Background: The importance of Notch and Fe65 function during development has been well recognized.
Results: Adaptor protein Fe65 attenuates Notch1 signaling via the accelerated degradation of the membrane-tethered Notch1.
Conclusion: Dual regulation of Notch1 signaling pathway by adaptor protein Fe65.
Significance: The findings of this study may begin to shed some light onto what may be a signal cross-talk mechanism of Notch1 signaling and the Fe65 adaptor protein.
Keywords: Adaptor Proteins, Notch, Protein Degradation, Protein-Protein Interactions, Ubiquitination, Fe65, Itch E3 Ligase
Abstract
Notch1 receptor functions as a critical controller of cell fate decisions and also as a key regulator of cell growth, differentiation, and proliferation in invertebrates and vertebrates. In this study, we have demonstrated that the adaptor protein Fe65 attenuates Notch1 signaling via the accelerated degradation of the membrane-tethered Notch1 in the cytoplasm. Fe65 also suppresses Notch1 transcriptional activity via the dissociation of the Notch1-IC-recombining binding protein suppressor of hairless (RBP)-Jk complex within the nucleus. Fe65 is capable of forming a trimeric complex with Itch and membrane-tethered Notch1, and Fe65 enhances the protein degradation of membrane-tethered Notch1 via an Itch-dependent proteasomal pathway. Collectively, our results demonstrate that Fe65 carries out different functions depending on its location in the regulation of Notch1 signaling.
Introduction
Notch is a single-pass type I transmembrane receptor that performs a key role in the determination of cell fate, differentiation, cell proliferation, cell survival, and cell death (1–4). Notch1 receptor is produced by furin in the endoplasmic reticular Golgi apparatus (S1 cleavage) during transport to the cell surface, where it is expressed in heterodimeric form (5, 6). After binding to the specific ligands, Jagged and Delta, the transmembrane C-terminal fragment of Notch is generated via proteolytic cleavage by a disintegrin and metalloprotease domain metalloprotease (S2 cleavage) (7, 8). The cleavage of this fragment by γ-secretase (S3 cleavage) subsequently induces the release of the Notch intracellular domain from the membrane (9). Then, the Notch1 intracellular (Notch1-IC)2 domain is translocated into the nucleus and binds to the cofactors CBF1/suppressor of hairless/Lag-1 (CSL), MAML-1 (mastermind-like-1), and p300/CBP, leading to the transcriptional activation of downstream target genes such as Hes1, Hes5, Hes7, Hey1, Hey2, and HeyL (8, 10–14).
After the transcriptional regulation of the target genes, Notch1-IC undergoes proteasomal degradation in the nucleus via the ubiquitin-proteasome system (15–20). Several E3 ubiquitin ligases have been implicated in the half-life of Notch1-IC, including Fbw7, which promotes proline-, glutamate-, serine-, threonine-rich domain-dependent Notch1 intracellular degradation in the nucleus (15, 17–20). Additionally, the results of a recent genetic study demonstrated that Itch regulates the PEST-independent degradation of cytoplasmic Notch protein (21). The E3 protein ubiquitin ligase (E3) Itch, a novel E3 Ub ligase, or atrophin-1 interacting protein 4 (AIP4, hereafter referred to as Itch), is absent in the non-agouti-lethal 18H or Itchy mice (22). The Itch gene encodes for 854 amino acids with a relative molecular weight of 113 kDa (23). Itch is a monomeric protein that belongs to the homologous to E6-AP carboxyl terminus-type family of E3s, whose modular structural organization consists of four WW domains and a unique proline-rich motif (22). In Drosophila, genetic evidence suggests that the homologous to E6-AP C terminus domain harboring the E3 ubiquitin ligase suppressor of deltex negatively regulates Notch receptor signaling (24). The results of a recent study have demonstrated that Itch ubiquitinates membrane-tethered Notch1 under both in vivo and in vitro conditions (21). They concluded that the Notch-IC domain functions as a direct substrate for Itch (21). However, the step regulated by Itch has yet to be clearly elucidated.
Fe65 is a neuronal adaptor protein that mediates the assembly of multimolecular complexes through a variety of protein-protein interaction domains: the WW domain, which binds to proline-rich sequences, and two C-terminal phosphotyrosine-binding domains (25, 26). Fe65 is a brain-enriched protein of unknown function that binds to the cytoplasmic domain of amyloid precursor protein (27). We also demonstrated previously that Notch1-IC inhibits AICD transcriptional activity via physical binding with amyloid precursor protein intracellular domain, Fe65, and Tip60 (28). Fe65 is localized within the cytoplasm and nucleus of the cell (29), although moderate amounts of Fe65 are tethered to membranes via binding with APP (30, 31). Fe65 expression can lead to the stabilization and nuclear translocation of AICD, where it may induce apoptosis through Tip60 (32). The functions of Fe65 remain to be clearly elucidated, but it is found within both the cytoplasm and nucleus and has been shown to play roles in cell motility and nuclear signaling. Whereas the importance of Notch and Fe65 function during development has been well recognized, the molecular and biochemical mechanisms via which Fe65 exerts its regulatory effect on Notch signaling remain incompletely understood.
Herein, we have evaluated the mechanism underlying the Fe65-mediated dual regulation of Notch1 signaling. Our data show that Fe65 inhibits the transcriptional activity of Notch1 via an induced reduction in the protein stability of membrane-tethered Notch1, and that the level of the membrane-tethered Notch1 protein was markedly down-regulated in the presence of Fe65 via the proteasomal degradation of membrane-tethered Notch1 through Itch in the cytoplasm. Additionally, Fe65 interacts physically with Notch1-IC and disrupts the Notch1-IC- recombining binding protein suppressor of hairless (RBP)-Jk transcription complex within the nucleus.
EXPERIMENTAL PROCEDURES
Cell Culture and Transfection
HEK293 cells and NIH3T3 cells were cultured in DMEM (Invitrogen) containing 10% fetal bovine serum and 1% penicillin/streptomycin in a humidified incubator with an atmosphere containing 5% CO2. The cultured cells were transiently transfected via the calcium phosphate method for human embryonic kidney 293 cells. For plasmid DNA transfection, the cells were grown to ∼80% confluence and transfected with the plasmids (18, 33).
Luciferase Reporter Assay
Human embryonic kidney 293 cells were cotransfected with 4XCSL-Luc (a four-time repeating section of the RBP-Jk target sequence, CGTGGGAA, with the luciferase gene), Hes1-Luc, Hes5-Luc, and β-galactosidase coupled with the indicated vector constructs. After 48 h of transfection, the cells were lysed in chemiluminescent lysis buffer (18.3% of 1 m K2HPO4, 1.7% of 1 m KH2PO4, 1 mm phenylmethylsulfonyl fluoride, 1 mm dithiothreitol) and analyzed with a luminometer (Berthold) for the luciferase assays. The luciferase reporter activity in each sample was normalized in relation to the β-galactosidase activity in the same lysate (34).
In Vitro Binding Assay
The recombinant GST, GST-Notch1-IC, and GST-Fe65 proteins were expressed in the Escherichia coli BL21 strain using the pGEX system as indicated (34). The GST fusion protein was then purified using glutathione-agarose beads (Sigma) in accordance with the manufacturer's instructions. An equal quantity of GST or GST-Notch1-IC or GST-Fe65 fusion protein was incubated with the lysates of the HEK293 cells, which were transfected for 3 h with combinations of expression vectors at 4 °C, with rotation. After incubation, the beads were washed three times in ice-cold phosphate-buffered saline and boiled with 20 μl of Laemmli sample buffer. The precipitates were then separated via SDS-PAGE, and the pull-down proteins were detected via immunoblotting with specific antibodies.
