNA end resection, which is in contrast to the genetic yeast data.
FIGURE 7.
Mre11 does not mediate the dissociation of Ku from DSBs in vivo. YFP-tagged Ku80-expressing U2-OS cells were transfected with control or Mre11 siRNA for 72 h and microirradiated. Kinetics of relative fluorescence intensity of YFP-Ku80 at DSBs was assessed after microirradiation in Mre11 knockdown cells or mock siRNA-treated cells. Error bars represent S.D.
DISCUSSION
One key unresolved question is how cells choose between NHEJ and HR for the repair of DSBs in eukaryotic cells. Substantial evidence shows that NHEJ and HR may not only compete but also collaborate to repair DSBs. Collaboration between NHEJ and HR to promote genome stability was supported by embryonic lethality and severe genomic instability of mice with double knockouts of the HR factor Rad54, and the NHEJ factor Ku80, which is in contrast with the viability of mice with a single knockdown of either protein (23). Conversely, competition between the pathways was illustrated as an increase in HR frequencies and even DNA resection intermediates following the deletion of single NHEJ factors (Ku, DNA-PKcs, and DNA ligase IV) (24–26). Likewise, deletion of CtIP in mammalian cells and either Sae2 null or Sae2-S267A mutant in yeast cause increased NHEJ frequencies (27, 28). It is not clear how the two pathways compete or coordinate to repair DSB, but one important factor regulating pathway choice is DNA end resection. In S/G2 phases of the cell cycle, the DNA end resection machinery quickly forms ssDNA ends, which initiate HR. Ku has weakened affinity for ssDNA compared with dsDNA and it has been suggested that resection likely pushes the pathway choice from NHEJ to HR due to this reduced affinity of Ku to bind to DNA ends (29). Previous data shows that Ku localizes to DSBs in S phase (30) and recent data generated in our laboratory shows the accumulation and dissociation kinetics/dynamics of Ku at DSBs is the same in S and non-S phase suggesting that a simple competition model for DNA ends is likely not correct (34).
As Ku is bound at DSBs in S phase and likely actively displaced for HR to occur, it is critical to uncover the mechanisms mediating the dissociation of Ku from DNA ends to free them for DNA end resection and thus DSB repair mediated by HR. In yeast, Ku accumulates at HO-induced DSB sites in the absence of Mre11 (7, 8) and blocks Exo1-mediated DNA resection. Deletion of Ku partially recovers DNA resection and cell survival upon IR in Mre11 mutant yeast cells suggesting the freeing of ends of Ku allows HR-mediated repair even in the absence of Mre11 (6, 31). An in vitro resection assay using a long linear DNA (4.4 kb) showed that purified yKu blocked Exo1-mediated DNA resection, which could be alleviated by the addition of MRX to the reaction (7). This data suggests that competition between Mre11 and Ku for DNA ends and active displacement of Ku from DNA ends by Mre11 may push the choice between HR and NHEJ for DSBs in favor of HR in yeast. It is of great interest to know whether human Mre11, Mre11/Rad50, or Mre11/Rad50/Nbs1 function similarly as the yeast proteins do in preventing Ku accumulation and/or mediating its dissociation from DNA ends. We established an in vitro DNA resection system with purified human proteins and a one-directional forked DNA substrate to clarify biochemically whether Exo1-dependent DNA resection was blocked by Ku and whether Mre11 or Mre11/Rad50 suppressed the ability of Ku to block processing of DNA ends. The importance of using a one-directional forked DNA substrate instead of a 4.4-kb piece of linear DNA is that the stoichiometrical loading of Ku onto the DNA substrate can be optimized with the forked DNA. Furthermore, as Ku has the propensity to slide along DNA, DNA end resection results using a long piece of DNA may just show the ability of Ku to slide along the DNA and not its active displacement. Using a biochemically defined in vitro system, we found Exo1-mediated DNA end resection was blocked by Ku, which was in agreement with previous yeast data (Fig. 2).
