Background: Half-life extension has become increasingly important for therapeutic proteins.
Results: Fusion of different bacterial immunoglobulin-binding domains to small recombinant antibodies prolonged their half-life to varying extents.
Conclusion: Fusion of domain C3 of streptococcal protein G showed the best effects, thus representing a promising module for half-life extension of small-sized therapeutics.
Significance: This study further established immunoglobulin-binding domains as suitable half-life extension modules.
Keywords: Antibody Engineering, Fusion Protein, Pharmacokinetics, Protein-Protein Interactions, Streptococcus, Half-life Extension, Immunoglobulin-binding Domain, Plasma Half-life, Recombinant Antibodies
Abstract
Many therapeutic proteins possessing a small size are rapidly cleared from circulation. Half-life extension strategies have therefore become increasingly important to improve the pharmacokinetic and pharmacodynamic properties of protein therapeutics. Here, we performed a comparative analysis of the half-life extension properties of various bacterial immunoglobulin-binding domains (IgBDs) derived from Staphylococcus protein A (SpA), Streptococcus protein G (SpG), and Finegoldia (formerly Peptostreptococcus) protein L (PpL). These domains, composed of 50–60 amino acid residues, were fused to the C terminus of a single-chain Fv and a bispecific single-chain diabody, respectively. All fusion proteins were produced in mammalian cells and retained their antigen-binding properties. The half-lives of the antibody molecules were prolonged to varying extents for the different IgBDs. The strongest effects in mice were observed for domain C3 of SpG (SpGC3) followed by domains B and D of SpA, suggesting that SpGC3 is particularly useful to extend the plasma half-life of small proteins.
Introduction
With a growing number of small protein therapeutics being developed, half-life extension strategies have become increasingly important to improve their pharmacokinetic and pharmacodynamic properties (1, 2). These strategies include approaches to increase the hydrodynamic radius of the protein, e.g. by hyperglycosylation, coupling to polyethylene glycol (PEG) chains or other hydrophilic polymers, and fusion to PEG-mimetic polypeptide chains or long circulation plasma protein and fragments thereof. Fusion to the Fc region of IgG molecules or to serum albumin has been shown to prolong the half-life of protein therapeutics, e.g. growth factors, hormones, and cytokines, and several of these molecules are already approved or in clinical development (2–4). Additionally, noncovalent interaction with plasma proteins has emerged as a half-life extension strategy. Here, most studies have focused so far on albumin-binding moieties, including peptides, antibody fragments, single-domain antibodies, and bacterial albumin-binding domains derived from protein A, fused or conjugated to a therapeutic compound (1).
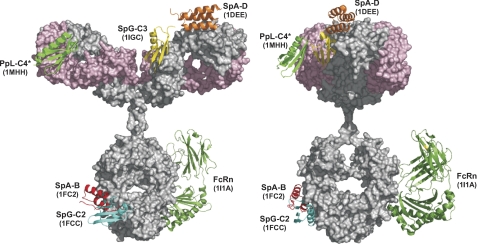
Recently, we have shown that fusion of an scDb2 to the immunoglobulin-binding domain B (IgBD) from protein A (SpAB) also results in a prolonged plasma half-life (5). Immunoglobulin-binding domains (IgBDs) are known from various bacterial proteins, including staphylococcal protein A (SpA), streptococcal protein G (SpG), and peptostreptococcal protein L (PpL) (6, 7). These IgBDs are composed of 50–60 amino acid residues forming either a 3-α-helix bundle or a compact structure composed of a 4-stranded β-sheet packed against a central α-helix (6). Primary binding sites of the domains from SpA and SpG on IgG have been localized at the CH2-CH3 interface of IgG heavy chain Fc fragments, overlapping with the binding site of FcRn (8). Furthermore, SpA and SpG but also PpL interact with different regions of Fab fragments (Fig. 1). Thus, binding to variable heavy chain (VH) domains belonging to the VH3 family was shown for the domain D of SpA, and this site is structurally distinguishable from its Fc-binding site (9). Domain C4 from PpL is capable of binding to certain subgroups of Vκ light chain domains (10, 11), and domain C3 of SpG has been shown to bind also to the CH1 domain of IgG molecules (12).
FIGURE 1.
Summary of IgBD bound to human IgG1. IgBDs from protein A (SpAB, SpAD), protein G (SpGC2, SpGC3), and protein L (PpLC4*) in complex with IgG were visualized on a human IgG1 model (29). In addition, the extracellular region of FcRn bound to the Fc region was included (8). Protein Data Bank entries are indicated for each IgBD and the FcRn. The structures were visualized with PyMOL (30). The IgG1 heavy chain is shown in gray, the light chain in light pink.
Here, we performed a comparative analysis of the capacity of various IgBDs derived from SpA, SpG, and PpL to extend the half-life of small recombinant antibodies, a scDb possessing a molecular mass of 55 kDa and a single-chain Fv fragment (scFv) with a molecular mass of 28 kDa. We show that half-lives in mice are prolonged to various extents depending on the applied IgBDs. The longest terminal half-life was seen for the fusion proteins containing domain C3 of SpG opening new applications as half-life extension module.
