Background: The catalytic subunit of the PP2A phosphatase interacts with structural and regulatory proteins and can be regulated post-translationally by phosphorylation and methylation.
Results: A novel alternatively spliced isoform of the PP2A catalytic subunit has been identified.
Conclusion: The alternatively spliced isoform shows a specific interaction profile and no phosphatase activity.
Significance: The alternatively spliced isoform may represent a novel mechanism of PP2A regulation.
Keywords: Alternative splicing, Gene expression, Protein-protein interactions, Serine threonine protein phosphatase, Signal transduction
Abstract
PP2A is the main serine/threonine-specific phosphatase in animal cells. The active phosphatase has been described as a holoenzyme consisting of a catalytic, a scaffolding, and a variable regulatory subunit, all encoded by multiple genes, allowing for the assembly of more than 70 different holoenzymes. The catalytic subunit can also interact with α4, TIPRL (TIP41, TOR signaling pathway regulator-like), the methyl-transferase LCMT-1, and the methyl-esterase PME-1. Here, we report that the gene encoding the catalytic subunit PP2Acα can generate two mRNA types, the standard mRNA and a shorter isoform, lacking exon 5, which we termed PP2Acα2. Higher levels of the PP2Acα2 mRNA, equivalent to the level of the longer PP2Acα mRNA, were detected in peripheral blood mononuclear cells that were left to rest for 24 h. After this time, the peripheral blood mononuclear cells are still viable and the PP2Acα2 mRNA decreases soon after they are transferred to culture medium, showing that generation of the shorter isoform depends on the incubation conditions. FLAG-tagged PP2Acα2 expressed in HEK293 is catalytically inactive. It displays a specific interaction profile with enhanced binding to the α4 regulatory subunit, but no binding to the scaffolding subunit and PME-1. Consistently, α4 out-competes PME-1 and LCMT-1 for binding to both PP2Acα isoforms in pulldown assays. Together with molecular modeling studies, this suggests that all three regulators share a common binding surface on the catalytic subunit. Our findings add important new insights into the complex mechanisms of PP2A regulation.
Introduction
Reversible phosphorylation is the most common mechanism of signal transduction in eukaryotic cells. Phosphatases play a pivotal role in this process, allowing a fine tuning of the activity, localization, and half-life of regulatory proteins involved in specific biochemical pathways (1, 2).
Type 2A phosphatases (PP2A,5 PP4, and PP6) belong to the PPP subfamily and act on a variety of substrates and cellular processes, including DNA replication, transcriptional regulation, translation, signal transduction, and apoptosis (1–3). PP2A is the major soluble serine/threonine-specific phosphatase in animal cells and one of the most conserved eukaryotic proteins (1, 2). The active PP2A phosphatase has been described as a holoenzyme consisting of a catalytic subunit (C), a scaffolding (PR65/A), and one of the several variable regulatory subunit (B). The diversity of regulatory subunits allows tight regulation of its substrate specificity and subcellular localization, and additional cellular and viral proteins can also associate with PP2A and regulate its activity (1, 2, 4). PP2A is a tumor suppressor, and its inactivation by mutations or viral proteins is a major mechanism of cellular transformation (5–8).
The combination of different types of catalytic, scaffolding, and regulatory subunits results in more than 70 different PP2A holoenzymes, each one exhibiting different specificity. However, this diversity relies mostly on the variable regulatory subunits, whereas the catalytic and scaffolding subunits display limited variability. Holoenzyme assembly is also under post-translational regulation by modifications such as phosphorylation or methylation of the catalytic subunit. Reversible methylation of PP2Ac C-terminal is mediated by leucine carboxy-methyl transferase 1 (LCMT-1) and protein phosphatase methyl esterase-1 (PME-1) (9, 10), which can form complexes with the catalytic subunit.
Tap42/α4 and Tip41/TIPRL are regulatory proteins that can associate with PP2A catalytic subunits as well as with PP4c and PP6c (11). Both can interact simultaneously constituting a trimeric complex (12). α4 has been recognized as a major regulator of PP2A complex assembly and PP2Ac degradation (13, 14). It suppresses the stress response factor c-Jun and represses p53-dependent transcription and apoptosis by maintaining both substrates in a low phosphorylation state (13). α4 deletion promotes apoptosis in a variety of cell lines and leads to progressive loss of all PP2A, PP4, and PP6 complexes. Association with α4 renders newly synthesized or old free catalytic subunits enzymatically inactive and resistant to proteasomal degradation until their assembly into functional phosphatase complexes (14). It was demonstrated that the ubiquitin-interacting motif of α4 mediates the interaction with monoubiquitinated forms of PP2Ac protecting them from polyubiquitination and degradation by the E3 ubiquitin ligase Mid1 (15). Although α4 C-terminal region interacts with E3 ubiquitin ligases, its association with PP2Ac is mediated by its N-terminal domain, whose structure reveals a tetratricopeptide repeat-like domain (16–18).
In human cells, so far only two PP2A catalytic subunits have been described, PP2Acα and PP2Acβ, which are encoded by different genes (1, 2). Here, we report the identification of a novel splice variant of PP2Acα in human cells. We show that the new variant is catalytically inactive and displays enhanced binding to the α4 regulatory subunit but does not bind to the PR65/A subunit. In addition, using recombinant proteins we found that the PP2A regulators PME-1 and LCMT-1 bind both PP2Ac isoforms with similar affinity, but their interaction is prevented by prior binding of α4 to the catalytic subunit. To our knowledge, this is the first case of alternative splicing described for PP2Acα, which might constitute a novel regulatory mechanism for this enzyme and provide new insights into the mechanism of PP2Ac association with α4.
EXPERIMENTAL PROCEDURES
Reagents and Antibodies
Rapamycin (LC Laboratories) was used at a final concentration of 200 nm. Phytohemagglutinin (PHA) (Sigma) was used in a final concentration of 2 μg/ml. The following antibodies were used in this study: rabbit antibodies for PP2Ac, TIPRL, phospho-RPS6, GAPDH and actin (Bethyl Laboratories), goat anti-α4 (Santa Cruz Biotechnology), mouse anti-PME-1 (Santa Cruz Biotechnology), mouse monoclonal anti-PR65/A (Invitrogen), mouse anti-FLAG M2 monoclonal antibody (Sigma), and anti-hexahistidine tag (Qiagen). Monoclonal anti-GST was obtained from a hybridoma (19).
