Abstract
Expression of the ISG15 specific protease USP18 is highly induced by type I interferons. The two main functions of USP18, i.e. its enzymatic activity and down-regulation of type I interferon signaling, are well characterized. However, to date all functional studies focused on full-length USP18. Here, we report that translation of human USP18 is initiated by a rare start codon (CUG). Usage of this non-canonical initiation site with its weak translation initiation efficiency promotes expression of an N-terminal truncated isoform (USP18-sf). In addition, an internal ribosome entry site (IRES) located in the 5′-coding region of USP18 also contributes to translation of USP18-sf. Functionally, both isoforms exhibit enzymatic activity and interfere with type I interferon signaling. However, USP18-sf shows different subcellular distribution compared with the full-length protein and enhanced deISGylation activity in the nucleus. Taken together, we report the existence of an N-terminal truncated isoform of USP18, whose expression is controlled on translational level by two independent mechanisms providing translational flexibility as well as cell type-specific resistance to inhibition of cap-dependent translation.
Keywords: Deubiquitination, Innate Immunity, Interferon, Signal Transduction, Translation Control, IRES, ISG15, USP18, Rare Start Codon
Introduction
Interferons (IFNs)2 are a group of small proteins that are produced intrinsically upon pathogen infections, genotoxic insults, or stress (1). They function as cytokines to stimulate the expression of hundreds of interferon-stimulated genes (ISGs) (2, 3). These ISGs regulate various aspects of cellular status, including proliferation, survival, and differentiation (4–6). We identified the interferon inducible gene USP18 (also known as UBP43) as a member of the ubiquitin-specific processing protease (UBP) family (7, 8). Further analysis of USP18 demonstrated that it is a bona fide ISG15-specific protease (9, 10). In addition, USP18 functions as a strong negative regulator of type I interferon-activated JAK/STAT signaling, which leads to hypersensitivity to IFN treatment in USP18-deficient cells (11–13). This regulatory effect is independent from its ISG15 isopeptidase activity and mediated through competition for interferon α/β receptor 2 (IFNAR2) binding with JAK1 (13). Accordingly, it was recently shown that USP18 is the key mediator of long-lasting refractoriness to continuous IFN-α treatment in the mouse liver (14).
Down-regulation of general protein translation, which helps to restrict production of viral proteins upon infection, is one of the characteristics of activated IFN signaling. Initiation of protein biosynthesis in Eukaryotes is mostly achieved through binding of the 40 S ribosomal subunit and related factors to the 5′-cap structure of the mRNA (15). Upon assembly the ribosome scans the mRNA from the 5′-end of the capped mRNA for the first AUG in good Kozak consensus context to start protein translation (16). The inhibitory effect of IFNs on protein translation is mediated by PKR induced phosphorylation of eukaryotic initiation factor 2α (eIF-2α), which blocks guanine nucleotide exchange factor eIF-2B activity (17–19), and also by caspase-3 induced cleavage of eIF4G, which inhibits formation of the eIF4F complex crucial for RNA cap structure binding (20, 21). Although both mechanisms are efficient for slowing down production of viral proteins they also affect translation of cellular RNAs. Thus, protein levels of negative regulators of IFN signaling which are crucial to control cellular levels of ISGs are also reduced. Interestingly, some of these regulatory factors seem to circumvent the induced reduction of cap-dependent protein translation by utilizing a cap-independent mechanism of translation such as internal ribosome entry site (IRES). For instance, interferon regulatory factor 2 (IRF2) and NF-κB-repressing factor (NRF), two negative regulators of the innate immune response, were reported to be expressed in an IRES-dependent manner under stress conditions (22, 23). This cap-independent method of protein translation helps to maintain critical protein levels when the cell is under stress, e.g. during activated IFN signaling.
In this report, we show that instead of using the canonical start codon AUG, translation of full-length USP18 protein is initiated by the rare start codon CUG. The relatively low translation initiation efficiency of the CUG start codon and following frequent skip by the scanning ribosome promotes expression of an N-terminal truncated isoform, which we named USP18-sf. In addition, expression of USP18-sf is also driven by an IRES element in the 5′ region of the USP18 coding sequence. This IRES-mediated expression helps to maintain critical protein levels of USP18 in a cell type-specific manner when cap-dependent translation is impaired. Moreover, we show that the newly identified isoform USP18-sf is the major deISGylation enzyme for nuclear proteins.
