Background: Cardiolipin is required for maintaining optimal mitochondrial function.
Results: Cardiolipin depletion selectively obstructed proliferation, mitochondrial function, and morphology in germ cells.
Conclusion: The contribution of cardiolipin to mitochondrial function and morphology varies among the different cell types in vivo.
Significance: This provides a biological basis for understanding the different sensitivities of organelles to changes in the lipid environment.
Keywords: C. elegans, Cardiolipin, Cell Division, Mitochondria, Phospholipid, Cardiolipin Synthase, Germ Cell
Abstract
Cardiolipin (CL) is a major membrane phospholipid specifically localized in mitochondria. At the cellular level, CL has been shown to have a role in mitochondrial energy production, mitochondrial membrane dynamics, and the triggering of apoptosis. However, the in vivo role of CL in multicellular organisms is largely unknown. In this study, by analyzing deletion mutants of a CL synthase gene (crls-1) in Caenorhabditis elegans, we demonstrated that CL depletion selectively caused abnormal mitochondrial function and morphology in germ cells but not in somatic cell types such as muscle cells. crls-1 mutants reached adulthood but were sterile with reduced germ cell proliferation and impaired oogenesis. In the gonad of crls-1 mutants, mitochondrial membrane potential was significantly decreased, and the structure of the mitochondrial cristae was disrupted. Contrary to the abnormalities in the gonad, somatic tissues in crls-1 mutants appeared normal with respect to cell proliferation, mitochondrial function, and mitochondrial morphology. Increased susceptibility to CL depletion in germ cells was also observed in mutants of phosphatidylglycerophosphate synthase, an enzyme responsible for producing phosphatidylglycerol, a precursor phospholipid of CL. We propose that the contribution of CL to mitochondrial function and morphology is different among the cell types in C. elegans.
Introduction
Cardiolipin (CL)2 is an anionic phospholipid with a unique diphosphatidylglycerol structure containing four acyl groups (1–3). In eukaryotic cells, CL localizes predominantly to the mitochondrial inner membrane, where it serves specific roles in mitochondrial structure and function. CL biosynthesis occurs via three enzymatic reactions, all catalyzed by enzymes present in the mitochondria (Fig. 1A) (4). The first step, catalyzed by phosphatidylglycerophosphate (PGP) synthase, is the synthesis of PGP from cytidine diphosphate-diacylglycerol (CDP-DAG) and glycerol 3-phosphate. PGP phosphatase dephosphorylates PGP to phosphatidylglycerol (PG). In the final step, CL synthase catalyzes the formation of CL from PG and CDP-DAG. These enzymes are highly conserved in eukaryotes (Figs. 1A, 2B, and 9B) (5).
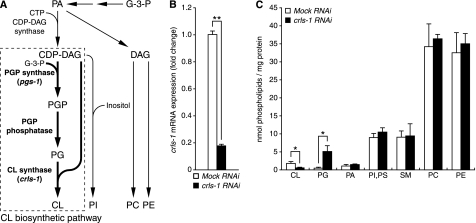
FIGURE 1.
CRLS-1 is the functional homologue of CL synthase in C. elegans. A, schematic summary of phospholipid biosynthesis. Phospholipid biosynthesis branches into two pathways utilizing phosphatidic acid (PA) as an intermediate molecule. One branch of the pathway converts phosphatidic acid to DAG, which eventually produces phosphatidylcholine (PC) and phosphatidylethanolamine (PE). The other branch of the pathway converts phosphatidic acid to CDP-DAG, which is catalyzed by CDP-DAG synthase. CL biosynthetic pathway is boxed with dashed lines. Gene names in parentheses represent putative homologues in C. elegans. In this study, we used crls-1 gene deletion mutants to reduce the content of CL, and pgs-1 gene deletion mutants to reduce the content of both CL and PG. B and C, CL content is decreased in crls-1(RNAi) worms. Alterations and the expression of crls-1 mRNA (B) and content of phospholipids (C) were examined in worms subjected to crls-1 or mock RNAi. The expression of the crls-1 gene was normalized to that of the eft-3 gene and is represented as a fold change over the mock RNAi control. G-3-P, glycerol 3-phosphate; PI, phosphatidylinositol; PS, phosphatidylserine; SM, sphingomyelin. The data represent the mean ± S.D. **, p < 0.01; *, p < 0.05.
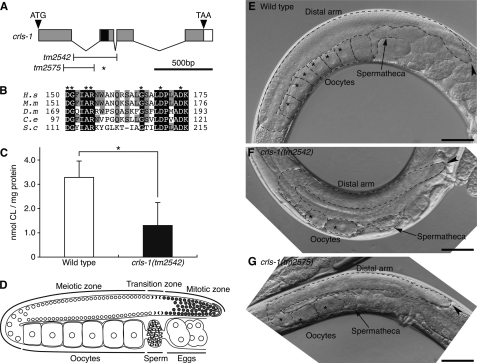
FIGURE 2.
Phenotypes of crls-1 mutants. A, genomic structure of crls-1. Gray and white boxes indicate exons and 3′-untranslated regions, respectively. The positions of the ATG start codon and TAA stop codon are shown. The black box indicates the DGXXARXXXXXXXXGXXXDXXXD sequence. T-bars below indicate the extent of deletion in crls-1(tm2542) and crls-1(tm2575). The deletion in crls-1(tm2575) causes a frameshift resulting in a premature stop codon (asterisk). B, multiple sequence alignment of the catalytic active motif of C. elegans CRLS-1 and homologous sequences in eukaryotes organized by ClustalW alignment. Residues identical in all five sequences are shaded in black, and residues identical in three or four proteins are shaded in gray. The accession numbers for the sequences used are as follows: (H.s) Homo sapiens, NP_061968; (M.s) Mus musculus, NP_001019556; (D.m) D. melanogaster, NP_733116; (C.e) C. elegans, NP_001022546; (S.c) Saccharomyces cerevisiae, NP_010139. C, content of CL was examined in wild type and crls-1(tm2542). The data represent the mean ± S.D. *, p < 0.05. D, schematic diagram of an adult hermaphrodite gonad. E–G, Nomarski images of the adult hermaphrodite gonads of wild type (E), crls-1(tm2542) (F), and crls-1(tm2575) (G) are shown. The gonads are outlined with the dashed lines. Asterisks indicate the germ nuclei at the diakinesis stage during oogenesis. Arrowheads indicate the distal end of each gonad. Scale bars, 50 mm.
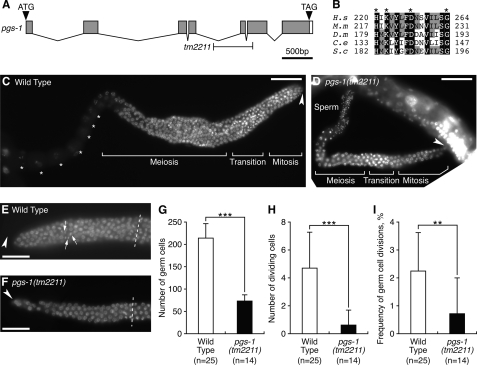
FIGURE 9.
pgs-1 is required for the mitotic proliferation of germ cells. A, genomic structure of pgs-1. Gray and white boxes indicate exons and 3′-untranslated regions, respectively. The positions of the ATG start codon and TGA stop codon are shown. T-bars below indicate the extent of deletion in pgs-1(tm2211). B, multiple sequence alignment of the catalytic active motif of C. elegans PGS-1 and homologous sequences in eukaryotes organized by ClustalW alignment. Residues identical in all five sequences are shaded in black, and residues identical in three or four proteins are shaded in gray. Asterisks indicate the HXKXXXXDXXXXXXG motif that is highly conserved in several hydrolases and phosphodiesterases. The accession numbers for the sequences used are as follows: (H.s) H. sapiens, NP_077733; (M.s) M. musculus, NP_598518; (D.m) D. melanogaster, NP_650751; (C.e) C. elegans, NP_490666; (S.c) S. cerevisiae, NP_009923. C and D, DAPI-stained images of adult hermaphrodite gonads of wild type (C) and pgs-1(tm2211) (D) are shown. The fluorescence signal from the sperm nuclei overlaps other portions of the specimen in C. Asterisks indicate the germ nuclei at the diakinesis stage during oogenesis. Arrowheads indicate the distal end of each gonad. Scale bars, 50 mm. E and F, DAPI-stained images of the mitotic zone in the adult hermaphrodite gonads of wild type (E) and pgs-1(tm2211) (F) are shown. Arrows indicate nuclei in M phase. Arrowheads indicate the distal end of each gonad. Dashed lines indicate the boundary between the mitotic and transition zones. Scale bars, 10 mm. G–I, quantitative analysis of germ cell mitosis in wild type and pgs-1(tm2211). The data represent the mean ± S.D. ***, p < 0.001; **, p < 0.01.