EXPERIMENTAL PROCEDURES
Materials
HRP-conjugated anti-His tag antibody was purchased from Santa Cruz Biotechnology (Santa Cruz, CA), and FITC-conjugated mouse anti-His tag antibody was from Dianova (Hamburg, Germany). Carcinoembryonic antigen (CEA) obtained from Europa Bioproducts (Cambridge, UK). Human and mouse serum IgG was purchased from Sigma. Human and mouse Fab and Fc fragments generated from serum IgG were purchased from EMELCA Bioscience (Breda, The Netherlands). The human colon adenocarcinoma cell line LS174T was purchased from ECACC (Wiltshire, UK) and cultured in RPMI 1640 medium, 5% FBS, 2 mm glutamine (Invitrogen). Jurkat cells were cultured in RPMI 1640 medium, 10% FBS, 2 mm glutamine. CD1 mice were purchased from Elevage Janvier (Le Genest St. Isle, France).
Construction and Production of scDb-IgBD and scFv-IgBD Fusion Proteins
DNA encoding the IgBDs (IgBD SpAB, SpAD, SpAEZ4, SpGC3, and PpLC4*), including a His6 tag at the C terminus, were synthesized by GeneArt (Regensburg, Germany) adding a NotI restriction site at the 5′ end and an EcoRI and XbaI restriction site at the 3′ end. IgBD SpAB was cloned into mammalian expression vector pSecTagA-scDb-CEACD3-ABD-L (13) cut with NotI and XbaI substituting the ABD-L domain and generating pSecTagA-scDb-CEACD3-SpAB. The IgBD SpAD, SpAEZ4, SpGC3, and PpLC4* were then cloned into pSecTagA-scDb-CEACD3-SpAB as NotI-XbaI fragments substituting the SpAB IgBD. ScFv-IgBD fusion proteins were generated cloning the IgBDs as NotI-EcoRI fragments into vector pSecTagA-scFv-CEA-4–1BBL (14). HEK293 cells were stably transfected with the plasmids, and the antibody-IgBD fusion proteins were purified from cell culture supernatant by IMAC essentially as described previously (15).
ELISA
CEA (3 μg/ml) or immunoglobulins (IgG at 1 μg/ml; IgM, IgA, and IgE at 10 μg/ml) were coated overnight at 4 °C, and the remaining binding sites were blocked with 2% (w/v) dry milk/PBS. Purified recombinant antibodies and serum samples were titrated in duplicate and incubated for 1 h at room temperature. Detection was performed with mouse HRP-conjugated anti-His tag antibody using TMB substrate (0.1 mg/ml TMB, 100 mm sodium acetate buffer, pH 6.0, 0.006% H2O2). The reaction was stopped with 50 μl of 1 m H2SO4. Absorbance was measured at 450 nm in an ELISA reader.
Flow Cytometry
Binding to CEA- and CD3-expressing cells was determined by flow cytometry (16). LS174T or Jurkat cells (2.5 × 105) were incubated with a dilution series of antibodies for 2 h at 4 °C. Cells were then washed with PBS, 2% FBS, 0.02% NaN3 (PBA), and bound antibodies were detected with FITC-conjugated mouse anti-His tag antibody. Data were fitted with Prism software (GraphPad, La Jolla, CA) from two independent binding curves.
Size Exclusion Chromatography
Purity and Stokes radii of the scDb-IgBD and scFv-IgBD fusions proteins were analyzed by HPLC size exclusion chromatography using a BioSuite 250 column (Waters Corporation, Milford, MA) and a flow rate of 0.5 ml/min. The following standard proteins were used: thyroglobulin, apoferritin, β-amylase, BSA, carbonic anhydrase, and cytochrome c.
Affinity Measurements
Affinities of IgBD fusion proteins for human and mouse serum IgG as well as Fab fragments thereof at neutral or acidic pH were determined by quartz crystal microbalance measurements (A-100 C-Fast system; Attana, Stockholm, Sweden). IgGs as well as Fab fragments were chemically immobilized on a low nonspecific binding carboxyl sensor chip according to the manufacturer's protocol at a density resulting in a signal increase of 65–95 Hz. Binding experiments were performed in PBST (0.1% Tween 20), pH 7.4 or pH 6.0, at a flow rate of 25 μl/min. The chip was regenerated with 25 μl of 10 mm glycine-HCl, pH 3.0. Before each measurement, a base line was measured which was subtracted from the binding curve. Data were collected by Attester 3.0 (version 3.1.1.8; Attana) and analyzed by Attache Office Evaluation Software (version 3.3.4; Attana), using a mass transport limited model for curve fitting.
Pharmacokinetics
Animal care and all experiments performed were in accordance with federal guidelines and have been approved by university and state authorities. CD1 mice (8–16 weeks, 30–40 g) received an intravenous injection of 25 μg of the IgBD fusion proteins in a total volume of 150 μl. In time intervals of 3 min, 30 min, 1 h, 2 h, 6 h, 1 day, 3 days, and 7 days, blood samples (50 μl) were taken from the tail and incubated on ice. Clotted blood was centrifuged at 13,000 × g for 30 min at 4 °C, and serum samples were stored at −20 °C. Serum concentrations of CEA-binding recombinant antibodies were determined by ELISA (as described above). For comparison, the first value (3 min) was set to 100%. Half-lives (t½α, t½β) and AUC were calculated with Excel. Terminal half-lives (t½β) were calculated using the last three or four serum concentrations shown in Fig. 1. For statistics, Student's t test was applied. Results are shown as mean value ± S.D.