DNA Cloning and Sequencing
The clone 3.4 cDNA from the novel PP2Acα2 isoform cloned into pACT2 and pGEX-5X-2, the α4 cDNA cloned into pET28a, and the TIPRL cDNA cloned into pET-TEV (a pET28a derivative) were already available in our laboratory from previous studies (12, 20). The PP2Acα cDNA (from the gene located on chromosome 5q31.1, GRCh37.p5, NC_000005.9) was amplified by RT-PCR from several immortalized human cell lines and from peripheral blood mononuclear cells (PBMCs) and cloned using the pGEM-T-easy cloning system (Promega) according to the manufacturer's instructions. The PME-1 and LCMT-1 cDNAs were amplified from a fetal brain cDNA library (Clontech) using oligonucleotides 5′-GGA TCC ATG GCC ACT AGG CAG AGG GAA TC-3′ (LCMT-1F), 5′-GCG GCC GCT TAA TAA GTT ATC TCC TTC AGC CCA AG-3′ (LCMT-1R), 5′-GGG AAT TCA TGT CGG CCC TCG AAA AGA GC-3′ (PME-1F), and 5′-GGG TCG ACT TAA CAG CCA GGA AAC ACA CAC TG-3′ (PME-1R) and cloned into pGEM-T vector (Promega). The cDNAs were subcloned into pET-TEV using BamHI and NotI restriction enzymes for LCMT-1, and EcoRI and SalI for PME-1. The cDNAs from the novel and standard PP2Ac isoforms were also subcloned into the mammalian expression vector pcDNA-FLAG, in-frame with the FLAG tag (Invitrogen), generating plasmids pcDNA-FLAG-PP2Acα and pcDNA-FLAG-PP2Acα2. Cloning procedures were carried out as described previously (21). cDNA sequences were verified by using an Applied Biosystems 3130 xl DNA analyzer.
Cell Culture and Handling
Buffy coat cells from healthy blood donors were obtained by centrifugation using triple top and bottom bag separation systems. The interface fraction (from here on called PBMCs) was harvested, washed three times in Hanks' salt solution, and transferred to culture flasks in RPMI 1640 medium (Invitrogen), with or without fetal calf serum (10%) and 2 mm l-glutamine. Cells were treated with rapamycin and/or PHA and kept in culture at 37 °C and 5% CO2. Finally, cells were harvested for RNA and protein extraction. For the experiments shown in the Fig. 3 (A and B), a Qiagen RNA blood mini kit was used to isolate total RNA, where erythrocytes were removed by lysis. The PBMCs were kept at room temperature in microcentrifuge tubes up to the moment of RNA extraction. Human immortalized cells HEK293, RAJI, and Jurkat were maintained in 100-mm dishes and submitted to starvation (medium without serum) and/or rapamycin treatment. HEK293 cells were cultured in minimal essential α medium (Invitrogen), and RAJI and Jurkat cells were cultured in RPMI 1640 medium.
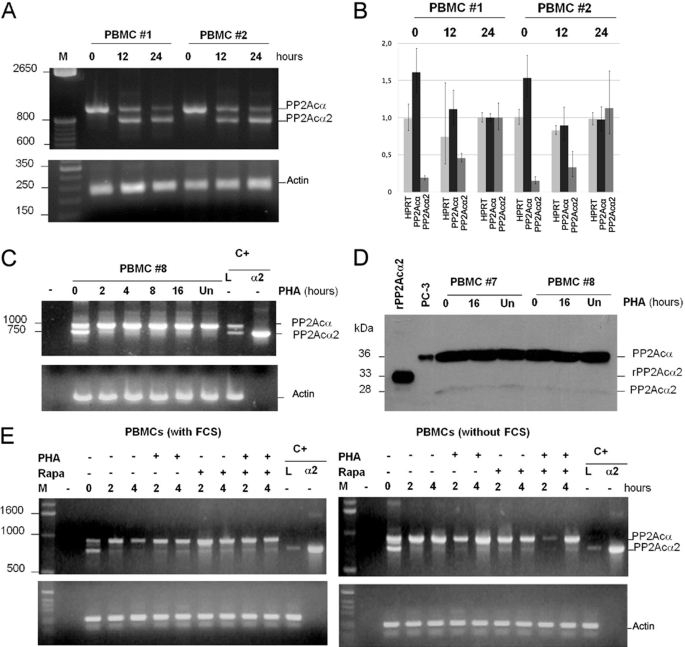
FIGURE 3.
Analysis of PP2Acα2 expression in PBMCs. A, analysis of the RT-PCR products of PP2Acα in PBMCs isolated from donors #1 and #2. RT-PCR was performed with RNA samples isolated at the moment of blood extraction (time 0) and 12 and 24 h after extraction. β-Actin was used as a control. B, real-time qPCR quantification of the samples from A. The numbers on the left indicate the -fold change relative to the sample taken from donor 1 (PBMC #1) at 24 h, which was used to normalize the other samples. Light gray: HPRT (control), medium gray: PP2Acα2, dark gray: ful-length PP2Acα. C, time course analysis of the RT-PCR products of PP2Acα in PBMC isolated from donor #8. After resting for 24 h, cells were transferred to culture followed by activation with PHA for the indicated times. Parallel amplification of a 300-bp actin cDNA was used as an internal control. D, Western blot analysis of the PP2Acα protein isoforms of PBMCs from donors #7 and #8. After resting for 24 h, cells were transferred to culture followed by treatment or not with phytohemagglutinin (PHA) for the indicated times. Recombinant PP2Acα2 was used as a reference in the Western blot. E, analysis of RT-PCR products of PP2Acα obtained from cDNAs of PBMCs. After resting for 24 h, cells were transferred to culture for the indicated times. +, indicates the cells treated with phytohemagglutinin (PHA) or rapamycin (Rapa). The two amplified bands correspond to the longer and shorter isoforms of PP2Acα (930 and 768 bp, respectively). L = positive control of cDNA amplification from leukocyte cDNA library; α2 = positive control of amplification of PP2Acα2 cDNA.
Whole cell extracts were prepared by suspending cells in high salt buffer supplemented with protease and phosphatase inhibitors from Calbiochem (250 mm NaCl, 50 mm Tris-HCl, pH 8.0, 5 mm EDTA, pH 8.0, 0.5% Nonidet P-40). After 30 min on ice, lysates were cleared by centrifugation (20,000 × g, 10 min, 4 °C).
To obtain stable clones expressing each PP2Ac isoform, ∼106 HEK293 cells were cultured in Dulbecco's modified Eagle's medium until they reached 60% confluency. The cells were transfected by electroporation with 20 μg of the constructs pcDNA-FLAGØ (empty vector), pcDNA-FLAG-PP2Acα, or pcDNA-FLAG-PP2Acα2 and submitted to selection with 1 mg/ml G-418 sulfate (Stratagene). Expression of FLAG-PP2Acα and FLAG-PP2Acα2 was analyzed by Western blot using an antibody against the FLAG tag (mouse anti-FLAG M2, Sigma).