So far, in vitro analysis has revealed that CL interacts with a number of mitochondrial proteins and is required for the optimal activity of several enzymes in the mitochondrial electron transport chain, including NADH dehydrogenase (complex I) (6), ubiquinol:cytochrome c reductase (complex III) (7–10), cytochrome c oxidase (complex IV) (11, 12), and ATP synthase (complex V) (13). Consistent with these observations, yeast CL synthase mutants (crd1 mutants), in which CL is not detected, show instability of electron transport chain supercomplexes, perturbation of coupling, and reduced mitochondrial membrane potential, leading to a decrease in the oxidative phosphorylation rate (14–19). crd1 mutants are viable under normal conditions; however, they show growth defects and mitochondrial DNA instability at high temperatures or under conditions of osmotic stress (14, 16). In contrast, knockdown of CL synthase in HeLa cells does not cause any defects in mitochondrial membrane potential, oxidative phosphorylation rate, or cell division under normal conditions (20, 21), although it is possible that residual CL may be sufficient to maintain normal mitochondrial function. In addition to the contribution of CL to mitochondrial oxidative phosphorylation, CL has also been shown to be involved in induction of apoptosis by interacting with pro-apoptotic proteins such as cytochrome c and tBid (22–24). As described above, the CL function has been analyzed at the cellular level using yeast and mammalian cells. However, the in vivo role of CL in multicellular organisms is largely unknown.
Caenorhabditis elegans is a suitable model organism to investigate the effects of gene depletion on individual cells or tissues because it is anatomically simple and has invariant lineage. In this study, we analyzed C. elegans mutants lacking the CL synthase gene (crls-1) and found that CL is required for germ cell proliferation. The number of germ cells was reduced in crls-1 mutants due to a reduction in the mitotic proliferation of germ cells. In the germ cells of crls-1 mutants, the mitochondrial membrane potential was significantly decreased and the structure of the mitochondrial cristae was disrupted. However, somatic cells in crls-1 mutants appeared normal with respect to cell proliferation, mitochondrial function, and mitochondrial morphology. These findings were confirmed by analysis of C. elegans mutants lacking a PGP synthase. This is the first study to show varying susceptibilities among different cell types to CL depletion in vivo.
EXPERIMENTAL PROCEDURES
General Methods and Strains
Maintenance and genetic manipulation of C. elegans were carried out as described previously (25). The Bristol strain N2 was used as the standard wild type strain. In addition, the following mutations, rearrangement, and transgenes were used: pgs-1(tm2211)I, bli-3(e767)I, rrf-3(pk1426)II, crls-1(tm2542)III, crls-1(tm2575)III, qC1 dpy-19(e1259) glp-1(q339)[qIs26]III, and wIs51[scm::gfp]. All strains were maintained at 20 °C. The deletion mutant strains crls-1(tm2542), crls-1(tm2575), and pgs-1(tm2211) were isolated as described previously (26). Each mutant was backcrossed seven times and was balanced with qC1 dpy-19(e1259) glp-1(q339)[qIs26] or bli-3(e767) for crls-1(tm2542) or pgs-1(tm2211), respectively.
Fluorescence and Nomarski Microscopy
Fluorescent images were obtained with an Axiovert 200 microscope (Carl Zeiss MicroImaging, Tokyo, Japan) equipped with a cooled AxioCam MRm CCD camera (Carl Zeiss MicroImaging) if not otherwise specified. Nomarski images were obtained with an IX71 microscope (Olympus, Tokyo, Japan) equipped with a cooled DS-5Mc CCD camera (Nikon, Tokyo, Japan).
Fixation and DAPI Staining
For gonad staining, young adult stage hermaphrodites were decapitated just below the pharyngeal bulb, allowing extrusion and extension of the gonad arms. The gonad was fixed with gonad fixative solution (3% formaldehyde, 75% methanol, and 6.2 mm K2HPO4 (pH 7.2)) for 10 min at room temperature. After washing twice with phosphate-buffered saline/Tween 20 (PBS-T, containing 0.1% Tween 20), the gonad was stained with 0.1 μg/ml DAPI in PBS-T for 10 min at room temperature. After washing twice with PBS-T, the gonad was mounted on a slide for observation.
Transgenic Rescue Analysis
DNA injection into the C. elegans germ line was carried out as described previously (59). A full-length cDNA of crls-1 was amplified by PCR with the following primers: 5′-GGA AGT GGT ACC ATG ATA GTA ACA TCG ATG TT-3′ and 5′-GCG AAA ACG AGC TCT AAT TAA ATT TTT TTG ATG G-3′ (the underlined sequences are KpnI and SacI sites, respectively). The amplified fragments were cloned into the pPD49.78 and pPD49.83 heat-shock vectors (a gift from A. Fire) at the KpnI/SacI sites for pST1 and pST2, respectively. Two fragments containing a point mutation to change Asp-116 to Ala were amplified by PCR with the following primers: 5′-GGA AGT GGT ACC ATG ATA GTA ACA TCG ATG TT-3′ and 5′-GAG AAG TTT ATC AGC GAC AGG AGC AAG CAC G-3′; 5′-AAA TCA CTG CTT GGC TCC GTG CTT GCT CCT-3′ and 5′-GCG AAA ACG AGC TCT AAT TAA ATT TTT TTG ATG G-3′ (italicized nucleotides represent the Asp-to-Ala mutation). The two overlapping fragments were fused by PCR with following primers: 5′-GGA AGT GGT ACC ATG ATA GTA ACA TCG ATG TT-3′ and 5′-GCG AAA ACG AGC TCT AAT TAA ATT TTT TTG ATG G-3′. The amplified fragments, forming a full-length crls-1D116A cDNA, were cloned into pPD49.78 and pPD49.83 at the KpnI/SacI sites for pST3 and pST4, respectively. The array Ex[hsp::crls-1] contained the plasmids pST1 (hsp16-41::crls-1), pST2 (hsp16-2::crls-1), and pRF4[rol-6(su1006)]. The array Ex[hsp::crls-1D116A] contained the plasmids pST3 (hsp16-41::crls-1D116A), pST4 (hsp16-2::crls-1D116A), and pRF4. For the experiments of transgenic rescue of crls-1 mutants, L1-staged hermaphrodites crls-1(tm2542), crls-1(tm2542);Ex[hsp::crls-1], and crls-1(tm2542); Ex[hsp::crls-1D116A] were subjected to heat shock at 37 °C for 45 min. The heat-shocked hermaphrodites were transferred to new nematode growth medium plates individually and were incubated at 20 °C until they finished egg laying.
Analysis of Germ Cell Proliferation
The frequency of germ cell division was scored in the DAPI-stained extruded gonad (27). The boundary between the mitotic and transition zones was determined by the presence of the characteristic crescent-shaped germ nuclei, and the number of germ cells in the mitotic zone was counted. To calculate the frequency of germ cell divisions, the number of germ cells in the mitotic phase showing condensed chromatin or dividing nuclei was counted and then divided by the total number of germ cells in the mitotic zone.
Visualization of Seam Cell Nuclei
Seam cell nuclei were visualized using the wIs51 transgene. wIs51[scm::gfp] expresses green fluorescent protein under a control of seam cell-specific promoters. Heterozygous crls-1(+/tm2542) or pgs-1(+/tm2211) worms, which carry a background transgenic insertion of wIs51, were generated. Gravid hermaphrodites were placed on nematode growth medium plates and incubated for 2 h to lay eggs. After removal of the parent hermaphrodites, the F1 progeny were incubated for 48 h. Young adult F1 hermaphrodites were mounted on a 4% agar pad and immobilized in 50 mm sodium azide. The number of seam cells on one lateral side was counted.
Determination of Mitochondrial Membrane Potential by DiSC3
L4-staged hermaphrodites were placed on nematode growth medium plates containing 0.075, 0.15, or 0.3 μm of DiSC3 (Sigma) and incubated for 24 h at 20 °C in the dark. Gonad arms were extruded and extended on a slide by decapitation just below the pharyngeal bulb. To avoid rupture of the extruded gonad, the samples were sealed with a piece of PCR-slide seal. The intensity of DiSC3 fluorescence was monitored on a BZ-8000 fluorescence microscope (Keyence, Tokyo, Japan) using a fixed exposure time and was quantified using ImageJ (National Institutes of Health, Bethesda). To determine the mean fluorescence values, the sum of the fluorescence values of the area corresponding to gonad or muscles was divided by the number of pixels.
