Abstract
Protein post-translational modification by S-nitrosylation conveys a ubiquitous influence of nitric oxide on signal transduction in eukaryotic cells. The wide functional purview of S-nitrosylation reflects in part the regulation by S-nitrosylation of the principal protein post-translational modifications that play a role in cell signaling, including phosphorylation, acetylation, ubiquitylation and related modifications, palmitoylation, and alternative Cys-based redox modifications. In this minireview, we discuss the mechanisms through which S-nitrosylation exerts its broad pleiotropic influence on protein post-translational modification.
Keywords: Nitric Oxide, Phosphorylation, Post-translational Modification, Redox Signaling, S-Nitrosylation, Sumoylation, Ubiquitylation, Acetylation, Palmitoylation
Introduction
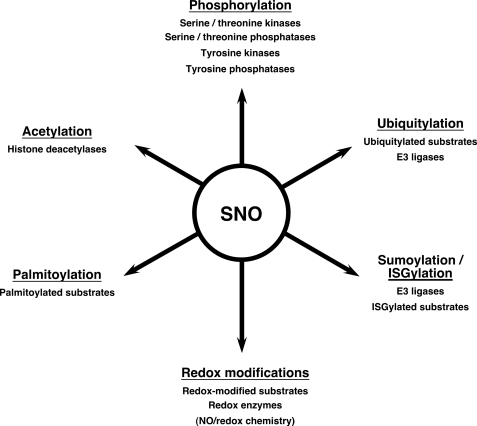
In eukaryotic cells, regulation of protein properties and function by post-translational modification is the central molecular mechanism that mediates signal transduction. It has been established over the past decade that S-nitrosylation, the addition of an NO group to a Cys thiol to form an S-nitrosoprotein (SNO-protein), regulates a broad spectrum of proteins in all functional protein classes and cell types examined (1–3). The literature now encompasses some 3000 S-nitrosoproteins (including those identified with exogenously administered, physiological, nitrosylating agents), which may represent only the tip of the iceberg in light of the emerging role of denitrosylases in determining detectable levels of protein S-nitrosylation (4, 5). As illustrated in this minireview, the picture that has formed indicates that the propagation or modulation of cell signals by S-nitrosylation often entails crosstalk with signaling modalities mediated by other principal mechanisms of post-translational modification. Indeed, it appears to date that, among post-translational modifications that convey cell signals, the breadth of the influence of S-nitrosylation may be comparable with that of phosphorylation and ubiquitylation, where signal crosstalk is established as a central operating principle (6). A number of recent reviews have emphasized the determinants of substrate specificity and the enzymatic mechanisms of S-nitrosylation/denitrosylation (1, 2, 4, 5, 7). We summarize here for the first time the role of S-nitrosylation as a pleiotropic regulator of protein post-translational modification (Fig. 1).
FIGURE 1.
Schematic summary of principal post-translational mechanisms regulated by S-nitrosylation and molecular loci of regulation.
The precis presented here raises issues of general relevance to the analysis of regulation by S-nitrosylation (as for other post-translational modifications). With regard to causality, it can be difficult to predict or assess the degree of modification required to exert a significant regulatory effect in vivo, in part because of the compartmentalized modification of substrate proteins involved (1, 8). Accordingly, only a small subset of proteins, such as those affiliated with a receptor or other partners (1, 8, 9), may convey the cell signals. Furthermore, the population stoichiometry of modification may be relatively unimportant in cases of gain of function, including induced protein-protein interactions and the activation of transcription factors or channels, where minor populations may convey biological activity. Nonetheless, S-nitrosylation and activation of a substantial proportion of a channel population have in fact been documented (10), and notably, a stoichiometry of 1 for S-nitrosylation has been observed in the case of inhibition of enzyme activity (mitochondrial caspase-3) (11, 12). Additional challenges are presented by the targeted S-nitrosylation of multiple, functionally related elements in signaling pathways because spatiotemporal analyses are largely beyond the state of the art. Nonetheless, mutation of identified sites of S-nitrosylation in concert with inhibition of stimulus-coupled NOS activity can provide strong evidence for a physiological role. An emerging understanding of the determinants of specificity of S-nitrosylation highlights the importance of direct binding of substrates by sources of NO groups, the reactivity of target Cys, and the nature of the nitrosylating agent, as illustrated by the newly appreciated roles for transnitrosylases (7).
Phosphorylation
Regulation by S-nitrosylation of both protein kinases and phosphatases influences a wide range of signal transduction pathways mediated by phosphorylation/dephosphorylation, and illustrative examples are provided here (Table 1). In many but not all cases, the effect of S-nitrosylation on kinase or phosphatase activity is inhibitory and is exerted through modification of a single Cys residue, and in the case of kinases, inhibition may be exerted directly through suppression of kinase activity or by modulating the interaction of kinase and substrate. These operating principles are well illustrated in the case of MAPK-mediated signaling (1) that mediates TNFα-induced apoptosis. Endogenous S-nitrosylation of the MAPK kinase kinase ASK1 (apoptosis signal-regulating kinase-1) at a single Cys inhibits the binding of ASK1 to its principal downstream effectors, MKK3 and MKK6 (13). S-Nitrosylation of a single Cys within JNK, the downstream MAPK target of MKK3/6, inhibits JNK-mediated phosphorylation and transactivation of c-Jun (14), at least in part by inhibiting binding of JNK and c-Jun (15). Thus, S-nitrosylation of specific regulatory Cys residues may exert a calibrated anti-apoptotic influence through modulation of sequential MAPK-mediated transduction stages. In addition to the MAPK subfamily of the CMCG family of kinases, S-nitrosylation acts on members of the cyclin-dependent kinase subfamily (16, 17). Endogenous S-nitrosylation of neuronal Cdk5 targets one or two Cys residues, including Cys-83, within the ATP-binding pocket, and the resultant modulation of kinase activity regulates dendritic growth, branching, and spine formation (17, 18). Excessive S-nitrosylation of Cdk5 is associated with dendritic spine loss in Alzheimer disease (18).
TABLE 1.