EXPERIMENTAL PROCEDURES
Plasmids and Antibodies
Full-length human USP18 sequence was amplified from cDNA of IFN-α treated HeLa cells. For cloning into pMSCV-puro (Clontech) BglII and HpaI restriction sites were introduced and for cloning into pCDNA3.1 HindIII and XhoI were introduced. USP18-sf fragment was also amplified from HeLa cells derived cDNA with BglII and HpaI restriction sites for cloning into pMSCV-puro. FLAG-tagged expression plasmids were generated by amplifying USP18 sequence starting from ATG1, CTG16, or ATG36 from pMSCV-puro-USP18 introducing HindIII and BamHI restriction sites and following subcloning into p3xFLAG (Sigma). Constructs for His-mISG15, FLAG-mUbcH8 and HA-hUBE1L were previously described (24). The discistronic constructs pR-ΔEnull-F and pR-EMCV-F were kindly provided by Dr. Peter Sarnow (Stanford University). 5′-region of human USP18 coding sequence (bp 1–105) was subcloned via EcoRI into pR-ΔENull-F to make pR-USP18-F. Promoterless monocistronic renilla reporter construct pRLnull (Promega) was digested with EcoRI and the above mentioned EcoRI-digested USP18 fragment subcloned into it to generate pR-USP18. All mutant constructs used in this report were generated by site-directed mutagenesis. For bacterial expression USP18-GCG36 and USP18-sf were amplified by PCR using p3xFLAG-hUSP18-GCG36 or p3xFLAG-hUSP18-sf as template and subsequently subcloned into pGEX4T-1 via BamHI and NotI restriction sites. All plasmids generated for this report were checked for point mutations or deletions by sequencing. Antibodies against V5 (Invitrogen), β-tubulin, FLAG (both Sigma), HA (Covance), GST (EMD), β-galactosidase, H4 (both Abcam) EGFR (Millipore), Parp, eIF2α, p-eIF2α, STAT1, and p-STAT1 (all Cell Signal) were purchased from indicated companies. Anti-hUSP18 antibody was previously described (13). Anti-mouse IgG Alexa Fluor 488 (Invitrogen) was used as secondary antibody for Fluorescence Microscopy.
Cell Lines and Transfection
HEK293T cells were cultured in Dulbecco's modified Eagle's medium (DMEM, Cellgro) with 10% bovine calf serum (BCS) and 2 mm l-glutamine. HeLa, MCF-7, HACAT, and A549 cells were cultured in DMEM with 10% fetal bovine serum (FBS) and 2 mm l-glutamine. KT-1, Jurkat, and Daudi cells were cultured in RPMI medium (Cellgro) with 10% FBS and 2 mm l-glutamine. Human IFN-α2a (Roche) was used at 10 ng/ml for induction of USP18 expression or in STAT1 phosphorylation experiments. For all transfections, cells were grown on 6-well plates and transfected using PEI (polyethyleneimine). For tunicamycin treatment, cells were incubated in presence of 2.5 μg/ml of tunicamycin (Calbiochem) for 14 h. Stable pools of MCF-7, Jurkat, Daudi, and HACAT cells were made after retroviral infection with the respective constructs by selection with 1–2.5 μg/ml of puromycin (Calbiochem). For RNA transfections, RNA was transcribed in vitro with mMESSAGE mMACHINE (Ambion) and purified RNAs transfected into HeLa cells with Lipofectamine 2000 (Invitrogen).
Western Blotting
Cells were lysed in 150 mm NaCl, 20 mm Tris pH 7.5, 0.5% Nonidet P-40, 1 mm EDTA, 1 mm DTT and protease inhibitor mixture (complete, Roche). In phosphorylation experiments phosphatase inhibitor (PhoSTOP, Roche) was added to lysis buffer. Proteins were electroblotted onto nitrocellulose membranes (HyBond, Amersham Biosciences Inc.) and fluorophore-conjugated secondary antibodies (Li-Cor) used for detection with Odyssey system (Li-Cor).
Fluorescence Microscopy and Subcellular Fractionation Assays
HeLa and MCF-7 cells were grown on glass slides and transfected with FLAG-hUSP18 or FLAG-hUSP18-sf expression constructs. 24 h later, cells were fixed in ice-cold methanol, incubated with corresponding primary and secondary antibodies and analyzed with a fluorescence microscope (Leica Microsystems). For subcellular fractionation experiments, nuclear fractions from IFN-α-treated cells were isolated using a standard two-step hypotonic/high salt buffer procedure.
Bicistronic and Monocistronic Reporter Assay
Transfected HeLa cells were lysed and protein lysates analyzed using the Dual-Luciferase® reporter system (Promega). Firefly and Renilla luciferase activity was measured in a luminometer (BD Biosciences).
Assay for Ubiquitin-specific Protease Activity
The assay for a ubiquitin-specific protease to deubiquitinate a ubiquitin-β-galactosidase fusion protein (Ub-Met-β-gal) has been previously described (25). Briefly, BL21(DE3) cells were co-transformed with GST-USP18 isoforms or GST-DUB2 together with a construct encoding a ubiquitin-β-galactosidase fusion protein. Cell lysates were prepared, and lysates analyzed by Western blotting using anti-β-galactosidase antibody.
RT-PCR
RNAs from transfected HeLa cells were isolated using RNeasy Mini kit (Qiagen). Extracted RNAs were then used for cDNA synthesis with qScript cDNA SuperMix (Quanta Biosciences). Oligos used to amplify different regions of dicistronic constructs:
P1(RLuc_forward): TCTTGACGAGCAATCCTAGAGC; P2(FLuc_reverse1): TGCAACTCCGATAAATAACGCG; P3(FLuc_forward): AGCTATGAAACGATATGGGCTG; P4(FLuc_reverse2): AGAGCGACACCTTTAGGCAG.