Immunoblot Analysis
After 48 h of transfection, the cultured cells were harvested and lysed in radioimmune precipitation assay buffer (50 mm Tris-HCl (pH 7.5), 150 mm NaCl, 1% Nonidet P-40, 0.5% sodium deoxycholate, 0.1% SDS, 1 mm PMSF, 1 mm DTT, 2 μg/ml each of leupeptin and aprotinin) for 30 min at 4 °C. The cell lysates were then subjected to 20 min of centrifugation at 12,000 × g at 4 °C. The resultant soluble fraction was boiled in Laemmli buffer and subjected to SDS-PAGE. After gel electrophoresis, the separated proteins were transferred via electroblotting onto PVDF membranes (Millipore). The membranes were then blocked with Tris-buffered saline solution (pH 7.4) containing 0.1% Tween 20 and 5% nonfat milk. The blotted proteins were subsequently probed with anti-Myc antibody (9E10), anti-HA (12CA5) antibody, or anti-FLAG M2 antibody (Sigma), followed by incubation with anti-mouse horseradish peroxidase-conjugated secondary antibodies (Amersham Biosciences, Inc.). The blots were then developed via ECL.
Coimmunoprecipitation
After 48 h of transfection, the cells were lysed for 10 min in 1 ml of radioimmune precipitation assay lysis buffer at room temperature. After 20 min of centrifugation at 12,000 × g, the supernatants were subjected to immunoprecipitation with specific antibodies. After overnight incubation, protein A-agarose was added, and the samples were incubated for 3 h at 4 °C on the rotator. The beads were then again washed three times in ice-cold PBS, and any proteins that remained bound to the beads were eluted by boiling in 5× protein sample buffer. The samples were separated via SDS-PAGE, and visualized by immunoblotting.
Protein Accumulation Assay
Cells were treated with the proteasomal inhibitor MG-132 (Sigma-Aldrich), or the translational inhibitor cycloheximide (Sigma-Aldrich). MG-132 was used at 0, 5, and 10 μm for 6 h for the dosage assay of the proteasomal inhibitors. Half-life experiments employing the cycloheximide-mediated inhibition of protein synthesis were conducted as described previously (18). Cycloheximide was used at 100 μm for 0, 1, 2, 4, and 6 h for the time-course assay of the translational inhibitors. Protein levels were analyzed via immunoblotting.
Ubiquitination Assay
The HEK293 cells were transfected with indicated expression vectors and then harvested 48 h after transfection. After 42 h of transfection, the cells were treated with 10 μm of MG132 for 6 h, and the cells were lysed for 10 min in 1 ml of radioimmune precipitation assay lysis buffer at room temperature. After 20 min of centrifugation at 12,000 × g, the supernatants were subjected to immunoprecipitation with Notch antibody. After overnight incubation, protein A-agarose was added, and the samples were incubated for 3 h at 4 °C on the rotator. The beads were then again washed three times in ice-cold PBS, and any proteins that remained bound to the beads were eluted by boiling in 5× protein sample buffer. The precipitates were separated via SDS-PAGE and visualized by immunoblotting with HA antibody (35).
Knockdown of Fe65 and Itch
Knockdown study of siRNA were designed against human Fe65 genes and human Itch genes and subcloned into the pGPU/GFP/Neo vector. The nucleotide sequences for siRNA targeting human Fe65 were as follows. Fe65, 5′-CUUAAUGCAUCUAUACUCUdTdT-3′ (upper strand) and 3′-dTdTGAAUUACGUAGAUAUGAGA-5′ (lower strand) (GenePharma). The nucleotide sequences for siRNA targeting human Itch were as follows. Itch, 5′-AAGGAGCAACAUCUGGAUUAAUAdTdT-3′ (upper strand) and 3′-dTdTUUCCUCGUUGUAGACCUAAUUAU-5′ (lower strand) (GenePharma). siRNA was transfected into HEK293 cells using Lipofectamine according to the supplier's manual.
RESULTS
Fe65 Suppresses Notch1 Transcriptional Activity
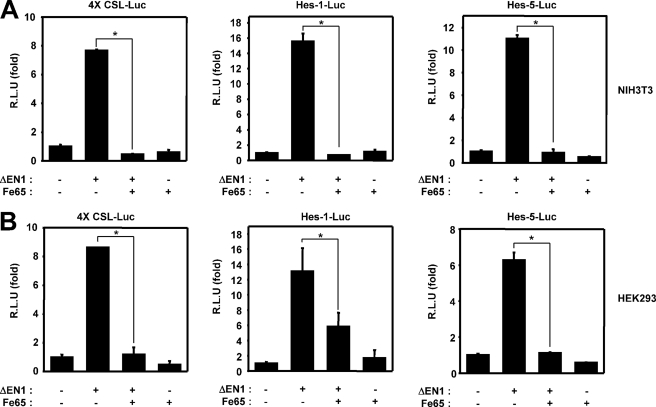
To evaluate the possible functions of Fe65 in Notch1 signaling, a reporter assay was conducted with NIH3T3 and HEK293 cells using luciferase reporter genes. The NIH3T3 cells or HEK293 cells were transfected with 4XCSL-Luc or Hes1-Luc or Hes5-Luc as well as either the active Notch1 mutant ΔEN1 (membrane-tethered Notch1) or an empty vector. As anticipated, ΔEN1-mediated transcription activity was found to have increased in these samples. We determined that Fe65 attenuated the ability of membrane-tethered Notch1 to stimulate transcription (Fig. 1, A and B). The basic helix-loop-helix proteins Hes1 and Hes5, both of which harbor multiple RBP-Jk-binding DNA sequences on their promoters, were identified as essential targets of Notch 1 in epithelial cells (36). Therefore, we confirmed the effects of Fe65 on the Notch1 signaling pathway using the Hes1 or Hes5 reporter systems. ΔEN1 expression significantly induced the activation of the Hes1 or Hes5 reporter systems (Fig. 1, A and B). Fe65 coexpression inhibits ΔEN1-induced natural Hes1 or Hes5 promoter transcriptional activity (Fig. 1, A and B).
FIGURE 1.
Fe65 suppresses the Notch1 signaling pathway. A, NIH3T3 cells were transfected with expression vectors for 4XCSL-Luc, Hes1-Luc, Hes5-Luc, and β-galactosidase, along with ΔEN1 and Fe65, as indicated. B, HEK293 cells were transfected with expression vectors for 4XCSL-Luc, Hes1-Luc, Hes5-Luc, and β-galactosidase, along with ΔEN1 and Fe65, as indicated. A and B, after 48 h of transfection, the cells were lysed, and the luciferase activity was determined. The data were normalized with β-galactosidase. These results represent the mean ± S.D. of three independent experiments. R.L.U., relative luciferase units. The data we evaluated for significant difference by Student's t test. *, p < 0.01 (analysis of variance).
Tip60 Does Not Affect Fe65-mediated Inhibition of Notch1 Signaling Pathway
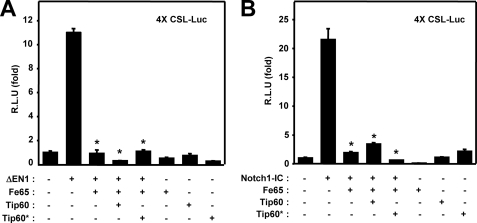
Previous reports have demonstrated that the Fe65 adaptor protein forms a complex with the Tip60 histone acetyltransferase (32, 34). We demonstrated previously that the expression of Tip60 down-regulates Notch1-IC-mediated transcriptional activity through Tip60-dependent acetylation of Notch1-IC (34). To determine the function of Tip60 in the Fe65-mediated suppression of Notch signaling, we introduced a Tip60 mutant (Tip60*) that cannot bind to Fe65. ΔEN1 expression was found to significantly induce the activation of the 4XCSL-Luc reporter system (Fig. 2A). When Fe65 was transiently coexpressed in HEK293 cells, constitutively active ΔEN1-mediated transcriptional activity was suppressed to a substantial degree (Fig. 2A). However, the forced expression of Tip60* exerted no effect on the negative role of Fe65 in ΔEN1-mediated transcriptional activity. We also confirmed the effects of Fe65 on the ΔEN1-mediated transcriptional activity using the Hes1 or Hes5 reporter systems (data not shown). Moreover, we also corroborated the effects of Tip60 on the Fe65-induced suppression of Notch signaling, using Notch1-IC. When Fe65 was transiently coexpressed in HEK293 cells, Notch1-IC-mediated transcriptional activity was apparently suppressed (Fig. 2B). However, coexpression of Tip60* exerted no effect on the negative role of Fe65 in Notch1-IC-mediated transcriptional activity (Fig. 2B). These results show that Fe65 involved in the regulation of Notch1 signaling in a Tip60-independent manner.
FIGURE 2.