RNA Isolation and cDNA Amplification by RT-PCR
Total RNA was isolated from PBMCs and cell lines using the TRIzol reagent (Invitrogen) or the QIAamp RNA Blood Mini kit (Qiagen) according to the manufacturer's instructions. RNA quality and concentration were determined by spectrophotometry. Total cDNAs were synthesized using the QuantiTect Reverse Transcription system (Qiagen) according to the supplier's instructions or using the MMLV reverse transcriptase (Invitrogen). For MMLV-reverse transcription reactions, initially 500 ng from total RNA, 10 mm dNTPs and10 μm oligo(dT) were incubated at 65 °C for 5 min. Subsequently, the reverse transcriptase (MMLV-RT) and the first strand buffer 5× (250 mm Tris-HCl, pH 8.3, 375 mm KCl, 15 mm MgCl2) were added to the reaction, which was incubated at 37 °C for 50 min and at 70 °C for 15 min. Amplification of the PP2Acα cDNAs by PCR was performed using the primers 5′-GAA TTC CAT ATG GAC GAG AAG GTG TTC AC-3′ and 5′-GGA TCC TTA CAG GAA GTA GTC TGG GGT AC-3′. β-Actin partial cDNA amplification was performed as a control using the primers 5′-GAA ACT ACC TTC AAC TCC ATC-3′ and 5′-CGA GGC CAG GAT GGA GCC GCC-3′. Amplicons were analyzed on 2% agarose gel electrophoresis.
Real-time PCR
To analyze the variation of the total amount of RNA in the experiment shown in Fig. 3A, the same samples were subjected to real-time qPCR amplification. The reaction mix contained 12.5 μl of SYBR Green PCR Master Mix 2× (Applied Biosystems), 1 μl of each primer, 1 μl of cDNA, and 10.5 μl of sterile water. The primers were designed to detect either the standard (Forward1: 5′-GAT CTT CTG TCT ACA TGG TGG TCT C-3′ and Reverse1: 5′-ACA CAT TGG ACC CTC ATG GGG AA-3′) or the novel isoform (Forward1 and Reverse2: 5′-CCA GTT ATA TCC CTC ATG GGG AAC-3′). Primers for detection of HPRT1 (Forward: 5′-TGA CAC TGG CAA AAC AAT GCA GA-3′ and Reverse: 5′-GGT CCT TTT CAC CAG CAA GCT-3′) were used as endogenous control. qPCR assays were performed on an ABI® PRISM® 7500 Sequence Detection System (Applied Biosystems). To ensure experimental accuracy, all reactions were performed in triplicate. Gene expression was quantified by the comparative CT method, normalizing CT values to the housekeeping gene GAPDH (forward primer: 5′-TGC ACC ACC AAC TGC TTA GC-3′; reverse primer: 5′-GGC ATG GAC TGT GGT CAT GAG-3′) and calculating relative expression values.
GST Pulldown Assays
GST-PP2Acα, GST-PP2Acα2, or GST alone were coexpressed in Escherichia coli BL21(DE3) with histidine-tagged TIPRL, PME-1, LCMT-1, or α4. Protein expression was induced with isopropyl 1-thio-β-d-galactopyranoside (0.5 mm), and the cells were incubated at 16 °C for 16 h or at 25 °C for 4 h. Cell lysis and interaction assays were performed as described previously (12).
PP2A Activity Assay
HEK293 cells stably transfected with pcDNA-FLAG, pcDNA-FLAG-PP2Acα, or pcDNA-FLAG-PP2Acα2 were suspended in lysis buffer (150 mm NaCl, 30 mm Tris-HCl, pH 8.0, 3 mm EDTA, pH 8.0, 0.3% Nonidet P-40), incubated on ice for 15 min, and centrifuged at 20,000 × g for 10 min at 4 °C. The cleared lysates were separated in triplicates (300 μl each), which were incubated with 1.5 μg of anti-FLAG M2 monoclonal antibody (Sigma) and 15 μl of Protein A/G plus beads for 16 h at 4 °C. The beads were harvested by centrifugation at 550 × g for 1 min at 4 °C and washed three times with lysis buffer. To perform the phosphatase assay, 40 μl of PP2A assay buffer (50 mm Tris-HCl, pH 8.5, 20 mm MgCl2, 1 mm DTT, 14 mm p-nitrophenyl phosphate) was added to the beads, and the reactions were incubated at 30 °C for 30 min. PP2A activity was measured as the absorbance of the supernatants at 405 nm using an EnSpire plate reader (Perkin-Elmer). The beads were subsequently analyzed by Western blot.
Molecular Docking
The PDB files corresponding to α4 N-terminal domain (PDB: 3QC1 (18)) and PP2A catalytic subunit (PDB: 3DW8, chain C (22)) were submitted to automated docking using the ClusPro 2.0 server (available on-line (23–26)). Each protein was analyzed both as receptor and ligand. The five highest scoring models were analyzed using PyMOL (available on-line (27)).
RESULTS
Identification of a Novel Splicing Isoform of PP2Acα
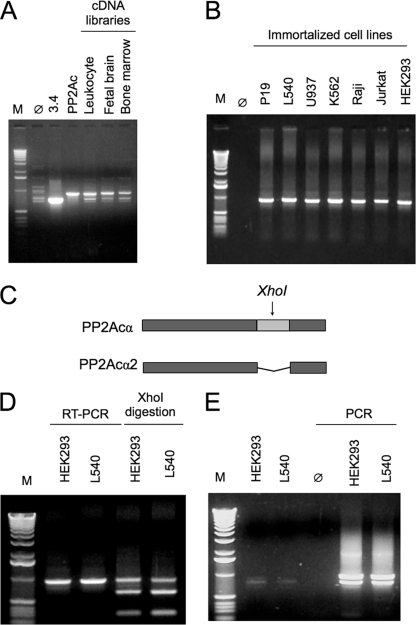
In a previous study, we have reported the interaction of type 2A phosphatase catalytic subunits with TIPRL. The interactions were initially identified in a yeast two-hybrid screen using TIPRL as bait to screen a human leukocyte cDNA library (12). One of the clones isolated in that screen, then denominated clone 3.4, corresponded to a novel variant of PP2Acα lacking an internal segment. Analysis of the PP2Acα gene structure revealed that the missing part corresponds exactly to the 5th exon, suggesting that the novel variant could result from alternative splicing (Fig. 1 and supplemental Fig. S1). Investigation of PP2Ac cDNAs from human cDNA libraries (leukocyte, fetal brain, and bone marrow) confirmed the presence of two distinct bands, one corresponding to the expected molecular weight of the standard PP2Acα cDNA and another to the shorter isoform (Fig. 1A), which we will refer to as PP2Acα2 from here on.
FIGURE 1.