RESULTS
ScDb-IgBD and scFv-IgBD fusion proteins were generated by fusing IgBDs from SpA, SpG, or PpL to the C terminus of a bispecific single-chain diabody directed against CEA and CD3 (scDb CEACD3) or a single-chain Fv directed against CEA (scFv CEA) (17) (Fig. 2a). From SpA we used domain B (18), domain D (9), and a modified version of domain E, which was described to exhibit increased thermal stability (19). From SpG we used domain C3 (12), and from PpL we used a mutated version of domain C4 (C4*) carrying mutations disrupting the second binding site for Fab fragments (10).
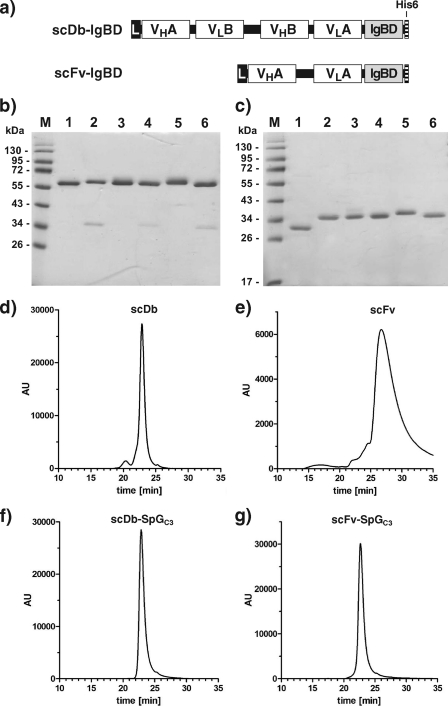
FIGURE 2.
Construction of scDb-IgBDs and scFv-IgBDs. a, composition of the scDb-IgBD and scFv-IgBD fusion protein. IgBDs are fused to the C terminus of a bispecific scDb or a scFv. b, SDS-PAGE analysis of purified scDb-CEACD3 (lane 1), scDb-SpAB (lane 2), scDb-SpAD (lane 3), scDb-SpAE4 (lane 4), scDb-SpGC3 (lane 5), and scDb-PpLC4* (lane 6) under reducing conditions. c, SDS-PAGE analysis of purified anti-CEA scFv (lane 1), scFv-SpAB (lane 2), scFv-SpAD (lane 3), scFv-SpAE4 (lane 4), scFv-SpGC3 (lane 5), and scFv-PpLC4* (lane 6) under reducing conditions. Two micrograms of protein were analyzed per lane, and the gel was stained with Coomassie Brilliant Blue G-250. Lane M, molecular mass standards. d–g, purified scDb, scFv as well as the scDb-SpGC3 and scFv-SpGC3 fusion proteins were analyzed by size exclusion chromatography.
The scDb-IgBD and scFv-IgBD fusion proteins were produced in stably transfected HEK293 cells and purified from the supernatant by immobilized metal affinity chromatography. Yields were in the range of 2–22 mg/liter supernatant. Purity of the fusion proteins was confirmed by SDS-PAGE analysis (Fig. 2, b and c). The scDb-IgBD fusion proteins migrated with apparent molecular masses of around 60 kDa and the scFv-IgBD fusion proteins with apparent molecular masses of ∼35 kDa, corresponding to the calculated molecular masses (Table 1). Purity was further confirmed by size exclusion chromatography. All fusion proteins showed a single peak corresponding to monomeric molecules (Fig. 2, d–g). The measured Stokes radii of the fusion proteins were in the range of 2.2–2.7 nm. Interestingly, the Stokes radii of the scFv-IgBD fusion proteins were similar to those of the scDb-IgBD fusion proteins, whereas the unmodified scFv had a Stokes radius of 1.2 nm (Table 1).
TABLE 1.
Stokes radii and pharmacokinetic properties
Data are shown as mean ± S.D.