Tip60 does not affect Fe65-mediated inhibition of Notch1 signaling pathway. A, HEK293 cells were transfected with expression vectors for 4XCSL-Luc and β-galactosidase, along with ΔEN1, Fe65, Tip60, and the Tip60* mutant as indicated. B, HEK293 cells were transfected with expression vectors for 4XCSL-Luc and β-galactosidase, along with Notch1-IC, Fe65, Tip60, and the Tip60* mutant as indicated. A and B, after 48 h of transfection, the cells were lysed, and the luciferase activity was determined. The data were normalized with β-galactosidase. These results represent the mean ± S.D. of three independent experiments. R.L.U., relative luciferase units. We assessed the data for significant differences via Student's t test compared with ΔEN1 or Notch1-IC. *, p < 0.01 (analysis of variance).
Fe65 Facilitates the Degradation of Membrane-tethered Notch1 by the Proteasome-dependent Pathway, but Notch1-IC Does not Affect Fe65-mediated Degradation
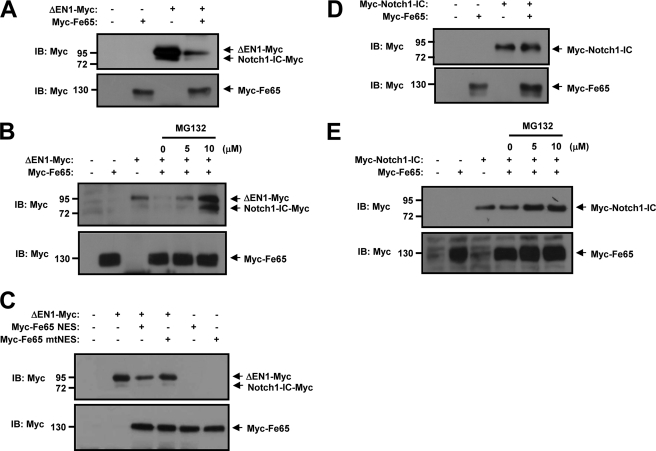
We carried out immunoblot analysis on HEK293 cells to determine whether Fe65 performs a function in the regulation of the ΔEN1 (membrane-tethered Notch1) protein level. Cells were cotransfected with ΔEN1-Myc and Myc-Fe65. We found that the ΔEN1 protein level (membrane-tethered Notch1) was reduced upon cotransfection of Fe65 (Fig. 3A). To determine whether the degradation of ΔEN1 protein level is mediated by the proteasome pathway, the proteasome inhibitor MG132 was employed for the treatment of ΔEN1-expressing and Fe65-expressing cells. MG132 can reversibly block all activities of the 26 S proteasome (37). The cells were treated with proteasome inhibitor for 6 h, and ΔEN1 protein was detected via an immunoblot assay. The ΔEN1 protein level was reduced in the presence of Fe65 but was restored to a significant extent by treatment with MG132 (Fig. 3B). Next, we transfected two expression vectors encoding for Fe65 fused at the COOH terminus to the nuclear export signal (NES) of the MAPKK or Fe65 fused to a mutated, nonfunctional version of this NES. The transfection of the Fe65-NES had the same effect as the wild-type Fe65, whereas the loss of function mutant NES conjugated Fe65 (Fe65-mNES) has no effect on the ΔEN1 protein level (Fig. 3C). To determine the possible role of Fe65 in the regulation of Notch1-IC protein stability, HEK293 cells were transfected with Myc-Notch1-IC and Myc-Fe65. Our studies showed that Fe65 could not regulate the steady-state level of Notch1-IC proteins (Fig. 3, D and E). We determined that the ΔEN1 protein level was reduced upon cotransfection of Fe65, but Notch1-IC protein was not reduced upon cotransfection of Fe65. These results showed that the stability of the ΔEN1 protein was down-regulated by Fe65 via the proteasome-dependent pathway. Our results demonstrate that Fe65 carries out different functions in Notch1 signaling, depending on its location.
FIGURE 3.
Fe65 facilitates degradation of membrane-tethered Notch1 by the proteasome-dependent pathway. A, HEK293 cells were transfected for 48 h with the indicated combinations of expression vectors encoding for ΔEN1-Myc and Myc-Fe65. B, HEK293 cells were transfected for 42 h with the indicated combinations of expression vectors encoding for ΔEN1-Myc and Myc-Fe65. The cells were then treated with the indicated quantity of MG132 for 6 h. C, HEK293 cells were transfected for 48 h with the indicated combinations of expression vectors encoding for ΔEN1-Myc, Myc-Fe65 NES, and Myc-Fe65 mNES. D, HEK293 cells were transfected for 48 h with the indicated combinations of expression vectors encoding for Myc-Notch1-IC and Myc-Fe65. E, HEK293 cells were transfected for 42 h with the indicated combinations of expression vectors encoding for Myc-Notch1-IC and Myc-Fe65. The cells were then treated with the indicated amounts of MG132 for 6 h. A–E, the cell lysates were also subjected to immunoblotting (IB) analysis with the indicated antibodies.
Fe65 Disrupts the Binding of Notch1-IC to RBP-Jk
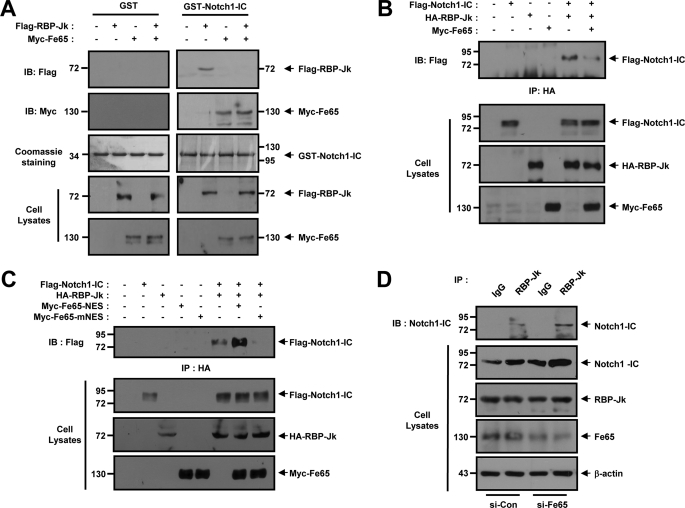
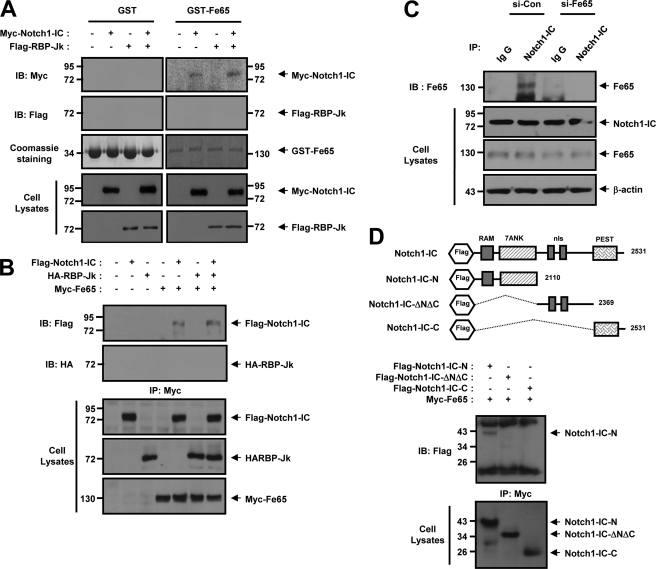
We attempted to evaluate the effects of Fe65 on Notch1-IC and RBP-Jk binding both in vitro and in intact cells. In the in vitro binding studies, purified GST or GST-Notch1-IC proteins were immobilized onto GSH-agarose. FLAG-RBP-Jk and Myc-Fe65 cells expressing the cell lysates were incubated with either GST or with GST-Notch1-IC, both of which were immobilized onto GSH-agarose. The interaction between GST-Notch1-IC and RBP-Jk was verified previously on bead complexes in the absence of Fe65 (Fig. 4A). The interaction GST-Notch1-IC and Fe65 has been detected on bead complexes in the absence or presence of RBP-Jk (Fig. 4A). The formation of the GST-Notch1-IC and RBP-Jk complexes was suppressed substantially as the result of Fe65 expression under in vitro conditions. To determine the effects of Fe65 on the molecular interactions occurring between Notch1-IC and RBP-Jk, coimmunoprecipitation was conducted with HEK293 cells via the cotransfection of FLAG-Notch1-IC, HA-RBP-Jk, and Myc-Fe65. The transfected cell lysates were immunoprecipitated using anti-HA antibody and immunoblotted with anti-FLAG antibody. The results showed that Fe65 interrupts the physical association between Notch1-IC and RBP-Jk (Fig. 4B). Furthermore, we also attempted to confirm the effects of Fe65-NES and Fe65-mNES on the physical association occurring between Notch1-IC and RBP-Jk (Fig. 4C). HEK293 cells were transfected with FLAG-Notch1-IC, HA-RBP-Jk, Myc-Fe65-NES, and Myc-Fe65-mNES. The cells were subjected to immunoprecipitation with anti-HA antibody and immunoblotted with anti-FLAG antibody (Fig. 4C). The results indicated that the loss of function mutant NES conjugated Fe65 (Fe65-mNES) interrupts the physical association between Notch1-IC and RBP-Jk (Fig. 4C), whereas NES-conjugated Fe65 (Fe65-NES) improves the physical association between Notch1-IC and RBP-Jk. We next attempted to ascertain whether Fe65 was able to interrupt the association between Notch1-IC and RBP-Jk in intact cells. HEK293 cells were transfected with Fe65 siRNA. The cells were subjected to immunoprecipitation with anti-RBP-Jk, and the resultant immunoprecipitates were examined via immunoblotting analysis with anti-Notch1 antibody. These results imply that Fe65 negatively regulated the transactivation of Notch1-IC target genes via the suppression of the interaction between Notch1-IC and RBP-Jk in intact cells (Fig. 4D).