Summary of illustrative examples of regulation by S-nitrosylation of protein post-translational modification
Exemplary instances of regulation by S-nitrosylation of serine/threonine kinases outside the MAPK family are provided by Akt kinase (PKB) and G protein-coupled receptor kinase (GRK).2 S-Nitrosylation of a single Cys within Akt inhibits insulin-stimulated Akt catalytic activity, and Akt S-nitrosylation is enhanced in diabetic mouse models (19, 20). Enhanced S-nitrosylation of Akt is also associated with aging-related deficits in skeletal muscle (21). S-Nitrosylation of a single Cys within the activation loop of GRK2, induced by stimulation of β2-adrenergic receptors (β2-ARs) that are coupled to endothelial NOS (eNOS; NOS3), inhibits ligand-stimulated activation of GRK2 and thus β2-AR phosphorylation and thereby suppresses receptor desensitization and down-regulation that are associated with heart failure and asthma (22). S-Nitrosylation also inhibits the activity of protein kinase C (23) as well as other serine/threonine kinases, including IκB kinase and the insulin receptor kinase, as discussed below.
Protein-tyrosine kinases (PTKs) function in a multiplicity of signaling pathways, and S-nitrosylation has been shown to regulate the function of both receptor and non-receptor PTKs. S-Nitrosylation of one or two Cys residues within the epidermal growth factor receptor, a prototypical receptor PTK, inhibits its auto(trans)phosphorylation and downstream signaling (24). Downstream signaling by receptor PTKs is often mediated by phosphorylation and activation of non-receptor PTKs, the prototype of which is c-Src. S-Nitrosylation of a single Cys within the catalytic domain of c-Src (which is conserved among other members of the Src family of non-receptor PTKs) activates rather than inhibits c-Src catalytic activity as assessed by tyrosine autophosphorylation, and c-Src activation by β-estradiol, an important step in cancer cell invasion, was shown to depend upon c-Src S-nitrosylation (25). An additional example of kinase activation by S-nitrosylation is provided by the (non-protein) kinase glucokinase. S-Nitrosylation of a single Cys residue (Cys-371) consequent upon activation of neuronal NOS (nNOS; NOS1) by insulin in pancreatic beta cells disrupts the binding of glucokinase and nNOS, thereby disinhibiting glucokinase to promote insulin secretion (notably, insulin is a prominent example of a receptor PTK ligand) (26).
Both protein-tyrosine phosphatases (PTPs) and serine/threonine phosphatases are regulated by S-nitrosylation. Members of the PTP superfamily utilize a catalytic Cys, and oxidative inhibition of PTPs by endogenously produced hydrogen peroxide, which may be reversible or irreversible, is well known. S-Nitrosylation inhibits the prototypical PTP, PTP1B, by targeting the active site Cys, and S-nitrosylation protected PTP1B from irreversible oxidation by hydrogen peroxide (27). Cellular irradiation induced transient S-nitrosylation of the active site Cys and thereby inactivated the SH2 (Src homology 2) domain-containing non-receptor tyrosine phosphatases SHP-1 and SHP-2 (28). Furthermore, PTP1B, SHP-1, and SHP-2 are S-nitrosylated consequent upon insulin stimulation of eNOS, which results in enhanced tyrosine phosphorylation of the insulin receptor and downstream signaling elements, including the Akt kinase, and thereby enhanced insulin responsiveness (29). The chemokine stromal cell-derived factor-1α (SDF-1α) signals through a MAPK cascade involving JNK3, and JNK activity is regulated by MKP7 (MAPK phosphatase 7). Activation of JNK3 by SDF-1α was shown to depend upon activation of eNOS, and an essential role was demonstrated for S-nitrosylation and inhibition of MKP7, resulting in enhanced JNK3 phosphorylation (30). Thus, S-nitrosylation of MKP7 is a crucial link in SDF-1α-regulated endothelial cell migration and angiogenesis (30).
Recent results demonstrated that PTEN (phosphatase and tensin homolog) is also regulated by S-nitrosylation. The gene encoding PTEN was identified as a tumor suppressor that is mutated in many cancers. Although the catalytic domain of PTEN is similar to that of dual specificity PTPs, its principal substrates are phosphoinositides and, in particular, phosphatidylinositol 3,4,5-trisphosphate. Thus, PTEN negatively regulates the phosphoinositide 3-kinase/Akt signaling pathway. S-Nitrosylation of a single Cys within PTEN inhibits phosphatidylinositol-3,4,5-trisphosphate phosphatase activity, and notably, the site of S-nitrosylation (Cys-83) is allosteric and distinct from the active site Cys (Cys-124) that is the target of oxidative inactivation of PTEN (31). S-Nitrosylation of PTEN enhances Akt signaling (phosphorylation of Akt substrates), and S-nitrosylation of PTEN is enhanced in ischemic brain tissue (31, 32), consistent with the role of Akt in promoting cell survival. Enhanced S-nitrosylation of PTEN was also observed in the brains of patients diagnosed with Alzheimer disease (33).
Acetylation
Nitric oxide plays a broad role in regulating the expression of eukaryotic genes, and this influence is exerted at least in part through S-nitrosylation (34, 35). S-Nitrosylation of regulatory binding partners of transcription factors can exert an extranuclear influence on transcription factor activation, stability, and/or nuclear targeting, as in the case of hypoxia-inducible factor-1α (HIF-1α) and p53 (see below) (1), and S-nitrosylation of transcription factors at DNA-binding or allosteric sites can directly regulate gene transcription in the nucleus, as in the case of NF-κB, where S-nitrosylation of critical redox-sensitive Cys residues inhibits NF-κB DNA binding and promoter activity and thereby NF-κB-dependent gene transcription (36). In addition, it has emerged that S-nitrosylation may also operate in the nucleus on epigenetic mechanisms of transcriptional regulation, in particular through regulation of histone acetylation/deacetylation (Table 1).
HDAC2 (histone deacetylase 2) is modified directly by S-nitrosylation. In neurons, S-nitrosylation of HDAC2 coupled to stimulation of nNOS by BDNF was localized to two Cys residues, Cys-262 and Cys-274, which are conserved across other Class I histone deacetylases (37). S-Nitrosylation did not affect deacetylase activity, but it induced HDAC2 release from chromatin and consequently enhanced acetylation of histones and transcriptional activation, in particular of cAMP response element-regulated genes. At least one function of this chromatin remodeling was to control the activation of genes that regulate dendritic growth and branching (37). In skeletal muscle, HDAC2 expression is up-regulated in the MDX mouse model of muscular dystrophy, which is also characterized by dysregulated NOS activity in dystrophic muscle (38). Either down-regulation of HDAC2 or repletion of NOS ameliorates the pathophysiological MDX muscle phenotype, and repletion of NOS results in S-nitrosylation of HDAC2 (38).