Identification and cloning of a novel splicing isoform of the PP2A catalytic subunit α. A, PCR amplification of PP2Ac cDNAs from three human cDNA libraries (leukocyte, fetal brain, and bone marrow) results in two distinct bands, corresponding to the canonical and to a novel shorter isoform (PP2Acα2). Clone 3.4 and PPA2cα indicate the control reactions containing the short and the canonical cDNA forms, respectively. B, RT-PCR amplification of PP2Ac cDNAs from immortalized cell lines results in a single band corresponding to the canonical isoform. C, diagram of the PP2Acα cDNAs showing the unique XhoI restriction site located at exon 5, which is missing in PP2Acα2. D and E, analysis of PP2Acα cDNA amplification by RT-PCR from two immortalized cell lines (HEK293 and L540). Following one round of PCR amplification, the PCR product was digested with XhoI to deplete the samples from the predominant PP2Acα amplification product. The cDNA containing exon 5 is digested with XhoI, producing two fragments (638 and 292 bp), whereas the shorter cDNA is not digested. E, the undigested PCR product was extracted from the gel and purified (left) and used as template in a second round of PCR amplification using PP2Acα-specific primers. This amplification resulted in two bands of similar intensity. The shorter band was cloned into pGEM-T easy vector and sequenced, which confirmed its identity as the shorter PP2Acα2 isoform.
In an attempt to verify the existence of this novel isoform in cultured cell lines, we amplified the PP2Ac cDNA by RT-PCR from several immortalized human cell lines, mostly from hematopoietic origin. For all the cell lines tested (P19, L450, U937, K562, RAJI, Jurkat, and HEK293), there apparently was amplification of only one PCR product that corresponded to the molecular weight of the standard PP2Acα cDNA (Fig. 1B). However, this observation needed confirmation. Therefore, to determine whether permanent cells lines express the PP2Acα2 mRNA, we relied on the existence of a unique XhoI restriction site within exon 5 of PP2Acα (Fig. 1C), which allows for selective digestion of the PP2Acα cDNA. The RT-PCR products from two representative cell lines, one adherent (HEK293) and one non-adherent (L540), were digested with XhoI to selectively digest the PP2Acα cDNA, so that it would not be amplified in subsequent PCR reactions (Fig. 1D). The faint bands corresponding to the shorter isoform were purified from gels and used as templates in a second round of PCR amplification, which resulted in two distinct bands (Fig. 1E). The shorter PCR product was purified from the gel and cloned into the pGEM-T vector for sequencing analysis, which confirmed that it was indeed the cDNA of the PP2Acα2 isoform.
The results described above suggested that PP2Acα2 mRNA exists in several immortalized human cell lines, however at very low expression levels. We, therefore, asked whether its expression might be induced under specific conditions. As a first attempt to identify such conditions, we tested serum starvation or addition of rapamycin to block cell growth. In RAJI and Jurkat cells, these treatments resulted in a detectable although rather modest increase in PP2Acα2 mRNA levels (Fig. 2A). This effect could also be observed at the protein level as the Western blot analysis revealed that the PP2Acα2 isoform is indeed expressed also as a protein in both cell lines (Fig. 2B).
FIGURE 2.
Analysis of the PP2Acα2 protein expression in RAJI and Jurkat cells. A, analysis of RT-PCR products of PP2Acα obtained from the cDNAs of RAJI and Jurkat cells. The two bands detected correspond to the longer and shorter PP2Ac isoforms (930 and 768 bp, respectively). L = positive control of cDNA amplification from a leukocyte cDNA library; 3.4 = positive control of amplification of PP2Acα2 cDNA. Parallel amplification of a 300-bp actin cDNA was used as an internal control. B, Western blot analysis of the protein isoforms of PP2Acα isolated from lysates of RAJI and Jurkat cells. +, indicates cells treated with either rapamycin (Rapa) or fetal bovine serum (FCS, 10%) for 48 h, respectively. Actin was used as a loading control in the Western blots.
PP2Acα2 Is Expressed in Resting Human Peripheral Blood Mononuclear Cells but Inhibited in Cultured Cells
Because the PP2Acα2 mRNA was expressed at relatively high levels in cDNA libraries obtained from human tissues, including leukocytes, but was barely detectable in immortalized cell lines, we decided to analyze its expression levels in primary PBMCs. In freshly isolated PBMCs from healthy donors; only the standard PP2Acα mRNA could be detected by RT-PCR (Fig. 3A). Interestingly, when the cells are left to rest at room temperature, the alternatively spliced PP2Acα2 mRNA is detected, being as abundant as that of the PP2Acα mRNA after 12–24 h at room temperature (Fig. 3A). A real-time PCR quantification of the samples analyzed in Fig. 3A showed that the abundance of PP2A mRNA decreased by 30% after 24 h, whereas the shorter isoform mRNA showed a 5-fold increase (Fig. 3B). The PBMCs are still viable after this period of incubation at room temperature as they recover following transfer to culture conditions. The PP2Acα2 mRNA decreases soon after the PBMCs are transferred to culture medium. A time course experiment of PBMCs that have been left to rest for 24 h showed that expression of PP2Acα2 mRNA was strongly inhibited in the first 2 h of culture, decreasing to a barely detectable level after 16 h (Fig. 3C). The PP2Acα2 protein was detected by Western blot, although the ratio between PP2Acα/PP2Acα2 proteins in PBMCs was not equivalent to the PP2Acα/PP2Acα2 mRNA ratio (Fig. 3D). Changes in culture conditions by addition of PHA, rapamycin, or serum starvation did not affect the levels of the PP2Acα2 isoform after the cells are transferred to culture medium (Fig. 3E). This shows that generation of the PP2Acα2 isoform takes place only under very stringent conditions that might involve reduced nutrient levels, which is the case when the PBMCs are stored for relatively long periods.
PP2Acα2 Is Catalytically Inactive, Shows Enhanced Binding of α4, and Does Not Interact with the Scaffolding Subunit PR65/A
After confirming the existence of a novel splicing variant of PP2Ac that is expressed as a protein, although at low levels compared with the standard isoform, the next step in our investigation consisted of analyzing the possible functional differences between these two PP2Acα isoforms. For this purpose, we generated stably transfected HEK293 cell lines expressing FLAG-tagged PP2Acα (clones A4 and C5) or PP2Acα2 (clone 3.4), as well as a control cell line transfected with the empty vector, and analyzed protein expression, enzyme activity, and interaction profiles.
Cells expressing FLAG-PP2Acα and FLAG-PP2Aα2 showed proliferation rates similar to the control cells transfected with the empty vector (not shown), indicating that they are not toxic to the cell at these expression levels. The cells transfected with pcDNA-FLAG-PP2Acα showed a corresponding band detectable by Western blotting using an anti-PP2Ac antibody (Fig. 4A), whereas in cells transfected with pcDNA-FLAG-PP2Acα2, the short isoform could be detected only by immunoprecipitation with anti-FLAG antibody (Fig. 4C). In addition, the cells transfected with pcDNA-FLAG-PP2Acα expressing the standard isoform showed a reduction in the levels of the endogenous PP2Acα, whereas no changes were observed in cells transfected with the empty vector and FLAG-PP2Acα2 (Fig. 4A). Transfection with FLAG-PP2Acα2 resulted also in a significant increase in the protein levels of the PP2Ac regulator α4, as well as in a decrease in the levels of the PP2A scaffolding subunit PR65/A, as seen by Western blot analysis (Fig. 4A).