FIGURE 4.
Fe65 disrupts the binding of Notch1-IC to RBP-Jk. A, HEK293 cells were transfected for 48 h with the indicated combinations of expression vectors encoding for FLAG-RBP-Jk and Myc-Fe65, as indicated. Cell lysates were then subjected to GST pull-down experiments with immobilized GST and GST-Notch1-IC. Proteins bound to GST or GST-Notch1-IC were analyzed via immunoblotting (IB) with anti-FLAG and anti-Myc antibody. GST or GST-Notch1-IC proteins were visualized via staining with Coomassie Brilliant Blue. B, HEK293 cells were transfected with expression vectors encoding for FLAG-Notch1-IC, HA-RBP-Jk, and Myc-Fe65, as indicated. After 48 h of transfection, the cell lysates were immunoprecipitated with anti-HA antibody, and the immunoprecipitates (IP) were immunoblotted with anti-FLAG antibody. C, HEK293 cells were transfected with expression vectors encoding for FLAG-Notch1-IC, HA-RBP-Jk, Myc-Fe65-NES, and Myc-Fe65-mNES as indicated. After 48 h of transfection, the cell lysates were immunoprecipitated with anti-HA antibody, and the immunoprecipitates were immunoblotted with anti-FLAG antibody. D, HEK293 cells were transfected for 48 h with the vector for siControl or siFe65. Cell lysates were then subjected to immunoprecipitation with anti-RBP-Jk or IgG antibody, and the resultant precipitates were subjected to immunoblotting analysis with anti-Notch1. Antibody to β-actin was used as a loading control. A–D, the cell lysates were also subjected to immunoblotting analysis with the indicated antibodies.
Notch1-IC Interacts Directly with Fe65 in Intact Cells
During our previous investigations, we have noticed that Fe65 may interact with Notch1-IC (28). In an effort to delineate more precisely the manner in which Fe65 prevents Notch1-IC and RBP-Jk-mediated transcription, we conducted a series of in vitro binding and coimmunoprecipitation experiments. In the in vitro binding studies, purified GST or GST-Fe65 proteins were immobilized onto GSH-agarose. FLAG-RBP-Jk and Myc-Notch1-IC expressing the cell lysates were incubated with either GST or GST-Fe65, both of which had been immobilized onto GSH-agarose. The interaction between GST-Fe65 and Notch1-IC has been detected on bead complexes in the absence of RBP-Jk (Fig. 5A). However, despite the performance of repeated experiments, no interaction was determined to have occurred between GST-Fe65 and RBP-Jk (Fig. 5A). To characterize the physical interaction occurring between Fe65 and Notch1-IC or RBP-Jk, coimmunoprecipitation was performed with HEK293 cells via the cotransfection of FLAG-Notch1-IC, HA-RBP-Jk, and Myc-Fe65. The transfected cell lysates were immunoprecipitated with anti-Myc antibody and immunoblotted with anti-FLAG or anti-HA antibody (Fig. 5B). The results indicated that Fe65 interacts with Notch1-IC but not with RBP-Jk, and no trimeric complex was detected in this case (Fig. 5B).
FIGURE 5.
Notch1-IC interacts directly with Fe65 in intact cells. A, HEK293 cells were transfected for 48 h with the indicated combinations of expression vectors encoding for Myc-Notch1-IC and FLAG-RBP-Jk, as indicated. Cell lysates were then subjected to GST pull-down experiments with immobilized GST and GST-Fe65. Proteins bound to GST or GST-Fe65 were analyzed via immunoblotting (IB) with anti-Myc or anti-FLAG antibody. GST or GST-Fe65 proteins were visualized via staining with Coomassie Brilliant Blue. B, HEK293 cells were transfected with expression vectors encoding for FLAG-Notch1-IC, HA-RBP-Jk, and Myc-Fe65, as indicated. After 48 h of transfection, the cell lysates were immunoprecipitated (IP) with anti-Myc antibody, and the immunoprecipitates were immunoblotted with anti-FLAG or anti-HA antibody. Cell lysates were also subjected to immunoblotting analysis with the indicated antibodies. C, HEK293 cells were transfected for 48 h with the vector for siControl or siFe65. Cell lysates were then subjected to immunoprecipitation with anti-Notch1-IC or IgG antibody, and the resultant precipitates were subjected to immunoblotting analysis with anti-Fe65. Cell lysates were also subjected to immunoblotting analysis with the indicated antibodies. Antibody to β-actin was used as a loading control. D, HEK 293 cells were transfected for 48 h with the indicated combination of expression vectors encoding for FLAG-Notch1-IC-N, FLAG-Notch1-IC-ΔNΔC, FLAG-Notch1-IC-C, along with Myc-Fe65. Cell lysates were then subjected to immunoprecipitation with anti-Myc, and the resultant precipitates were subjected to immunoblotting analysis with anti-FLAG antibody. Cell lysates were also subjected to immunoblotting analysis with the indicated antibodies. RAM, RBP-J association module of Notch-IC; PEST, proline-, glutamate-, serine-, threonine-rich domain of Notch-IC; nls, nuclear localization signals of Notch-IC.
Next, we attempted to ascertain whether Fe65 was capable of physically associating with Notch1-IC in intact cells. HEK293 cells were transfected with Fe65 siRNA. The cells were subjected to immunoprecipitation with anti-Notch1-IC, and the resultant immune pellets were examined via immunoblotting analysis with anti-Fe65 antibody. The immunoblot data demonstrated that the pellets interacted directly with endogenous Notch1-IC in the intact cells (Fig. 5C). These findings strongly indicate that a physical interaction between the two endogenous proteins Fe65 and Notch1 does indeed occur in intact cells. Notch1-IC harbors a CDC domain that harbors a RBP-J association module domain, seven ankyrin repeats, an opa-repeat region domain, and a PEST domain within its structure. We attempted to determine which, if any, of these domains might be involved in the interaction between Notch1-IC and Fe65. We utilized a variety of FLAG-Notch1 deletion mutants: Notch1-IC-N, Notch1-IC-ΔNΔC, and Notch1-IC-C. We conducted coimmunoprecipitation assays using the same three Notch1-IC deletion mutants and Myc-Fe65. As shown in Fig. 5D, the RAM-ankyrin repeat domains of Notch1-IC interact with Fe65. These data, therefore, suggest that the RAM-ANK domain of Notch1-IC was critically important to the interaction of Fe65 with Notch1-IC.