RESULTS
Human Cell Lines Express Two USP18 Isoforms upon IFN-α Treatment
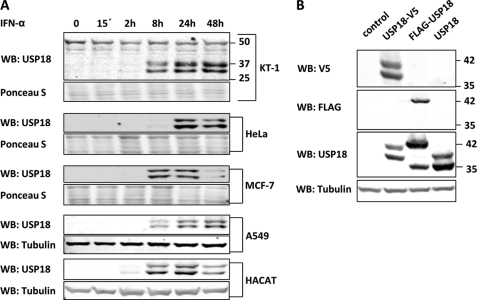
Several IFN responsive human cell lines were treated with recombinant hIFN-α to compare induction and expression pattern of USP18. In all cell lines tested the USP18 antibody detects two specific interferon inducible bands (Fig. 1A). The time point of highest expression as well as ratio of the two USP18-specific bands vary among cell lines. For instance, MCF-7 and HACAT cells show highest levels between 8–24 h after IFN treatment whereas expression in KT-1 and A549 cells peak between 24 and 48 h. Furthermore, both HeLa and HACAT cells have significantly higher amounts of the faster migrating band at all times.
FIGURE 1.
Expression pattern of human USP18 in mammalian cell lines. A, IFN-responsive cell lines were treated with 10 ng/ml hIFN-α. Cells were harvested at indicated time points, lysed, and cell lysates subjected to USP18 Western blotting. B, 293T cells were transfected with expression plasmids coding for human untagged, N-terminal FLAG-tagged or C-terminal V5-tagged USP18. Transfection of empty vector was used as negative control. After 36 h, cell lysates were prepared and probed for FLAG, V5, USP18, and tubulin by Western blotting
To further examine the pattern of USP18 protein expression, we performed transfection studies with either N-terminal or C-terminal-tagged USP18 proteins. Exogenous expression of USP18 with C-terminal V5 tag led to detection of two specific bands whereas N-terminal FLAG-tagged USP18 showed a single band by Western blotting (Fig. 1B), suggesting that the two USP18 isoforms share the C-terminal part but differ in the N-terminal region. An expression plasmid coding for untagged USP18 was included as a control. The observed difference between FLAG- and V5-tagged USP18 when probed with anti-USP18 antibody is due to the different molecular weights of the produced fusion proteins. The faster migrating band in the FLAG-USP18 lane detected with anti-USP18 antibody does not have the N-terminal FLAG tag and migrates at the same position as the lower band in the USP18 lane. This indicates that the faster migrating protein in the FLAG-USP18 transfectant lacks the N-terminal region of USP18 and is likely the same as the faster migrating protein in the untagged USP18 sample. Furthermore, using the full-length coding sequence of USP18 ruled out that the lower band is expressed due to alternative mRNA splicing indicating another mechanism, such as an alternative translational start site or proteolytic cleavage as reported by Potu et al. (26), to produce two isoforms of USP18. Interestingly, C-terminal-tagged murine Usp18 also shows two specific bands when overexpressed suggesting a conserved mechanism between the two species (data not shown).
Expression of Full-length USP18 Is Initiated by the Rare Start Codon CUG, Which Promotes Expression of an N-terminal Truncated Isoform
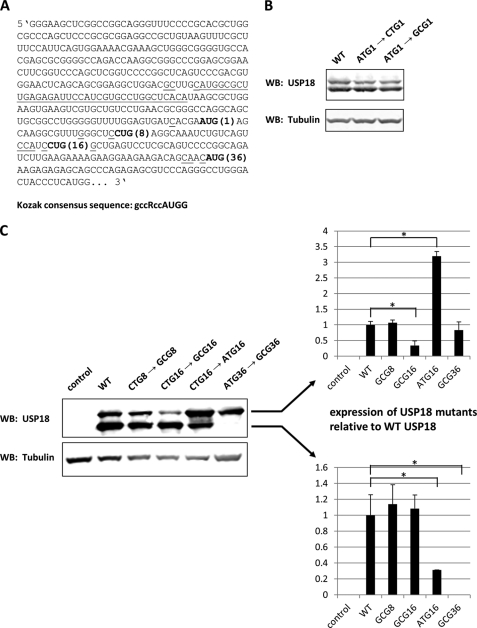
Next, we wanted to address the question whether the faster migrating USP18 specific band observed by Western blotting is due to proteolytic cleavage. In this case elimination of the full-length protein should also eliminate the smaller band. To investigate this we mutated the reported start codon (ATG1) in the USP18 coding sequence (Fig. 2A). Surprisingly, none of the introduced mutations changed the expression pattern compared with the wild type sequence (Fig. 2B). This finding, though unexpected, can be explained by the weak Kozak consensus of ATG1 and suggests the existence of another primary translation initiation site further downstream.
FIGURE 2.
Translation of human USP18 is initiated by the non-canonical start codon CUG promoting expression of an N-terminal truncated isoform. A, 5′ region of human USP18 mRNA including the 5′-UTR is shown. In-frame AUG and CUG codons are in bold letters. Underlined nucleotides indicate Kozak consensus, and the dotted line marks a uORF in the 5′-UTR of USP18. B, 293T cells were transfected with plasmids coding for wild type USP18 or USP18 mutants disrupting start codon ATG1. Whole cell lysates were analyzed for USP18 and tubulin expression by Western blotting. C, 293T cells were transfected with plasmids coding for different USP18 mutants and whole cell lysates were probed for USP18 by Western blotting. Empty vector was added as negative control and tubulin was used as loading control. Expression levels of USP18 isoforms normalized to tubulin from three independent transfections were quantified with Li-Cor Odyssey system. Asterisks indicate statistical significance (t test) of p < 0.05.