| Construct | Molecular mass | Sr | t½α | t½β | AUC |
|---|---|---|---|---|---|
| kDa | nm | h | h | %h | |
| scDb | 54.5 | 2.6 | 0.2 ± 0.1 | 1.3 ± 0.2 | 52 ± 16 |
| scDb-SpAB | 59.9 | 2.6 | 2.2 ± 0.5 | 11.8 ± 1.6 | 1,407 ± 352 |
| scDb-SpAD | 60.1 | 2.2 | 2.0 ± 0.8 | 9.0 ± 4.6 | 1,042 ± 403 |
| scDb-SpAEZ4 | 59.7 | 2.4 | 1.0 ± 0.3 | 4.2 ± 0.6 | 435 ± 79 |
| scDb-SpGC3 | 59.6 | 2.7 | 2.8 ± 1.2 | 23.3 ± 5.9 | 1,879 ± 279 |
| scDb-PpLC4* | 60.3 | 2.3 | 0.8 ± 0.1 | 2.4 ± 0.6 | 233 ± 136 |
| scFv | 26.7 | 1.2 | 0.1 ± 0.01 | 0.6 ± 0.2 | 16 ± 4 |
| scFv-SpAB | 33.6 | 2.7 | 1.0 ± 0.1 | 4.4 ± 0.7 | 377 ± 200 |
| scFv-SpAD | 33.8 | 2.3 | 1.1 ± 0.3 | 4.9 ± 1.0 | 434 ± 222 |
| scFv-SpAEZ4 | 33.5 | 2.4 | 0.4 ± 0.2 | 1.1 ± 0.4 | 63 ± 29 |
| scFv-SpGC3 | 33.4 | 2.7 | 0.8 ± 0.1 | 20.8 ± 11.0 | 1,040 ± 589 |
| scFv-PpLC4* | 34.0 | 2.5 | 0.6 ± 0.2 | 2.7 ± 1.0 | 207 ± 128 |
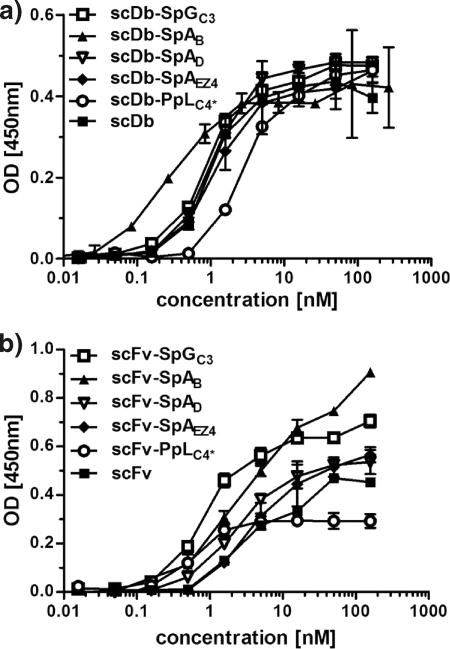
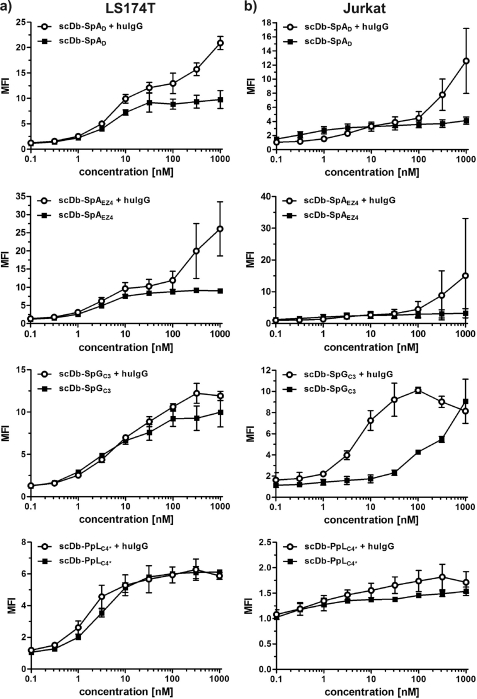
All fusion proteins showed binding to CEA in ELISA (Fig. 3) and to CEA-expressing LS174T in flow cytometry studies, with EC50 values in the low nanomolar range (Fig. 4). The bispecific scDb fusion proteins showed also binding to CD3-expressing Jurkat cells (Fig. 4). In the flow cytometry experiments, no reduction in binding was observed in the presence of excess amounts of human IgG. In contrast, an increased binding was seen, indicating that binding of the IgBD fusion protein to IgG increases sensitivity, presumably due to avidity effects.
FIGURE 3.
Binding of scDb-IgBD and scFv-IgBD fusion proteins to CEA in ELISA. Increasing concentrations of the scDb-IgBD (a) or scFv-IgBD (b) fusion proteins were analyzed for binding to immobilized CEA.
FIGURE 4.
Cell binding of scDb-IgBD fusion proteins. Binding of scDb-IgBD fusion proteins to CD3-positive Jurkat cells and CEA-expressing LS174T cells was analyzed in the absence or presence of human IgG (100 μg/ml IgG), respectively. Binding was detected with a FITC-labeled anti-His tag antibody. Binding is shown as mean fluorescence intensity (MFI) calculated from MFI of cells incubated with fusion protein/MFI of cells alone (background).