Important new insight into the regulation of nuclear S-nitrosylation has been provided recently by Snyder and co-workers (39). These investigators established a role for the enzyme GAPDH in conveying S-nitrosylation-based signals from the cytosol to the nucleus: GAPDH that is S-nitrosylated at the single (active site) Cys-150, by NO generated endogenously in the context of apoptotic signaling, binds the E3 ubiquitin ligase Siah1 and is thereby co-translocated to the nucleus (40). Subsequently, they reported that S-nitrosylation of GAPDH promoted its binding to nuclear substrates, including HDAC2 and the Class III deacetylase sirtuin-1, followed by transnitrosylative transfer of the NO group from SNO-GAPDH to binding partners (39). Furthermore, trans-S-nitrosylation inhibited sirtuin-1 activity and its effects on transcription. Thus, SNO-GAPDH functions as a nuclear S-nitrosylase to facilitate histone acetylation and thereby gene transcription (7, 39). Most recently, this group has shown that histone acetylation is also facilitated through degradation by Siah1, co-translocated to the nucleus with SNO-GAPDH, of the histone methyltransferase SUV39H1 (suppressor of variegation 3–9 homologue 1) (41). Although lysine acetylation is best characterized in the context of nuclear histones, proteomic analysis has revealed that the mammalian acetylome comprises at least 1700 proteins involved in a broad range of cellular functions, most of which are non-nuclear (42). The role of S-nitrosylation in regulating extranuclear lysine acetylation remains unexplored.
Long-chain Fatty Acid S-Acylation (S-Palmitoylation)
Cys residues within proteins may be subject to covalent lipid modification by thioether-linked polyisoprenylation or thioester-linked S-palmitoylation. S-Palmitoylation is unique among Cys-directed and other covalent lipid modifications of proteins in that it is, at least in many cases, reversible and dynamic. Members of the DHHC family (characterized by an aspartate-lysine-lysine-cysteine active site) serve as S-palmitoylating enzymes, and two classes of depalmitoylating enzymes have been identified (43). S-Palmitoylation has been shown to influence a broad range of protein properties and functions, including subcellular localization and stability, protein-protein interaction, and signal propagation (43). As in the case of S-nitrosylation, only one or a few Cys residues are S-palmitoylated within most modified proteins. To the extent that the molecular determinants that target S-palmitoylation and S-nitrosylation are shared, S-palmitoylation and S-nitrosylation might operate antiphasically. A mutually competitive relationship between S-palmitoylation and S-nitrosylation was suggested initially on the basis of decreased palmitoylation of multiple neuronal proteins (including GAP-43 and SNAP-25) in the presence of nitric oxide donors as assessed by metabolic labeling (incorporation of [3H]palmitate) (44). Subsequent studies, also employing metabolic labeling, reported inhibition by S-nitrosylating agents of S-palmitoylation of the mammalian β2-AR, transferrin receptor, and caveolin (45, 46) as well as the coronavirus spike protein (47).
A direct demonstration of mutually competitive S-nitrosylation and S-palmitoylation in neurons under physiological conditions was provided recently by Ho et al. (48). PSD-95 (postsynaptic density protein of 95 kDa), which serves as the principal scaffolding protein of the post-synaptic density, is known to be dynamically S-palmitoylated at Cys-3 and Cys-5 (49). Ho et al. (48) showed that these residues are also subject to endogenous S-nitrosylation and that stimulation of S-nitrosylation decreased palmitoylation as assessed by metabolic labeling as well as by acyl-biotin exchange (which directly indicates levels of S-palmitoylated substrate). Conversely, inhibition of S-palmitoylation, either pharmacologically in cultured cells or by genetic knock-out of the relevant DHHC in intact mice, enhanced endogenous S-nitrosylation. The physiological relevance of this dynamic reciprocity was demonstrated by the finding that synaptic clustering of PSD-95, known to require Cys-3/Cys-5 palmitoylation and to play an essential role in the organization of glutamatergic receptors that are scaffolded by PSD-95 (49), was decreased by nNOS activation consequent upon stimulation of NMDA receptors. It is of note that, although as yet unexplored, S-nitrosylation may potentially inhibit protein S-palmitoylation by targeting the reactive Cys residue in CoA and thereby suppressing formation of fatty acyl-CoA, the precursor for S-acylation, or the reactive Cys residue at the active site of the DHHC enzymes that is required for their S-palmitoylating function. In view of the hundreds of proteins (distributed across essentially all functional classes) that are known to be modified endogenously by S-nitrosylation and/or S-palmitoylation, the implications of their interaction are profound (Table 1).
Ubiquitylation, SUMOylation, and ISGylation
Ubiquitylation is the ligation by the 8.5-kDa protein ubiquitin of a target protein Lys residue (monoubiquitylation), which may be followed by formation of ubiquitin chains through attachment of additional ubiquitin moieties to one or more of the seven Lys residues within conjugated ubiquitin (polyubiquitylation). Activation of ubiquitin by an E1 ubiquitin-activating enzyme results in the thioester linkage of ubiquitin to the E1 active site Cys thiol, followed by transfer of ubiquitin via transthioesterification to the active site Cys thiol of an E2 ubiquitin-conjugating enzyme and finally by formation of an isopeptide bond between a target protein lysine and the C-terminal glycine of ubiquitin catalyzed by an E3 ubiquitin ligase that can interact with both E2 and substrate. Ubiquitylation was initially shown to govern protein degradation through polyubiquitylation that targets modified substrates to the proteasome. However, it is now well established that the form of ubiquitylation (monoubiquitylation versus polyubiquitylation and the type of inter-ubiquitin linkages in polyubiquitin chains) dictates disparate fates that are associated with a broad range of cellular processes and signaling events (including, for example, receptor and membrane trafficking and gene transcription) (Table 1).