Electron Microscopy
Transmission electron microscopy was performed by the Hanaichi Ultrastructure Research Institute Co. (Okazaki, Japan). Young adult hermaphrodites were fixed with 4% paraformaldehyde and 1% glutaraldehyde in 100 mm PBS for 1 h at 20 °C, followed by further fixation with 2% paraformaldehyde and 2% glutaraldehyde in 100 mm PBS for more than 24 h at 4 °C. The samples were post-fixed with 2% osmium tetroxide in 100 mm phosphate buffer for 4 h at 4 °C, followed by dehydration in an ethanol series, substitution with propylene oxide, and infiltration with Q651 resin (Mitsui Chemicals, Tokyo, Japan). Transverse sections were post-stained with uranyl acetate and lead citrate. The specimens were observed with a JEM-200EX electron microscope (JEOL, Tokyo, Japan).
Rhythmic Behavioral Analysis
Young adult hermaphrodites were placed in drops of PBS, and simple rhythmic thrashing swimming was measured. Swimming was scored as the number of seconds taken for 30 thrashes and calculated backward to the swimming rate. Each worm was scored twice.
RNAi
RNAi against the crls-1 gene was performed by the feeding of double-stranded RNA as described previously (28). Escherichia coli strain HT115 (DE3) was transformed with the pPD129.36 L4440 DoubleT-7scriptII vector (a gift from A. Fire, Stanford University School of Medicine, Stanford, CA) containing the full-length cDNA region of crls-1. An empty pPD129.36 plasmid was used for a mock RNAi control. Single colonies of transformed E. coli were cultured in LB medium containing 100 μg/ml ampicillin overnight at 37 °C. The pre-cultured E. coli was diluted 1:100 and incubated for 6 h at 37 °C. After that, E. coli was seeded on nematode growth medium plates containing 1 mm isopropyl β-d-thiogalactopyranoside and further incubated overnight at room temperature to induce the expression of double-stranded RNA. Late L4-staged hermaphrodites of rrf-3 mutants were placed on the feeding RNAi plates and incubated overnight at 20 °C to lay eggs (29). The F1 progeny that were laid during this period were incubated for 48 h at 25 °C. Young adult F1 hermaphrodites were collected and used for biochemical analyses.
Total RNA Isolation and Quantitative Real Time PCR
Total RNA was extracted from young adult worms with Isogen (Nippongene, Toyama, Japan). cDNA was synthesized from the RNA using Moloney murine leukemia virus reverse transcriptase (Invitrogen). Quantitative real time PCR was performed using a CFX96 real time detection system with SsoFast EvaGreen Supermix (Bio-Rad). The sequences of the oligonucleotide primers used were as follows: 5′-CTC GGC TGT ATC ACA GGA TTC ACC A-3′ and 5′-ACG AGG AGG ATC ACA GTC TAG CGA-3′ for crls-1; 5′-GGC CGT CCC AGG AGA CAA CG-3′ and 5′-GAC CTG GGC GTG GAA GGT GC-3′ for eft-3. eft-3 was amplified as an internal control, and crls-1 gene expression was normalized against eft-3 expression.
Lipid Analysis
Approximately 10,000 young adult worms were collected, and the total phospholipids were extracted using the method of Bligh and Dyer (30). Phospholipids were separated by one-dimensional thin layer chromatography on Silica Gel 60 TLC plates (Merck) in chloroform/methanol/acetic acid (65:25:13, v/v/v). Phospholipids were identified by comigration with known standards. The position of each spot was visualized by staining with 0.001% primuline in acetone/H2O (4:1, v/v) under ultraviolet light. The area of silica gel corresponding to each phospholipid was scraped off and subjected to phosphorus quantification (31).
RESULTS
Identification of C. elegans CL Synthase crls-1
CL synthase catalyzes the formation of CL using PG and CDP-DAG as substrates (Fig. 1A) (1). A data base search of the complete C. elegans genome sequence revealed the presence of a single CL synthase homologue, F23H11.9, which we named crls-1 (cardiolipin synthase homologue-1). The crls-1 gene product, CRLS-1, consists of 246 amino acid residues and shows 45% identity with the human CL synthase. CRLS-1 has a CDP-alcohol phosphatidyltransferase motif that is highly conserved in the CRLS family proteins (32). In particular, the “DGXXARXXXXXXXXGXXXDXXXD” sequence, which is essential for the enzyme activity of CL synthase, is completely conserved within the phosphatidyltransferase motif of C. elegans CRLS-1 (Fig. 2B). To confirm that CRLS-1 contributes to CL synthesis in C. elegans, we determined the CL content in worms subjected to crls-1 RNAi (Fig. 1, B and C). In crls-1 RNAi worms, the expression of crls-1 mRNA and the CL content were decreased to 18 and 39%, respectively, of those in mock RNAi worms (CL content: 1.94 nmol/mg protein in mock RNAi worms versus 0.76 nmol/mg protein in crls-1 RNAi worms). Concurrently, the content of PG, which is the precursor of CL, increased 8-fold in crls-1 RNAi worms compared with mock RNAi worms (0.64 nmol/mg protein in mock RNAi worms versus 5.26 nmol/mg protein in crls-1 RNAi worms). However, the amount of other phospholipids was not substantially affected in crls-1 RNAi worms. These data indicate that CRLS-1 is a functional homologue of CL synthase in C. elegans.
crls-1 Is Required for Germ Cell Proliferation
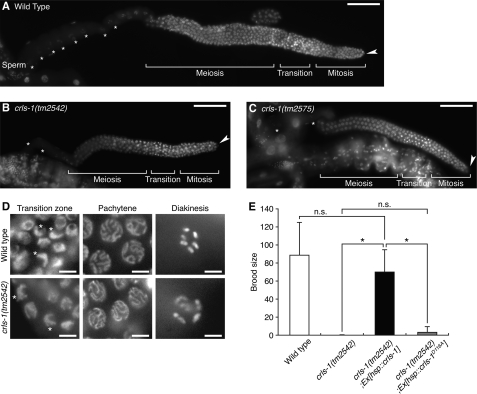
To investigate the physiological role of CL in vivo, we examined the phenotypes of two different deletion alleles of crls-1; tm2542 and tm2575 (Fig. 2A). Both alleles lack the DGXXARXXXXXXXXGXXXDXXXD sequence that is essential for the enzyme activity of CL synthase, suggesting that the products are functionless (Fig. 2A) (33). Consistent with the crls-1 RNAi worms, the CL content was significantly decreased in crls-1(tm2542) (Fig. 2C). Both the crls-1(tm2542) and crls-1(tm2575) mutants were viable and reached adulthood; however, they showed extremely reduced brood size compared with the wild type (Table 1). In C. elegans, the gonadal arm of an adult hermaphrodite is U-shaped, with a distal arm containing a syncytium of germ line nuclei and a proximal arm containing oocytes and fertilized eggs (Fig. 2, D and E). Germ cells at the distal end of the gonad proliferate by mitosis, and as they migrate proximally, they initiate meiosis at the transition zone (34). In crls-1 mutants, the distal gonadal arm was reduced in size, and the number of oocytes was decreased (Fig. 2, F and G). DAPI staining revealed that the number of germ cells in the distal arm was reduced in crls-1 mutants (Fig. 3, A–C). The reduced germ cell number was not due to enhanced apoptosis of germ cells because a mutation in ced-3, an essential gene for apoptotic cell death, did not restore normal germ cell number in crls-1 mutants (data not shown). Although the number of germ cells was decreased, meiosis appeared to occur normally in crls-1 mutants, i.e. crls-1 mutant gonads contained crescent-shaped nuclei (leptotene/zygotene stage) in the transition zone, thread-like chromosomes (pachytene stage) in the meiotic zone, and highly condensed chromosomes (diakinesis stage) in the oocytes in a manner similar to those in wild type gonads (Fig. 3D). The reduced brood size of crls-1 mutants was rescued by expression of crls-1 under the control of a heat shock promoter, whereas expression of a catalytically inactive mutant crls-1 (crls-1D116A) failed to rescue the defect in crls-1 mutants (Fig. 3E) (20, 32). Thus, crls-1 is required for reproductive function through its enzymatic activity.
TABLE 1.