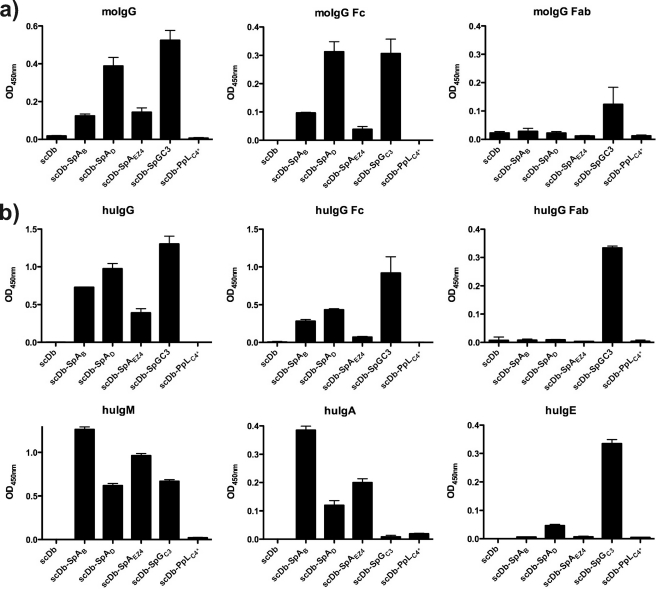
Binding to various extent of the scDb-IgBD fusion proteins to human and mouse immunoglobulins was demonstrated in ELISA (Fig. 5). Except for the PpLC4* fusion protein, all other IgBD fusion proteins showed binding to mouse and human serum IgG. Strongest binding to mouse and human IgG was seen for SpGC3 followed by SpAD and SpAB. This pattern was also seen using Fc fragments of mouse and human serum IgG. In contrast, binding to Fab fragments of mouse and human serum IgG was observed only for scDb-SpGC3, demonstrating that SpGC3 can bind to the Fc and Fab fragments of human and mouse IgG. In these assays, binding of SpGC3 to human and mouse Fab fragments was weaker than binding to Fc fragments. Furthermore, the SpA- and SpG-IgBD fusion proteins bound to human IgM. Binding to human IgA was observed for the three SpA-IgBD fusion proteins, whereas some weak binding to human IgE was observed for scDb-SpGC3 and to some extent for scDb-SpAD, presumably through binding to the Fc region. Unmodified scDb included as negative control did not reveal any binding to the various Ig preparations.
FIGURE 5.
Binding of scDb-IgBD to IgG, Fab, and Fc analyzed by ELISA. Human and mouse IgG as well as Fab and Fc fragments thereof were immobilized, and binding of the scDb-IgBD fusion proteins was determined at a concentration of 100 nm. Furthermore, the scDb-IgBD fusion proteins were analyzed for binding to human IgM, human IgA, and human IgE.
In further experiments, we measured binding to mouse and human serum IgG and its Fab fragments by quartz crystal microbalance (Table 2). Affinities of the SpA- and SpG-IgBD fusion proteins for human IgG were in the low nanomolar range. Because of the heterogeneity of the IgG preparations composed of different IgG subclasses the obtained affinities represent most like average values. Strongest binding was observed for the SpGC3 fusion proteins. The measured affinities for mouse IgG were 2–10-fold weaker. In these experiments, we could demonstrate a weak binding of the scDb-PpLC4* fusion protein to human and mouse IgG as well as human Fab fragments. An affinity of 160 nm for human IgG was described for this mutated domain, similar to the affinity of the wild-type domain (130 nm) (20). We measured a much lower affinity of only 1.5 μm for the scDb-PpLC4* fusion protein, indicating that fusion to a protein negatively influences interaction with IgG. An affinity for Fab fragments of 190 nm was determined for scDb-SpGC3, and some very weak binding to human Fab with an KD of 3 μm was measured for scDb-PpLC4*. We further analyzed binding of scDb-SpGC3 to human serum IgG Fc fragments and to a humanized IgG1 antibody (Trastuzumab) (Table 2). At neutral pH, the fusion protein showed an affinity of 10 nm for Fc fragments and 5 nm for human IgG1. These experiments confirmed a high affinity of scDb-SpGC3 for human IgG Fc and further demonstrate that SpGC3 is capable of binding to IgG Fab and Fc fragments. At acidic pH, the affinity of SpGC3 was unaltered or even slightly increased, whereas SpAB and SpAD exhibited reduced affinities at pH 6, in accordance with data described for protein A and protein G (21) (Table 2).
TABLE 2.
Affinities of scDb-IgBD fusion proteins for human and mouse IgG and Fab fragments thereof at neutral or acidic pH determined by quartz crystal microbalance (QCM) measurements
| Construct | Ig | Human KD |
Mouse KD |
||
|---|---|---|---|---|---|
| pH 7.4 | pH 6.0 | pH 7.4 | pH 6.0 | ||
| nm | nm | nm | nm | ||
| SpAB | IgG | 9 | 350 | 380 | 15,000 |
| SpAD | IgG | 22 | 148 | 50 | 773 |
| IgG Fab | —a | NDb | — | ND | |
| SpAEZ4 | IgG | 37 | 654 | 78 | 2,480 |
| IgG Fab | — | ND | — | ND | |
| SpGC3 | IgG | 6 | 5 | 67 | 46 |
| IgG Fab | 190 | 150 | 190 | 190 | |
| IgG Fc | 10 | 3.6 | ND | ND | |
| humanized IgG1c | 5 | 4.5 | |||
| PpLC4* | IgG | 1,510 | 1,130 | — | ND |
| IgG Fab | 3,100 | ND | — | ND | |
a —, no binding detected.
b N.D., not determined.
c Trastuzumab was used as IgG1 probe.
