Background: Whether AGR2 promotes adenocarcinoma growth as a secreted or ER-localized protein is not known.
Results: A unique carboxyl-terminal motif, KTEL, is required for AGR2 ER localization and function.
Conclusion: Not all ER localization motifs are interchangeable.
Significance: Specific ER localization signals may be required for protein function. AGR2's tumor-promoting effects are mediated from the ER and not as a secreted protein.
Keywords: Cancer Biology, Endoplasmic Reticulum (ER), Growth Factors, Protein Sorting, Signal Transduction
Abstract
Soluble proteins are enriched in the endoplasmic reticulum (ER) by retrograde transport from the Golgi that is mediated by the KDEL receptors. In addition to the classic carboxyl-terminal KDEL motif, a variety of sequence variants are also capable of receptor binding that result in ER localization. Although different ER localization signals that exhibit varying affinities for the KDEL receptors exist, whether there are functional implications was unknown. The present study determines whether AGR2 requires a specific ER localization signal to be functionally active. AGR2 is expressed in most human adenocarcinomas and serves a role in promoting growth and the transformed phenotype. Using two different cell lines in which AGR2 induces expression of either the EGFR ligand amphiregulin or the transcription factor CDX2, only the highly conserved wild-type carboxyl-terminal KTEL motif results in the appropriate outcome. Deletion of the KTEL motif results in AGR2 secretion and loss of AGR2 function. AGR2 function is also lost when ER residence is achieved with a carboxyl-terminal KDEL or KSEL instead of a KTEL motif. Thus variations in ER localization sequences may serve a specific functional role, and in the case of AGR2, this role is served specifically by KTEL.
Introduction
Soluble proteins are targeted to the endoplasmic reticulum (ER)2 by two major mechanisms. The first is signal peptide-directed translocation of newly translated proteins into the ER. Second, the proteins remain enriched in the ER by KDEL receptor-mediated retrieval of proteins that have entered the intermediate compartment or the Golgi apparatus (1–3). Whether a protein is retrieved back to the ER is determined by four carboxyl-terminal amino acids that mediate binding to one of three KDEL receptors in higher vertebrates. The classic endoplasmic reticulum localization motif in higher vertebrates is KDEL (4). Subsequent work led to a Prosite motif ((K/R/H/Q/S/A)(D/E/N/Q)EL) that encompasses a set of carboxyl-terminal peptide sequences that result in ER residence. The implications of why different carboxyl-terminal sequences capable of ER localization exist are not clear, but recent studies have demonstrated that affinity for the three known KDEL receptors is affected by which localization motif is employed (5, 6). Whether different ER localization motifs also carry functional significance is not known.
Anterior Gradient Homolog 2 (AGR2) encodes a 17-kDa protein that is highly conserved in vertebrates. AGR2 was first described in Xenopus laevis where its expression is responsible for the development of a glandular organ called the cement gland (7, 8). A significant role in tissue regeneration was established for AGR2 in salamanders where it functions in nerve-dependent limb regeneration (9, 10). AGR2 is also expressed by secretory cells in the normal murine intestine (11). In humans, enhanced AGR2 expression was first described in breast cancer, which was followed by similar observations in most human adenocarcinomas, including those derived from the esophagus, pancreas, lung, ovary, and prostate (12–19). Both in vitro and in vivo studies have demonstrated that AGR2 promotes tumor growth and metastasis (11, 14, 20).
Recent studies have provided insights into the AGR2 mechanism of action. AGR2 expression in esophageal and lung adenocarcinoma cells induces expression of the EGF receptor ligand amphiregulin (AREG), which is responsible for the transformed phenotype observed in cultured cells in vitro (21). In addition, AGR2 stimulation of AREG expression required activation of the Hippo signaling pathway co-activator, YAP1. Thus AGR2 expression promotes tumor growth and the transformed phenotype by affecting the Hippo and EGF signaling pathways.
The induction of AREG expression also provides a means to identify structural requirements for AGR2 activity, including protein domains that are essential for its biologic action. The AGR2 N terminus contains a sequence motif characteristic of signal peptides, which results in protein targeting to the secretory pathway of the cell. Indeed, several studies have proposed that AGR2 secretion from the cell is necessary for its action (7, 10, 14, 16). In addition, yeast two-hybrid screens identified AGR2 binding proteins that naturally occur on the cell surface (9, 22). Whether AGR2 binding to the identified receptors results in a biological response, however, has yet to be established.
Immunocytochemistry of AGR2-expressing cells, however, reveals an intracellular pattern that is most consistent with an ER distribution (11, 21). The carboxyl terminus of AGR2 contains a tetra-peptide sequence, KTEL, that is conserved in all vertebrates from Xenopus to humans (Treefam accession TF321449 (23)). Although the sequence does not agree with the Prosite consensus sequence for ER residence (4, 24), a recent study by Raykhel et al. (5) demonstrated that the KTEL motif does result in binding to the three known KDEL receptors, which results in ER localization. The study also demonstrated that the KTEL motif results in lower affinities for the three known KDEL receptors when compared with proteins terminating with a KDEL sequence.
This study addresses two questions concerning AGR2 biology and the functional significance of endoplasmic reticulum localization signals. The first is whether ER residence of AGR2 is necessary for its function. The second is whether endoplasmic retention by the KTEL sequence is absolutely required for AGR2 function, as suggested by its high conservation in all species where AGR2 is expressed, or whether other ER localization signals may serve a similar role.
EXPERIMENTAL PROCEDURES
Cell Lines
IEC-6, a rat small intestinal jejunal cell line (ATCC, Manassas, VA), was cultured in Dulbecco's modified Eagle's medium with 4 mm l-glutamine, 1.5 g/liter sodium bicarbonate, 4.5 g/liter glucose, and supplemented with 0.1 unit/ml bovine insulin and 10% fetal bovine serum (Hyclone, Thermo Fisher Scientific) (25). The IEC-6 stable cell lines expressing AGR2-KTEL, AGR2-KDEL, AGR2-KSEL, and AGR2-STOP were transfected with pcDNA3.1 expression vectors (Invitrogen) and cultured in the presence of 2 mg/ml of G418 (Mediatech, Inc., Manassas, VA). Human OE33 esophageal adenocarcinoma cells were obtained from Sigma-Aldrich and cultured in RPMI 1640 with 10% FBS. Transient transfection for different AGR2 mutants was used for OE33 cells. The GFP-KTEL and RFP-KTEL expression vectors were transiently transfected into both IEC-6 and OE33 cells.