Within the first 150 bp of the USP18 coding sequence we found a second in-frame ATG at codon position 36 (ATG36). Furthermore, we identified two in-frame CTGs at codon position 8 and 16, respectively (Fig. 2A). Although rare in mammalian cells the CUG codon was reported to function as a translation initiation site in some mammalian genes (27), and its translation efficiency is significantly enhanced in an optimal Kozak consensus context (28, 29). Therefore, we mutated both CTG8 and CTG16 to GCG to examine if one of the two codons can function as translation initiation site for USP18. The GCG8 mutation did not have a striking effect on expression of USP18 (Fig. 2C). This is not surprising considering its relatively weak Kozak consensus. Mutating CTG16 to GCG16, however, significantly reduced expression of the upper band suggesting that full-length USP18 starts from CTG16. To test if inefficient translation initiation at position CUG16 and the resulting frequent skip by the scanning ribosome, a mechanism also referred to as leaky scanning, promote translation of an N-terminal truncated isoform further downstream, we mutated CTG16 to ATG16 to optimize translation initiation efficiency at this site. The introduced mutation elevated expression of full-length USP18 and reduced expression of the smaller isoform as determined by quantification with the Li-Cor Odyssey system (Fig. 2C, right panel). This suggests that the weak translation initiation efficiency of CUG16 promotes expression of an N-terminal truncated isoform from an alternative translational start side further downstream. Taken together, these data show that the CUG codon at position 16 is the primary translation initiation start site for USP18 and that its low translation initiation efficiency leads to a frequent skip by the scanning ribosome, which in turn contributes to translation initiation at an alternative start site producing an N-terminal truncated USP18 isoform. In fact, mutating ATG36 to GCG completely eliminated the faster migrating band showing that an N-terminal truncated isoform is expressed because of an alternative translational start site (Fig. 2C).
From this point, on we name the N-terminal truncated isoform USP18-sf and refer to USP18 protein starting from CUG16 as USP18. Supplemental Fig. S1A depicts the deduced protein sequence of USP18 including conserved domains of the UBP family. Interestingly, sequence alignment of human USP18 with other species showed that the two in-frame CTGs and ATGs as well as the corresponding Kozak consensuses are quite conserved (supplemental Fig. S1B).
IRES Element in the 5′ Region of USP18 Coding Sequence Contributes to USP18-sf Expression
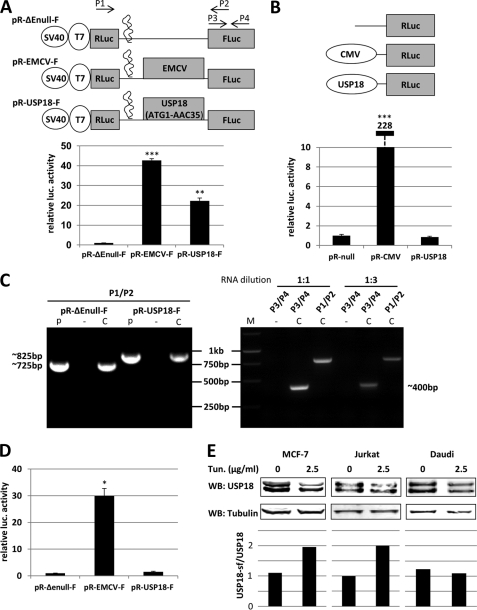
In the previous section we showed that expression of USP18-sf is due to weak translation initiation efficiency of CUG16. In fact, ∼70% of USP18-sf levels are lost when CTG16 is mutated to ATG16 (Fig. 2C). However, this optimized translation initiation site did not completely eliminate USP18-sf expression suggesting the existence of an additional mechanism of alternative translation initiation. A plausible explanation for this finding could be existence of an IRES or cryptic promoter activity upstream of ATG36. Because most cellular IRES elements are believed to be ≥100 bp, the 5′ region spanning from ATG1 to ATG36 that encompasses 105 nucleotides could be sufficient (30). To test if the region from ATG1 to ATG36 has IRES activity we cloned the nucleotide sequence from ATG1 to AAC35 into a dicistronic luciferase construct (Fig. 3A). The highly efficient IRES from encephalomyocarditis virus (EMCV) was used as a positive control. Because ribosomal read-through can produce false-positive luciferase readings all of these constructs contain a RNA hairpin loop downstream of the Renilla reporter gene. This secondary RNA structure is capable of inhibiting ribosomal read-through (31, 32). The results of the luciferase assay showed that the 5′ region of USP18 coding sequence leads to a 22-fold increase of luciferase activity compared with the negative control pR-ΔEnull-F (Fig. 3A). Cap-dependent Renilla activity was similar throughout all samples. These results suggest the existence of an IRES element in the 5′ coding region of USP18 mRNA.
FIGURE 3.