Viability and fertility of crls-1 mutants
Adult hermaphrodites of the crls-1 heterozygote were allowed to lay eggs for overnight at 20 °C, and the F1 progeny were scored for embryonic lethality and brood size. The brood sizes were determined by placing individual worms on seeded plates and allowing self-fertilization at the indicated temperature. The P0 worms were then transferred to a fresh plate at 24-h intervals for the next 4 days. The total F1 progenies on the plates were counted. Genotype of the animals were examined by genomic PCR. The data represent the mean ± S.D.
| Genotype | Embryonic lethal | n | Brood size | n |
|---|---|---|---|---|
| % | ||||
| Wild type | 0 | >500 | 320.94 ± 39.39 | 63 |
| crls-1(tm2542) | 0 | 450 | 6.12 ± 2.71 | 24 |
| crls-1(tm2575) | 0 | 435 | 3.76 ± 2.43 | 21 |
FIGURE 3.
crls-1 is required for germ line development. A–C, DAPI-stained images of adult hermaphrodite gonads of wild type (A), crls-1(tm2542) (B), and crls-1(tm2575) (C) are shown. The fluorescence signal from the sperm nuclei overlaps other portions of the specimen in B and C. Asterisks indicate the germ nuclei at the diakinesis stage during oogenesis. Arrowheads indicate the distal end of each gonad. Scale bars, 50 mm. D, DAPI-stained images of germ cell nuclei in adult hermaphrodite of the transition zone, pachytene stage, and diakinesis stage are shown. Asterisks indicate the typical crescent-shaped nuclei in the transition zone of germ cells. Scale bars, 5 mm. E, transgenic worms expressing the CRLS-1 or CRLS-1D116A protein under the control of a heat shock promoter were produced as described under “Experimental Procedures.” The brood sizes of wild type, crls-1(tm2542), and crls-1(tm2542) expressing exogenous CRLS-1 or CRLS-1D116A were examined. Wild type and crls-1(tm2542) were derived from the same crls-1(+/tm2542) heterozygous mother. The data represent the mean ± S.D. *, p < 0.05; n.s., not significant.
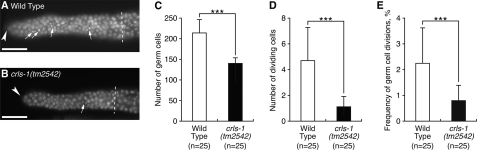
We further examined the mitotic capacity of germ cells by measuring the frequency of germ cell divisions, because division frequency often correlates with germ cell number (Fig. 4) (35, 36). In wild type gonads, more than 200 germ cells were present at the distal mitotic zone, and 2% of these germ cells had dividing nuclei as reported previously (Fig. 4, A and C–E) (27, 37). In contrast, the mitotic zone of the crls-1 mutant gonads contained fewer germ cells, and dividing germ cells were found at lower frequency (Fig. 4, B–E). These data indicate that crls-1 is required for the mitotic proliferation of germ cells in C. elegans.
FIGURE 4.
crls-1 is required for mitotic proliferation of germ cells. A and B, DAPI-stained images of the mitotic zone in the adult hermaphrodite gonads of wild type (A) and crls-1(tm2542) (B) are shown. Arrows indicate nuclei in mitotic phase (M phase), showing condensed chromatin or dividing nuclei. Arrowheads indicate the distal end of each gonad. Dashed lines indicate the boundary between the mitotic and transition zones. Scale bars, 10 mm. C–E, quantitative analysis of germ cell mitosis in wild type and crls-1(tm2542). The data represent the mean ± S.D. ***, p < 0.001.
Somatic Cell Proliferation Occurs Normally in crls-1 Mutants
Next, we examined somatic cell proliferation in crls-1 mutants. In C. elegans, 556 somatic cells are generated from one fertilized egg during embryogenesis (38). A time-sequential observation revealed that somatic cells, such as intestinal cells and epithelial cells, proliferated and differentiated normally through somatic cell divisions in crls-1 mutant embryos (data not shown). Consistent with this observation, crls-1 mutants did not show any embryonic lethality (Table 1).
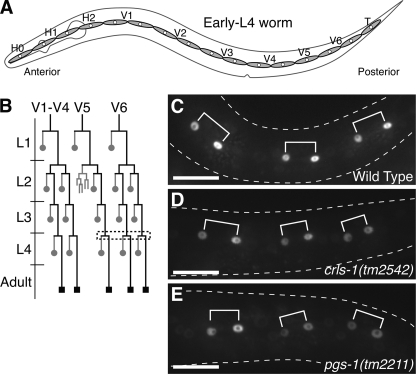
During the post-embryonic development of C. elegans, certain types of somatic cells, such as vulval cells and seam cells, proliferate to form somatic organs (the egg-laying apparatus and lateral epithelia, respectively) (38). During vulval development, three vulval precursor cells divide to produce 22 somatic vulval cells, forming the vulva at the adult stage (Table 2; Fig. 5A). In crls-1 mutants, 22 vulval cells were presented in the normal configuration, and the morphology of the mature vulva was indistinguishable from that of the wild type, indicating that vulval cell divisions are not affected in crls-1 mutants (Fig. 5, B and C). In addition to the vulval cells, seam cell divisions also occurred normally in crls-1 mutants, i.e. all seam cells divided along the anterior-posterior axis to produce two daughter cells in a manner similar to that in the wild type (Fig. 6). Consistent with this, the seam cell number at the adult stage was the same as in the wild type (Table 2). Taken together, these data indicate that the somatic cell proliferation occurs normally throughout development in crls-1 mutants.
TABLE 2.
Average number of vulval cells and seam cells in crls-1 and pgs-1 mutants
| Genotype | No. of vulval cells | n | No. of seam cells | n |
|---|---|---|---|---|
| Wild type | 22 ± 0 | 16 | 16.03 ± 0.44 | 90 |
| crls-1(tm2542)a | 22 ± 0 | 8 | 16.09 ± 0.67 | 40 |
| pgs-1(tm2211)b | 22 ± 0 | 15 | 16.02 ± 0.12 | 51 |
a crls-1(tm2542) homozygous mutants were derived from crls-1(tm2542)/qC1 mother.
b pgs-1(tm2211) homozygous mutants were derived from pgs-1(tm2211)/bli-3(e767) mother. The data represent the mean ± S.D.
FIGURE 5.
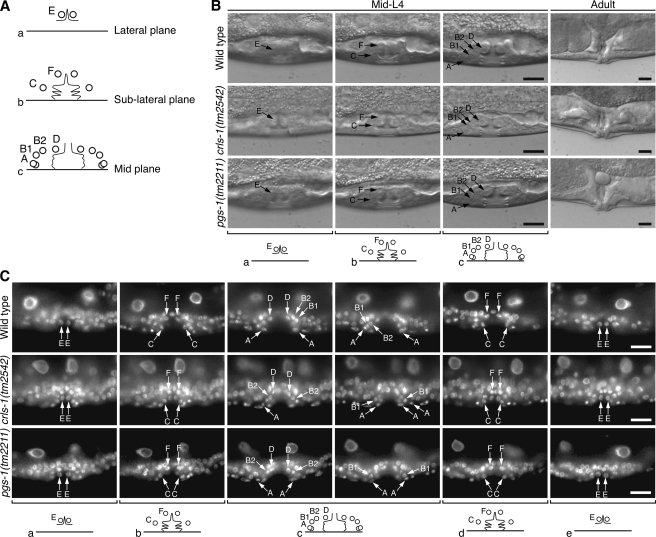
Vulval cell divisions are normal in crls-1 and pgs-1 mutants. A, schematic diagram of vulval cell positions in mid-L4 larva. Circles, thin lines, and thick horizontal lines indicate nuclei, cell boundaries, and the ventral cuticle, respectively. Anterior is left and dorsal is top. The letters A, B1, and B2, for example, correspond to the names of the vulval cells vulA, vulB1, and vulB2. B and C, positions of the vulval cells and the morphology of the mature vulva were observed by Nomarski microscopy (B) and DAPI staining (C). The top, middle, and bottom panels show wild type, crls-1(tm2542), and pgs-1(tm2211), respectively. Schematics corresponding to each focal plane are shown below the images. Arrows indicate the nuclei of each vulval cell lineage. Scale bars, 10 mm.
FIGURE 6.