In eukaryotes, polyubiquitylation-mediated degradation of some proteins is governed by the N-end rule, according to which an N-terminal Asn, Gln, or Cys is converted to Arg to allow recognition by an E3 ligase (50). Whereas Asn or Gln is enzymatically deamidated to Asp or Glu prior to conjugation with Arg, arginylation of N-terminal Cys is mediated through a modification, assumed to be S-nitrosylation, followed by O2-dependent oxidation to cysteine sulfonic acid, as exemplified in the case of the RGS (regulator of G protein signaling) proteins (50). Thus, S-nitrosylation presumably serves as a necessary step in implementing the ubiquitin-dependent N-end mechanism of protein degradation and thereby governs the turnover of multiple substrates.
S-Nitrosylation has been shown to inhibit RING finger E3 ligase activity. S-Nitrosylation of the neuronal RING finger E3 ligase parkin, which targets Cys within BIR (baculoviral inhibitor of apoptosis repeat motif) and RING domains, is enhanced in rodent parkinsonian models and brains of human patients with sporadic Parkinson disease, and the consequent inhibition of parkin activity (which may be preceded by activation (50)) has been implicated in protein accumulation and aggregation that characterizes Parkinson (and other neurodegenerative) disease (51, 52). In the case of the RING finger E3 ligase XIAP (X-linked inhibitor of apoptosis protein), S-nitrosylation that targets Cys residues within RING and BIR domains, respectively, inhibits ubiquitin-mediated degradation of pro-apoptotic caspase cysteine proteases (caspase-3) and releases and disinhibits XIAP-bound caspase-3 and thereby promotes cell death (53), and elevated levels of SNO-XIAP have been detected in the brains of patients with any of a number of neurodegenerative diseases (53, 54). As indicated above in the case of the N-end rule pathway, S-nitrosylation of substrates may regulate their propensity to undergo ubiquitylation. S-Nitrosylation of a single Cys within the Fe2+-dependent degradation sequence of IRP2 (iron regulatory protein 2) was reported to enhance its ubiquitylation and proteasomal degradation (55). In contrast, endogenous S-nitrosylation of a pair of Cys residues within the key apoptosis regulatory protein Bcl-2 inhibits its ubiquitylation and proteasomal degradation and suppresses apoptosis (56), and similarly, S-nitrosylation of a pair of Cys residues within the anti-apoptotic FLICE inhibitory protein FLIP, which is down-regulated in association with apoptotic signaling, inhibits its ubiquitylation and exerts an anti-apoptotic effect (57). In the case of the “tumor suppressor” transcription factor p53, ubiquitylation by the RING finger E3 ligase HDM2 mediates rapid turnover by proteasomal degradation, and S-nitrosylation of a single Cys within HDM2 inhibits p53 binding and thereby stabilizes p53 and activates p53-dependent transcription (58, 59).
Additional examples illustrate control of ubiquitylation by S-nitrosylation via regulation of alternative post-translational modifications. The transcription factor NF-κB is complexed with and sequestered in the cytoplasm by IκB (inhibitor of NF-κB) proteins, and phosphorylation of IκB by the IκB kinase complex (which contains the catalytic IκB kinase (IKK) α and IKKβ subunits) induces IκB ubiquitylation and degradation to allow translocation of NF-κB to the nucleus and DNA binding (60). Reynaert et al. (61) showed that endogenous S-nitrosylation of Cys-179 within IKKβ inhibits IκB phosphorylation, thereby regulating the proteasomal targeting of IκB and NF-κB-dependent transcription. In the case of HIF-α, hydroxylation of HIF-α proline residues facilitates binding of the von Hippel-Lindau protein, pVHL, which functions as the substrate recognition module of an E3 ubiquitin ligase. Because hydroxylation is O2-dependent, HIF-α is stabilized by hypoxia. However, it was observed that exogenous S-nitrosylating agents as well as endogenous NOS activity could stabilize HIF-1α at normoxia (58, 62), and it was later demonstrated that S-nitrosylation of a single Cys within the oxygen-dependent degradation domain of HIF-1α could inhibit normoxic pVHL binding and thus HIF-1α ubiquitylation and degradation (63). Normoxic S-nitrosylation of HIF-1α is enhanced in mice with a targeted deletion of the denitrosylase S-nitrosoglutathione reductase, and the effects of ischemic myocardial infarction are ameliorated in these animals in association with up-regulation by HIF-1α of vascular endothelial growth factor and enhanced angiogenesis (64). In addition, as described above, ubiquitylation of nuclear proteins is subserved in part by binding and co-translocation to the nucleus of GAPDH and the E3 ligase Siah1, which are dependent upon GAPDH S-nitrosylation (40). Binding of SNO-GAPDH stabilizes Siah1 and thereby enhances degradation of nuclear proteins (40).
There exist a number of ubiquitin-like proteins, most prominently the SUMO family of small ubiquitin-like modifiers and ISG15, the product of interferon-stimulated gene 15, that modify protein Lys residues in a fashion similar to ubiquitylation to regulate a wide range of protein functions (58, 59). S-Nitrosylation of a single Cys within the SUMO E3 ligase Pias3 (protein inhibitor of activated STAT3) promoted its interaction with and ubiquitylation by the ubiquitin E3 ligase Trim32 (tripartite motif-containing 32), resulting in enhanced degradation of Pias3 and suppression of sumoylation (65). ISG15 is modified directly by S-nitrosylation at a single Cys that participates in homodimerization, and S-nitrosylation results in enhanced ISGylation (66).
There are several hundred mammalian E3 ligases, which are distributed among at least four mechanistically distinct classes (67). Little is known about the susceptibility to and potential functional influence of S-nitrosylation across E3 enzymes (or for that matter, E1 and E2 enzymes, which possess reactive, active site Cys thiols). The possible roles of S-nitrosylation in regulating ubiquitylation-mediated signaling that is at least partly independent of substrate turnover also remain unexplored.