Antibodies
Mouse monoclonal anti-CDX2 antibodies were obtained from Biogenex (San Ramon, CA). A mouse monoclonal anti-GRP78 (HNGC symbol HSPA5) was obtained from Enzo Life Sciences (Farmingdale, NY). The antibody was generated against the peptide SEKDEL derived from HSPA5 (also known as GRP78), which reacts against all KDEL proteins, and is thus referred to as an anti-KDEL antibody. Rabbit anti-human AGR2 antisera were produced as previously described (21).
Expression Vectors
All of the recombinant DNA constructs utilized the pcDNA3.1 expression vector (Invitrogen), and stable expression was achieved using G418 selection. The AGR2 constructs utilized the human AGR2 gene (GenBankTM accession number NM_006408). In vitro mutagenesis was performed using the QuikChange® site-directed mutagenesis kit (Stratagene, Santa Clara, CA). Dr. Erik Snapp (Albert Einstein College of Medicine, Bronx, NY) graciously provided the GFP- and RFP-KDEL expression vectors (26). The KDEL terminus was then mutated to KTEL for the purposes of this study.
Quantitative Real Time PCR
RNA levels were quantified using real time PCR. Quantitative PCR primers included: rat Agr2, 5′-TCAGTCTCGGCAATCCTGCTTCTT-3′ and 5′-TCTTTAAAGCTTGA-3′; rat Cdx2, 5′-GCGGCGAAACCTTTGTGAATGGAT-3′ and 5′-ACTCAGCTTTCCTCCTGATGGTGA-3′; rat β-actin, 5′-TGAACACGGCATTGTCACCAACTG-3′ and 5′-ATACAGGGACAACACAGCCTGGAT-3′; human AGR2, 5′-ATGAGTGCCCACACAGTCAA-3′ and 5′-GGACATACTGGCCATCAGGA-3′; human AREG, 5′-GTGGTGCTGTCGCTCTTGATA-3′ and 5′-ACTCACAGGGGAAATCTCACT-3′; and human β-actin, 5′-GAGCGCGGCTACAGCTT-3′ and 5′-TCCTTAATGTCACGCACGATTT-3′.
Total RNA was isolated from cells using TRIzol reagent (Invitrogen). First strand cDNA was synthesized from total RNA using Superscript II reverse transcriptase (Invitrogen) with random hexamer primers. Real time PCRs were performed using IQSYBR Green Supermix and the iCycler iQTM detection system (Bio-Rad).
Protein Immunoblotting
The protein concentrations of cell lysates were spectrophotometrically determined (NanoDrop 2000; Thermo Fischer Scientific, Wilmington, DE). Protein samples were resolved using 4–12% Bis-Tris gel (Invitrogen) and transferred to PVDF membranes (Millipore Corp., Bedford, MA). The membranes were blocked for 1 h using 5% nonfat milk in buffer containing 20 mm Tris, 137 mm NaCl, 0.1% Tween 20, pH 7.6, followed by incubation with primary antibodies (anti-AGR2, 1:2000; anti-CDX2, 1:50; anti-β-actin, 1:5000; anti-KDEL, 1:1000). Detection was achieved with the appropriate secondary antibodies followed by chemiluminescence (GE Healthcare). For detection of AGR2 in the culture media, equal numbers of cells were plated in regular culture media. After 1 day, the media were changed to serum-free media. Twenty-four hours later, the media were collected and concentrated using a spin column that retains 90–95% of a 17-kDa protein (MICROCON YM-30; Millipore), followed by immunoblotting for secreted AGR2. The concentrated media were stained with colloidal Coomassie Brilliant Blue R-250 (Invitrogen) to normalize for the amount of protein loaded for each cell line. All of the immunoblots were quantified using a flatbed scanner and ImageJ.
Immunocytochemistry
25,000 cells were plated in 8-well chamber slides (Applied Scientific South San Francisco, CA). The cells were fixed in 4% paraformaldehyde for 10 min, followed by permeabilization using 0.1% saponin, 10% FBS in PBS for 20 min. PBS with 10% FBS was then used for blocking for 20 min. Primary antibodies were diluted in PBS with 0.1% saponin 10% FBS and incubated for 2 h (anti-AGR2 1:300; anti-cdx2 1:5, anti-KDEL 1:50) at room temperature. Anti-mouse or anti-rabbit IgG conjugated to either Alexa 594 or Alexa 488 (Invitrogen) was used as a secondary antibody. Imaging was achieved with a Nikon TS-1 microscope equipped for confocal laser scanning fluorescence microscopy (Nikon C1 system) or with standard fluorescence with a Nikon Eclipse E600 microscope.
Miscellaneous Methods
Secreted AREG in solution was measured using the Amphiregulin Duo-set ELISA kit (DY262; R & D Systems, Minneapolis, MN.). Quantification of immunofluorescent images was achieved using ImageJ. The image threshold before counting cells utilized the algorithm by Li (40).
RESULTS
Expression of Secreted and ER Retained AGR2 Isoforms