Seam cell divisions are not affected in crls-1 and pgs-1 mutants. A, schematic diagram of seam cells on each side in an early L4 larva. B, post-embryonic division pattern of V seam cells. The expression pattern of scm::gfp, which is specifically expressed in the nuclei of seam cells, is indicated by black lines. The dashed box marks the lineages observed in C–E. Gray circles represent anterior daughters that fuse with the epithelial syncytium, hyp7, and black squares denote seam cells. C–E, fluorescence images of scm::gfp just after the divisions at the L4 stage in wild type (C), crls-1(tm2542) (D), and pgs-1(tm2211) (E) are shown. The shapes of the worms are outlined with dashed lines. Three pairs of daughter cells are shown with brackets. Scale bars, 20 mm.
crls-1 Is Required for the Maintenance of Mitochondrial Function and Morphology in Germ Cells but Not in Muscle Cells
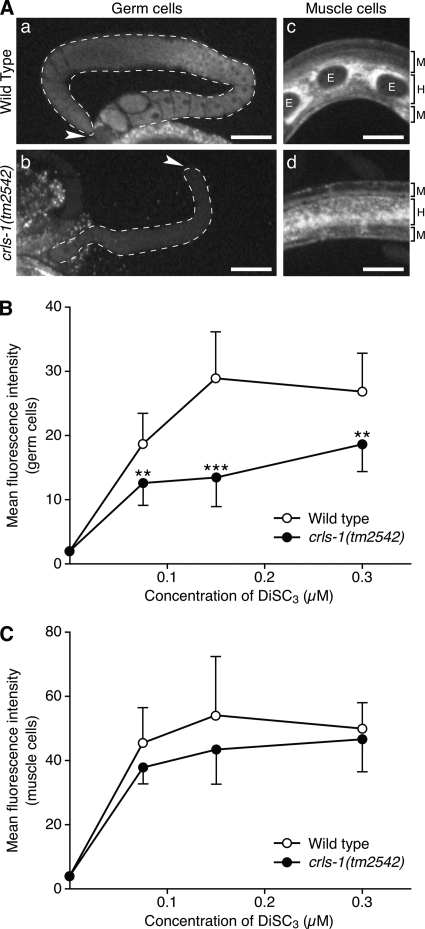
Previous studies using yeast CL synthase mutants showed that a lack of CL causes a reduction in mitochondrial membrane potential (14–16, 18, 19, 39). Thus, we first stained crls-1 mutants with DiSC3, a membrane potentially sensitive mitochondrial dye (40), to evaluate the function of the mitochondria. The level of DiSC3 fluorescence intensity indicated a significant reduction in mitochondrial membrane potential in crls-1 germ cells (Fig. 7, A and B). However, DiSC3 staining was not significantly affected in crls-1 muscle cells (Fig. 7, A and C). Consistent with this observation, muscle function appeared normal in crls-1 mutants as judged by the thrashing body movement in water (123.17 cycles/min in wild type versus 113.09 cycles/min in crls-1 mutants).
FIGURE 7.
Mitochondrial membrane potential is decreased in crls-1 germ cells. A, representative fluorescence images of germ cells (panels a and b) and muscle cells (panels c and d) in the wild type and crls-1(tm2542) stained with 0.15 μm DiSC3 are shown. The gonads are outlined with the dashed lines. Arrowheads indicate the distal end of each gonad. Brackets on the right-hand side of the images (panels c and d) indicate the layers of muscle cells (M) and hypodermis (H). E, embryos. Scale bars, 50 mm. B and C, quantification of the fluorescence intensity of DiSC3 in germ cells (B) and muscle cells (C) of wild type and crls-1(tm2542). The data represent the mean ± S.D. ***, p < 0.001; **, p < 0.01.
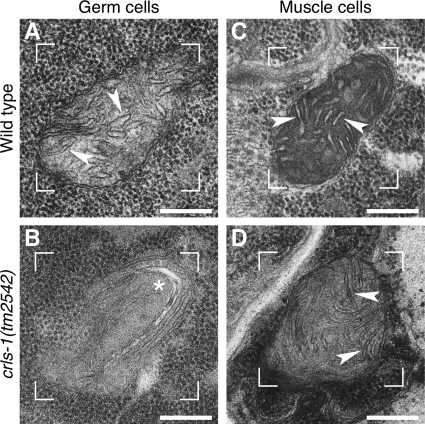
We next examined the ultrastructural morphology of mitochondria in crls-1 mutants by transmission electron microscopy (Fig. 8). In wild type germ cells, the mitochondria displayed many typical cristae that randomly crossed the mitochondrial matrix (arrowheads in Fig. 8A). However, in crls-1 germ cells, cristae were not observed in the mitochondrial matrix, and elongated inner membranes were located along the edge of the mitochondria (asterisk in Fig. 8B). In contrast to the germ cells, mitochondrial morphology appeared normal in muscle cells in crls-1 mutants (arrowheads in Fig. 8, C and D). These data indicate that C. elegans crls-1 is required for the maintenance of mitochondrial function and morphology in germ cells but not in muscle cells.
FIGURE 8.
Mitochondrial cristae structures are disrupted in crls-1 germ cells. Transmission electron micrographs of germ cells (A and B) and muscle cells (C and D) in wild type and crls-1(tm2542) are shown. In the mitochondrial matrix of crls-1 germ cells, elongated inner membranes were located along the edge of the mitochondria, instead of the obvious cristae structures (asterisk in B). Representative mitochondria are enclosed by brackets. Arrowheads indicate the cristae structures. Scale bars, 200 nm.
Accumulation of PG Content Is Not Responsible for the Germ Cells Defects in crls-1 Mutants
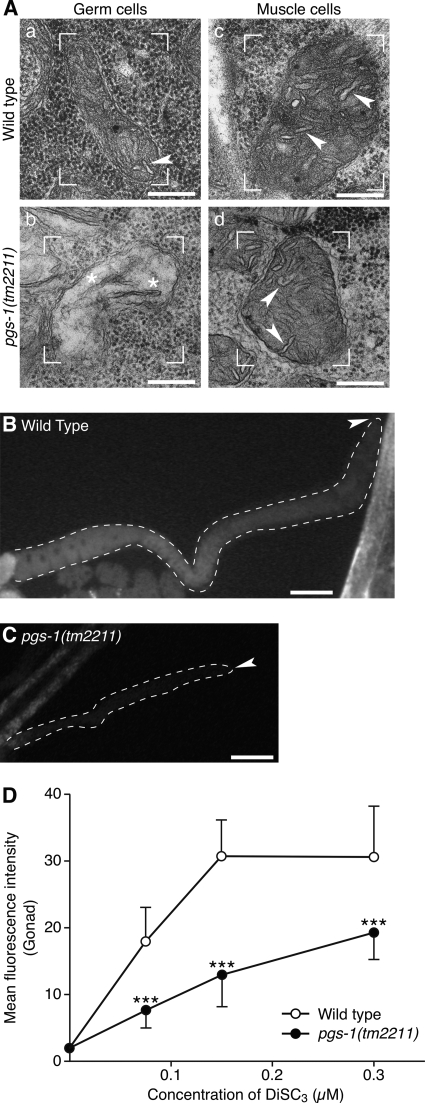
To rule out the possibility that accumulation of PG causes the germ cell defects in crls-1 mutants, we examined the phenotypes of pgs-1(tm2211), a deletion mutant lacking the phosphatidylglycerophosphate synthase gene (Fig. 9A). PGP synthase catalyzes the formation of PGP, a precursor of PG and CL, using CDP-DAG and glycerol 3-phosphate as substrates (Figs. 1A and 9B) (41, 42). Thus, lack of PGP synthase results in depletion of both PG and CL. In pgs-1 mutants, germ cell divisions occurred infrequently, and the number of germ cells was decreased (Fig. 9, C–I) in a manner similar to crls-1 mutants. In addition, deficiency of pgs-1 caused the appearance of several elongated cristae structures in the mitochondrial matrix (Fig. 10A, panels a and b), and reduced mitochondrial membrane potential in germ cells (Fig. 10, B–D). In contrast, pgs-1 mutants did not show obvious defects in somatic tissues, as judged by the divisions of vulval cells and seam cells (Table 2; Figs. 5 and 6), and mitochondrial morphology in muscle cells (Fig. 10A, panels c and d). These data indicate that loss of CL, but not accumulation of PG, causes the germ cell defects in crls-1 mutants.