Cys-based Redox Modifications (Glutathionylation, Sulfhydration, and Alternative Cys Oxidations)
In addition to S-nitrosylation, Cys residues in proteins may undergo a variety of redox-based modifications and electrophilic substitutions (68, 69). There is strong evidence and a structural rationale to suggest that oxidative modifications of thiol are not in general functionally interchangeable (8, 69), and a molecular code for redox based-regulation has been entertained (70). The extent to which particular modifications convey physiological signals is an area of active investigation (69). To date, physiological roles for the modification of Cys residues by the formation of sulfinic acid and glutathione mixed disulfide are best established in the context of cellular proliferation and differentiation, acting through inhibition of phosphatases (69, 71), but recent examples of regulation of members of other classes of proteins, as in activation of the ATM (ataxia telangiectasia mutated) protein kinase by hydrogen peroxide-mediated disulfide-linked dimerization (72), may portend a far wider purview. Crosstalk among S-nitrosylation and alternative redox modifications may be predicted because 1) in general, S-nitrosylation will prevent further oxidation of protein thiols (27, 73); 2) S-nitrosylation may catalyze disulfide formation between vicinal thiols within or between proteins at active or allosteric sites (74), although the physiological relevance of accelerated disulfide formation is not well established; and 3) S-nitrosylation may enhance glutathionylation (75, 76) or sulfhydration of the S-nitrosylated Cys in specific instances where structural features orient the S-NO to favor attack on sulfur. In addition, S-nitrosylation will predictably regulate the activity of oxidases and reductases that employ active site or allosteric thiols to control both the production of redox-active second messengers and the redox modifications of proteins that may subserve signaling. One notable example is the inhibitory S-nitrosylation of peroxiredoxin 2 (77); the peroxiredoxins have a recognized role in PTK-mediated signaling through regulation of PTP activity (71). Exploration of interplay and crosstalk between S-nitrosylation and alternative redox-based and electrophilic modifications of protein Cys residues will assume increasing importance with the establishment of physiological roles for alternative modifications in cell signaling (Table 1).
This work was supported, in whole or in part, by National Institutes of Health Grants R01HL059130, R01HL095463, R01HL091876, and P01HL75443. This work was also supported by Defense Advanced Research Projects Agency Grant N66001-10-C-2015. This is the third article in the Thematic Minireview Series on Redox Sensing and Regulation.
- GRK
- G protein-coupled receptor kinase
- β2-AR
- β2-adrenergic receptor
- eNOS
- endothelial NOS
- PTK
- protein-tyrosine kinase
- nNOS
- neuronal NOS
- PTP
- protein-tyrosine phosphatase
- SDF-1α
- stromal cell-derived factor-1α
- HIF-1α
- hypoxia-inducible factor-1α
- IKK
- IκB kinase.
REFERENCES
- 1. Hess D. T., Matsumoto A., Kim S. O., Marshall H. E., Stamler J. S. (2005) Protein S-nitrosylation: purview and parameters. Nat. Rev. Mol. Cell Biol. 6, 150–166 [DOI] [PubMed] [Google Scholar]
- 2. Foster M. W., McMahon T.J., Stamler J. S. (2003) S-Nitrosylation in health and disease. Trends Mol. Med. 9, 160–168 [DOI] [PubMed] [Google Scholar]
- 3. Sen N., Snyder S. H. (2010) Protein modifications involved in neurotransmitter and gasotransmitter signaling. Trends Neurosci. 33, 493–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Benhar M., Forrester M. T., Stamler J. S. (2009) Protein denitrosylation: enzymatic mechanisms and cellular functions. Nat. Rev. Mol. Cell Biol. 10, 721–732 [DOI] [PubMed] [Google Scholar]
- 5. Seth D., Stamler J. S. (2011) The SNO-proteome: causation and classifications. Curr. Opin. Chem. Biol. 15, 129–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. López-Otín C., Hunter T. (2010) The regulatory crosstalk between kinases and proteases in cancer. Nat. Rev. Cancer 10, 278–292 [DOI] [PubMed] [Google Scholar]
- 7. Stamler J. S., Hess D. T. (2010) Nascent nitrosylases. Nat. Cell Biol. 12, 1024–1026 [DOI] [PubMed] [Google Scholar]
- 8. Woo H. A., Yim S. H., Shin D. H., Kang D., Yu D. Y., Rhee S. G. (2010) Inactivation of peroxiredoxin 1 by phosphorylation allows localized H2O2 accumulation for cell signaling. Cell 140, 517–528 [DOI] [PubMed] [Google Scholar]
- 9. Ozawa K., Whalen E. J., Nelson C. D., Mu Y., Hess D. T., Lefkowitz R. J., Stamler J. S. (2008) S-Nitrosylation of β-arrestin regulates β-adrenergic receptor trafficking. Mol. Cell 31, 395–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Eu J. P., Sun J., Xu L., Stamler J. S., Meissner G. (2000) The skeletal muscle calcium release channel: coupled O2 sensor and NO signaling functions. Cell 102, 499–509 [DOI] [PubMed] [Google Scholar]
- 11. Mannick J. B., Schonhoff C., Papeta N., Ghafourifar P., Szibor M., Fang K., Gaston B. (2001) S-Nitrosylation of mitochondrial caspases. J. Cell Biol. 154, 1111–1116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Mannick J. B., Hausladen A., Liu L., Hess D. T., Zeng M., Miao Q. X., Kane L. S., Gow A. J., Stamler J. S. (1999) Fas-induced caspase denitrosylation. Science 284, 651–654 [DOI] [PubMed] [Google Scholar]