FIGURE 4.
Characterization of PP2Acα2 stably expressed in HEK293 cells. A, Western blot analysis of the whole cell lysates obtained from HEK293 cells transfected with empty pcDNA-FLAG vector (vector), FLAG-PP2Acα2 (3.4), or FLAG-PP2Acα (A4 and C5) using antibodies to detect the catalytic subunit of PP2A. Phosphorylation of the ribosomal protein RPS6 is slightly reduced in clone C5 and unchanged in clones 3.4 and A4. Expression of FLAG-PP2Acα2 (clone 3.4) results in increased expression of α4 and reduced expression of PR65/A. B, phosphatase assays using FLAG immunoprecipitates from the four cell lineages described above. PP2Acα2 is catalytically inactive. C, Western blot analysis of the FLAG immunoprecipitates assayed in B to detect the presence of the FLAG-tagged proteins (using and anti-FLAG antibody) and their interaction with endogenous proteins using antibodies specific to α4, PR65/A, and TIPRL. PP2Acα2 precipitates higher amounts of α4 and does not precipitates PR65/A.
An in vitro activity assay with p-nitrophenyl phosphate as substrate and the immunopurified FLAG-tagged PP2Acα isoforms showed that PP2Acα2 does not display any detectable enzymatic activity (Fig. 4B). If this was due only to a defect in the active site, it would be expected that incorporation of the defective enzyme into heterotrimeric PP2A complexes might lead to a dominant negative phenotype. However, no enhanced phosphorylation of RPS6, a known PP2A target, was detected in cellular extracts expressing PP2Acα2, indicating that the existence of a catalytically inactive variant of PP2Ac does not suppress the activity of the endogenous enzyme (Fig. 4). This explains also why the levels of the endogenous PP2Acα levels are not affected in the cells transfected with pcDNA-FLAG-PP2Acα2.
The FLAG immunoprecipitates used in the phosphatase activity assay were analyzed by Western blot to detect interactions of the two PP2Ac isoforms with endogenous proteins. PP2Acα2 precipitated a significantly higher amount of endogenous α4 than PP2Acα, whereas the interaction with TIPRL remained unchanged. However, a striking finding was that the interaction of PP2Acα2 with the PR65/A subunit was completely abolished (Fig. 4C). This finding indicates that PP2Acα2 is not incorporated into the mature PP2A heterotrimeric complexes.
Binding of α4 to PP2Acα2 Prevents Its Interaction with PME-1 and LCMT-1
An examination of the position of exon 5 in the three-dimensional structure of PP2Ac heterotrimeric complex with PR65/A (28, 29) shows that it is close to the active site (supplemental Fig. S2). This might explain the loss of catalytic activity by PP2Acα2, but the analysis of the structure also indicates that the residues encoded by exon 5 are distant from the site of interaction with PR65/A, suggesting that PP2Acα2 interaction with this subunit should not be directly affected by absence of the exon 5 amino acid residues. Another possibility to be considered is that the preferential binding of PP2Acα2 to α4 is responsible for its reduced interaction with PR65/A, however, this implies that exon 5 amino acids participate in the interaction interface with α4. The interaction site for α4 on PP2Ac is still elusive due to the lack of structural data and conflicting information obtained from biochemical experiments. Mutation analysis suggested that α4 and PR65/A compete for PP2Ac binding, indicating that these two subunits share a site of interaction on the side opposite the catalytic site of the phosphatase (30). However, the observation that okadaic acid treatment induces the dissociation of PP2Ac and α4 suggested that α4 might occlude the active site of PP2Ac (31).
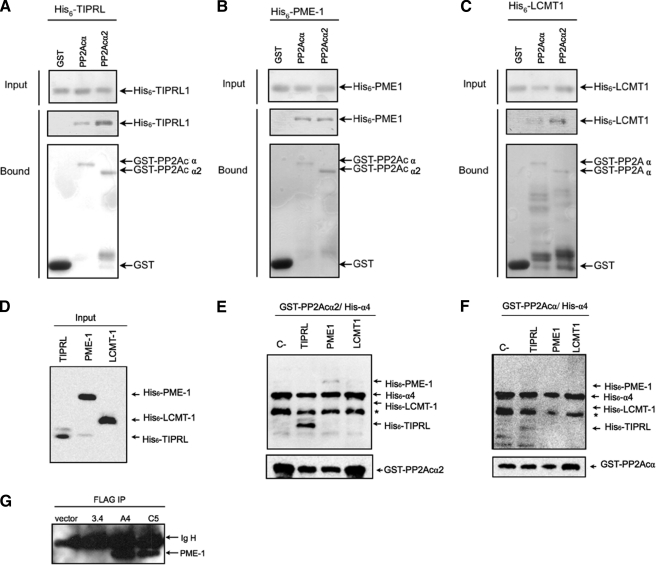
The PP2A regulators PME-1 and LCMT-1 catalyze, respectively, methylation and demethylation of the C-terminal Leu309 residue of PP2Ac, which is located near the catalytic site in the three-dimensional structure of the enzyme (9, 10). An examination of the crystal structures of PP2Ac in complex with these two proteins showed that the amino acids encoded by exon 5 participate extensively in these contacts (supplemental Fig. S2). We, therefore, sought to determine how this might affect the interactions of PME-1 and LCMT-1 with the novel PP2Acα2 isoform. We co-expressed recombinant GST-tagged PP2Acα and PP2Acα2 with histidine-tagged PME-1 or LCMT-1 in E. coli and performed in vitro pulldown assays. Histidine-tagged TIPRL was used as a parallel control. This assay showed that PME-1, TIPRL, and LCMT-1 all bind to both PP2Acα isoforms, with TIPRL and LCMT-1 showing a slight preference for PP2Acα2 (Fig. 5, A–C). We used PME-1 or LCMT-1 to obtain an indirect indication of the site of interaction of α4 on PP2A, by performing in vitro binding assays to the preassembled PP2Ac·α4 complex. GST-tagged PP2Acα or PP2Acα2 were co-expressed in E. coli with histidine-tagged α4, and the lysates of these co-expressions were mixed with lysates from cells expressing histidine-tagged TIPRL, PME-1, or LCMT-1. The GST-tagged proteins were isolated using glutathione-Sepharose beads, and the interacting proteins were analyzed by Western blot (Fig. 5, D–F). Our previous results showed that TIPRL binds to PP2Ac in the presence of α4 and that residues 210–309 of PP2Ac are sufficient to interact with TIPRL (12). Consistently, TIPRL was detected in association with the PP2Ac·α4 complex in the present assay. However, neither PME-1 nor LCMT-1 was detected in GST-PP2Ac·α4 pulldown assays, suggesting that α4 physically blocks the binding sites for PME-1 and LCMT-1 on both PP2Acα isoforms. The same result was observed in a similar experiment using purified proteins instead of cell lysates (not shown). This finding is in agreement with the fact that the residues encoded by exon 5 of PP2Ac affect its affinity for binding to α4. An analysis of the FLAG immunoprecipitations shown in Fig. 4C with an antibody directed to PME-1 showed that it binds only to PP2Acα, and not to PP2Acα2 (Fig. 5G), which is in line with our result described above (Fig. 4C) of the lack of interaction of PP2Acα2 with PR65/A.