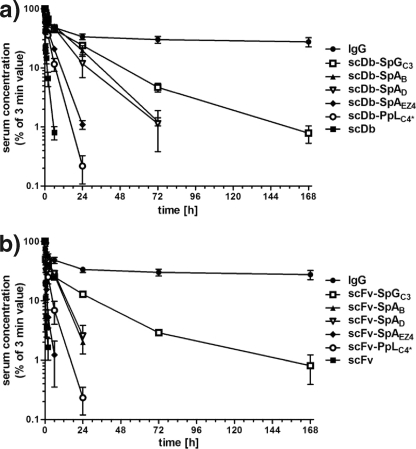
Half-lives of scDb-IgBD and scFv-IgBD fusion proteins were analyzed in CD1 mice receiving a single intravenous injection of the IgBD fusion proteins. Serum concentrations were determined by ELISA, i.e. detecting molecules with CEA-binding activity. The unmodified antibodies were rapidly cleared from circulation with terminal half-lives of 1.3 h for the scDb and 0.6 h for the scFv. The half-lives of the antibody IgBD fusion proteins were prolonged to varying extent (Fig. 6 and Table 1). The longest half-life was found for the SpGC3 fusion proteins (terminal half-life of 23.3 and 20.8 h for scDb-SpGC3 and scFv-SpGC3, respectively). In contrast, only a marginal improvement was observed for the SpAEZ4 and the PpLC4* fusion proteins. The SpAB and SpAD fusion proteins showed an intermediate extension of the half-life (terminal half-life of 9–11.8 h for the scDb fusion proteins and 4.4–4.9 h for the scFv fusion proteins, respectively) (Table 1). The extended half-life of the IgBD fusion proteins also resulted in an increased AUC. The strongest increase was determined for the SpGC3 fusion proteins (36-fold for scDb-SpGC3 and 65-fold for scFv-SpGC3). The AUCs of the SpAB and SpAD fusion proteins were increased 20–27-fold.
FIGURE 6.
Plasma half-life of scDb-IgBD and scFv-IgBD fusion proteins compared with unmodified proteins (scDb, scFv) and a chimeric IgG. scDb-IgBD (a) and scFv-IgBD (b) fusion proteins were injected intraperitoneally into CD1 mice (25 μg/animal), and serum concentrations of the antibody molecules were determined at different time points by ELISA. Data were normalized considering maximal concentration at the first time point (3 min).
DISCUSSION
In this study, we have generated a panel of novel IgBD fusion proteins using IgBDs from various bacterial immunoglobulin receptors. Although all fusion proteins retained their antigen-binding activity, differences were observed in their capacity to interact with immunoglobulins.
The unmodified antibodies, an scDb of 55 kDa and an scFv of 28 kDa, were rapidly cleared from circulation with terminal half-lives of 1.3 h for the scDb and 0.6 h for the scFv. In a previous study of the same scDb used here, we showed that this is mainly due to rapid renal clearance (22), reflected also by the Stokes radius of only 2.6 nm, which allows almost unhindered passage through the glomerular filtration barrier (23). Only marginal effects on the half-life was seen for the SpAEZ4 and the PpLC4* fusion proteins with 2–3-fold increased half-lives. Fusion of SpAD to the scDb resulted in a similar half-life extension (7-fold prolonged terminal half-life) observed in a previous study with an scDb-SpAB fusion protein (5), which was also seen to a similar extent for the two scFv fusion proteins. Compared with the SpAB and SpAD fusion proteins, we found that the SpGC3 fusion proteins exhibited even further extended half-lives, which was 2.5-fold increased for the scDb fusion protein and ∼5-fold increased for the scFv fusion protein, corresponding to 2–3-fold increased AUCs. These findings illustrate that the IgBDs are capable of binding to serum immunoglobulins in vivo and that the IgBDs exhibit different capacities to extend the plasma half-life of two small recombinant antibodies.
The half-life of recombinant proteins bound to serum immunoglobulins is influenced by several factors, including (i) the selectivity for long circulating immunoglobulins; (ii) the stability of the complexes in plasma, i.e. at neutral pH; (iii) the stability of the complex in the sorting endosome, i.e. at acidic pH; and (iv) interference of bound IgBD fusion proteins with recycling of the complexes by the FcRn.
A reduced binding at acidic pH might result in dissociation of the IgBD fusion proteins from bound immunoglobulins in the acidic environment of the early and recycling endosomes (pH ∼6.3–6.8) (24) leading to intracellular degradation of the fusion proteins. Of note, all SpA-IgBD fusion proteins showed a markedly reduced affinity at pH 6, whereas the SpGC3 fusion protein exhibited an unaltered or even increased affinity. This indicates that the SpGC3-Ig complexes are stable in the acidic environment of the sorting endosome enabling recycling into the bloodstream.
All IgBDs from SpA and SpG bind to the Fc region of IgG and are also described to be capable of binding to Fab fragments, although in our studies we could not detect binding of the SpA-IgBD fusion proteins to human or mouse Fab fragments generated from serum IgG, indicating a rather low affinity for Fab fragments. This is in accordance with data obtained for the individual IgBDs from SpA where affinities for VH3-containing scFv fragments of 2.6–14 μm were determined by BIAcore measurements (25). Furthermore, they differ in their binding sites on Fab arms. Whereas SpA domains bind to the variable heavy chain domains (VH) belonging to the VH3 germ line family, the SpG domain binds to the constant heavy chain domain 1 (CH1) at a region highly conserved between the different human IgG subclasses. Thus, interaction with the VH domains results in binding to all immunoglobulin classes, whereas binding to CH1 should be rather selective for IgG molecules. Because the different immunoglobulin classes exhibit different half-lives, selective binding of the IgBD fusion proteins to the Fab arms can directly influence half-life.