Fe65 Negatively Regulates Notch1 Signaling via an E3 Ligase, Itch
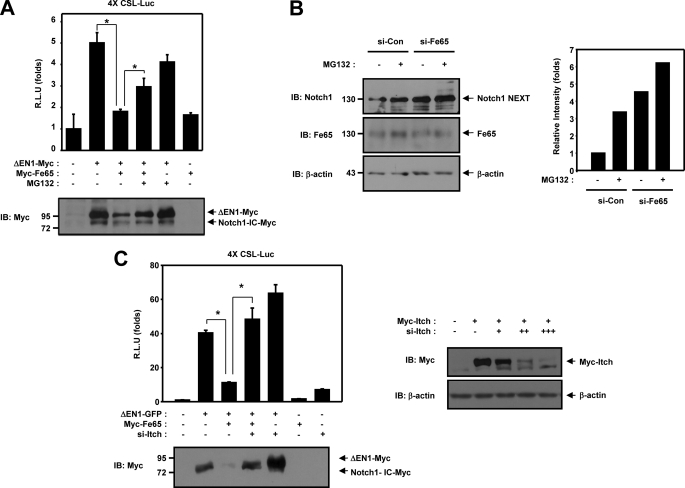
We evaluated the involvement of Fe65 in the membrane-tethered Notch1 proteasome-dependent degradation pathway by conducting luciferase reporter gene assays and Western blot analysis. The transcriptional activity of ΔEN1 was inhibited by Fe65 but recovered in the presence of MG132 (Fig. 6A). The ΔEN1 protein level was reduced in the presence of Fe65 but was restored significantly by treatment with MG132 (Fig. 6A). After ligand binding, it is cleaved (S2 cleavage) by tumor necrosis factor α-converting enzyme to yield a membrane-tethered intermediate fragment referred to as Notch1 extracellular truncation (NEXT). This fragment is then cleaved by presenilin-dependent γ-secretase to release the intracellular domain from the membrane. Moreover, the endogenous Notch1 NEXT protein level was lower in the wild-type cells than in the Fe65 siRNA-transfected cells (Fig. 6B, first and third lanes), and treatment with MG132 enhanced the endogenous Notch1 NEXT protein level by inhibiting proteasomal degradation (Fig. 6B, second and fourth lanes). These results showed that the stability of the Notch1 NEXT protein was down-regulated by Fe65 via the proteasome-dependent pathway. Generally, the ubiquitination of proteins results in their rapid degradation, and membrane-tethered Notch1 is degraded by the ubiquitin-proteasome system in the cytoplasm (16). Membrane-tethered Notch1 is ubiquitinated by Itch (24). We expected that Itch might function as a mediator for the negative regulation of membrane-tethered Notch1 by Fe65. Therefore, we evaluated the involvement of Itch using the Itch siRNA. When Itch siRNA was cotransfected with ΔEN1 and Fe65, the transcriptional activation of ΔEN1 was increased (Fig. 6C). According to Western blot analysis, the membrane-tethered Notch1 protein level was shown to have been reduced by Fe65 and was restored markedly by the cotransfection of Itch siRNA (Fig. 6C). These results demonstrated that Itch siRNA could recover and enhance the transcriptional activity and protein level of membrane-tethered Notch1 in the presence of Fe65.
FIGURE 6.
Fe65 negatively regulates Notch1 signaling via an E3 ligase Itch. A, HEK293 cells were transfected with expression vectors for 4XCSL-Luc and β-galactosidase, along with ΔEN1 and Fe65 as indicated. After 42 h of transfection, the cells were treated with 10 μm of MG132 for 6 h, after which the cells were lysed, and the luciferase activity was determined. B, HEK293 cells were transfected for 48 h with the vector for siControl or siFe65. After 42 h of transfection, the cells were treated with 10 μm of MG132 for 6 h and the cell lysates were immunoblotted (IB) with anti-Notch1 or anti-Fe65 antibody. Antibody to β-actin was used as a loading control. C, HEK293 cells were transfected with expression vectors for 4XCSL-Luc and β-galactosidase, along with ΔEN1, Fe65 and Itch siRNA as indicated. After 48 h of transfection, the cells were lysed, and the luciferase activity was determined. HEK293 cells were transfected for 48 h with the indicated combinations of expression vectors encoding for Myc-Itch and Itch siRNA. The cell lysates were immunoblotted with anti-Myc or anti-β-actin antibody. Antibody to β-actin was used as a loading control. A and C, the data were normalized with β-galactosidase. The results were expressed as the mean ± S.E. of three independent experiments. R.L.U., relative luciferase units. The cell lysates were immunoblotted with anti-Myc antibody. The data we assessed for significant differences via Student's t test. *, p < 0.01 (analysis of variance).
Unlike the levels observed for the control short interference plasmid-transfected cells, the cells transfected with short interference Itch harbored low levels of Itch (Fig. 6C). Accordingly, we suggest that Fe65 negatively regulates membrane-tethered Notch1 through Itch.
Fe65 Facilitates Stable Association between Membrane-tethered Notch1 and E3 Ligase Itch via the Formation of a Trimeric Complex
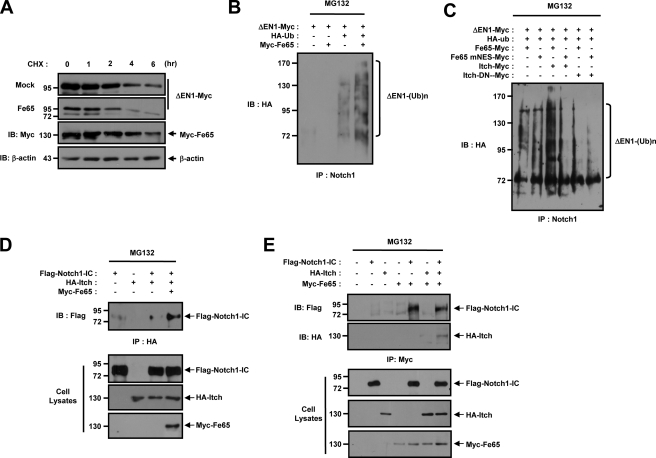
We attempted to determine whether ΔEN1 could be subjected to proteasome-mediated proteolysis, as reported previously. We transfected the HEK293 cells with ΔEN1-Myc and Myc-Fe65, and the quantity of remaining ΔEN1 was evaluated after various periods of cycloheximide treatment. We determined the protein stability of ΔEN1 in HEK293 cells by cycloheximide treatment with or without Fe65. Cycloheximide interacts with the translocase enzyme and blocks protein synthesis in eukaryotic cells. After cycloheximide treatment, the level of ΔEN1 protein declined gradually, with approximately half of the protein degraded after 3 h without Fe65 (Fig. 7A). Upon cycloheximide treatment, the reduced level of ΔEN1 protein declined rapidly, with approximately half of the protein being degraded after 1.5 h in the presence of Fe65 (Fig. 7A). No reduction was noted in the level of actin employed as a control (Fig. 7A). This result demonstrates that ΔEN1 is rapidly turned over in the presence of Fe65. To characterize the ubiquitination of ΔEN1-Myc, HA-Ub and Myc-Fe65 were cotransfected in HEK293 cells. Precipitation with anti-Notch1 antibody and immunoblotting with ΔEN1 demonstrated that the polyubiquitinated ΔEN1 appeared as bands. The ubiquitinated ΔEN1 levels increased with the coexpression of Fe65 (Fig. 7B). We also attempted to determine whether Fe65 could regulate the level of ΔEN1 ubiquitination through Itch. Immunoblot analysis of the ΔEN1 precipitated with anti-Notch1 antibody showed that the polyubiquitinated ΔEN1 levels were increased with the coexpression of Itch with wild-type Fe65 (Fig. 7C). However, the ubiquitination of ΔEN1 was decreased substantially by coexpression of Itch and Fe65-mNES (Fig. 7C).
FIGURE 7.