USP18 coding sequence shows IRES activity, which helps maintain USP18-sf protein levels when cap-dependent translation is down-regulated. A, schematic representation of the dicistronic plasmids used in transient transfections to analyze USP18 IRES activity. Indicated dicistronic plasmids were transiently transfected into HeLa cells. 24 h post-transfection, respective luciferase activities corresponding to firefly luciferase (FLuc) and Renilla luciferase (RLuc) were measured. Shown are mean FLuc readings of three independent experiments normalized to RLuc. Asterisks indicate statistical significance compared with control plasmid (t test) of p < 0.05. B, USP18 sequence used in A was cloned into promoterless Renilla reporter construct to analyze cryptic promoter activity. Shown RLuc constructs were co-transfected with CMV driven firefly reporter gene construct into HeLa cells. 24 h later luciferase activities corresponding to FLuc and RLuc were determined. FLuc activity was used to compensate for differences in transfection efficiency. Asterisks indicate statistical significance compared with control plasmid (t test) of p < 0.05. C, HeLa cells were transfected with dicistronic luciferase plasmids to confirm presence of intact dicistronic transcripts and absence of monocistronic FLuc transcripts by RT-PCR. Oligos used for amplification are depicted in A. Left panel: integrity of dicistronic transcripts produced by the indicated plasmids was verified by amplification of the full intercistronic region RLuc-insert-FLuc as compared with plasmid DNA control. Right panel: analysis of pR-USP18-F using different dilution of input RNA and two different primer combinations as indicated above the lanes. Expected size of amplified fragments are shown next to the gel pictures. (p) = plasmid DNA, (−) = noRT, (C) = cDNA, (M) = Marker. D, HeLa cells were transfected with in vitro transcribed, capped dicsitronic RNAs as indicated. 8 h later FLuc activity and RLuc activity was determined. Shown are mean FLuc values normalized to RLuc of three independent experiments. Asterisks indicate statistical significance compared with control plasmid (t test) of p < 0.05. E, MCF-7, Jurkat and Daudi cells stably expressing USP18 were treated with 2.5 μg/ml tunicamycin or carrier. 14 h later cells were harvested and whole cell lysates probed for USP18. USP18-sf and USP18 levels were quantified with the Li-Cor Odyssey system and difference in protein levels shown as USP18-sf/USP18 ratio.
However, in some cases using the Renilla luciferase (RLuc)/firefly luciferase (FLuc) dicistronic reporter system was reported to cause false positive FLuc readings due to formation of aberrant monocistronic transcripts produced by cryptic promoter activity or RNA splicing (33, 34). To examine if the nucleotide sequence from ATG1-AAC35 of USP18 exhibits cryptic promoter activity we cloned this region into promoterless pRLnull vector in front of the Renilla reporter gene. The same construct with functional CMV promoter upstream of the Renilla reporter was used as positive control. The plasmids were transfected into HeLa cells together with a CMV driven firefly reporter construct for normalization of transfection efficiency. In this assay the USP18 sequence did not show any induction of Renilla luciferase activity, which indicates that it does not possess cryptic promoter activity (Fig. 3B). Next, we verified the integrity of the produced dicistronic mRNAs by RT-PCR. The used primers span the entire intercistronic region and are depicted in Fig. 3A. For both pR-ΔEnull-F and pR-USP18-F a single transcript, which corresponds to the PCR product amplified from plasmid DNA, was produced (Fig. 3C, left panel). This finding confirmed the presence of intact dicistronic RNA. DNase I treatment and NoRT control proved that the RT-PCR products were derived from mRNAs and not from transfected plasmid DNA. Additionally, we investigated the potential existence of shorter monocistronic FLuc RNAs. For this purpose, we performed RT-PCR analysis with different dilutions of input RNA from pR-USP18-F-transfected cells and different oligo sets. In this experiment primer set P1/P2 would amplify FLuc insert-RLuc, whereas the primer set P3/P4 would amplify a region of the FLuc gene only. In the case of aberrant splice sites within the inserted USP18 sequence, the amount of amplified product by P3/P4 would be significantly higher than that of P1/P2. However, in our assay no significant difference between the amplified products was observed (Fig. 3C, right panel).
Another way to exclude both cryptic promoter activity and aberrant splicing is the transfection of dicistronic RNAs to assess IRES activity. However, a drawback of this method is that cellular IRESs cannot associate with nuclear IRES trans-acting factors (ITAFs), which may be required for activity of some IRESs (35–37). We used the plasmids shown in Fig. 3A for in vitro transcription of capped, dicistronic RNAs. These RNAs were then transfected into HeLa cells and cell lysates analyzed for RLuc and FLuc activity. In this experiment, however, USP18 sequence only led to a 2-fold increase of FLuc activity compared with empty control vector, which may be attributed to the mentioned lack of association with crucial nuclear ITAFs (Fig. 3D).