FIGURE 10.
pgs-1 is required for the maintenance of mitochondrial morphology and function in germ cells. A, mitochondrial cristae structures are disrupted in pgs-1 germ cells. Transmission electron micrographs of germ cells (panels a and b) and muscle cells (panels c and d) in wild type and pgs-1(tm2211) are shown. In pgs-1 germ cells, the mitochondria contained several elongated cristae structures in the mitochondrial matrix (asterisk in panel c). Representative mitochondria are enclosed by brackets. Arrowheads indicate the cristae structures. Scale bars, 200 nm. B–D, mitochondrial membrane potential is decreased in pgs-1 germ cells. Representative fluorescence images of adult hermaphrodite germ cells in wild type (B) and pgs-1(tm2211) (C) stained with 0.15 μm DiSC3 are shown. The gonads are outlined with the dashed lines. Arrowheads indicate the distal end of each gonad. Scale bars, 50 mm. D, quantification of the fluorescence intensity of DiSC3 in germ cells of wild type and pgs-1(tm2211). The data represent the mean ± S.D. ***, p < 0.001.
DISCUSSION
Previous studies using yeast showed that inhibition of CL synthase results in the instability of electron transport chain supercomplexes, a reduction in mitochondrial membrane potential, and a decrease in the oxidative phosphorylation rate (14, 16, 18, 19, 39, 43). In this study, by analyzing deletion mutants of the CL synthase gene (crls-1) in the simple multicellular organism C. elegans, we demonstrated that CL depletion selectively affects mitochondrial function and morphology in germ cells (Figs. 7 and 8). Furthermore, we found that CL is required for mitotic proliferation of germ cells but not that of somatic cells (Table 2; Figs. 4–6).
It is interesting that germ cells are more susceptible to CL depletion than somatic cells. Previous studies have indicated the mitochondrial heterogeneity in cell types with respect to morphological diversity (44, 45), steroidogenic capacity (46), protein profiles (47, 48), and lipid profiles (49, 50). Based on the existence of such mitochondrial heterogeneity, we propose two models that could explain the hypersensitivity of the germ cells to CL depletion. One possibility is that in vivo, CL is selectively required for germ cell-specific mitochondrial proteins. Using the genome-wide microarray data base generated by Reinke et al. (51), we found several germ cell-enriched genes encoding putative mitochondrial proteins, such as asb-1, ucr-2.3, bath-43, and hmg-5 (Table 3). At this time, in vivo functions of most of these genes are largely unknown; however, asb-1, which encodes a germ cell-specific isoform of mitochondrial ATP synthase b subunit, has been reported to be required for germ line development in C. elegans (52). CL may be needed to maintain the function and/or stability of such a germ cell-specific mitochondrial protein. The other possibility is that physical properties of the CL-depleted membrane vary between germ cells and somatic cells because of the difference in the mitochondrial lipid composition between these cell types (49, 50). In somatic cells such as muscle cells, mitochondrial lipids may compensate for a CL-depleted membrane environment by creating suitable microdomains for mitochondrial proteins, and thus, mitochondrial function and morphology are maintained in somatic cells. Recently, Drosophila mutants of a CL synthase gene were reported to exhibit abnormal mitochondrial morphology and function in muscle cells (53). The difference in the mitochondrial phenotype of muscle cells between C. elegans and Drosophila melanogaster may also be caused by the difference in the mitochondrial lipid composition between these species. For example, D. melanogaster lacks long chain polyunsaturated fatty acids that confer membrane fluidity (54).
TABLE 3.
Germ cell-enriched genes encoding putative mitochondrial proteins
| Sequence name | Gene name | Predicted protein localizationa,b | Brief description | Ref. |
|---|---|---|---|---|
| F28F8.2 | acs-2 | Mito, Ex | Long chain fatty acid acyl-CoA ligase | 60 |
| T05F1.2 | Mito, Ex | Unnamed protein | ||
| T22F3.4 | rpl-11.1 | Mito, Ex | Large ribosomal subunit L11 protein | |
| R09E10.6 | Mito, Cyto | Unnamed protein | ||
| F26H9.4 | Mito, Cyto | Nicotinamide mononucleotide adenylyltransferase | ||
| W01A11.7 | Mito, Nuc | Unnamed protein | ||
| F10D11.1 | sod-2 | Mito, Cyto | Manganese superoxide dismutase | 61 |
| T24D1.3 | Mito, Nuc | Unnamed protein | ||
| K08F4.11 | gst-3 | Mito, Cyto, Nuc | Glutathione S-transferase | |
| F35G12.10 | asb-1 | Mito, Pero, Ex | Mitochondrial F1F0-ATP synthase b subunit | 52 |
| T24C4.1 | ucr-2.3 | Mito, Nuc, Ex | Ubiquinol cytochrome c reductase complex core protein 2 | |
| T16H12.5 | bath-43 | Mito, Cyto, Nuc | Speckle-type POZ protein SPOP and related proteins | |
| F45E4.9 | hmg-5 | Mito, Cyto, Nuc | HMG box-containing protein | 62 |
| C49C3.7 | Mito, Cyto, Nuc | Unnamed protein | ||
| T26A5.2 | Mito, Nuc, Cyto, Ex | Unnamed protein | ||
| K04C2.3 | Mito, Cyto, Nuc, Ex | Unnamed protein |
a Protein localization was examined by WoLF PSORT, a computer program for the prediction of subcellular localization sites of proteins based on their amino acid sequences.
b The abbreviations used are as follows: Mito, mitochondria; Nuc, nucleus; Cyto, cytosol; Ex, extracellular.
In eukaryotes, cell division is regulated by cyclin and cyclin-dependent kinase. Inhibition of these general cell cycle regulators, such as C. elegans cyclin E (cye-1) and cyclin-dependent kinase-1 (cdk-1), reduces cell division of both germ cells and somatic cells, including the vulval cells and seam cells (55, 56), and thus, it causes lethality at a high frequency. In contrast, crls-1 mutants grow to adulthood and exhibit defects in cell division specifically in germ cells (Table 1; Fig. 4). We found no defects in the cell division of somatic cells (Table 2; Figs. 5 and 6). These observations indicate that CL is not involved in the general cell division machinery. In C. elegans, germ cells are significantly more proliferative than somatic cells (34), which make the germ line a highly energy-consuming organ. It would appear that in crls-1 mutants, the decrease in mitochondrial membrane potential in germ cells causes reduced energy production, which leads to inhibition of germ cell division. In the germ line of C. elegans hermaphrodite, sperm are produced during the late larval stage, and thereafter, oocytes are produced at the adult stage. Because the sperm and oocytes are produced from a common pool of progenitor germ cells, we assume that the reduced germ cell division of crls-1 mutants affects not only oocyte production but also sperm production. This would lead to the striking reduction of brood size in crls-1 mutants, the reduction occurring to a greater extent than would be expected from the reduction in germ cell division (Table 1; Fig. 4). To support this idea, crosses of crls-1 mutant males to wild type hermaphrodites resulted in smaller brood sizes than those using wild type males and hermaphrodites (474.00 broods by wild type male versus 159.75 broods by crls-1 mutant male).
This study demonstrates for the first time that the contribution of CL to mitochondrial function and morphology varies among the different cell types in vivo, raising the question of its generality. As mentioned above, Drosophila mutants of a CL synthase gene show abnormal mitochondrial morphology and function in muscle cells (53). In addition, disturbance of the CL remodeling causes abnormal mitochondrial morphology and function in muscle cells, such as the human Barth syndrome and the calcium-independent phospholipase A2γ knock-out mice (57, 58). To address this question, generation of CL synthase knock-out mice and subsequent analysis of their susceptibility to CL depletion in various tissues need to be carried out. In addition, crls-1 mutants still contained residual CL, although crls-1 is the single homologue of CL synthase in C. elegans (Fig. 2C). These data suggest that C. elegans has a bypass mechanism to produce CL except for the de novo synthetic pathway. Nonetheless, our study suggests the existence of a complementary system of the biomembrane in some cell types, at least in C. elegans. Further genetic analysis using crls-1 mutants may reveal the molecular mechanisms underlying the different sensitivities of organelles to the changes in the lipid environment.
Acknowledgments
We greatly thank A. Takimoto and M. Ikuta for excellent technical assistance; Drs. K. Gengyo-Ando (Saitama University) and S. Mitani (Tokyo Women's Medical University School of Medicine) for providing alleles of crls-1 and pgs-1; Dr. A. Fire (Stanford University School of Medicine) for convenient vectors; and Dr. T. Inoue (National University of Singapore) for helpful discussions and comments on this study. Some strains were provided by the Caenorhabditis Genetics Center (University of Minnesota, Minneapolis), which is funded by the National Institutes of Health, NCRR.
This work was supported by Grant-in-aid for Scientific Research (C) 22590072 from the Japan Society for the Promotion of Science (to Y. N.) and by a Kitasato University research grant for young researchers (to T. S.).