Fe65 facilitates stable association between Notch1 and E3 ligase Itch through the formation of a trimeric complex. A, HEK293 cells were transfected for 48 h with the indicated combinations of expression vectors encoding for ΔEN1-Myc and Myc-Fe65. The cells were then treated with 100 μm of cycloheximide (CHX) for the indicated time periods. Antibody to β-actin was used as a loading control. B, HEK293 cells were transfected with expression vectors encoding for ΔEN1-Myc, HA-Ub, and Myc-Fe65 in the indicated combinations. After 42 h of transfection, the cells were treated with 10 μm of MG132 for 6 h, the cell lysates were immunoprecipitated (IP) with anti-Notch1 antibody, and the immunoprecipitates were immunoblotted (IB) with anti-HA antibody. C, HEK293 cells were transfected with expression vectors for ΔEN1-Myc, HA-Ub, Itch-Myc, Itch-DN-Myc, Myc-Fe65, and Myc-Fe65 mNES, as indicated. After 42 h of transfection, the cell lysates were treated with 10 μm of MG132 for 6 h, the cell lysates were immunoprecipitated with anti-Notch1, antibody and the immunoprecipitates were immunoblotted with anti-HA antibody. Cell lysates were also subjected to immunoblotting analysis with the indicated antibodies. D, HEK293 cells were transfected with expression vectors encoding for FLAG-Notch1-IC, HA-Itch, and Myc-Fe65 in the indicated combinations. After 48 h of transfection, the cells were lysed, the cell lysates were immunoprecipitated with anti-HA antibody, and the immunoprecipitates were immunoblotted with anti-FLAG antibody. The cell lysates were also subjected to immunoblotting analysis with the indicated antibodies. E, HEK293 cells were transfected with expression vectors encoding for FLAG-Notch1-IC, HA-Itch, and Myc-Fe65 in the indicated combinations. After 48 h of transfection, the cells were lysed, the cell lysates were immunoprecipitated with anti-Myc antibody, and the immunoprecipitates were immunoblotted with anti-FLAG or anti-HA antibody. Cell lysates were also subjected to immunoblotting analysis with the indicated antibodies.
Previous reports have shown that Numb binds to the intracellular domain of Notch1 and then recruits the Itch to the complex (21, 38–40). Itch polyubiquitinates Notch1-IC, leading to its degradation in a proteasome-dependent manner (21, 39, 41–43). We also attempted to determine whether Fe65 could regulate the physical interaction between Itch and Notch1-IC or form a trimeric complex. We subsequently evaluated the involvement of Fe65 in the physical association between Itch and Notch1-IC via a coimmunoprecipitation experiment. HEK293 cells were cotransfected with vectors encoding for Flag-Notch1-IC, HA-Itch, and Myc-Fe65, and were then subjected to coimmunoprecipitation analysis. Immunoblot analysis using the anti-FLAG antibody of anti-HA antibody immunoprecipitates from the transfected cells demonstrated that Fe65 facilitates the physical association between Itch and Notch1-IC in the cells (Fig. 7D). These results revealed that the down-regulation of the membrane-tethered Notch1 protein by Fe65 occurred via an Itch-dependent pathway. At this point, we evaluated the formation of a trimeric complex between Fe65 and Notch1-IC or Itch in an effort to define more precisely the role of Fe65 in the negative regulation of Notch1 signaling. We detected binding between Notch1-IC and Fe65 but not between Fe65 and Itch, although the trimeric complex was detected in this case (Fig. 7E). Therefore, the results demonstrated that Fe65 interacts with Itch in the presence of Notch1-IC, thereby forming a trimeric complex.
Fe65 Prevent Notch1-IC-inhibited Cell Death and Notch1-Ic-mediated Signaling
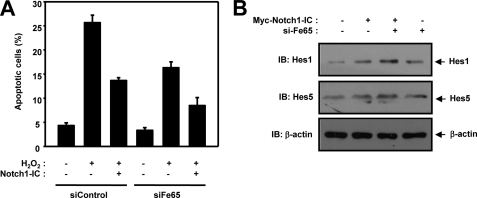
Next we evaluated the effect of Fe65 on Notch1-IC-inhibited cell death. H2O2 induced incremental cell death in HEK293 cells, and this cell death might be repressed by Notch1-IC and Fe65 siRNA, as shown by DAPI staining (Fig. 8A). These results suggest that Fe65 prevents Notch1-IC-inhibited cell death.
FIGURE 8.
Fe65 prevents Notch1-IC-inhibited cell death and Notch1-Ic-mediated signaling. A, HEK293 cells were transfected for 48 h with expression vector encoding GFP, Notch1-IC, siControl, or siFe65. The transfected cells were treated with the indicated quantity of H2O2 for 20 min. The cells were stained with DAPI, and TUNEL and GFP-positive cells were scored for apoptotic nuclei with a fluorescence microscope. Data are mean ± S.D. of values from three independent experiments. B, HEK293 cells were transfected with expression vectors encoding for Myc-Notch1-IC and Fe65 siRNA. The cell lysates were also subjected to immunoblotting (IB) analysis with anti-Hes1 and anti-Hes5 antibody. Antibody to β-actin was used as a loading control.
Furthermore, we also attempted to confirm the effects of Fe65 on Notch1-mediated Notch target gene expression (Fig. 8B). HEK293 cells were transfected with Myc-Notch1-IC and Fe65 siRNA. The cells were immunoblotted with anti-Hes1 and anti-Hes5 antibody (Fig. 8B). We found that Fe65 siRNA increased the Notch target gene expression. These results revealed that Fe65 negatively regulated the transactivation of Notch1-IC target genes via the suppression of the interaction between Notch1-IC and RBP-Jk.
DISCUSSION
In this study, we have demonstrated that Fe65 promotes the degradation of membrane-tethered Notch1. The nuclear localization of Fe65 inhibits Notch1 transcription activity via the dissociation of the Notch1-IC and RBP-Jk complex. Furthermore, Notch1-IC is capable of forming a trimeric complex with Fe65 and Itch. Fe65 thereby enhanced the protein degradation of membrane-tethered Notch1 via an Itch-dependent proteasomal pathway.
We demonstrated previously that the expression of Notch1-IC down-regulates AICD-Fe65-Tip60 complex-mediated transcriptional activity, reactive oxygen species generation, and cell death through physical binding with AICD, Fe65, and Tip60 (28). The neuronal adaptor protein Fe65 consists of several protein interaction domains, including one WW and two phosphotyrosine interaction/binding domains. Fe65 interacts with APP and nuclear protein (32). Fe65 expression is also developmentally regulated, with levels declining after embryonic day 15 and increasing again progressively from postnatal day 10 to adulthood (44). Previous reports have suggested the possibility of cross-talk between the Fe65 and Notch1 signaling pathway (28, 34). Fe65 belongs to a family of WW domain proteins, among which many have been shown to be involved in transcriptional regulation (45, 46). Our results demonstrated that Notch1 transcriptional activity was attenuated by the presence of Fe65, which indicates that Fe65 may also involve the suppression of Notch1 transcriptional activity. We also found that Fe65 is involved in the regulation of Notch1 signaling in a Tip60-independent manner.
The nuclear localization of Fe65 has also been reported (47), and several studies have suggested that one potential function for Fe65 in the nucleus may involve gene transactivation (32, 47, 48). However, the precise function of Fe65 in the nucleus and the mechanisms that regulate the translocation of Fe65 into the nucleus remain unclear. We have determined that Notch1-IC interacts directly with Fe65. Intriguingly, our results showed that the inhibitory mechanism functioned via the suppression of the interaction of Notch1-IC and RBP-Jk.