If expression of USP18-sf is not only driven by inefficient translation initiation at CUG16 but also by IRES a reduction of cap-dependent translation in vivo should have a bigger effect on protein levels of USP18 than USP18-sf. Tunicamycin is known to cause endoplasmic reticulum stress, which leads to reduction of general protein translation by phosphorylation of eIF2α and cleavage of eIF4G (38–40). Cleavage of eIF4G specifically inhibits cap-dependent translation initiation by inactivating the eIF4F complex with respect to its ability to recognize capped mRNAs. These characteristics of tunicamycin make it a suitable reagent to study if IRES mediated cap-independent expression of a target gene is induced or maintained under stress condition in vivo (22, 41, 42). However, activity of some viral IRESs, such as EMCV and polio, have been shown to be sensitive to Tunicamycin treatment, which results in a down-regulation of activity (43). To test if USP18 IRES activity is affected by tunicamycin induced translational stress we performed a dicistronic luciferase assay in the presence of tunicamycin (supplemental Fig. S2). Compared with EMCV IRES USP18 IRES activity exhibits relative resistance to tunicamycin treatment. Therefore we used tunicamycin as a model reagent to investigate if IRES-mediated translation leads to an increase of USP18-sf levels as compared with USP18. Cells were treated with tunicamycin for 14 h and whole cell lysates analyzed for USP18 protein levels. In MCF-7 and Jurkat cells the treatment led to an increase of USP18-sf/USP18 ratios whereas no difference was observed in Daudi cells, suggesting cell type-specific IRES activity (Fig. 3E). In MCF-7 and Jurkat cells quantification with the Li-Cor Odyssey system showed approximately twice as much USP18-sf after tunicamycin treatment, which indicates that production of USP18-sf is not completely halted due to cap-independent translation. Taking all the presented results together, we conclude that the 5′ region of USP18 mRNA shows IRES activity, which accounts for up to 30% of expressed USP18-sf, and that this cap-independent mechanism of translation initiation is cell-type dependent and requires a yet unknown nuclear ITAF.
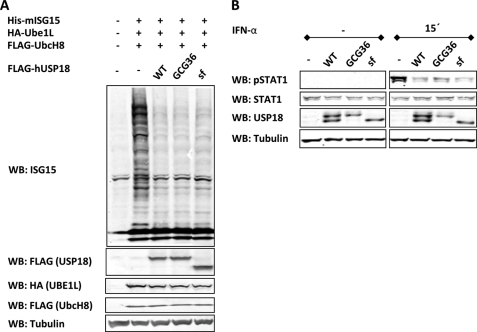
Both USP18 Isoforms Exhibit Similar Enzymatic Activity and Interfere with Type I IFN Signaling
We next examined if USP18-sf behaves the same way as the full-length protein regarding its known functions. So far, two main functions of USP18 have been reported; the ISG15 specific enzyme activity and the negative regulation of the type I IFN signaling pathway (9, 13). To investigate the ISG15 specific enzymatic activity of the USP18 isoforms we used our previously established transient transfection ISGylation system (24). DNA constructs expressing components of the ISGylation system (ISG15, E1, and E2) and USP18 (expressing both isoforms), USP18-GCG36 (only expressing full-length USP18) or USP18-sf were transiently transfected into 293T cells. In this assay both USP18 isoforms exhibit enzymatic activity and reduce total levels of ISGylated protein to the same extent (Fig. 4A). Next, we generated HACAT cells stably expressing USP18, USP18-GCG36, or USP18-sf to analyze interference with type I IFN-activated JAK/STAT signaling for each isoform separately. Stable cell lines were treated with IFN-α for 15min and phosphorylation of STAT1 was determined by Western blotting. Fig. 4B shows that both USP18 isoforms reduce phosphorylation of STAT1 by the same amount. A similar effect in MCF-7 cells stably expressing USP18 isoforms was also observed (supplemental Fig. S3).
FIGURE 4.
Both USP18 isoforms show enzyme activity and inhibit type I interferon induced JAK/STAT signaling. A, 293T cells were transfected with ISGylation system (FLAG-UBE1L, HA-UbcH8, and His-ISG15) with or without USP18 isoforms. 36 h after transfection whole cell lysates were probed for ISG15 as well as components of the ISGylation system by Western blotting. Tubulin was used as loading control. B, HACAT cells stably expressing USP18, USP18-GCG36, or USP18-sf were treated with 10 ng/ml hIFN-α for 15min. Whole cell lysates were then subjected to USP18, STAT1, and phospho-STAT1 Western blot. Tubulin was used as loading control.
Because USP18 belongs to the ubiquitin specific protease family, we also examined the possibility of isoform specific protease activity toward ubiquitin conjugates. For this purpose, GST-tagged USP18 isoforms were co-expressed together with ubiquitin-β-galactosidase fusion protein in Escherichia coli. Cleavage of ubiquitin from the fusion protein was monitored by anti-galactosidase Western blotting. Only the known ubiquitin protease DUB2 (44) showed significant cleavage of ubiquitin even though expression level was much lower compared with the USP18 isoforms (supplemental Fig. S4A). These results indicate that none of the isoforms are good enzymes toward ubiquitin conjugates.
Recently, a report suggested that USP18 is a regulator of epidermal growth factor receptor (EGFR) expression on translational level (45) and we aimed to examine if there is an isoform specific effect on EGFR protein levels. However, in our system we did not see any effect of USP18 on EGFR protein levels (supplemental Fig. S4B).