FIGURE 5.
GST pulldown assays using recombinant proteins. A–C, interaction assays of TIPRL, PME-1, and LCMT-1. Fusions of the indicated phosphatase catalytic subunits were co-expressed in E. coli with histidine-tagged TIPRL, PME-1, or LCMT1. GST was used as a negative control. GST fusion proteins were isolated from extracts by binding to glutathione-Sepharose beads. Bound proteins were resolved by SDS-PAGE and detected by immunoblotting with anti-His or anti-GST primary antibodies. Both PP2Ac isoforms interacted specifically with the regulatory proteins tested. D, Western blot analysis using anti-hexahistidine antibody of E. coli extracts expressing histidine-tagged TIPRL, PME-1, and LCMT-1. E and F, interaction assays of the PP2Ac·α4 complexes and TIPRL, PME-1, or LCMT1. GST fusions of the indicated phosphatase catalytic subunits were co-expressed in E. coli with histidine-tagged α4, and the extracts were mixed with the extracts shown in D and incubated for 2 h as 4 °C. Bound proteins were resolved by SDS-PAGE and detected by immunoblotting with anti-hexahistidine antibody (upper panels) or anti GST (lower panels). Only TIPRL interacted with PP2Ac in the presence of α4. The asterisk indicates a proteolysis product of α4. G, Western blot analysis of the FLAG immunoprecipitates shown in Fig. 4C to detect the presence of endogenous PME-1 using anti PME-1 antibody.
Structural Model of the α4·PP2Ac Complex
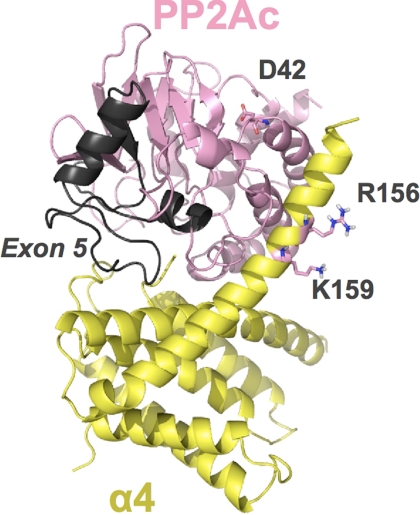
The striking differences observed in the activity and interactions of the two PP2Acα isoforms led us to investigate what are the possible structural explanations for these differences. After several unsuccessful attempts to crystallize the α4·PP2Acα2 complex, we performed an automated docking of PP2Ac to the recently published crystal structure of the N-terminal domain of α4 (α4ΔC) using the Cluspro server. Both proteins were analyzed as receptor and ligand. The five highest scored models from each docking were analyzed using PyMOL, and, although slightly different from each other, these models displayed a consistent pattern of binding of α4 close to the active site of PP2Ac (supplemental Figs. S3 and S4). The highest scoring model obtained using α4 as receptor and PP2Ac as ligand, depicted in Fig. 6, suggests that helixes 5 and 6 of α4 bind to PP2Ac in a structure that resembles a clamp. Mutation of aspartate 42 of PP2Ac and arginine 156 and lysine 159 of α4 are sufficient to disrupt their interaction (18, 30). Interestingly, these residues appear in close proximity in our model, suggesting that they might form salt bridges that stabilize the complex. In this model, the loop formed by amino acids encoded by exon 5 of PP2Ac is part of the interaction interface with α4, which is consistent with the results shown in the previous section. The fact that PP2Acα2, which lacks exon 5, binds to α4 with higher affinity than PP2Acα suggests that this is a destabilizing interface, possibly required for the regulated dissociation of the many complexes that PP2Acα forms in vivo.
FIGURE 6.
Schematic representation of the highest scoring model obtained using α4 (PDB: 3QC1) as receptor and PP2Ac (PDB: 3DW8, chain C) as ligand. α4 is colored in yellow, and PP2Ac is colored in pink. Exon 5 of PP2Ac is colored in gray. Residues Asp42 of PP2Ac and Arg156 and Lys159 of α4 are represented as sticks. The model was obtained from automated docking using ClusPro 2.0 (available on-line) and rendered using PyMOL (available on-line).
DISCUSSION
The catalytic subunit of PP2A can associate with a variety of proteins and dephosphorylate innumerous substrates, acting like a hub in cellular signaling networks and controlling innumerous cellular processes (1–4). Such important tasks need many forms of fine regulation. Here, we describe a novel isoform of PP2Acα generated by exon 5 skipping. The finding that the shorter PP2Acα2 isoform is found in PBMCs that are maintained in conditions similar to those used for storage of transfusion blood is intriguing and reveals a mechanism of alternative splicing that mediates a novel mode of PP2Acα regulation. This mechanism seems to be associated to nutrient restrictions, or it might also be possible that it is a mechanism found in resting cells, which would explain why the levels of the shorter PP2Acα2 isoform decrease soon after transfer to culture medium. Despite many attempts to detect endogenous PP2Acα2 in cultured cell lines, it could only be found in trace amounts relative to the standard isoform and under specific conditions, such as starvation of RAJI and Jurkat. Whether this isoform might represent a conspicuous protein in any cell type or condition still remains an open question.
To characterize the catalytic activity and interaction profile of this new PP2Acα isoform, we generated stable HEK293 cell lines expressing FLAG-tagged PP2Acα or PP2Acα2 and also expressed recombinant GST-tagged proteins in E. coli. Interestingly, no significant differences in binding affinity for the PP2A regulators TIPRL, α4, PME-1, and LCMT-1 were noticed when comparing the two GST-tagged PP2Acα isoforms expressed in bacteria. However, when we analyzed the same proteins expressed in mammalian cells, we found that PP2Acα2 precipitated at least twice as much α4 as compared with the standard 309-residue isoform, whereas the interaction with the PR65/A subunit virtually disappeared while the interaction with TIPRL remained unchanged. Changes in the expression levels of α4 and PR65/A caused by expression of PP2Acα2 were consistent with these interaction profiles, with a marked reduction in PR65/A, and an increase in α4 levels. It is possible that PR65/A is destabilized by the unbalance caused by expression of the PP2Acα2 isoform and is targeted for degradation, whereas α4 is stabilized by its enhanced interaction with the PP2Acα2 isoform.