- CL
- cardiolipin
- CDP-DAG
- cytidine diphosphate-diacylglycerol
- DiSC3
- 3,3′-dipropylthiocarbocyanine iodide
- PG
- phosphatidylglycerol
- PGP
- phosphatidylglycerophosphate.
REFERENCES
- 1. Schlame M., Rua D., Greenberg M. L. (2000) The biosynthesis and functional role of cardiolipin. Prog. Lipid Res. 39, 257–288 [DOI] [PubMed] [Google Scholar]
- 2. Houtkooper R. H., Vaz F. M. (2008) Cardiolipin, the heart of mitochondrial metabolism. Cell. Mol. Life Sci. 65, 2493–2506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Joshi A. S., Zhou J., Gohil V. M., Chen S., Greenberg M. L. (2009) Cellular functions of cardiolipin in yeast. Biochim. Biophys. Acta 1793, 212–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Schlame M., Haldar D. (1993) Cardiolipin is synthesized on the matrix side of the inner membrane in rat liver mitochondria. J. Biol. Chem. 268, 74–79 [PubMed] [Google Scholar]
- 5. Osman C., Voelker D. R., Langer T. (2011) Making heads or tails of phospholipids in mitochondria. J. Cell Biol. 192, 7–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Fry M., Green D. E. (1981) Cardiolipin requirement for electron transfer in complex I and III of the mitochondrial respiratory chain. J. Biol. Chem. 256, 1874–1880 [PubMed] [Google Scholar]
- 7. Yu C. A., Yu L. (1980) Structural role of phospholipids in ubiquinol-cytochrome c reductase. Biochemistry 19, 5715–5720 [DOI] [PubMed] [Google Scholar]
- 8. Hayer-Hartl M., Schägger H., von Jagow G., Beyer K. (1992) Interactions of phospholipids with the mitochondrial cytochrome c reductase studied by spin-label ESR and NMR spectroscopy. Eur. J. Biochem. 209, 423–430 [DOI] [PubMed] [Google Scholar]
- 9. Schägger H., Hagen T., Roth B., Brandt U., Link T. A., von Jagow G. (1990) Phospholipid specificity of bovine heart bc1 complex. Eur. J. Biochem. 190, 123–130 [DOI] [PubMed] [Google Scholar]
- 10. Gomez B., Jr., Robinson N. C. (1999) Phospholipase digestion of bound cardiolipin reversibly inactivates bovine cytochrome bc1. Biochemistry 38, 9031–9038 [DOI] [PubMed] [Google Scholar]
- 11. Robinson N. C., Zborowski J., Talbert L. H. (1990) Cardiolipin-depleted bovine heart cytochrome c oxidase. Binding stoichiometry and affinity for cardiolipin derivatives. Biochemistry 29, 8962–8969 [DOI] [PubMed] [Google Scholar]
- 12. Sedlák E., Robinson N. C. (1999) Phospholipase A(2) digestion of cardiolipin bound to bovine cytochrome c oxidase alters both activity and quaternary structure. Biochemistry 38, 14966–14972 [DOI] [PubMed] [Google Scholar]
- 13. Eble K. S., Coleman W. B., Hantgan R. R., Cunningham C. C. (1990) Tightly associated cardiolipin in the bovine heart mitochondrial ATP synthase as analyzed by 31P nuclear magnetic resonance spectroscopy. J. Biol. Chem. 265, 19434–19440 [PubMed] [Google Scholar]
- 14. Jiang F., Ryan M. T., Schlame M., Zhao M., Gu Z., Klingenberg M., Pfanner N., Greenberg M. L. (2000) Absence of cardiolipin in the crd1 null mutant results in decreased mitochondrial membrane potential and reduced mitochondrial function. J. Biol. Chem. 275, 22387–22394 [DOI] [PubMed] [Google Scholar]
- 15. Koshkin V., Greenberg M. L. (2000) Oxidative phosphorylation in cardiolipin-lacking yeast mitochondria. Biochem. J. 347, 687–691 [PMC free article] [PubMed] [Google Scholar]
- 16. Koshkin V., Greenberg M. L. (2002) Cardiolipin prevents rate-dependent uncoupling and provides osmotic stability in yeast mitochondria. Biochem. J. 364, 317–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zhang M., Mileykovskaya E., Dowhan W. (2002) Gluing the respiratory chain together. Cardiolipin is required for supercomplex formation in the inner mitochondrial membrane. J. Biol. Chem. 277, 43553–43556 [DOI] [PubMed] [Google Scholar]
- 18. Pfeiffer K., Gohil V., Stuart R. A., Hunte C., Brandt U., Greenberg M. L., Schägger H. (2003) Cardiolipin stabilizes respiratory chain supercomplexes. J. Biol. Chem. 278, 52873–52880 [DOI] [PubMed] [Google Scholar]
- 19. Zhang M., Mileykovskaya E., Dowhan W. (2005) Cardiolipin is essential for organization of complexes III and IV into a supercomplex in intact yeast mitochondria. J. Biol. Chem. 280, 29403–29408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Choi S. Y., Gonzalvez F., Jenkins G. M., Slomianny C., Chretien D., Arnoult D., Petit P. X., Frohman M. A. (2007) Cardiolipin deficiency releases cytochrome c from the inner mitochondrial membrane and accelerates stimuli-elicited apoptosis. Cell Death Differ. 14, 597–606 [DOI] [PubMed] [Google Scholar]
- 21. Huang Z., Jiang J., Tyurin V. A., Zhao Q., Mnuskin A., Ren J., Belikova N. A., Feng W., Kurnikov I. V., Kagan V. E. (2008) Cardiolipin deficiency leads to decreased cardiolipin peroxidation and increased resistance of cells to apoptosis. Free Radic. Biol. Med. 44, 1935–1944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Nomura K., Imai H., Koumura T., Kobayashi T., Nakagawa Y. (2000) Mitochondrial phospholipid hydroperoxide glutathione peroxidase inhibits the release of cytochrome c from mitochondria by suppressing the peroxidation of cardiolipin in hypoglycaemia-induced apoptosis. Biochem. J. 351, 183–193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Lutter M., Fang M., Luo X., Nishijima M., Xie X., Wang X. (2000) Cardiolipin provides specificity for targeting of tBid to mitochondria. Nat. Cell Biol. 2, 754–761 [DOI] [PubMed] [Google Scholar]
- 24. Gonzalvez F., Gottlieb E. (2007) Cardiolipin. Setting the beat of apoptosis. Apoptosis 12, 877–885 [DOI] [PubMed] [Google Scholar]
- 25. Brenner S. (1974) The genetics of Caenorhabditis elegans. Genetics 77, 71–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Gengyo-Ando K., Mitani S. (2000) Characterization of mutations induced by ethyl methanesulfonate, UV, and trimethylpsoralen in the nematode Caenorhabditis elegans. Biochem. Biophys. Res. Commun. 269, 64–69 [DOI] [PubMed] [Google Scholar]
- 27. Maciejowski J., Ugel N., Mishra B., Isopi M., Hubbard E. J. (2006) Quantitative analysis of germ line mitosis in adult C. elegans. Dev. Biol. 292, 142–151 [DOI] [PubMed] [Google Scholar]
- 28. Timmons L. (2004) Endogenous inhibitors of RNA interference in Caenorhabditis elegans. BioEssays 26, 715–718 [DOI] [PubMed] [Google Scholar]
- 29. Simmer F., Tijsterman M., Parrish S., Koushika S. P., Nonet M. L., Fire A., Ahringer J., Plasterk R. H. (2002) Loss of the putative RNA-directed RNA polymerase RRF-3 makes C. elegans hypersensitive to RNAi. Curr. Biol. 12, 1317–1319 [DOI] [PubMed] [Google Scholar]
- 30. Bligh E. G., Dyer W. J. (1959) A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 37, 911–917 [DOI] [PubMed] [Google Scholar]
- 31. Rouser G., Fkeischer S., Yamamoto A. (1970) Two-dimensional thin layer chromatographic separation of polar lipids and determination of phospholipids by phosphorus analysis of spots. Lipids 5, 494–496 [DOI] [PubMed] [Google Scholar]
- 32. Williams J. G., McMaster C. R. (1998) Scanning alanine mutagenesis of the CDP-alcohol phosphotransferase motif of Saccharomyces cerevisiae cholinephosphotransferase. J. Biol. Chem. 273, 13482–13487 [DOI] [PubMed] [Google Scholar]