- 13. Park H. S., Yu J. W., Cho J. H., Kim M. S., Huh S. H., Ryoo K., Choi E. J. (2004) Inhibition of apoptosis signal-regulating kinase 1 by nitric oxide through a thiol redox mechanism. J. Biol. Chem. 279, 7584–7590 [DOI] [PubMed] [Google Scholar]
- 14. Park H. S., Huh S. H., Kim M. S., Lee S. H., Choi E. J. (2000) Nitric oxide negatively regulates c-Jun N-terminal kinase/stress-activated protein kinase by means of S-nitrosylation. Proc. Natl. Acad. Sci. U.S.A. 97, 14382–14387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Park H. S., Mo J. S., Choi E. J. (2006) Nitric oxide inhibits an interaction between JNK1 and c-Jun through nitrosylation. Biochem. Biophys. Res. Commun. 351, 281–286 [DOI] [PubMed] [Google Scholar]
- 16. Kumar S., Barthwal M. K., Dikshit M. (2010) Cdk2 nitrosylation and loss of mitochondrial potential mediate NO-dependent biphasic effect on HL-60 cell cycle. Free Radic. Biol. Med. 48, 851–861 [DOI] [PubMed] [Google Scholar]
- 17. Zhang P., Yu P. C., Tsang A. H., Chen Y., Fu A. K., Fu W. Y., Chung K. K., Ip N. Y. (2010) S-Nitrosylation of cyclin-dependent kinase 5 (Cdk5) regulates its kinase activity and dendrite growth during neuronal development. J. Neurosci. 30, 14366–14370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Qu J., Nakamura T., Cao G., Holland E. A., McKercher S. R., Lipton S. A. (2011) S-Nitrosylation activates Cdk5 and contributes to synaptic spine loss induced by β-amyloid peptide. Proc. Natl. Acad. Sci. U.S.A. 108, 14330–14335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Yasukawa T., Tokunaga E., Ota H., Sugita H., Martyn J. A., Kaneki M. (2005) S-Nitrosylation-dependent inactivation of Akt/protein kinase B in insulin resistance. J. Biol. Chem. 280, 7511–7518 [DOI] [PubMed] [Google Scholar]
- 20. Carvalho-Filho M. A., Ueno M., Hirabara S. M., Seabra A. B., Carvalheira J. B., de Oliveira M. G., Velloso L. A., Curi R., Saad M. J. (2005) S-Nitrosation of the insulin receptor, insulin receptor substrate 1, and protein kinase B/Akt: a novel mechanism of insulin resistance. Diabetes 54, 959–967 [DOI] [PubMed] [Google Scholar]
- 21. Wu M., Katta A., Gadde M. K., Liu H., Kakarla S. K., Fannin J., Paturi S., Arvapalli R. K., Rice K. M., Wang Y., Blough E. R. (2009) Aging-associated dysfunction of Akt/protein kinase B: S-nitrosylation and acetaminophen intervention. PLoS ONE 4, e6430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Whalen E. J., Foster M. W., Matsumoto A., Ozawa K., Violin J. D., Que L. G., Nelson C. D., Benhar M., Keys J. R., Rockman H. A., Koch W. J., Daaka Y., Lefkowitz R. J., Stamler J. S. (2007) Regulation of β-adrenergic receptor signaling by S-nitrosylation of G-protein-coupled receptor kinase 2. Cell 129, 511–522 [DOI] [PubMed] [Google Scholar]
- 23. Choi H., Tostes R. C., Webb R. C. (2011) S-Nitrosylation inhibits protein kinase C-mediated contraction in mouse aorta. J. Cardiovasc. Pharmacol. 57, 65–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Murillo-Carretero M., Torroglosa A., Castro C., Villalobo A., Estrada C. (2009) S-Nitrosylation of the epidermal growth factor receptor: a regulatory mechanism of receptor tyrosine kinase activity. Free Radic. Biol. Med. 46, 471–479 [DOI] [PubMed] [Google Scholar]
- 25. Rahman M. A., Senga T., Ito S., Hyodo T., Hasegawa H., Hamaguchi M. (2010) S-Nitrosylation at cysteine 498 of c-Src tyrosine kinase regulates nitric oxide-mediated cell invasion. J. Biol. Chem. 285, 3806–3814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Rizzo M. A., Piston D. W. (2003) Regulation of β-cell glucokinase by S-nitrosylation and association with nitric-oxide synthase. J. Cell Biol. 161, 243–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Chen Y. Y., Chu H. M., Pan K. T., Teng C. H., Wang D. L., Wang A. H., Khoo K. H., Meng T. C. (2008) Cysteine S-nitrosylation protects protein-tyrosine phosphatase 1B against oxidation-induced permanent inactivation. J. Biol. Chem. 283, 35265–35272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Barrett D. M., Black S. M., Todor H., Schmidt-Ullrich R. K., Dawson K. S., Mikkelsen R. B. (2005) Inhibition of protein-tyrosine phosphatases by mild oxidative stresses is dependent on S-nitrosylation. J. Biol. Chem. 280, 14453–14461 [DOI] [PubMed] [Google Scholar]
- 29. Hsu M. F., Meng T. C. (2010) Enhancement of insulin responsiveness by nitric oxide-mediated inactivation of protein-tyrosine phosphatases. J. Biol. Chem. 285, 7919–7928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Pi X., Wu Y., Ferguson J. E., 3rd, Portbury A. L., Patterson C. (2009) SDF-1α stimulates JNK3 activity via eNOS-dependent nitrosylation of MKP7 to enhance endothelial migration. Proc. Natl. Acad. Sci. U.S.A. 106, 5675–5680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Numajiri N., Takasawa K., Nishiya T., Tanaka H., Ohno K., Hayakawa W., Asada M., Matsuda H., Azumi K., Kamata H., Nakamura T., Hara H., Minami M., Lipton S. A., Uehara T. (2011) On-off system for PI3 kinase-Akt signaling through S-nitrosylation of phosphatase with sequence homology to tensin (PTEN). Proc. Natl. Acad. Sci. U.S.A. 108, 10349–10354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Pei D. S., Sun Y. F., Song Y. J. (2009) S-Nitrosylation of PTEN involved in ischemic brain injury in rat hippocampal CA1 region. Neurochem. Res. 34, 1507–1512 [DOI] [PubMed] [Google Scholar]