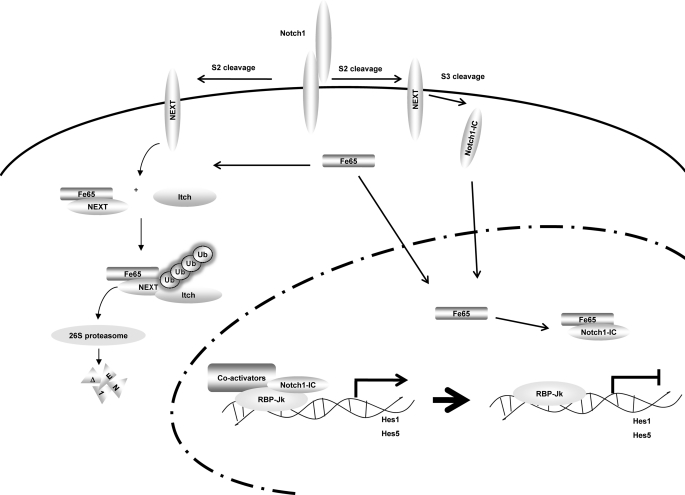
Itch belongs to the Nedd4/Rsp5p family of E3 ubiquitin ligases, which have been reported to control the down-regulation of both transmembrane proteins through mono-ubiquitination, and that of cytoplasmic proteins by polyubiquitination and proteasome degradation (39). E3 ubiquitin ligases that target membrane-associated Notch1 for ubiquitination have been described recently described (39). The RING finger containing E3 ligase Cbl regulates the ubiquitination of membrane-bound Notch1 and has also been reported to result in the lysosomal degradation of Notch1 (49). The HECT type E3 ligase Itch also targets a membrane-tethered form of Notch1 for ubiquitination (23). In this study, we determined that Fe65 stimulated the proteasomal degradation of membrane-tethered Notch1 via interaction with Itch. We have demonstrated that membrane-tethered Notch1 interacts with Itch, and that together Fe65 and Itch cooperate to increase the ubiquitination of Notch1. The Fe65 NES mutant does not enhance membrane-tethered Notch1 ubiquitination. Our results suggest that Fe65 may function as an adaptor for the recruitment of the E3 ligase Itch and components of the ubiquitination machinery to the membrane-tethered Notch1 receptor, thus promoting Notch1 ubiquitination and protein degradation (Fig. 9).
FIGURE 9.
Proposed model for the inhibition of Notch1 signaling pathway by Fe65. Fe65 suppresses Notch1 signaling via an acceleration of the degradation of NEXT in the cytoplasm. Notch1 is capable of forming a trimeric complex with Itch and Fe65. Fe65 enhanced the protein degradation of NEXT via the Itch-dependent proteasomal pathway. Additionally, Fe65 suppresses Notch1 transcriptional activity via the dissociation of the Notch1-IC-RBP-Jk complex in the nucleus.
In summary, the results of this study reveal that Fe65 functions as a negative regulator in Notch1 signaling via the promotion of membrane-tethered Notch1 protein degradation. Fe65 also dissociates from the Notch1-IC-RBP-Jk complex in the nucleus without affecting Notch1-IC protein turnover. Henceforth, the findings of this study may begin to shed some light onto what may be a signal cross-talk mechanism of Notch1 signaling and the Fe65 adaptor protein.
Acknowledgments
We thank Raphael Kopan (Washington University Medical School, St. Louis, MO) for the Notch1-related constructs, Tommaso Russo (Universita di Napoli Federico II, Italy) for the Fe65-related constructs, Thomas Südhof (University of Texas Southwestern, TX) for the Fe65 and Tip60 constructs, and Gerry Melino (University of Leicester, UK) for the Itch-related constructs.
This research was supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF), funded by Ministry of Education, Science and Technology Grants 2009-0071386 and 2011-0014796.
- Notch1-IC
- Notch1 intracellular
- CSL
- CBF1/suppressor of hairless/Lag-1
- NES
- nuclear export sequence
- NEXT
- Notch1 extracellular truncation.
REFERENCES
- 1. Artavanis-Tsakonas S., Matsuno K., Fortini M. E. (1995) Notch signaling. Science 268, 225–232 [DOI] [PubMed] [Google Scholar]
- 2. Egan S. E., St-Pierre B., Leow C. C. (1998) Notch receptors, partners and regulators. From conserved domains to powerful functions. Curr. Top Microbiol. Immunol. 228, 273–324 [DOI] [PubMed] [Google Scholar]
- 3. Lai E. C. (2004) Notch signaling. Control of cell communication and cell fate. Development 131, 965–973 [DOI] [PubMed] [Google Scholar]
- 4. Weinmaster G. (1998) Notch signaling. Direct or what? Curr. Opin. Genet. Dev. 8, 436–442 [DOI] [PubMed] [Google Scholar]
- 5. Lieber T., Kidd S., Young M. W. (2002) Kuzbanian-mediated cleavage of Drosophila Notch. Genes Dev. 16, 209–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Pan D., Rubin G. M. (1997) Kuzbanian controls proteolytic processing of Notch and mediates lateral inhibition during Drosophila and vertebrate neurogenesis. Cell 90, 271–280 [DOI] [PubMed] [Google Scholar]
- 7. Brou C., Logeat F., Gupta N., Bessia C., LeBail O., Doedens J. R., Cumano A., Roux P., Black R. A., Israël A. (2000) A novel proteolytic cleavage involved in Notch signaling. The role of the disintegrin-metalloprotease TACE. Mol. Cell 5, 207–216 [DOI] [PubMed] [Google Scholar]
- 8. Mumm J. S., Kopan R. (2000) Notch signaling. From the outside in. Dev. Biol. 228, 151–165 [DOI] [PubMed] [Google Scholar]
- 9. Mumm J. S., Schroeter E. H., Saxena M. T., Griesemer A., Tian X., Pan D. J., Ray W. J., Kopan R. (2000) A ligand-induced extracellular cleavage regulates γ-secretase-like proteolytic activation of Notch1. Mol. Cell 5, 197–206 [DOI] [PubMed] [Google Scholar]
- 10. Capell A., Steiner H., Romig H., Keck S., Baader M., Grim M. G., Baumeister R., Haass C. (2000) Presenilin-1 differentially facilitates endoproteolysis of the β-amyloid precursor protein and Notch. Nat. Cell Biol. 2, 205–211 [DOI] [PubMed] [Google Scholar]
- 11. De Strooper B., Annaert W., Cupers P., Saftig P., Craessaerts K., Mumm J. S., Schroeter E. H., Schrijvers V., Wolfe M. S., Ray W. J., Goate A., Kopan R. (1999) A presenilin-1-dependent γ-secretase-like protease mediates release of Notch intracellular domain. Nature 398, 518–522 [DOI] [PubMed] [Google Scholar]
- 12. Ray W. J., Yao M., Mumm J., Schroeter E. H., Saftig P., Wolfe M., Selkoe D. J., Kopan R., Goate A. M. (1999) Cell surface presenilin-1 participates in the γ-secretase-like proteolysis of Notch. J. Biol. Chem. 274, 36801–36807 [DOI] [PubMed] [Google Scholar]
- 13. Steiner H., Duff K., Capell A., Romig H., Grim M. G., Lincoln S., Hardy J., Yu X., Picciano M., Fechteler K., Citron M., Kopan R., Pesold B., Keck S., Baader M., Tomita T., Iwatsubo T., Baumeister R., Haass C. (1999) A loss of function mutation of presenilin-2 interferes with amyloid β-peptide production and notch signaling. J. Biol. Chem. 274, 28669–28673 [DOI] [PubMed] [Google Scholar]
- 14. Weinmaster G. (1997) The ins and outs of notch signaling. Mol. Cell Neurosci. 9, 91–102 [DOI] [PubMed] [Google Scholar]
- 15. Gupta-Rossi N., Le Bail O., Gonen H., Brou C., Logeat F., Six E., Ciechanover A., Israël A. (2001) Functional interaction between SEL-10, an F-box protein, and the nuclear form of activated Notch1 receptor. J. Biol. Chem. 276, 34371–34378 [DOI] [PubMed] [Google Scholar]
- 16. Lai E. C. (2002) Protein degradation. Four E3s for the notch pathway. Curr. Biol. 12, R74–78 [DOI] [PubMed] [Google Scholar]
- 17. Minella A. C., Clurman B. E. (2005) Mechanisms of tumor suppression by the SCF(Fbw7). Cell Cycle 4, 1356–1359 [DOI] [PubMed] [Google Scholar]
- 18. Mo J. S., Kim M. Y., Han S. O., Kim I. S., Ann E. J., Lee K. S., Seo M. S., Kim J. Y., Lee S. C., Park J. W., Choi E. J., Seong J. Y., Joe C. O., Faessler R., Park H. S. (2007) Integrin-linked kinase controls Notch1 signaling by down-regulation of protein stability through Fbw7 ubiquitin ligase. Mol. Cell. Biol. 27, 5565–5574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Oberg C., Li J., Pauley A., Wolf E., Gurney M., Lendahl U. (2001) The Notch intracellular domain is ubiquitinated and negatively regulated by the mammalian Sel-10 homolog. J. Biol. Chem. 276, 35847–35853 [DOI] [PubMed] [Google Scholar]