The most striking differences between the PP2Acα isoforms is the fact PP2Acα2, lacking amino acids encoded by exon 5, is catalytically inactive toward p-nitrophenyl phosphate and does not interact with the PR65/A subunit. However, the interaction analysis of the phosphatase regulator α4 also produced important findings for the understanding of the functional differences between the two PP2Acα isoforms. It has long been discussed whether α4 is a phosphatase inhibitor or an allosteric regulator; however, recent findings suggested that the primary function of α4 is to stabilize an immature form of the PP2A catalytic subunit and to promote its assembly into the canonical heterotrimeric complexes (14). According to this model, the α4·PP2Ac complex is catalytically inactive, and the function of α4 would be to stabilize and to direct the catalytic subunit to the correct complexes, where it would become active. Our findings agree with this model, because the higher affinity of the PP2Acα2 isoform for α4 favors the stabilization of a trapped intermediate complex in the maturing process of PP2A, which is unable to reach the final active state in the heterotrimeric complex.
In vitro binding assays using recombinant proteins showed that α4 prevents PME-1 and LCMT-1 from interacting with both PP2Acα2 and PP2Acα, suggesting that these proteins share a common interaction site on the catalytic subunit, which includes residues encoded by exon 5 and is close to the phosphatase active site (9, 10). Molecular modeling of the α4·PP2Ac complex supports this proposal while accommodating also previous information from point mutations (18, 30). Taken together, our data indicate that α4 blocks the PP2Ac active site, however, okadaic acid does not induce the dissociation of α4 from either one of the isoforms (Ref. 12 and data not shown). Incubation with a molar excess of PME-1 or LCMT-1, or even the drug metformin, which was recently reported to induce dissociation of this complex in HeLa cell extracts (32), were unable to dissociate α4 from PP2Ac in vitro (data not shown). Our results also suggested that PP2Acα2, but not PP2Acα, is unable to dissociate from α4, but due to the high stability of this complex it was not possible to study the dissociation process in vitro.
One important question to be answered is whether PP2Acα2 is catalytically inactive due to a defect in the active site or simply because it is prevented from being incorporated into the active holoenzyme, because it does not interact with the PR65/A subunit. It was not possible to test the activity of the recombinant protein expressed in E. coli, because even the standard isoform is inactive when expressed in bacteria (data not shown). Most probably, we observed a combination of these two factors, which raises an interesting question: could these effects be separated by generating a PP2Ac mutant, which is enzymatically inactive but still is incorporated into heterotrimeric complexes? In a recent work, a PP2Ac active site mutant (H118N) was found to completely lose its interaction with the PR65/A subunit, whereas the interaction with α4 remained unchanged (33). This suggests that the regulation of PP2Ac by α4 might be so robust that any modification that results in a defective enzyme is readily detected and sequestered in an inactive form.
In summary, the results presented in this work strongly suggest a new regulatory mechanism for PP2A which is, to our knowledge, the first case of alternative splicing described for PP2Acα. This work also sheds new light on the mechanism of regulation of the canonical PP2Acα isoform by α4.
Acknowledgments
We thank Maria Eugenia R. Camargo and Elaine Cristina Teixeira for technical assistance and Adriana Duarte from the Hemocentro, Universidade Estadual de Campinas, Campinas, for providing “buffy coat” samples.
This work was supported by Fundação de Amparo à Pesquisa do Estado São Paulo, Conselho Nacional de Desenvolvimento Científico e Tecnológico, and Laboratório Nacional de Biociências, Centro Nacional de Pesquisa em Energia e Materiais.

This article contains supplemental Figs. S1–S4.
- PP2A
- protein phosphatase 2A
- PME-1
- protein phosphatase methyl esterase-1
- LCMT-1
- leucine carboxy-methyl transferase 1
- RPS6
- ribosomal protein S6
- PBMC
- peripheral blood mononuclear cell
- PHA
- phytohemagglutinin
- HEK293
- human embryonic kidney 293
- TIPRL
- TOR signaling pathway regulator-like.
REFERENCES
- 1. Janssens V., Goris J. (2001) Protein phosphatase 2A. A highly regulated family of serine/threonine phosphatases implicated in cell growth and signaling. Biochem. J. 353, 417–439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Zolnierowicz S. (2000) Type 2A protein phosphatase, the complex regulator of numerous signaling pathways. Biochem. Pharmacol. 60, 1225–1235 [DOI] [PubMed] [Google Scholar]
- 3. Goldberg Y. (1999) Protein phosphatase 2A. Who shall regulate the regulator? Biochem. Pharmacol. 57, 321–328 [DOI] [PubMed] [Google Scholar]
- 4. Virshup D. M., Shenolikar S. (2009) From promiscuity to precision. Protein phosphatases get a makeover. Mol. Cell 33, 537–545 [DOI] [PubMed] [Google Scholar]
- 5. Eichhorn P. J., Creyghton M. P., Bernards R. (2009) Protein phosphatase 2A regulatory subunits and cancer. Biochim. Biophys. Acta 1795, 1–15 [DOI] [PubMed] [Google Scholar]
- 6. Junttila M. R., Puustinen P., Niemelä M., Ahola R., Arnold H., Böttzauw T., Ala-aho R., Nielsen C., Ivaska J., Taya Y., Lu S. L., Lin S., Chan E. K., Wang X. J., Grènman R., Kast J., Kallunki T., Sears R., Kähäri V. M., Westermarck J. (2007) CIP2A inhibits PP2A in human malignancies. Cell 130, 51–62 [DOI] [PubMed] [Google Scholar]
- 7. Mumby M. (2007) PP2A. Unveiling a reluctant tumor suppressor. Cell 130, 21–24 [DOI] [PubMed] [Google Scholar]
- 8. Van Hoof C., Goris J. (2004) PP2A fulfills its promises as tumor suppressor. Which subunits are important? Cancer Cell 5, 105–106 [DOI] [PubMed] [Google Scholar]