- 33. Jiang F., Rizavi H. S., Greenberg M. L. (1997) Cardiolipin is not essential for the growth of Saccharomyces cerevisiae on fermentable or nonfermentable carbon sources. Mol. Microbiol. 26, 481–491 [DOI] [PubMed] [Google Scholar]
- 34. Kimble J., Hirsh D. (1979) The postembryonic cell lineages of the hermaphrodite and male gonads in Caenorhabditis elegans. Dev. Biol. 70, 396–417 [DOI] [PubMed] [Google Scholar]
- 35. Ariz M., Mainpal R., Subramaniam K. (2009) C. elegans RNA-binding proteins PUF-8 and MEX-3 function redundantly to promote germ line stem cell mitosis. Dev. Biol. 326, 295–304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Michaelson D., Korta D. Z., Capua Y., Hubbard E. J. (2010) Insulin signaling promotes germ line proliferation in C. elegans. Development 137, 671–680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Crittenden S. L., Leonhard K. A., Byrd D. T., Kimble J. (2006) Cellular analyses of the mitotic region in the Caenorhabditis elegans adult germ line. Mol. Biol. Cell 17, 3051–3061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Sulston J. E., Horvitz H. R. (1977) Post-embryonic cell lineages of the nematode, Caenorhabditis elegans. Dev. Biol. 56, 110–156 [DOI] [PubMed] [Google Scholar]
- 39. Zhang M., Su X., Mileykovskaya E., Amoscato A. A., Dowhan W. (2003) Cardiolipin is not required to maintain mitochondrial DNA stability or cell viability for Saccharomyces cerevisiae grown at elevated temperatures. J. Biol. Chem. 278, 35204–35210 [DOI] [PubMed] [Google Scholar]
- 40. Gásková D., DeCorby A., Lemire B. D. (2007) DiS-C3(3) monitoring of in vivo mitochondrial membrane potential in C. elegans. Biochem. Biophys. Res. Commun. 354, 814–819 [DOI] [PubMed] [Google Scholar]
- 41. Chang S. C., Heacock P. N., Clancey C. J., Dowhan W. (1998) The PEL1 gene (renamed PGS1) encodes the phosphatidylglycero-phosphate synthase of Saccharomyces cerevisiae. J. Biol. Chem. 273, 9829–9836 [DOI] [PubMed] [Google Scholar]
- 42. Kawasaki K., Kuge O., Chang S. C., Heacock P. N., Rho M., Suzuki K., Nishijima M., Dowhan W. (1999) Isolation of a Chinese hamster ovary (CHO) cDNA encoding phosphatidylglycerophosphate (PGP) synthase, expression of which corrects the mitochondrial abnormalities of a PGP synthase-defective mutant of CHO-K1 cells. J. Biol. Chem. 274, 1828–1834 [DOI] [PubMed] [Google Scholar]
- 43. Zhong Q., Gohil V. M., Ma L., Greenberg M. L. (2004) Absence of cardiolipin results in temperature sensitivity, respiratory defects, and mitochondrial DNA instability independent of pet56. J. Biol. Chem. 279, 32294–32300 [DOI] [PubMed] [Google Scholar]
- 44. Munn E. A. (1974) The Structure of Mitochondria, pp. 1–69, Academic Press, New York [Google Scholar]
- 45. Kuznetsov A. V., Hermann M., Saks V., Hengster P., Margreiter R. (2009) The cell type specificity of mitochondrial dynamics. Int. J. Biochem. Cell Biol. 41, 1928–1939 [DOI] [PubMed] [Google Scholar]
- 46. Hanukoglu I. (1992) Steroidogenic enzymes. Structure, function, and role in regulation of steroid hormone biosynthesis. J. Steroid Biochem. Mol. Biol. 43, 779–804 [DOI] [PubMed] [Google Scholar]
- 47. Mootha V. K., Bunkenborg J., Olsen J. V., Hjerrild M., Wisniewski J. R., Stahl E., Bolouri M. S., Ray H. N., Sihag S., Kamal M., Patterson N., Lander E. S., Mann M. (2003) Integrated analysis of protein composition, tissue diversity, and gene regulation in mouse mitochondria. Cell 115, 629–640 [DOI] [PubMed] [Google Scholar]
- 48. Johnson D. T., Harris R. A., French S., Blair P. V., You J., Bemis K. G., Wang M., Balaban R. S. (2007) Tissue heterogeneity of the mammalian mitochondrial proteome. Am. J. Physiol. Cell. Physiol. 292, C689–C697 [DOI] [PubMed] [Google Scholar]
- 49. Courtade S., Marinetti G. V., Stotz E. (1967) The structure and abundance of rat tissue cardiolipins. Biochim. Biophys. Acta 137, 121–134 [DOI] [PubMed] [Google Scholar]
- 50. White D. A. (1973) in Form and Function of Phospholipids (Ansell G. B., Hawthorne J. N., Dawson R. M. C., eds) pp. 441–482, Elsevier Scientific Publishing Co., Inc., New York [Google Scholar]
- 51. Reinke V., Smith H. E., Nance J., Wang J., Van Doren C., Begley R., Jones S. J., Davis E. B., Scherer S., Ward S., Kim S. K. (2000) A global profile of germ line gene expression in C. elegans. Mol. Cell 6, 605–616 [DOI] [PubMed] [Google Scholar]
- 52. Kawasaki I., Hanazawa M., Gengyo-Ando K., Mitani S., Maruyama I., Iino Y. (2007) ASB-1, a germ line-specific isoform of mitochondrial ATP synthase b subunit, is required to maintain the rate of germ line development in Caenorhabditis elegans. Mech. Dev. 124, 237–251 [DOI] [PubMed] [Google Scholar]
- 53. Acehan D., Malhotra A., Xu Y., Ren M., Stokes D. L., Schlame M. (2011) Cardiolipin affects the supramolecular organization of ATP synthase in mitochondria. Biophys. J. 100, 2184–2192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Shen L. R., Lai C. Q., Feng X., Parnell L. D., Wan J. B., Wang J. D., Li D., Ordovas J. M., Kang J. X. (2010) Drosophila lacks C20 and C22 PUFAs. J. Lipid Res. 51, 2985–2992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Boxem M., Srinivasan D. G., van den Heuvel S. (1999) The Caenorhabditis elegans gene ncc-1 encodes a cdc2-related kinase required for M phase in meiotic and mitotic cell divisions, but not for S phase. Development 126, 2227–2239 [DOI] [PubMed] [Google Scholar]
- 56. Fay D. S., Han M. (2000) Mutations in cye-1, a Caenorhabditis elegans cyclin E homolog, reveal coordination between cell cycle control and vulval development. Development 127, 4049–4060 [DOI] [PubMed] [Google Scholar]
- 57. Acehan D., Vaz F., Houtkooper R. H., James J., Moore V., Tokunaga C., Kulik W., Wansapura J., Toth M. J., Strauss A., Khuchua Z. (2011) Cardiac and skeletal muscle defects in a mouse model of human Barth syndrome. J. Biol. Chem. 286, 899–908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Mancuso D. J., Sims H. F., Han X., Jenkins C. M., Guan S. P., Yang K., Moon S. H., Pietka T., Abumrad N. A., Schlesinger P. H., Gross R. W. (2007) Genetic ablation of calcium-independent phospholipase A2γ leads to alterations in mitochondrial lipid metabolism and function resulting in a deficient mitochondrial bioenergetic phenotype. J. Biol. Chem. 282, 34611–34622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Mello C. C., Kramer J. M., Stinchcomb D., Ambros V. (1991) Efficient gene transfer in C. elegans. Extrachromosomal maintenance and integration of transforming sequences. EMBO J. 10, 3959–3970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Van Gilst M. R., Hadjivassiliou H., Yamamoto K. R. (2005) A Caenorhabditis elegans nutrient response system partially dependent on nuclear receptor NHR-49. Proc. Natl. Acad. Sci. U.S.A. 102, 13496–13501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Hunter T., Bannister W. H., Hunter G. J. (1997) Cloning, expression, and characterization of two manganese superoxide dismutases from Caenorhabditis elegans. J. Biol. Chem. 272, 28652–28659 [DOI] [PubMed] [Google Scholar]
- 62. Sumitani M., Kasashima K., Matsugi J., Endo H. (2011) Biochemical properties of Caenorhabditis elegans HMG-5, a regulator of mitochondrial DNA. J. Biochem. 149, 581–589 [DOI] [PMC free article] [PubMed] [Google Scholar]