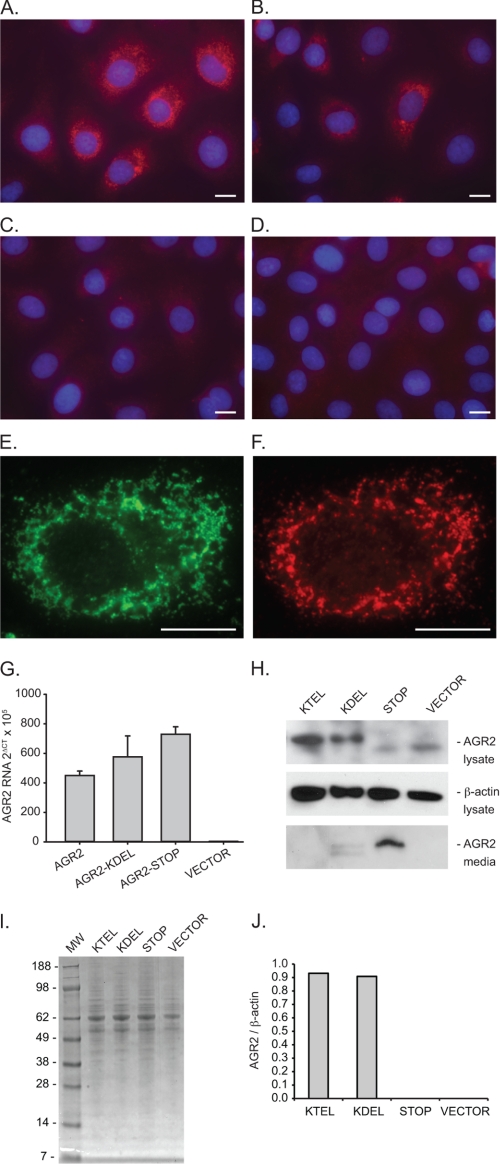
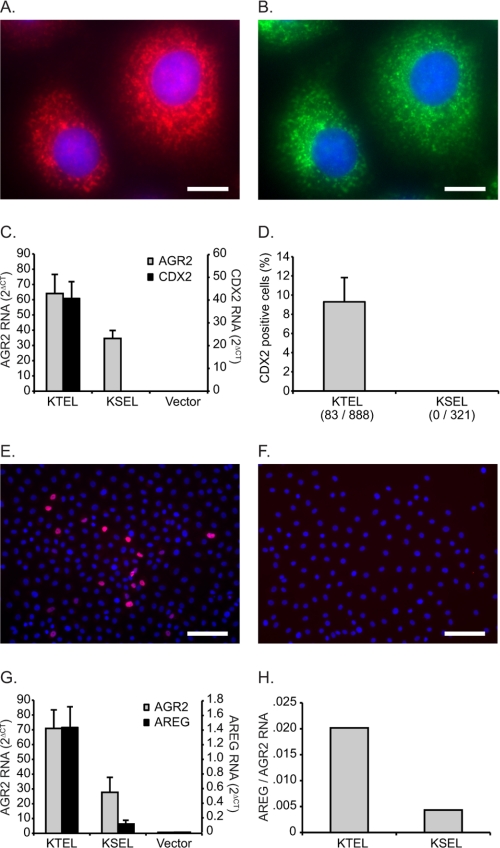
Our previous studies have utilized RNA interference to define the function of AGR2 in tumor cells. To explore the functional significance of the carboxyl-terminal sequence, a cell line with no AGR2 expression was used in this study. The presence of wild-type AGR2 containing a KTEL carboxyl-terminal domain may confound experiments when mutant proteins are expressed in the same cells. For the present study, function was prospectively monitored by the AGR2-induced appearance of new gene transcription, which provided a cleaner background to assess functional determinants. IEC-6 cells are derived from normal rat intestinal crypts and represent nontransformed cells that were first propagated as sustained primary cultures (25). Previous studies have shown that IEC-6 cells can be induced to differentiate through the expression of various developmental genes (27–30). No AGR2 expression is detected in wild-type IEC-6 cells when evaluated with quantitative real time PCR, protein immunoblotting, or immunocytochemistry (Fig. 1).
FIGURE 1.
AGR2 expression in rat intestinal IEC-6 cells. A–D, immunofluorescence images of IEC-6 cells transfected with AGR2 constructs terminating with a carboxyl-terminal KTEL (A), KDEL (B), or a STOP inserted before the wild-type KTEL (C) sequence. D, cells transfected with the expression vector alone. A–D, anti-AGR2 antibodies (red) and nuclear DAPI staining (blue). E and F, representative IEC-6 cell transfected with wild-type AGR2 and labeled with anti-KDEL (E, green) and anti-AGR2 (F, red) antibodies. The white bars in the images equal 10 μm. G, real time quantitative PCR determination of AGR2 RNA from transfected IEC-6 cells. The error bars represent S.D. H, protein immunoblots of transfected IEC-6 cell lysates (anti-AGR2 and anti-β-actin) or concentrated media (anti-AGR2). I, Coomassie Blue-stained SDS-PAGE of culture media depicted in H to show protein loading. J, graph of AGR2 protein levels as determined by densitometry of the immunoblot in H. The values are normalized by the corresponding β-actin band density.
Stable wild-type AGR2 expression was achieved in IEC-6 cells to produce IEC-6:AGR2-KTEL. In addition to wild-type AGR2, IEC-6 cells were also transfected with DNA constructs in which the carboxyl-terminal sequence was mutated from KTEL to KDEL (T173D, IEC-6:AGR2-KDEL) or a STOP was inserted before the KTEL (K172STOP, IEC-6:AGR2-STOP). The cells exhibited no gross phenotypic changes. Consistent with other proteins retained in the ER, immunofluorescence for AGR2 in IEC-6:AGR2-KTEL and IEC-6:AGR2-KDEL cells revealed punctate cytoplasmic staining consistent with an ER distribution (Fig. 1, A and B). ER localization was confirmed when the cells were also probed with an anti-KDEL antibody that binds HSPA5 (also known as GRP78), an ER resident protein, which revealed a similar staining pattern (Fig. 1, E and F). In contrast, IEC-6:AGR2-STOP cells (Fig. 1C) showed no cytoplasmic staining for AGR2 and were similar to IEC-6 cells transfected with the vector control (Fig. 1D). Protein immunoblotting of cell lysates also revealed the presence of AGR2 protein for IEC-6:AGR2-KTEL and IEC-6:AGR2-KDEL cells but not in the IEC-6:AGR2-STOP cells. In contrast, AGR2 was detected in the media only for the IEC-6:AGR2-STOP cells, revealing that it is secreted and not retained (Fig. 1H). Thus IEC-6:AGR2-KTEL and IEC-6:AGR2-KDEL cells exhibited ER localization in IEC-6 cells.
Only KTEL Retained AGR2 Induces CDX2 in IEC-6 Cells
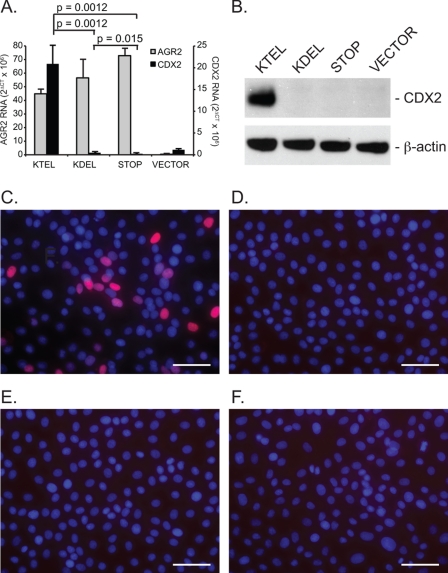
A screen for markers of intestinal development revealed that expression of the transcription factor CDX2 was markedly induced upon expression of wild-type AGR2 in IEC-6 cells. Real time quantitative PCR did not detect CDX2 expression in wild-type IEC-6 cells, but IEC-6:AGR2-KTEL cells showed a dramatic induction of CDX2 RNA (Fig. 2A). CDX2 protein production was confirmed with protein immunoblotting (Fig. 2B). Immunocytochemistry revealed the expected nuclear localization for CDX2, although the distribution among cells was heterogenous (Fig. 2C). Thus CDX2 expression in IEC-6 cells is a downstream result of AGR2 expression and can be used as a functional readout.
FIGURE 2.