Next, we tested whether human Mre11 or Mre11/Rad50 competed with Ku for DNA end binding or actively displaced Ku from DNA end in our in vitro system. A previous report showed that either Ku or RPA blocks digestion of the 3′ overhang DNA substrate by purified MRN complex (32). However, it is still not clear whether its blockage is due to interaction between Ku and MRN proteins or bidirectional loading of Ku-induced symmetric Ku and DNA complex at the 3′ overhang DNA. Therefore, Mre11 or Mre11/Rad50 were tested in both structural and functional aspects in the in vitro DNA assays with F-DNA. Direct competition between human Mre11 and Ku on DNA binding was tested first. We observed that Mre11 had much lower binding affinity to forked DNA than Ku and even excess Mre11 could not compete with Ku for DNA binding (Fig. 3). Furthermore, Mre11/Rad50 showed even lower DNA binding affinity than Mre11 and failed to compete with Ku for DNA binding. X-ray crystal structures of Mre11/Rad50 suggest that the ABC ATPase of Rad50 mediates ATP hydrolysis allowing an open-to-close conformational change that helps to load Mre11/Rad50 on pre-occupied DNA ends by Ku or other DNA adducts (18, 22). However, in our finding, Mre11/Rad50 failed to bind to DNA in the presence of Ku regardless of ATP (Fig. 4). Subsequently, we studied the effect of nuclease activity of Mre11 or Mre11/Rad50 on Ku binding in vitro and found that neither the endo- nor exonuclease activity of Mre11 or Mre11/Rad50 could actively displace Ku from DNA ends. Thus, lower DNA binding affinity of Mre11 or Mre11/Rad50 than Ku for dsDNA does not allow Ku to be dissociated from the DNA end and even allosteric changes in Mre11/Rad50 or nuclease activities cannot mediate the dissociation of human Ku from DNA ends.
DNA resection in yeast requires coordination of several proteins including Mre11/Rad50/Xrs2, Sae2, Sgs1, and Exo1. Among them, Exo1-dependent long range 5′ to 3′ DNA resection is promoted by Mre11/Rad50/Xrs2 (7, 33). Therefore, we wanted to test whether human Mre11 or Mre11/Rad50 collaboratively helped Exo1 to mediate DNA resection in the presence of Ku in vitro. We found that in our in vitro DNA resection system, as long as Ku was present, Exo1 could not resect DNA even with help from the enzymatically active Mre11 or Mre11/Rad50. Our in vitro studies show that neither Mre11 nor Mre11/Rad50 or their exonuclease or endonucleases activities are able to actively displace Ku from DSB ends. This data are in contrast to data generated in yeast that showed that Ku recruitment to DSBs was suppressed by Mre11/Rad50/Xrs2 and suggest that a more complex mechanism mediates the active displacement of human Ku from DNA ends (6, 7).
Supplementary Material
Acknowledgments
We are grateful to Dr. Guo-Min Li, Department of Toxicology and Biochemistry, University of Kentucky, College of Medicine, for kindly providing recombinant exonuclease 1 baculovirus for protein purification and Asaithamby Aroumougame for assisting in reading and editing the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grants CA50519, CA134991, and CA92584 and Cancer Research Institute of Texas Grant RP110465.

This article contains supplemental Figs. S1–S5.
- DSB
- DNA double strand break
- NHEJ
- non-homologous end-joining
- HR
- homologous recombination
- DNA-PKcs
- DNA-dependent protein kinase catalytic subunit
- Exo1
- exonuclease 1
- MR
- Mre11/Rad50
- MRN
- Mre11/Rad50/Nbs1
- MRX
- Mre11/Rad50/Xrs2
- yKu
- yeast Ku
- F-DNA
- forked DNA
- nt
- nucleotide(s).