USP18-sf Is the Major DeISGylation Enzyme for Nuclear Proteins
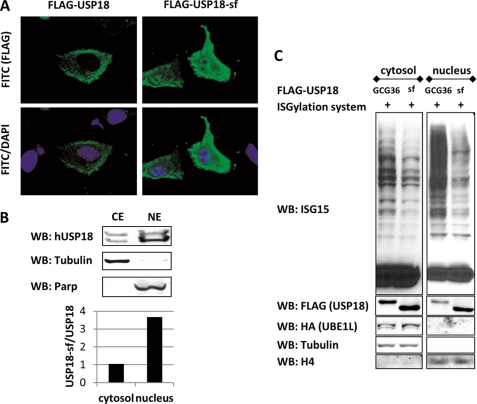
We used immunofluorescence microscopy to investigate the subcellular localization of both USP18 isoforms. HeLa cells were transfected with FLAG-tagged USP18 or USP18-sf construct and analyzed with a fluorescence microscope. We found that USP18 is mainly located in the cytosol whereas USP18-sf is evenly distributed between cytosol and nucleus (Fig. 5A). Because the polyethylenimine (PEI) transfections were performed in the absence of IFN and PEI was reported not to induce an IFN response, it is unlikely that other IFN-inducible factors are responsible for the exclusion of full-length USP18 from the nucleus (46). To further support this finding we aimed to determine the subcellular localization of endogenous USP18. Because the available antibody for human USP18 is not suitable for immunostaining we prepared cytosolic and nuclear extracts from IFN-α treated HeLa cells and probed for USP18. Again, we observed a significantly higher amount of USP18-sf in the nuclear fraction as quantified by the Li-Cor Odyssey system (Fig. 5B). The observed nuclear abundance of USP18-sf indicates that this isoform carries out most of USP18's functions in the nucleus.
FIGURE 5.
USP18 isoforms show different subcellular localization and USP18-sf transfected cells show reduced protein ISGylation in the nucleus. A, HeLa cells were transfected with FLAG-USP18 or FLAG-USP18-sf. Immunostaining of MeOH fixed cells shows that FLAG-USP18-sf is localized in the nucleus and the cytosol whereas FLAG-USP18 is almost exclusively located in the cytosol. B, HeLa cells were treated with 10 ng/ml hIFN-α for 36 h to induce expression of USP18. Cells were then lysed and nuclear/cytosolic fractions prepared. Each subcellular fraction was probed for USP18 as well as nuclear and cytosolic markers. USP18-sf/USP18 ratio was quantified with the Li-Cor Odyssey system. C, HeLa cells were transiently cotransfected with ISGylation system and either FLAG-USP18-GCG36 or FLAG-USP18-sf expression construct. 36 h after transfection cytosolic and nuclear extracts were prepared and probed for ISG15, USP18, UBE1L as well as nuclear and cytosolic markers.
Based on the observed differential localization between the two USP18 isoforms it is likely that USP18-sf is the major deISGylation enzyme for nuclear proteins. In fact, nuclear extracts of HeLa cells transfected with components of the ISGylation system and FLAG-USP18-GCG36 or FLAG-USP18-sf showed significantly lower levels of ISGylated protein in the nuclear fraction when USP18-sf was present (Fig. 5C). Accordingly, compared with FLAG-USP18-GCG36 transfected cells more FLAG-USP18-sf was detected in the nuclear fraction. MCF-7 cells showed the same difference in subcellular localization, and consequently reduced levels of ISGylated protein in the nucleus of FLAG-USP18-sf transfected cells were observed (supplemental Fig. S5).
DISCUSSION
IFN-responsive cell lines express two USP18 isoforms upon induction. Here, we show that translation of full-length USP18 is not initiated by the previously reported canonical AUG start codon but from a non-canonical CUG codon further downstream. Moreover, we present evidence that expression of an N-terminal truncated isoform is controlled by two independent mechanisms providing resistance to down-regulation of cap-dependent translation in a cell-type specific manner. Detailed functional characterization of this isoform, USP18-sf, revealed that it behaves like the full-length protein. However, different subcellular localization suggests more specialized functions.
Previous reports have shown that endogenous expression of USP18 with a C-terminal tag leads to detection of two specific bands by Western blotting but no effort was made to identify a regulating mechanism or a functional difference between the two (13, 26, 47). All IFN responsive human cell lines that we tested in this study express two USP18 isoforms with varying ratios (Fig. 1A). For instance, HeLa, A549, and HACAT cells show higher levels of USP18-sf at all times whereas KT-1 and MCF-7 cells show equal levels. Because we have shown that expression of USP18-sf is partially mediated by IRES, a cell type-specific difference in expression of specific ITAFs may contribute to the observed difference. In fact, approximately half of the cellular proteins that are translated in an IRES-dependent manner require ITAFs for efficient translation (48). In addition, it is possible that cell condition and/or cell state affects translation initiation efficiency at CUG16 or IRES-mediated expression of USP18-sf, which may contribute to the observed difference in USP18 expression pattern among the cell lines.
The initial transfection experiments using untagged as well as C- or N-terminal tagged versions of USP18 clearly ruled out the possibility that USP18-sf is produced by alternative mRNA splicing (Fig. 1B). Interestingly, the 5′-untranslated region (UTR) of USP18 contains an upstream open reading frame (uORF), a known translational regulatory element (Fig. 2A)(49). In case of short uORFs reinitiation of protein translation can control the choice of translational start sites and protein synthesis of functionally important isoforms (50, 51). However, regulatory elements located in the 5′-UTR do not seem to have a crucial effect on translation of the USP18 isoforms. The expression constructs used in this report only contain the coding sequence of USP18 but still produce both isoforms similar to the pattern seen in endogenous USP18 (see Figs. 1B or 2B). We cannot rule out, though, that the present uORF might contribute to skipping of ATG1 to some degree.