- 20. Wu G., Lyapina S., Das I., Li J., Gurney M., Pauley A., Chui I., Deshaies R. J., Kitajewski J. (2001) SEL-10 is an inhibitor of notch signaling that targets notch for ubiquitin-mediated protein degradation. Mol. Cell. Biol. 21, 7403–7415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Qiu L., Joazeiro C., Fang N., Wang H. Y., Elly C., Altman Y., Fang D., Hunter T., Liu Y. C. (2000) Recognition and ubiquitination of Notch by Itch, a hect-type E3 ubiquitin ligase. J. Biol. Chem. 275, 35734–35737 [DOI] [PubMed] [Google Scholar]
- 22. Perry W. L., Hustad C. M., Swing D. A., O'Sullivan T. N., Jenkins N. A., Copeland N. G. (1998) The itchy locus encodes a novel ubiquitin protein ligase that is disrupted in a18H mice. Nat. Genet. 18, 143–146 [DOI] [PubMed] [Google Scholar]
- 23. Liu Y. C. (2007) The E3 ubiquitin ligase Itch in T cell activation, differentiation, and tolerance. Semin. Immunol. 19, 197–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Cornell M., Evans D. A., Mann R., Fostier M., Flasza M., Monthatong M., Artavanis-Tsakonas S., Baron M. (1999) The Drosophila melanogaster Suppressor of deltex gene, a regulator of the Notch receptor signaling pathway, is an E3 class ubiquitin ligase. Genetics 152, 567–576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. McLoughlin D. M., Miller C. C. (2008) The FE65 proteins and Alzheimer's disease. J. Neurosci. Res. 86, 744–754 [DOI] [PubMed] [Google Scholar]
- 26. Russo T., Faraonio R., Minopoli G., De Candia P., De Renzis S., Zambrano N. (1998) Fe65 and the protein network centered around the cytosolic domain of the Alzheimer's β-amyloid precursor protein. FEBS Lett. 434, 1–7 [DOI] [PubMed] [Google Scholar]
- 27. Gandy S. (2005) The role of cerebral amyloid β accumulation in common forms of Alzheimer disease. J. Clin. Invest. 115, 1121–1129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kim S. Y., Kim M. Y., Mo J. S., Park H. S. (2007) Notch1 intracellular domain suppresses APP intracellular domain-Tip60-Fe65 complex mediated signaling through physical interaction. Biochim. Biophys. Acta 1773, 736–746 [DOI] [PubMed] [Google Scholar]
- 29. Nakaya T., Kawai T., Suzuki T. (2008) Regulation of FE65 nuclear translocation and function by amyloid β-protein precursor in osmotically stressed cells. J. Biol. Chem. 283, 19119–19131 [DOI] [PubMed] [Google Scholar]
- 30. Nakaya T., Suzuki T. (2006) Role of APP phosphorylation in FE65-dependent gene transactivation mediated by AICD. Genes Cells 11, 633–645 [DOI] [PubMed] [Google Scholar]
- 31. Scheinfeld M. H., Ghersi E., Laky K., Fowlkes B. J., D'Adamio L. (2002) Processing of β-amyloid precursor-like protein-1 and -2 by γ-secretase regulates transcription. J. Biol. Chem. 277, 44195–44201 [DOI] [PubMed] [Google Scholar]
- 32. Cao X., Südhof T. C. (2001) A transcriptionally [correction of transcriptively] active complex of APP with Fe65 and histone acetyltransferase Tip60. Science 293, 115–120 [DOI] [PubMed] [Google Scholar]
- 33. Chen C., Okayama H. (1987) High-efficiency transformation of mammalian cells by plasmid DNA. Mol. Cell. Biol. 7, 2745–2752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kim M. Y., Ann E. J., Kim J. Y., Mo J. S., Park J. H., Kim S. Y., Seo M. S., Park H. S. (2007) Tip60 histone acetyltransferase acts as a negative regulator of Notch1 signaling by means of acetylation. Mol. Cell. Biol. 27, 6506–6519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Mo J. S., Ann E. J., Yoon J. H., Jung J., Choi Y. H., Kim H. Y., Ahn J. S., Kim S. M., Kim M. Y., Hong J. A., Seo M. S., Lang F., Choi E. J., Park H. S. (2011) Serum- and glucocorticoid-inducible kinase 1 (SGK1) controls Notch1 signaling by downregulation of protein stability through Fbw7 ubiquitin ligase. J. Cell Sci. 124, 100–112 [DOI] [PubMed] [Google Scholar]
- 36. Kageyama R., Ohtsuka T. (1999) The Notch-Hes pathway in mammalian neural development. Cell Res. 9, 179–188 [DOI] [PubMed] [Google Scholar]
- 37. Rock K. L., Gramm C., Rothstein L., Clark K., Stein R., Dick L., Hwang D., Goldberg A. L. (1994) Inhibitors of the proteasome block the degradation of most cell proteins and the generation of peptides presented on MHC class I molecules. Cell 78, 761–771 [DOI] [PubMed] [Google Scholar]
- 38. Dho S. E., French M. B., Woods S. A., McGlade C. J. (1999) Characterization of four mammalian numb protein isoforms. Identification of cytoplasmic and membrane-associated variants of the phosphotyrosine binding domain. J. Biol. Chem. 274, 33097–33104 [DOI] [PubMed] [Google Scholar]
- 39. McGill M. A., McGlade C. J. (2003) Mammalian numb proteins promote Notch1 receptor ubiquitination and degradation of the Notch1 intracellular domain. J. Biol. Chem. 278, 23196–23203 [DOI] [PubMed] [Google Scholar]
- 40. Verdi J. M., Schmandt R., Bashirullah A., Jacob S., Salvino R., Craig C. G., Program A. E., Lipshitz H. D., McGlade C. J. (1996) Mammalian NUMB is an evolutionarily conserved signaling adapter protein that specifies cell fate. Curr. Biol. 6, 1134–1145 [DOI] [PubMed] [Google Scholar]
- 41. Chastagner P., Israël A., Brou C. (2008) AIP4/Itch regulates Notch receptor degradation in the absence of ligand. PLoS ONE 3, e2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. McGill M. A., Dho S. E., Weinmaster G., McGlade C. J. (2009) Numb regulates post-endocytic trafficking and degradation of Notch1. J. Biol. Chem. 284, 26427–26438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wakamatsu Y., Maynard T. M., Jones S. U., Weston J. A. (1999) NUMB localizes in the basal cortex of mitotic avian neuroepithelial cells and modulates neuronal differentiation by binding to NOTCH-1. Neuron 23, 71–81 [DOI] [PubMed] [Google Scholar]
- 44. Kesavapany S., Banner S. J., Lau K. F., Shaw C. E., Miller C. C., Cooper J. D., McLoughlin D. M. (2002) Expression of the Fe65 adapter protein in adult and developing mouse brain. Neuroscience 115, 951–960 [DOI] [PubMed] [Google Scholar]
- 45. Sudol M., Hunter T. (2000) NeW wrinkles for an old domain. Cell 103, 1001–1004 [DOI] [PubMed] [Google Scholar]
- 46. Sudol M., Sliwa K., Russo T. (2001) Functions of WW domains in the nucleus. FEBS Lett. 490, 190–195 [DOI] [PubMed] [Google Scholar]
- 47. von Rotz R. C., Kohli B. M., Bosset J., Meier M., Suzuki T., Nitsch R. M., Konietzko U. (2004) The APP intracellular domain forms nuclear multiprotein complexes and regulates the transcription of its own precursor. J. Cell Sci. 117, 4435–4448 [DOI] [PubMed] [Google Scholar]
- 48. Telese F., Bruni P., Donizetti A., Gianni D., D'Ambrosio C., Scaloni A., Zambrano N., Rosenfeld M. G., Russo T. (2005) Transcription regulation by the adaptor protein Fe65 and the nucleosome assembly factor SET. EMBO Rep. 6, 77–82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Diévart A., Beaulieu N., Jolicoeur P. (1999) Involvement of Notch1 in the development of mouse mammary tumors. Oncogene 18, 5973–5981 [DOI] [PubMed] [Google Scholar]