AGR2 expression in IEC-6 cells induces CDX2 expression and requires a carboxyl-terminal KTEL sequence. A, real time quantitative PCR determination of AGR2 RNA (gray columns, left ordinate) and CDX2 RNA (black columns, right ordinate) levels in transfected IEC-6 cells. The error bars represent S.D. (n = 2). B, protein immunoblots of IEC-6 cell lysates for CDX2 and β-actin. C–F, immunofluorescence images of IEC-6 cells transfected with the same AGR2 constructs as depicted in Fig. 1A but probed with an anti-CDX2 antibody (red) or DAPI nuclear stain (blue). C, IEC-6:AGR2-KTEL; D, IEC-6:AGR2-KDEL; E, IEC-6:AGR2-STOP; F, IEC-6:VECTOR. The white bars represent 50 μm.
In contrast to the IEC-6:AGR2-KTEL cells, the IEC-6:AGR2-KDEL and IEC-6:AGR2-STOP cells did not show significant CDX2 RNA or protein expression (Fig. 2), despite the presence of AGR2 RNA (Fig. 2A) and protein (Fig. 1, H and J). Thus secreted AGR2 (IEC-6:AGR2-STOP) was not able to induce CDX2 expression. In addition, only ER localization by KTEL resulted in CDX2 expression. When a single amino acid substitution was performed changing KTEL to KDEL, CDX2 expression was not induced despite the presence of AGR2 in the ER.
KTEL and KDEL carboxyl-terminal Proteins Share the Same Receptor
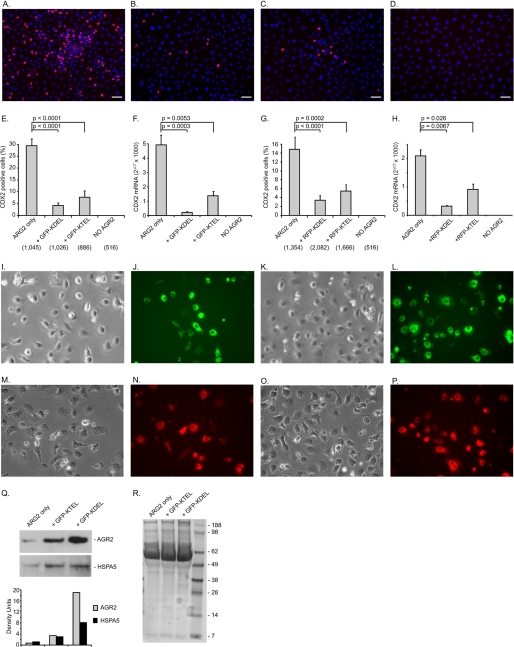
Whether wild-type AGR2 is being retained in the ER by the same mechanism as other KDEL-retained ER resident proteins was explored using competition experiments. Previous studies in yeast revealed that overexpression of a carboxyl-terminated HDEL protein results in secretion of ER resident proteins through competition with the ERD2 receptor (1, 31). The HDEL receptor (ERD2) represents the sole determinant for ER localization in yeast, because there is only one HDEL receptor. IEC-6:AGR2-KTEL cells that express CDX2 were transiently transfected with a previously described GFP-KDEL construct. The GFP-KDEL construct (also known as ER-GFP) is an inert protein with no interacting partners and does not undergo glycosylation or form disulfide bridges (26). Transient transfection of GFP-KDEL in IEC-6:AGR2-KTEL cells resulted in a 7-fold reduction in CDX2 expression as determined by the fraction of cells that express nuclear CDX2 expression on immunofluorescence (Fig. 3, B and E). Expression of a KTEL carboxyl-terminated GFP, GFP-KTEL, also induced a 3.8-fold decrease in CDX2 nuclear expression (Fig. 3, C and E). Consistent with the decrease in CDX2 protein detected by immunofluorescence, a concomitant decrease in CDX2 RNA was also observed using real time quantitative PCR of cells transfected with either GFP-KDEL or GFP-KTEL (Fig. 3F). To ensure that the observed results were not due to the GFP protein itself, RFP-tagged proteins were similarly expressed in the IEC-6:AGR2-KTEL cells, resulting in a similar outcome as measured by immunocytochemistry or CDX2 RNA levels (Fig. 3, G and H).
FIGURE 3.
CDX2 induction is mediated by binding to the KDEL receptors. Shown are the effects of expression with GFP and RFP proteins terminated at the carboxyl terminus with KDEL or KTEL on CDX2 expression in IEC-6:AGR2-KTEL cells. A–D show anti-CDX2 (red) and DAPI (blue) staining in IEC-6 cells transfected with AGR2 alone (A), AGR2 + GFP-KDEL (B), AGR2 + GFP-KTEL (C), and expression vector alone and no AGR2 (D). The white bars represent 50 μm. E, proportion of CDX2-positive cells for the constructs shown in A–D as quantified with ImageJ software. The total number of cells counted as determined by DAPI staining is listed in parentheses below the abscissa. F, real time quantitative PCR of CDX2 RNA normalized to β-actin for the same cells shown in E. G, proportion of CDX2-positive cells for similar constructs as shown in E except that RFP was substituted for GFP. H, real time quantitative PCR of CDX2 RNA normalized to β-actin for the same cells shown in G. I–L, controls for GFP transfection efficiency (400× magnification). Phase contrast (I and K) and GFP fluorescence (J and L) of IEC-6-AGR2 cells transfected with GFP-KDEL (I and J) or GFP-KTEL (K and L) are shown. M–P, controls for RFP transfection efficiency. Phase contrast (M and O) and RFP fluorescence (N and P) of IEC-6-AGR2 cells transfected with RFP-KDEL (M and N) or RFP-KTEL (O and P) are shown. Q, culture media of the cells transfected with the GFP constructs were evaluated with protein immunoblotting with anti-AGR2 and anti-KDEL (HSPA5) antibodies. Below each lane is a column graph depicting the density of the bands. R, Coomassie Blue staining of equal amounts of culture media as a loading control.
As observed for other ER resident proteins, overexpression of GFP-KDEL is expected to displace AGR2 if they share the same KDEL receptor. With the expression of either GFP-KDEL or GFP-KTEL, greater amounts of AGR2 are secreted into the culture media (Fig. 3Q). Protein immunoblotting of the culture media also showed increased secretion of soluble HSPA5 ER proteins as detected with anti-KDEL antibodies (Fig. 3Q). The results indicate that both KTEL- and KDEL-terminated proteins bind to the same receptors. The loss of function observed with KDEL-terminated AGR2 is not due to binding a different receptor.
Expression of KTEL-terminated AGR2 Is Required for Secretion of Amphiregulin in Human Esophageal Adenocarcinoma Cells
AGR2 is normally expressed in human esophageal adenocarcinoma OE33 cells where it induces the expression of the EGF receptor ligand, amphiregulin (AREG) (21). When AGR2 is overexpressed in OE33 cells, AREG expression is also induced. Thus OE33 cells provided an opportunity to study the role of the AGR2 carboxyl-terminal KTEL sequence in a tumor cell line that normally expresses the protein.