Finally, efficient FcRn-mediated recycling requires that the IgBDs do not interfere with binding of IgG to the FcRn. The Fc binding sites of the IgBDs analyzed in this study are located at the CH2-CH3 domain interface of the Fc region and are therefore overlapping with the binding site for FcRn (8, 26, 27) (see also Fig. 1). Our binding studies indicate that the interaction of the SpA-IgBD fusion proteins, and also the SpGC3 fusion proteins, with IgG is dominated by binding to the Fc region. Previously, it was shown that a genetically engineered functional analog of SpAB (domain Z) inhibits binding of IgG to FcRn (28), suggesting that the SpA-IgBD fusion proteins interfere strongly with FcRn recycling of the fusion proteins. However, the SpGC3 fusion proteins do not reach the long half-life of IgG, which is in the range of 1–2 weeks in mice, indicating that also the Fc-binding site of SpGC3 interferes with recycling by the FcRn, leaving room for further improvements.
Overall, we established for the different IgBD fusion proteins differences in the pH stability of binding and the selectivity for different immunoglobulin regions, which might explain the observed differences in half-lives between the SpA-IgBD and the SpGC3 fusion proteins. The remarkably long half-life mediated by SpGC3 makes this domain a promising module for half-life extension of small sized therapeutics.
Acknowledgment
We thank Prof. Hans-Heinrich Heidtmann (St. Joseph Hospital, Bremerhaven, Germany) for providing Trastuzumab.
This work was supported by Deutsche Forschungsgemeinschaft Grant Ko1461/5.
- scDb
- single-chain diabody
- CEA
- carcinoembryonic antigen
- AUC
- area-under-the-curve
- FcRn
- neonatal Fc receptor
- IgBD
- immunoglobulin-binding domain
- PpL
- Peptostreptococcus protein L
- scFv
- single-chain fragment variable
- SpA
- Staphylococcus protein A
- SpG
- Streptococcus protein G.
REFERENCES
- 1. Kontermann R. E. (2009) Strategies to extend plasma half-lives of recombinant antibodies. BioDrugs 23, 93–109 [DOI] [PubMed] [Google Scholar]
- 2. Kontermann R. E. (2011) Strategies for extended serum half-life of protein therapeutics. Curr. Opin. Biotechnol. 22, 868–876 [DOI] [PubMed] [Google Scholar]
- 3. Huang C. (2009) Receptor-Fc fusion therapeutics, traps, and MIMETIBODY technology. Curr. Opin. Biotechnol. 20, 692–699 [DOI] [PubMed] [Google Scholar]
- 4. Chuang V. T., Kragh-Hansen U., Otagiri M. (2002) Pharmaceutical strategies utilizing recombinant human serum albumin. Pharm. Res. 19, 569–577 [DOI] [PubMed] [Google Scholar]
- 5. Unverdorben F., Schwarz A., Richter F., Hutt M., Kontermann R. E. (2012) Half-life extension of a single-chain diabody by fusion to domain B of staphylococcal protein A. Protein Eng. Design Sel., in press [DOI] [PubMed] [Google Scholar]
- 6. Tashiro M., Montelione G. T. (1995) Structures of bacterial immunoglobulin-binding domains and their complexes with immunoglobulins. Curr. Opin. Struct. Biol. 5, 471–481 [DOI] [PubMed] [Google Scholar]
- 7. Sidorin E. V., Solov'eva T. F. (2011) IgG-binding proteins of bacteria. Biochemistry (Mosc.) 76, 363–378 [DOI] [PubMed] [Google Scholar]
- 8. Burmeister W. P., Huber A. H., Bjorkman P. J. (1994) Crystal structure of the complex of rat neonatal Fc receptor with Fc. Nature 372, 379–383 [DOI] [PubMed] [Google Scholar]
- 9. Graille M., Stura E. A., Corper A. L., Sutton B. J., Taussig M. J., Charbonnier J. B., Silverman G. J. (2000) Crystal structure of a Staphylococcus aureus protein A domain complexed with the Fab fragment of a human IgM antibody: structural basis for recognition of B-cell receptors and superantigen activity. Proc. Natl. Acad. Sci. U.S.A. 97, 5399–5404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Graille M., Harrison S., Crump M. P., Findlow S. C., Housden N. G., Muller B. H., Battail-Poirot N., Sibaï G., Sutton B. J., Taussig M. J., Jolivet-Reynaud C., Gore M. G., Stura E. A. (2002) Evidence for plasticity and structural mimicry at the immunoglobulin light chain-protein L interface. J. Biol. Chem. 277, 47500–47506 [DOI] [PubMed] [Google Scholar]
- 11. Akerström B., Björck L. (1989) Protein L: an immunoglobulin light chain-binding bacterial protein. Characterization of binding and physicochemical properties. J. Biol. Chem. 264, 19740–19746 [PubMed] [Google Scholar]