REFERENCES
- 1. Lindahl T., Prigent C., Barnes D. E., Lehmann A. R., Satoh M. S., Roberts E., Nash R. A., Robins P., Daly G. (1993) DNA joining in mammalian cells. Cold Spring Harb. Symp. Quant. Biol. 58, 619–624 [DOI] [PubMed] [Google Scholar]
- 2. Burma S., Chen B. P., Chen D. J. (2006) Role of non-homologous end joining (NHEJ) in maintaining genomic integrity. DNA Repair 5, 1042–1048 [DOI] [PubMed] [Google Scholar]
- 3. Pastink A., Eeken J. C., Lohman P. H. (2001) Genomic integrity and the repair of double strand DNA breaks. Mutat. Res. 480, 37–50 [DOI] [PubMed] [Google Scholar]
- 4. Sun H., Treco D., Szostak J. W. (1991) Extensive 3′-overhanging, single-stranded DNA associated with the meiosis-specific double strand breaks at the ARG4 recombination initiation site. Cell 64, 1155–1161 [DOI] [PubMed] [Google Scholar]
- 5. White M. B., Word C. J., Humphries C. G., Blattner F. R., Tucker P. W. (1990) Immunoglobulin D switching can occur through homologous recombination in human B cells. Mol. Cell Biol. 10, 3690–3699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Mimitou E. P., Symington L. S. (2010) Ku prevents Exo1- and Sgs1-dependent resection of DNA ends in the absence of a functional MRX complex or Sae2. EMBO J. 29, 3358–3369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Shim E. Y., Chung W. H., Nicolette M. L., Zhang Y., Davis M., Zhu Z., Paull T. T., Ira G., Lee S. E. (2010) Saccharomyces cerevisiae Mre11/Rad50/Xrs2 and Ku proteins regulate association of Exo1 and Dna2 with DNA breaks. EMBO J. 29, 3370–3380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wu D., Topper L. M., Wilson T. E. (2008) Recruitment and dissociation of nonhomologous end-joining proteins at a DNA double strand break in Saccharomyces cerevisiae. Genetics 178, 1237–1249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Sonoda E., Hochegger H., Saberi A., Taniguchi Y., Takeda S. (2006) Differential usage of non-homologous end-joining and homologous recombination in double strand break repair. DNA Repair 5, 1021–1029 [DOI] [PubMed] [Google Scholar]
- 10. Siede W., Friedl A. A., Dianova I., Eckardt-Schupp F., Friedberg E. C. (1996) The Saccharomyces cerevisiae Ku autoantigen homologue affects radiosensitivity only in the absence of homologous recombination. Genetics 142, 91–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Aravind L., Koonin E. V. (2000) SAP, a putative DNA-binding motif involved in chromosomal organization. Trends Biochem. Sci. 25, 112–114 [DOI] [PubMed] [Google Scholar]
- 12. Feldmann H., Driller L., Meier B., Mages G., Kellermann J., Winnacker E. L. (1996) HDF2, the second subunit of the Ku homologue from Saccharomyces cerevisiae. J. Biol. Chem. 271, 27765–27769 [DOI] [PubMed] [Google Scholar]
- 13. Uematsu N., Weterings E., Yano K., Morotomi-Yano K., Jakob B., Taucher-Scholz G., Mari P. O., van Gent D. C., Chen B. P., Chen D. J. (2007) Autophosphorylation of DNA-PKcs regulates its dynamics at DNA double strand breaks. J. Cell Biol. 177, 219–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. So S., Davis A. J., Chen D. J. (2009) Autophosphorylation at serine 1981 stabilizes ATM at DNA damage sites. J. Cell Biol. 187, 977–990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Walker J. R., Corpina R. A., Goldberg J. (2001) Structure of the Ku heterodimer bound to DNA and its implications for double strand break repair. Nature 412, 607–614 [DOI] [PubMed] [Google Scholar]
- 16. Yoo S., Kimzey A., Dynan W. S. (1999) Photocross-linking of an oriented DNA repair complex. Ku bound at a single DNA end. J. Biol. Chem. 274, 20034–20039 [DOI] [PubMed] [Google Scholar]