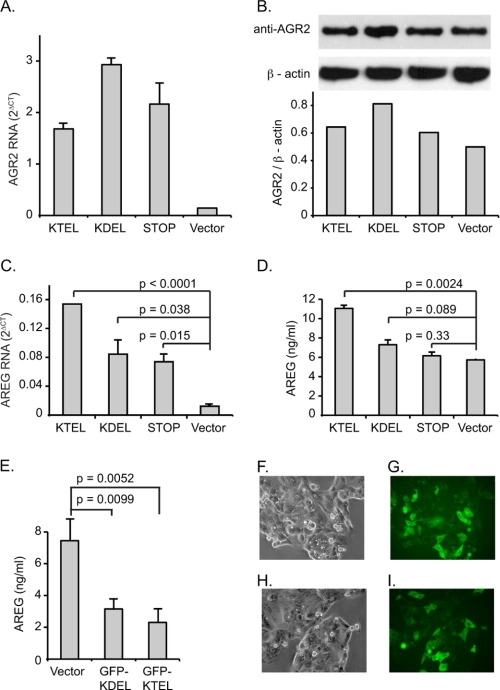
The same AGR2 DNA constructs previously used with the IEC-6 cells were also transiently transfected into OE33 cells to determine whether ER localization by the carboxyl-terminal KTEL sequence was similarly required for efficient AREG induction. Expression of wild-type KTEL carboxyl-terminated AGR2 enhanced AREG RNA and protein secretion by 12.5- and 1.9-fold, respectively (Fig. 4, C and D). Wild-type AGR2 overexpression resulted in higher levels of AREG secreted into the culture media (Fig. 4D). Expression of KDEL carboxyl-terminated AGR2, however, induced AREG RNA expression and protein secretion by only 4- and 1.3-fold, respectively, whereas the AGR2 (STOP) construct resulted in 4.7- and 1.1-fold differences from the control. Thus the functional significance of the carboxyl-terminal ER localization motif is conserved in a tumor cell line in which AGR2 expression is normally observed.
FIGURE 4.
AGR2 induction of AREG in OE33 esophageal adenocarcinoma cells requires a carboxyl-terminal KTEL sequence. A, real time quantitative PCR of total AGR2 RNA in OE33 cells transiently transfected with AGR2 constructs terminating with a carboxyl-terminal KTEL, KDEL, or a STOP sequence (before the wild-type KTEL). Vector designates transfection with the expression vector alone. B, immunoblots of cell lysates for total AGR2 and β-actin of transfected cells. The AGR2 antisera recognizes both the endogenous and transiently expressed constructs of AGR2. Under each lane is a column graph depicting the total AGR2/β-actin ratio determined by densitometry. C, real time quantitative PCR of total AREG RNA in the same cells as in A. D, determination of AREG protein levels in the culture media by ELISA. E, determination of AREG concentration in the media of wild-type OE33 cells that have been transiently transfected with GFP-KDEL and GFP-KTEL constructs. F–I, controls for GFP transfection efficiency. Phase contrast (F and H) and GFP fluorescence (G and I) of OE33 cells transfected with GFP-KDEL (F and G) or GFP-KTEL (H and I) are shown. The error bars for all of the column graphs represent S.D.
Whether the AGR2 induction of AREG expression also requires binding to the KDEL receptors was evaluated in OE33 cells through overexpression of the GFP-KDEL and GFP-KTEL constructs. Constructs terminating in either KDEL or KTEL motifs resulted in a decrease in AREG protein secretion of 2.4- and 3.2-fold, respectively (Fig. 4E).
ER Localization Signals with Similar Physical Characteristics as KTEL Cannot Support AGR2 Function
The substitution of KDEL for KTEL is predicted to result in a significant change in physical characteristics that may affect AGR2 function. Thus an alternative construct was evaluated in which a carboxyl-terminal sequence of KSEL replaced the KTEL ER localization motif. Previous work demonstrated that KSEL exhibits affinity for the KDEL receptors and results in ER localization (5). In addition, the substitution of serine for threonine is predicted to be the least disruptive. AGR2-KSEL expression in IEC-6 cells resulted in an ER distribution similar to that observed with anti-KDEL antibodies (Fig. 5, A and B). KSEL-terminated AGR2, however, did not induce CDX2 RNA or protein (Fig. 5, C–F). When AGR2-KSEL is expressed in OE33 cells, there is 4.7-fold less induction of AREG RNA compared with transfection with AGR2-KTEL (Fig. 5, G and H).
FIGURE 5.
Mutation of KTEL to KSEL results in compromised AGR2 induction of CDX2 and AREG. A and B, immunofluorescence labeling of AGR2-KSEL-expressing IEC-6 cells with anti-AGR2 (A) and anti-KDEL (B) antibodies. The white bars represent 10 μm. C, quantified RNA levels determined by real time quantitative PCR for AGR2 (left ordinate) and CDX2 (right ordinate) for AGR2-KTEL- and AGR2-KSEL-transfected IEC-6 cells. D, proportion of CDX2 positive cells in IEC-6 cells transfected with the AGR2-KTEL or AGR2-KSEL constructs. E and F, representative images from IEC-6 cells transfected with the AGR2-KTEL (E) or AGR2-KSEL (F) constructs and stained for CDX2 (red) and DAPI (blue). The white bars represent 100 μm. G, quantified RNA levels determined by real time quantitative PCR for AGR2 (left ordinate) and AREG (right ordinate) derived from AGR2-KTEL- and AGR2-KSEL-transfected OE33 cells. H, OE33 data from G that depict AREG expression normalized to total AGR2 expression. The error bars on all graphs represents S.D.
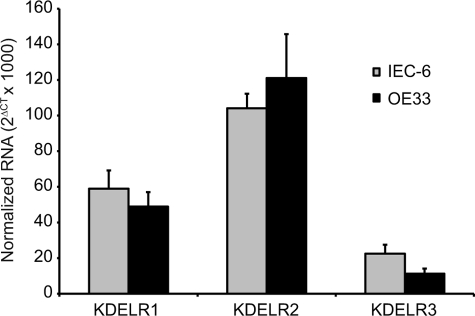
Both IEC-6 and OE33 Cells Express All Three Known KDEL Receptors
To determine which KDEL receptors were being affected by the competition experiments, real time PCR was performed on both cell lines to determine the relative levels of the three known KDEL receptors, KDELR1, KDELR2, and KDELR3. Both IEC-6 and OE33 cells expressed all three receptors, although in different proportions (Fig. 6). Similar to what has been previously reported for HeLa cells, RNA for KDELR2 was most abundant compared with the other two receptors.