- 12. Derrick J. P., Wigley D. B. (1994) The third IgG-binding domain from streptococcal protein G: an analysis by x-ray crystallography of the structure alone and in a complex with Fab. J. Mol. Biol. 243, 906–918 [DOI] [PubMed] [Google Scholar]
- 13. Hopp J., Hornig N., Zettlitz K. A., Schwarz A., Fuss N., Müller D., Kontermann R. E. (2010) The effects of affinity and valency of an albumin-binding domain (ABD) on the half-life of a single-chain diabody-ABD fusion protein. Protein Eng. Des. Sel. 23, 827–834 [DOI] [PubMed] [Google Scholar]
- 14. Müller D., Frey K., Kontermann R. E. (2008) A novel antibody-4-1BBL fusion protein for targeted costimulation in cancer immunotherapy. J. Immunother. 31, 714–722 [DOI] [PubMed] [Google Scholar]
- 15. Müller D., Karle A., Meissburger B., Höfig I., Stork R., Kontermann R. E. (2007) Improved pharmacokinetics of recombinant bispecific antibody molecules by fusion to human serum albumin. J. Biol. Chem. 282, 12650–12660 [DOI] [PubMed] [Google Scholar]
- 16. Benedict C. A., MacKrell A. J., Anderson W. F. (1997) Determination of the binding affinity of an anti-CD34 single-chain antibody using a novel, flow cytometry-based assay. J. Immunol. Methods 201, 223–231 [DOI] [PubMed] [Google Scholar]
- 17. Chester K. A., Begent R. H., Robson L., Keep P., Pedley R. B., Boden J. A., Boxer G., Green A., Winter G., Cochet O., Hawkins R. E. (1994) Phage libraries for generation of clinically useful antibodies. Lancet 343, 455–456 [DOI] [PubMed] [Google Scholar]
- 18. Gouda H., Torigoe H., Saito A., Sato M., Arata Y., Shimada I. (1992) Three-dimensional solution structure of the B domain of staphylococcal protein A: comparisons of the solution and crystal structures. Biochemistry 31, 9665–9672 [DOI] [PubMed] [Google Scholar]
- 19. Meininger D. P., Rance M., Starovasnik M. A., Fairbrother W. J., Skelton N. J. (2000) Characterization of the binding interface between the E-domain of staphylococcal protein A and an antibody Fv fragment. Biochemistry 39, 26–36 [DOI] [PubMed] [Google Scholar]
- 20. Beckingham J. A., Bottomley S. P., Hinton R., Sutton B. J., Gore M. G. (1999) Interactions between a single immunoglobulin-binding domain of protein L from Peptostreptococcus magnus and a human κ light chain. Biochem. J. 340, 193–199 [PMC free article] [PubMed] [Google Scholar]
- 21. Akerström B., Björck L. (1986) A physicochemical study of protein G, a molecule with unique immunoglobulin G-binding properties. J. Biol. Chem. 261, 10240–10247 [PubMed] [Google Scholar]
- 22. Stork R., Campigna E., Robert B., Müller D., Kontermann R. E. (2009) Biodistribution of a bispecific single-chain diabody and its half-life extended derivatives. J. Biol. Chem. 284, 25612–25619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Haraldsson B., Sörensson J. (2004) Why do we not all have proteinuria? An update of our current understanding of the glomerular barrier. News Physiol. Sci. 19, 7–10 [DOI] [PubMed] [Google Scholar]
- 24. Jovic M., Sharma M., Rahajeng J., Caplan S. (2010) The early endosome: a busy sorting station for proteins at the crossroads. Histol. Histopathol. 25, 99–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Jansson B., Uhlén M., Nygren P. A. (1998) All individual domains of staphylococcal protein A show Fab binding. FEMS Immunol. Med. Microbiol. 20, 69–78 [DOI] [PubMed] [Google Scholar]
- 26. Deisenhofer J. (1981) Crystallographic refinement and atomic models of a human Fc fragment and its complex with fragment B of protein A from Staphylococcus aureus at 2.9- and 2.8-Å resolution. Biochemistry 20, 2361–2370 [PubMed] [Google Scholar]
- 27. Kim J. R., Tsen M. F., Ghetie V., Ward E. S. (1994) Localization of the site of the murine IgG1 molecule that is involved in binding to the intestinal Fc receptor. Eur. J. Immunol. 24, 2429–2434 [DOI] [PubMed] [Google Scholar]
- 28. Raghavan M., Chen M. Y., Gastinel L. N., Bjorkman P. J. (1994) Investigation of the interaction between the class I MHC-related Fc receptor and its immunoglobulin G ligand. Immunity 1, 303–315 [DOI] [PubMed] [Google Scholar]
- 29. Clark M. (1997) IgG effector mechanisms. Chem. Immunol. 65, 88–110 [PubMed] [Google Scholar]
- 30. DeLano W. L. (2010) The PyMOL Molecular Graphics System, version 1.3r1, Schrödinger, LLC, New York [Google Scholar]