- 17. Llorente B., Symington L. S. (2004) The Mre11 nuclease is not required for 5′ to 3′ resection at multiple HO-induced double strand breaks. Mol. Cell Biol. 24, 9682–9694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Williams G. J., Williams R. S., Williams J. S., Moncalian G., Arvai A. S., Limbo O., Guenther G., SilDas S., Hammel M., Russell P., Tainer J. A. (2011) ABC ATPase signature helices in Rad50 link nucleotide state to Mre11 interface for DNA repair. Nat. Struct. Mol. Biol. 18, 423–431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Langerak P., Mejia-Ramirez E., Limbo O., Russell P. (2011) Release of Ku and MRN from DNA ends by Mre11 nuclease activity and Ctp1 is required for homologous recombination repair of double strand breaks. PLoS Genet. 7, e1002271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Buis J., Wu Y., Deng Y., Leddon J., Westfield G., Eckersdorff M., Sekiguchi J. M., Chang S., Ferguson D. O. (2008) Mre11 nuclease activity has essential roles in DNA repair and genomic stability distinct from ATM activation. Cell 135, 85–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Paull T. T., Gellert M. (1998) The 3′ to 5′ exonuclease activity of Mre 11 facilitates repair of DNA double strand breaks. Mol. Cell 1, 969–979 [DOI] [PubMed] [Google Scholar]
- 22. Lim H. S., Kim J. S., Park Y. B., Gwon G. H., Cho Y. (2011) Crystal structure of the Mre11-Rad50-ATPγS complex: understanding the interplay between Mre11 and Rad50. Genes Dev. 25, 1091–1104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Couëdel C., Mills K. D., Barchi M., Shen L., Olshen A., Johnson R. D., Nussenzweig A., Essers J., Kanaar R., Li G. C., Alt F. W., Jasin M. (2004) Collaboration of homologous recombination and nonhomologous end-joining factors for the survival and integrity of mice and cells. Genes Dev. 18, 1293–1304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Allen C., Kurimasa A., Brenneman M. A., Chen D. J., Nickoloff J. A. (2002) DNA-dependent protein kinase suppresses double strand break-induced and spontaneous homologous recombination. Proc. Natl. Acad. Sci. U.S.A. 99, 3758–3763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kass E. M., Jasin M. (2010) Collaboration and competition between DNA double strand break repair pathways. FEBS Lett. 584, 3703–3708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Pierce A. J., Hu P., Han M., Ellis N., Jasin M. (2001) Ku DNA end-binding protein modulates homologous repair of double strand breaks in mammalian cells. Genes Dev. 15, 3237–3242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Bennardo N., Cheng A., Huang N., Stark J. M. (2008) Alternative NHEJ is a mechanistically distinct pathway of mammalian chromosome break repair. PLoS Genet 4, e1000110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Huertas P., Cortés-Ledesma F., Sartori A. A., Aguilera A., Jackson S. P. (2008) CDK targets Sae2 to control DNA-end resection and homologous recombination. Nature 455, 689–692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Falzon M., Fewell J. W., Kuff E. L. (1993) EBP-80, a transcription factor closely resembling the human autoantigen Ku, recognizes single to double strand transitions in DNA. J. Biol. Chem. 268, 10546–10552 [PubMed] [Google Scholar]
- 30. Kim J. S., Krasieva T. B., Kurumizaka H., Chen D. J., Taylor A. M., Yokomori K. (2005) Independent and sequential recruitment of NHEJ and HR factors to DNA damage sites in mammalian cells. J. Cell Biol. 170, 341–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Tomita K., Matsuura A., Caspari T., Carr A. M., Akamatsu Y., Iwasaki H., Mizuno K., Ohta K., Uritani M., Ushimaru T., Yoshinaga K., Ueno M. (2003) Competition between the Rad50 complex and the Ku heterodimer reveals a role for Exo1 in processing double strand breaks but not telomeres. Mol. Cell. Biol. 23, 5186–5197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Paull T. T., Gellert M. (1999) Nbs1 potentiates ATP-driven DNA unwinding and endonuclease cleavage by the Mre11/Rad50 complex. Genes Dev. 13, 1276–1288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Fiorentini P., Huang K. N., Tishkoff D. X., Kolodner R. D., Symington L. S. (1997) Exonuclease I of Saccharomyces cerevisiae functions in mitotic recombination in vivo and in vitro. Mol. Cell. Biol. 17, 2764–2773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Shao Z., Davis A. J., Fattah K. R., So S., Sun J., Lee K., Harrison L., Yang J., Chen D. J. (2012) Persistently bound Ku at DNA ends attenuates DNA end resection and homologous recombination. DNA Repair doi: 10.1016/j.dnarep.2011.12.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.