FIGURE 6.
IEC-6 and OE33 cells express all three KDEL receptor isoforms. Shown is real time quantitative PCR for the three KDEL receptors (KDELR1, KDELR2, and KDELR3) in human OE33 and rat IEC-6 cells. The ordinate represents RNA levels normalized to β-actin. The error bars represent S.D. of three determinations.
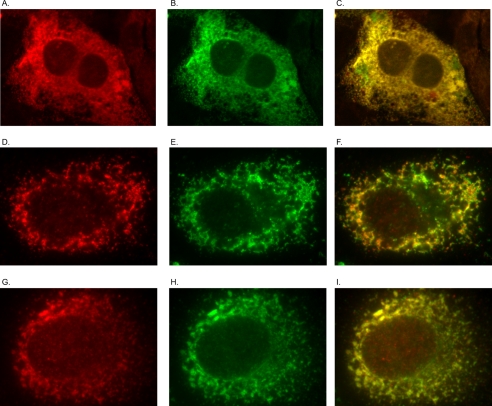
KTEL Carboxyl-terminated Proteins Sort to Same ER Compartment as KDEL-terminated Proteins
Additional experiments were performed to evaluate sorting by the KTEL motif independent of the AGR2 protein. GFP and RFP with either a carboxyl-terminal KTEL or KDEL were expressed together by co-transfection in CHO cells. Subsequent confocal immunocytochemistry revealed that both motifs resulted in co-localization of the GFP and RFP signals in a pattern consistent with the ER (Fig. 7, A–C).
FIGURE 7.
Carboxyl-terminal KDEL and KTEL proteins are both sorted to the same compartment. A–C, confocal imaging of CHO cells transfected with RFP-KDEL (A) and GFP-KTEL (B) in CHO cells. C, merged RFP/GFP image. D–I, IEC-6 cells transfected with AGR2-KTEL (D–F) or AGR2-KDEL (G–I) and probed with anti-AGR2 (D and G) or anti-KDEL (E and H) antibodies. F and I, merged images of the preceding two panels.
Additional experiments were also performed using IEC-6 cells to determine whether AGR2 that is terminated with either KTEL or KDEL will co-localize with other ER resident proteins as determined using the anti-KDEL antibody. In both scenarios, IEC-6 cells that express either AGR2 with a KTEL or KDEL carboxyl terminus co-localized with ER resident proteins as determined with the anti-KDEL antibody (Fig. 7, D–I). Thus ER localization by KTEL is required for AGR2 function, and a preferential distribution within the ER was not discernible.
DISCUSSION
Previous work established that AGR2 expression in esophageal and lung adenocarcinoma cell lines induces expression of the EGF receptor ligand, AREG. For the present study, AGR2 was discovered to induce the de novo expression of CDX2 in IEC-6 cells. AREG and CDX2 expression thus provided an opportunity to evaluate the protein domains necessary for AGR2 function. Published studies to date have reported an extracellular role for AGR2 as a secreted protein as well as an intracellular role as a protein chaperone or stress response factor (7, 10, 14, 16, 32, 33). Among the factors favoring an intracellular role based in the ER is the presence of a carboxyl-terminal motif, KTEL, that is conserved from Xenopus to humans.
Using immunofluorescence studies of Myc-tagged AGR2, Raykhel et al. (5) previously demonstrated that AGR2 is retained in the ER. The same study utilized biomolecular fluorescence complementation to show that AGR2 binds to all three isoforms of the KDEL receptor, although with lower affinity than controls possessing a KDEL carboxyl-terminal sequence. Although a variety of different carboxyl-terminal motifs bind to the three KDEL receptor isoforms and exhibit different affinities, whether there are functional consequences remained unknown.
The present study demonstrates that AGR2 residence within the ER is required for its function as reflected by AREG expression in OE33 adenocarcinoma cells or CDX2 expression by IEC-6 intestinal cells. Converting AGR2 to a secretory protein by deleting the KTEL terminus nullified the functional readout in both cell lines. The results are consistent with experiments in which application of recombinant AGR2 to the culture media was not able to induce either AREG or CDX2 expression.3
ER residence alone, however, is not sufficient for AGR2 function because conversion of the KTEL sequence to the classic KDEL motif failed to induce AREG or CDX2 expression. The results demonstrate that differences in ER localization signals may serve an essential functional role.
The results of the competition experiments are consistent with AGR2 ER localization that is mediated by the same receptors as those responsible for retaining KDEL-terminated proteins. Previous studies revealed that the single yeast counterpart of the KDEL receptor, ERD2, can be saturated by high expression of a HDEL-terminated ligand, resulting in secretion of ER resident proteins such as GRP78 (HSPA5) (1). In addition, saturation of the yeast ERD2 receptor with HDEL-terminated ligand inhibited yeast growth (31), a finding also observed in the present study. Similar to the yeast studies, overexpression of an independent protein that terminates in either KDEL or KTEL compromised AGR2 function and resulted in its secretion. Consistent with the previous work of Raykhel et al. (5), where higher affinity for the KDEL receptors was exhibited by KDEL compared with KTEL-terminated fluorescent proteins, a consistent effect was observed in the inhibition of AGR2 function or promotion of AGR2 secretion (Fig. 3, E–G and Q).
The distribution of KTEL-terminated proteins, including AGR2, was similar to that of other ER resident proteins as determined using the anti-KDEL antibodies. When considered in the context of the competition experiments, the data would support involvement of the previously described KDEL receptors. Because the generation of isoform-specific anti-KDEL receptor antibodies have been unsuccessful (5), whether AGR2 preferentially associates with a particular isoform cannot be determined. Given that the KDEL receptors were identified by sequence homology to the yeast ERD2 gene or to each other, it is possible that another receptor exists that is capable of binding both KDEL- and KTEL-terminated proteins and is required for AGR2 function. If this is true, however, the distribution of such a receptor must result in a distribution for AGR2 that is similar to the other ER proteins retrieved by the KDEL receptors.
Several possibilities may account for the diversity of ER localization signals. One hypothesis is that KDEL receptor binding influences AGR2 function by affecting its subcellular location or promoting a protein complex that includes the receptors. The major difference between the various ER localization sequences described to date is the different affinities displayed for the three KDEL receptors. Recent studies have suggested that different affinities for the KDEL receptor may result in changes in subcellular distribution that are dynamic and dependent on situational conditions such as the cell density or growth media (6).
Protein complexes that include chaperones bound to the KDEL receptors have also been described that may mediate a function other than the retrograde transport of proteins back to the ER (34). Of particular interest are studies showing that ligand-bound KDEL receptors also bind p38 MAPKs and JNKs, which suggests potential involvement in physiological and pathological processes (35). High throughput binding assays have also revealed additional associations between the KDEL receptors and proteins not known to serve a role in retrograde transport, but the functional implications are currently unclear (36–38).