- 33. Kwak Y. D., Ma T., Diao S., Zhang X., Chen Y., Hsu J., Lipton S. A., Masliah E., Xu H., Liao F. F. (2010) NO signaling and S-nitrosylation regulate PTEN inhibition in neurodegeneration. Mol. Neurodegener. 5, 49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Bogdan C. (2001) Nitric oxide and the regulation of gene expression. Trends Cell Biol. 11, 66–75 [DOI] [PubMed] [Google Scholar]
- 35. Illi B., Colussi C., Grasselli A., Farsetti A., Capogrossi M. C., Gaetano C. (2009) NO sparks off chromatin: tales of a multifaceted epigenetic regulator. Pharmacol. Ther. 123, 344–352 [DOI] [PubMed] [Google Scholar]
- 36. Kelleher Z. T., Matsumoto A., Stamler J. S., Marshall H. E. (2007) NOS2 regulation of NF-κB by S-nitrosylation of p65. J. Biol. Chem. 282, 30667–30672 [DOI] [PubMed] [Google Scholar]
- 37. Nott A., Watson P. M., Robinson J. D., Crepaldi L., Riccio A. (2008) S-Nitrosylation of histone deacetylase 2 induces chromatin remodeling in neurons. Nature 455, 411–415 [DOI] [PubMed] [Google Scholar]
- 38. Colussi C., Mozzetta C., Gurtner A., Illi B., Rosati J., Straino S., Ragone G., Pescatori M., Zaccagnini G., Antonini A., Minetti G., Martelli F., Piaggio G., Gallinari P., Steinkuhler C., Clementi E., Dell'Aversana C., Altucci L., Mai A., Capogrossi M. C., Puri P. L., Gaetano C. (2008) HDAC2 blockade by nitric oxide and histone deacetylase inhibitors reveals a common target in Duchenne muscular dystrophy treatment. Proc. Natl. Acad. Sci. U.S.A. 105, 19183–19187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kornberg M. D., Sen N., Hara M. R., Juluri K. R., Nguyen J. V., Snowman A. M., Law L., Hester L. D., Snyder S. H. (2010) GAPDH mediates nitrosylation of nuclear proteins. Nat. Cell Biol. 12, 1094–1100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Hara M. R., Agrawal N., Kim S. F., Cascio M. B., Fujimuro M., Ozeki Y., Takahashi M., Cheah J. H., Tankou S. K., Hester L. D., Ferris C. D., Hayward S. D., Snyder S. H., Sawa A. (2005) S-Nitrosylated GAPDH initiates apoptotic cell death by nuclear translocation following Siah1 binding. Nat. Cell Biol. 7, 665–674 [DOI] [PubMed] [Google Scholar]
- 41. Sen N., Snyder S. H. (2011) Neurotrophin-mediated degradation of histone methyltransferase by S-nitrosylation cascade regulates neuronal differentiation. Proc. Natl. Acad. Sci. U.S.A. 108, 20178–20183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Choudhary C., Kumar C., Gnad F., Nielsen M. L., Rehman M., Walther T. C., Olsen J. V., Mann M. (2009) Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science 325, 834–840 [DOI] [PubMed] [Google Scholar]
- 43. Salaun C., Greaves J., Chamberlain L. H. (2010) The intracellular dynamic of protein palmitoylation. J. Cell Biol. 191, 1229–1238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Hess D. T., Patterson S. I., Smith D. S., Skene J. H. (1993) Neuronal growth cone collapse and inhibition of protein fatty acylation by nitric oxide. Nature 366, 562–565 [DOI] [PubMed] [Google Scholar]
- 45. Adam L., Bouvier M., Jones T. L. (1999) Nitric oxide modulates β2-adrenergic receptor palmitoylation and signaling. J. Biol. Chem. 274, 26337–26343 [DOI] [PubMed] [Google Scholar]
- 46. Baker T. L., Booden M. A., Buss J. E. (2000) S-Nitrosocysteine increases palmitate turnover on Ha-Ras in NIH 3T3 cells. J. Biol. Chem. 275, 22037–22047 [DOI] [PubMed] [Google Scholar]
- 47. Akerström S., Gunalan V., Keng C. T., Tan Y. J., Mirazimi A. (2009) Dual effect of nitric oxide on SARS-CoV replication: viral RNA production and palmitoylation of the S protein are affected. Virology 395, 1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ho G. P., Selvakumar B., Mukai J., Hester L. D., Wang Y., Gogos J. A., Snyder S. H. (2011) S-Nitrosylation and S-palmitoylation reciprocally regulate synaptic targeting of PSD-95. Neuron 71, 131–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. El-Din El-Husseini A., Bredt D. S. (2002) Protein palmitoylation: a regulator of neuronal development and function. Nat. Rev. Neurosci. 3, 791–802 [DOI] [PubMed] [Google Scholar]
- 50. Hu R. G., Sheng J., Qi X., Xu Z., Takahashi T. T., Varshavsky A. (2005) The N-end rule pathway as a nitric oxide sensor controlling the levels of multiple regulators. Nature 437, 981–986 [DOI] [PubMed] [Google Scholar]
- 51. Yao D., Gu Z., Nakamura T., Shi Z. Q., Ma Y., Gaston B., Palmer L. A., Rockenstein E. M., Zhang Z., Masliah E., Uehara T., Lipton S. A. (2004) Nitrosative stress linked to sporadic Parkinson disease: S-nitrosylation of parkin regulates its E3 ubiquitin ligase activity. Proc. Natl. Acad. Sci. U.S.A. 101, 10810–10814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Chung K. K., Thomas B., Li X., Pletnikova O., Troncoso J. C., Marsh L., Dawson V. L., Dawson T. M. (2004) S-Nitrosylation of parkin regulates ubiquitination and compromises parkin's protective function. Science 304, 1328–1331 [DOI] [PubMed] [Google Scholar]
- 53. Nakamura T., Wang L., Wong C. C., Scott F. L., Eckelman B. P., Han X., Tzitzilonis C., Meng F., Gu Z., Holland E. A., Clemente A. T., Okamoto S., Salvesen G. S., Riek R., Yates J. R., 3rd, Lipton S. A. (2010) Transnitrosylation of XIAP regulates caspase-dependent neuronal cell death. Mol. Cell 39, 184–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Tsang A. H., Lee Y. I., Ko H. S., Savitt J. M., Pletnikova O., Troncoso J. C., Dawson V. L., Dawson T. M., Chung K. K. (2009) S-Nitrosylation of XIAP compromises neuronal survival in Parkinson disease. Proc. Natl. Acad. Sci. U.S.A. 106, 4900–4905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Kim S., Wing S. S., Ponka P. (2004) S-Nitrosylation of IRP2 regulates its stability via the ubiquitin-proteasome pathway. Mol. Cell. Biol. 24, 330–337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Azad N., Vallyathan V., Wang L., Tantishaiyakul V., Stehlik C., Leonard S. S., Rojanasakul Y. (2006) S-Nitrosylation of Bcl-2 inhibits its ubiquitin-proteasomal degradation. A novel anti-apoptotic mechanism that suppresses apoptosis. J. Biol. Chem. 281, 34124–34134 [DOI] [PubMed] [Google Scholar]