- 9. Xing Y., Li Z., Chen Y., Stock J. B., Jeffrey P. D., Shi Y. (2008) Structural mechanism of demethylation and inactivation of protein phosphatase 2A. Cell 133, 154–163 [DOI] [PubMed] [Google Scholar]
- 10. Stanevich V., Jiang L., Satyshur K. A., Li Y., Jeffrey P. D., Li Z., Menden P., Semmelhack M. F., Xing Y. (2011) The structural basis for tight control of PP2A methylation and function by LCMT-1. Mol. Cell 41, 331–342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chen J., Peterson R. T., Schreiber S. L. (1998) α4 associates with protein phosphatases 2A, 4, and 6. Biochem. Biophys. Res. Commun. 247, 827–832 [DOI] [PubMed] [Google Scholar]
- 12. Smetana J. H., Zanchin N. I. (2007) Interaction analysis of the heterotrimer formed by the phosphatase 2A catalytic subunit, α4 and the mammalian ortholog of yeast Tip41 (TIPRL). FEBS J. 274, 5891–5904 [DOI] [PubMed] [Google Scholar]
- 13. Kong M., Fox C. J., Mu J., Solt L., Xu A., Cinalli R. M., Birnbaum M. J., Lindsten T., Thompson C. B. (2004) The PP2A-associated protein α4 is an essential inhibitor of apoptosis. Science 306, 695–698 [DOI] [PubMed] [Google Scholar]
- 14. Kong M., Ditsworth D., Lindsten T., Thompson C. B. (2009) α4 is an essential regulator of PP2A phosphatase activity. Mol. Cell 36, 51–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. McConnell J. L., Watkins G. R., Soss S. E., Franz H. S., McCorvey L. R., Spiller B. W., Chazin W. J., Wadzinski B. E. (2010) α4 is a ubiquitin-binding protein that regulates protein serine/threonine phosphatase 2A ubiquitination. Biochemistry 49, 1713–1718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. McConnell J. L., Gomez R. J., McCorvey L. R., Law B. K., Wadzinski B. E. (2007) Identification of a PP2A-interacting protein that functions as a negative regulator of phosphatase activity in the ATM/ATR signaling pathway. Oncogene 26, 6021–6030 [DOI] [PubMed] [Google Scholar]
- 17. McDonald W. J., Sangster S. M., Moffat L. D., Henderson M. J., Too C. K. (2010) α4 phosphoprotein interacts with EDD E3 ubiquitin ligase and poly(A)-binding protein. J. Cell Biochem. 110, 1123–1129 [DOI] [PubMed] [Google Scholar]
- 18. LeNoue-Newton M., Watkins G. R., Zou P., Germane K. L., McCorvey L. R., Wadzinski B. E., Spiller B. W. (2011) The E3 ubiquitin ligase- and protein phosphatase 2A (PP2A)-binding domains of the α4 protein are both required for α4 to inhibit PP2A degradation. J. Biol. Chem. 286, 17665–17671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Assmann E. M., Alborghetti M. R., Camargo M. E., Kobarg J. (2006) FEZ1 dimerization and interaction with transcription regulatory proteins involves its coiled-coil region. J. Biol. Chem. 281, 9869–9881 [DOI] [PubMed] [Google Scholar]
- 20. Smetana J. H., Oliveira C. L., Jablonka W., Aguiar Pertinhez T., Carneiro F. R., Montero-Lomeli M., Torriani I., Zanchin N. I. (2006) Low resolution structure of the human α4 protein (IgBP1) and studies on the stability of α4 and of its yeast ortholog Tap42. Biochim. Biophys. Acta 1764, 724–734 [DOI] [PubMed] [Google Scholar]
- 21. Ausubel F., Brent R., Kingston R., Moore D. D., Seidman J. G., Smith J. A., Struhl K. (1998) Current Protocols in Molecular Biology, John Wiley and Sons, New York [Google Scholar]
- 22. Xu Y., Chen Y., Zhang P., Jeffrey P. D., Shi Y. (2008) Structure of a protein phosphatase 2A holoenzyme. Insights into B55-mediated Tau dephosphorylation. Mol. Cell 31, 873–885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kozakov D., Hall D. R., Beglov D., Brenke R., Comeau S. R., Shen Y., Li K., Zheng J., Vakili P., Paschalidis I. Ch., Vajda S. (2010) Achieving reliability and high accuracy in automated protein docking. ClusPro, PIPER, SDU, and stability analysis in CAPRI rounds 13–19. Proteins 78, 3124–3130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kozakov D., Brenke R., Comeau S. R., Vajda S. (2006) PIPER. An FFT-based protein docking program with pairwise potentials. Proteins 65, 392–406 [DOI] [PubMed] [Google Scholar]
- 25. Comeau S. R., Gatchell D. W., Vajda S., Camacho C. J. (2004) ClusPro. An automated docking and discrimination method for the prediction of protein complexes. Bioinformatics 20, 45–50 [DOI] [PubMed] [Google Scholar]
- 26. Comeau S. R., Gatchell D. W., Vajda S., Camacho C. J. (2004) ClusPro. A fully automated algorithm for protein-protein docking. Nucleic Acids Res. 32, W96–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. DeLano W. L. (2002) The PyMOL Molecular Graphics System, DeLano Scientific, San Carlos, CA [Google Scholar]
- 28. Xu Y., Xing Y., Chen Y., Chao Y., Lin Z., Fan E., Yu J. W., Strack S., Jeffrey P. D., Shi Y. (2006) Structure of the protein phosphatase 2A holoenzyme. Cell 127, 1239–1251 [DOI] [PubMed] [Google Scholar]
- 29. Cho U. S., Xu W. (2007) Crystal structure of a protein phosphatase 2A heterotrimeric holoenzyme. Nature 445, 53–57 [DOI] [PubMed] [Google Scholar]
- 30. Prickett T. D., Brautigan D. L. (2004) Overlapping binding sites in protein phosphatase 2A for association with regulatory A and α-4 (mTap42) subunits. J. Biol. Chem. 279, 38912–38920 [DOI] [PubMed] [Google Scholar]
- 31. Kloeker S., Reed R., McConnell J. L., Chang D., Tran K., Westphal R. S., Law B. K., Colbran R. J., Kamoun M., Campbell K. S., Wadzinski B. E. (2003) Parallel purification of three catalytic subunits of the protein serine/threonine phosphatase 2A family (PP2A(C), PP4(C), and PP6(C)) and analysis of the interaction of PP2A(C) with α4 protein. Protein Expr. Purif. 31, 19–33 [DOI] [PubMed] [Google Scholar]
- 32. Kickstein E., Krauss S., Thornhill P., Rutschow D., Zeller R., Sharkey J., Williamson R., Fuchs M., Köhler A., Glossmann H., Schneider R., Sutherland C., Schweiger S. (2010) Biguanide metformin acts on tau phosphorylation via mTOR/protein phosphatase 2A (PP2A) signaling. Proc. Natl. Acad. Sci. U.S.A. 107, 21830–21835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lizotte D. L., Blakeslee J. J., Siryaporn A., Heath J. T., DeLong A. (2008) A PP2A active site mutant impedes growth and causes misregulation of native catalytic subunit expression. J. Cell Biochem. 103, 1309–1325 [DOI] [PubMed] [Google Scholar]