We hypothesize that the ER localization motif serves a biofunctional role, which accounts for the diversity in motifs observed. The carboxyl-terminal domain is required for both ER localization and enzymatic function. KDEL receptor binding of soluble lumenal ER proteins such as the thioredoxins is pH-dependent, where the higher acidity of the Golgi apparatus results in receptor association. Once the receptor transports its bound ligand back to the ER where the pH is less acidic, the ligand disassociates and becomes a soluble protein (39). Similar to other ER thioredoxins, soluble AGR2 is likely to bind to substrate(s) that are currently being actively pursued. We hypothesize that the ER localization signal is critical for conformation of AGR2 and substrate binding in the ER as a soluble protein. Thus carboxyl-terminal alteration to KDEL or KSEL retains ER localization, but disrupts the function of AGR2 as an enzyme in the ER. The highly conserved amino acid sequence of the AGR2 ER localization signal in all species supports an important functional role.
In summary, the present work establishes that AGR2 localization in the ER by KTEL is required for its ability to induce AREG or CDX2 expression. ER residence alone, however, is not sufficient, because a specific localization signal is required for AGR2 function. Thus the diversity in ER localization signals serves an essential functional role and provides a rationale for the highly conserved carboxyl-terminal motif of AGR2. Because AGR2 expression is enhanced in almost all human adenocarcinomas, the current findings provide insights into potential mechanisms, the subcellular location required for AGR2 function, and potential therapeutic strategies for cancer.
Acknowledgments
We appreciate the technical assistance provided by May Tun and Dariusz Wodziak's critical reading of the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grants DK063624 (to A. W. L.) and DK56339 (to the Stanford University Digestive Disease Center). This work was also supported by the Stanford Cancer Center.
A. Gupta, A. Dong, and A. W. Lowe, personal observations.
- ER
- endoplasmic reticulum
- Bis-Tris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- RFP
- red fluorescent protein.
REFERENCES
- 1. Dean N., Pelham H. R. (1990) Recycling of proteins from the Golgi compartment to the ER in yeast. J. Cell Biol. 111, 369–377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Lewis M. J., Sweet D. J., Pelham H. R. (1990) The ERD2 gene determines the specificity of the luminal ER protein retention system. Cell 61, 1359–1363 [DOI] [PubMed] [Google Scholar]
- 3. Semenza J. C., Hardwick K. G., Dean N., Pelham H. R. (1990) ERD2, a yeast gene required for the receptor-mediated retrieval of luminal ER proteins from the secretory pathway. Cell 61, 1349–1357 [DOI] [PubMed] [Google Scholar]
- 4. Munro S., Pelham H. R. (1987) A C-terminal signal prevents secretion of luminal ER proteins. Cell 48, 899–907 [DOI] [PubMed] [Google Scholar]
- 5. Raykhel I., Alanen H., Salo K., Jurvansuu J., Nguyen V. D., Latva-Ranta M., Ruddock L. (2007) A molecular specificity code for the three mammalian KDEL receptors. J. Cell Biol. 179, 1193–1204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Alanen H. I., Raykhel I. B., Luukas M. J., Salo K. E., Ruddock L. W. (2011) Beyond KDEL. The role of positions 5 and 6 in determining ER localization. J. Mol. Biol. 409, 291–297 [DOI] [PubMed] [Google Scholar]
- 7. Aberger F., Weidinger G., Grunz H., Richter K. (1998) Anterior specification of embryonic ectoderm. The role of the Xenopus cement gland-specific gene XAG-2. Mech. Dev. 72, 115–130 [DOI] [PubMed] [Google Scholar]
- 8. Sive H. L., Hattori K., Weintraub H. (1989) Progressive determination during formation of the anteroposterior axis in Xenopus laevis. Cell 58, 171–180 [DOI] [PubMed] [Google Scholar]
- 9. Kumar A., Godwin J. W., Gates P. B., Garza-Garcia A. A., Brockes J. P. (2007) Molecular basis for the nerve dependence of limb regeneration in an adult vertebrate. Science 318, 772–777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kumar A., Delgado J. P., Gates P. B., Neville G., Forge A., Brockes J. P. (2011) The aneurogenic limb identifies developmental cell interactions underlying vertebrate limb regeneration. Proc. Natl. Acad. Sci. U.S.A. 108, 13588–13593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wang Z., Hao Y., Lowe A. W. (2008) The adenocarcinoma-associated antigen, AGR2, promotes tumor growth, cell migration, and cellular transformation. Cancer Res. 68, 492–497 [DOI] [PubMed] [Google Scholar]
- 12. Hao Y., Triadafilopoulos G., Sahbaie P., Young H. S., Omary M. B., Lowe A. W. (2006) Gene expression profiling reveals stromal genes expressed in common between Barrett's esophagus and adenocarcinoma. Gastroenterology 131, 925–933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lowe A. W., Olsen M., Hao Y., Lee S. P., Taek Lee K., Chen X., van de Rijn M., Brown P. O. (2007) Gene expression patterns in pancreatic tumors, cells and tissues. PLoS One 2, e323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ramachandran V., Arumugam T., Wang H., Logsdon C. D. (2008) Anterior gradient 2 is expressed and secreted during the development of pancreatic cancer and promotes cancer cell survival. Cancer Res. 68, 7811–7818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Thompson D. A., Weigel R. J. (1998) hAG-2, the human homologue of the Xenopus laevis cement gland gene XAG-2, is coexpressed with estrogen receptor in breast cancer cell lines. Biochem. Biophys. Res. Commun. 251, 111–116 [DOI] [PubMed] [Google Scholar]
- 16. Zhang J. S., Gong A., Cheville J. C., Smith D. I., Young C. Y. (2005) AGR2, an androgen-inducible secretory protein overexpressed in prostate cancer. Genes Chromosomes Cancer 43, 249–259 [DOI] [PubMed] [Google Scholar]
- 17. Fritzsche F. R., Dahl E., Dankof A., Burkhardt M., Pahl S., Petersen I., Dietel M., Kristiansen G. (2007) Expression of AGR2 in non small cell lung cancer. Histol. Histopathol. 22, 703–708 [DOI] [PubMed] [Google Scholar]