- 57. Chanvorachote P., Nimmannit U., Wang L., Stehlik C., Lu B., Azad N., Rojanasakul Y. (2005) Nitric oxide negatively regulates Fas CD95-induced apoptosis through inhibition of ubiquitin-proteasome-mediated degradation of FLICE inhibitory protein. J. Biol. Chem. 280, 42044–42050 [DOI] [PubMed] [Google Scholar]
- 58. Brüne B., von Knethen A., Sandau K. B. (2001) Transcription factors p53 and HIF-1α as targets of nitric oxide. Cell. Signal. 13, 525–533 [DOI] [PubMed] [Google Scholar]
- 59. Schonhoff C. M., Daou M. C., Jones S. N., Schiffer C. A., Ross A. H. (2002) Nitric oxide-mediated inhibition of Hdm2-p53 binding. Biochemistry 41, 13570–13574 [DOI] [PubMed] [Google Scholar]
- 60. Karin M., Ben-Neriah Y. (2000) Phosphorylation meets ubiquitination: the control of NF-κB activity. Annu. Rev. Immunol. 18, 621–663 [DOI] [PubMed] [Google Scholar]
- 61. Reynaert N. L., Ckless K., Korn S. H., Vos N., Guala A. S., Wouters E. F., van der Vliet A., Janssen-Heininger Y. M. (2004) Nitric oxide represses inhibitory κB kinase through S-nitrosylation. Proc. Natl. Acad. Sci. U.S.A. 101, 8945–8950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Palmer L. A., Gaston B., Johns R. A. (2000) Normoxic stabilization of hypoxia-inducible factor-1 expression and activity: redox-dependent effect of nitrogen oxides. Mol. Pharmacol. 58, 1197–1203 [DOI] [PubMed] [Google Scholar]
- 63. Li F., Sonveaux P., Rabbani Z. N., Liu S., Yan B., Huang Q., Vujaskovic Z., Dewhirst M. W., Li C. Y. (2007) Regulation of HIF-1α stability through S-nitrosylation. Mol. Cell 26, 63–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Lima B., Lam G. K., Xie L., Diesen D. L., Villamizar N., Nienaber J., Messina E., Bowles D., Kontos C. D., Hare J. M., Stamler J. S., Rockman H. A. (2009) Endogenous S-nitrosothiols protect against myocardial injury. Proc. Natl. Acad. Sci. U.S.A. 106, 6297–6302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Qu J., Liu G. H., Wu K., Han P., Wang P., Li J., Zhang X., Chen C. (2007) Nitric oxide destabilizes Pias3 and regulates sumoylation. PLoS ONE 2, e1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Okumura F., Lenschow D. J., Zhang D. E. (2008) Nitrosylation of ISG15 prevents the disulfide bond-mediated dimerization of ISG15 and contributes to effective ISGylation. J. Biol. Chem. 283, 24484–24488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Nakayama K. I., Nakayama K. (2006) Ubiquitin ligases: cell cycle control and cancer. Nat. Rev. Cancer 6, 369–381 [DOI] [PubMed] [Google Scholar]
- 68. Finkel T. (2011) Signal transduction by reactive oxygen species. J. Cell Biol. 194, 7–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Janssen-Heininger Y. M., Mossman B. T., Heintz N. H., Forman H. J., Kalyanaraman B., Finkel T., Stamler J. S., Rhee S. G., van der Vliet A. (2008) Redox-based regulation of signal transduction: principles, pitfalls, and promises. Free Radic. Biol. Med. 45, 1–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Kim S. O., Merchant K., Nudelman R., Beyer W. F., Jr., Keng T., DeAngelo J., Hausladen A., Stamler J. S. (2002) OxyR: a molecular code for redox-related signaling. Cell 109, 383–396 [DOI] [PubMed] [Google Scholar]
- 71. Rhee S. G., Kang S. W., Jeong W., Chang T. S., Yang K. S., Woo H. A. (2005) Intracellular messenger function of hydrogen peroxide and its regulation by peroxiredoxins. Curr. Opin. Cell Biol. 17, 183–189 [DOI] [PubMed] [Google Scholar]
- 72. Guo Z., Kozlov S., Lavin M. F., Person M. D., Paull T. T. (2010) ATM activation by oxidative stress. Science 330, 517–521 [DOI] [PubMed] [Google Scholar]
- 73. Murphy E., Kohr M., Sun J., Nguyen T., Steenbergen C. (2012) S-Nitrosylation: a radical way to protect the heart. J. Mol. Cell. Cardiol., in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Arnelle D. R., Stamler J. S. (1995) NO+, NO, and NO− donation by S-nitrosothiols: implications for regulation of physiological functions by S-nitrosylation and acceleration of disulfide formation. Arch. Biochem. Biophys. 318, 279–285 [DOI] [PubMed] [Google Scholar]
- 75. Baba S. P., Wetzelberger K., Hoetker J. D., Bhatnagar A. (2009) Post-translational glutathiolation of aldose reductase (AKR1B1): a possible mechanism of protein recovery from S-nitrosylation. Chem. Biol. Interact. 178, 250–258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. West M. B., Hill B. G., Xuan Y. T., Bhatnagar A. (2006) Protein glutathiolation by nitric oxide: an intracellular mechanism regulating redox protein modification. FASEB J. 20, 1715–1717 [DOI] [PubMed] [Google Scholar]
- 77. Fang J., Nakamura T., Cho D. H., Gu Z., Lipton S. A. (2007) S-Nitrosylation of peroxiredoxin 2 promotes oxidative stress-induced neuronal cell death in Parkinson disease. Proc. Natl. Acad. Sci. U.S.A. 104, 18742–18747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Erwin P. A., Lin A. J., Golan D. E., Michel T. (2005) Receptor-regulated dynamic S-nitrosylation of endothelial nitric-oxide synthase in vascular endothelial cells. J. Biol. Chem. 280, 19888–19894 [DOI] [PubMed] [Google Scholar]