- 18. Zhu H., Lam D. C., Han K. C., Tin V. P., Suen W. S., Wang E., Lam W. K., Cai W. W., Chung L. P., Wong M. P. (2007) High resolution analysis of genomic aberrations by metaphase and array comparative genomic hybridization identifies candidate tumour genes in lung cancer cell lines. Cancer Lett. 245, 303–314 [DOI] [PubMed] [Google Scholar]
- 19. Edgell T. A., Barraclough D. L., Rajic A., Dhulia J., Lewis K. J., Armes J. E., Barraclough R., Rudland P. S., Rice G. E., Autelitano D. J. (2010) Increased plasma concentrations of anterior gradient 2 protein are positively associated with ovarian cancer. Clin. Sci. 118, 717–725 [DOI] [PubMed] [Google Scholar]
- 20. Liu D., Rudland P. S., Sibson D. R., Platt-Higgins A., Barraclough R. (2005) Human homologue of cement gland protein, a novel metastasis inducer associated with breast carcinomas. Cancer Res. 65, 3796–3805 [DOI] [PubMed] [Google Scholar]
- 21. Dong A., Gupta A., Pai R. K., Tun M., Lowe A. W. (2011) The human adenocarcinoma-associated gene, AGR2, induces expression of amphiregulin through Hippo pathway co-activator YAP1 activation. J. Biol. Chem. 286, 18301–18310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Fletcher G. C., Patel S., Tyson K., Adam P. J., Schenker M., Loader J. A., Daviet L., Legrain P., Parekh R., Harris A. L., Terrett J. A. (2003) hAG-2 and hAG-3, human homologues of genes involved in differentiation, are associated with oestrogen receptor-positive breast tumours and interact with metastasis gene C4.4a and dystroglycan. Br. J. Cancer 88, 579–585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ruan J., Li H., Chen Z., Coghlan A., Coin L. J., Guo Y., Hériché J. K., Hu Y., Kristiansen K., Li R., Liu T., Moses A., Qin J., Vang S., Vilella A. J., Ureta-Vidal A., Bolund L., Wang J., Durbin R. (2008) TreeFam: 2008 Update. Nucleic Acids Res. 36, D735–D740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Pelham H. R. (1990) The retention signal for soluble proteins of the endoplasmic reticulum. Trends Biochem. Sci. 15, 483–486 [DOI] [PubMed] [Google Scholar]
- 25. Quaroni A., Wands J., Trelstad R. L., Isselbacher K. J. (1979) Epithelioid cell cultures from rat small intestine. Characterization by morphologic and immunologic criteria. J. Cell Biol. 80, 248–265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Snapp E. L., Sharma A., Lippincott-Schwartz J., Hegde R. S. (2006) Monitoring chaperone engagement of substrates in the endoplasmic reticulum of live cells. Proc. Natl. Acad. Sci. U.S.A. 103, 6536–6541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Suh E., Traber P. G. (1996) An intestine-specific homeobox gene regulates proliferation and differentiation. Mol. Cell. Biol. 16, 619–625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Soubeyran P., André F., Lissitzky J. C., Mallo G. V., Moucadel V., Roccabianca M., Rechreche H., Marvaldi J., Dikic I., Dagorn J. C., Iovanna J. L. (1999) Cdx1 promotes differentiation in a rat intestinal epithelial cell line. Gastroenterology 117, 1326–1338 [DOI] [PubMed] [Google Scholar]
- 29. Yoshida S., Kajimoto Y., Yasuda T., Watada H., Fujitani Y., Kosaka H., Gotow T., Miyatsuka T., Umayahara Y., Yamasaki Y., Hori M. (2002) PDX-1 induces differentiation of intestinal epithelioid IEC-6 into insulin-producing cells. Diabetes 51, 2505–2513 [DOI] [PubMed] [Google Scholar]
- 30. Escaffit F., Paré F., Gauthier R., Rivard N., Boudreau F., Beaulieu J. F. (2006) Cdx2 modulates proliferation in normal human intestinal epithelial crypt cells. Biochem. Biophys. Res. Commun. 342, 66–72 [DOI] [PubMed] [Google Scholar]
- 31. Townsley F. M., Frigerio G., Pelham H. R. (1994) Retrieval of HDEL proteins is required for growth of yeast cells. J. Cell Biol. 127, 21–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zhao F., Edwards R., Dizon D., Afrasiabi K., Mastroianni J. R., Geyfman M., Ouellette A. J., Andersen B., Lipkin S. M. (2010) Disruption of Paneth and goblet cell homeostasis and increased endoplasmic reticulum stress in Agr2−/− mice. Dev. Biol. 338, 270–279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Park S. W., Zhen G., Verhaeghe C., Nakagami Y., Nguyenvu L. T., Barczak A. J., Killeen N., Erle D. J. (2009) The protein disulfide isomerase AGR2 is essential for production of intestinal mucus. Proc. Natl. Acad. Sci. U.S.A. 106, 6950–6955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Capitani M., Sallese M. (2009) The KDEL receptor. New functions for an old protein. FEBS Lett. 583, 3863–3871 [DOI] [PubMed] [Google Scholar]
- 35. Yamamoto K., Hamada H., Shinkai H., Kohno Y., Koseki H., Aoe T. (2003) The KDEL receptor modulates the endoplasmic reticulum stress response through mitogen-activated protein kinase signaling cascades. J. Biol. Chem. 278, 34525–34532 [DOI] [PubMed] [Google Scholar]
- 36. Ewing R. M., Chu P., Elisma F., Li H., Taylor P., Climie S., McBroom-Cerajewski L., Robinson M. D., O'Connor L., Li M., Taylor R., Dharsee M., Ho Y., Heilbut A., Moore L., Zhang S., Ornatsky O., Bukhman Y. V., Ethier M., Sheng Y., Vasilescu J., Abu-Farha M., Lambert J. P., Duewel H. S., Stewart I. I., Kuehl B., Hogue K., Colwill K., Gladwish K., Muskat B., Kinach R., Adams S. L., Moran M. F., Morin G. B., Topaloglou T., Figeys D. (2007) Large-scale mapping of human protein-protein interactions by mass spectrometry. Mol. Syst. Biol. 3, 89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Miller J. P., Lo R. S., Ben-Hur A., Desmarais C., Stagljar I., Noble W. S., Fields S. (2005) Large-scale identification of yeast integral membrane protein interactions. Proc. Natl. Acad. Sci. U.S.A. 102, 12123–12128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Hardwick K. G., Boothroyd J. C., Rudner A. D., Pelham H. R. (1992) Genes that allow yeast cells to grow in the absence of the HDEL receptor. EMBO J. 11, 4187–4195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Wilson D. W., Lewis M. J., Pelham H. R. (1993) pH-dependent binding of KDEL to its receptor in vitro. J. Biol. Chem. 268, 7465–7468 [PubMed] [Google Scholar]
- 40. Li C. H., Tam P. K. S. (1998) An iterative algorithm for minimum cross entropy thresholding. Pattern Recognition Letters 18, 771–776 [Google Scholar]