Although common in viruses and bacteria alternative translation initiation codons are rarely found in mammalian cells. However, in recent years a growing number of non-AUG initiated translation of mammalian genes has been described (27). The general belief is that translation initiation at non-AUG start codons is mediated by “wobble” pairing of the methionine preloaded initiator tRNA with the non-AUG start codon (52). However, Schwab et al. reported that in case of CUG-initiated generation of cryptic peptides for immune surveillance the CUG codon can also be decoded with a leucine (28). In this report, we present evidence that the CUG codon at position 16 is the major translation initiation site for expression of full-length USP18 and that AUG36 is the translational start site of USP18-sf. The finding that the proposed primary translational start site AUG1 is not capable of initiating protein translation, as shown by transfection experiments with several USP18 mutants, was very surprising (Fig. 2B). This inability to commence translation is most likely due to a very weak Kozak consensus sequence at position AUG1 (Fig. 2A). Further analysis showed that CUG16 is the primary translational start site of USP18. Mutating CTG16 to GCG significantly reduced USP18 protein levels detected by Western blotting (Fig. 2C). However, this mutation did not completely eliminate expression of USP18. Approximately 25% of the full-length protein was still detected. This interesting result raises the possibility that even CUG8 might function as a translation initiation start site despite the weaker Kozak consensus. In addition, an ∼70% drop of USP18-sf levels in the ATG16 mutant showed that expression of the short isoform is mainly promoted by leaky scanning, which is a common mechanism of translational control in mammalian cells (16). For instance, the human c-MYC gene produces three different proteins from the same mRNA transcript via leaky scanning in a context dependent manner. Similar to USP18, translation of the full-length protein is initiated by a CUG codon which leads to expression of two isoforms downstream (53). Expression of these two N-terminal truncated c-Myc isoforms is completely eliminated when the CTG codon is mutated to ATG, which makes leaky scanning the exclusive regulating mechanism (54). In contrast, mutating CTG16 to ATG in USP18 did not completely eliminate expression of USP18-sf, which rules out leaky scanning as the sole molecular mechanism (Fig. 2C). This finding is further supported by the fact that anti-USP18 antibody detects USP18-sf in FLAG-USP18 transfectants despite translation initiation at the FLAG coding sequence, which precedes the USP18 sequence and shows perfect Kozak consensus (Fig. 1B). Together these findings strongly suggested an additional way of translational control such as IRES or cryptic promoter.
Cap-independent translation was first observed in RNA genomes of members of the picornavirus family (55). Although the canonical cap-dependent model of cap recognition and ribosomal scanning is the major translational mechanism in eukaryotes several cellular mRNAs have been reported to possess IRES mediated translation (56). For instance, under stress condition translation of IRF2 and NRF, both of which negatively regulate interferon signaling, was reported to be driven by an IRES element located in their 5′-UTR region (22, 23). Our detailed analysis of the potential USP18 IRES activity clearly ruled out existence of a cryptic promoter in the 5′ nucleotide sequence of USP18 (Fig. 3B), which left a functional IRES within the coding region as the most likely explanation. A number of independently performed experiments provided sufficient evidence to support the existence of a functional IRES in the 5′ region of the USP18 coding sequence (Fig. 3). However, transfection of dicistronic RNAs did not show significant IRES activity of the cloned USP18 sequence. It is well-known, though, that RNA transfections prevent cellular IRESs to associate with nuclear ITAFs, which may be required for activity of some IRESs (57, 58). Based on our results, it is highly likely that the USP18 IRES requires association with a nuclear ITAF to exhibit significant activity. Furthermore, our data suggest that the required nuclear ITAF must be expressed in a cell type-specific manner since not all cell lines tested showed an increase in USP18-sf/USP18 ratio upon inhibition of cap-dependent translation (Fig. 3E). Isoform-specific differences in protein stability or increased levels of phosphorylated eIF2α, which was reported to promote ribosomal bypass in suboptimal sequence context for translation initiation (59), are not likely to contribute to the observed increase in USP18-sf levels further supporting the existence of a functional IRES (supplemental Fig. S6). In addition, the observed IRES activity is not dependent on IFN because overexpression of FLAG-tagged USP18 or untagged USP18-ATG16 in the absence of IFN showed significant levels of USP18-sf (Figs. 1B and 2C). This non-canonical mechanism of translation initiation helps to sustain levels of USP18-sf when cap-dependent translation is reduced (Fig. 3E). Interestingly, the two cytokine inhibitory factors, IRF2 and the N terminal truncated SOCS3 isoform, show stable or even increased protein levels upon induction of ER stress (22, 60). This might be an indication that IRES-dependent translation is a common characteristic of some IFN regulatory factors to maintain required levels under stress conditions.
Functionally, we looked at all reported functions of USP18, including the enzyme activity and modulation of IFN signaling, but the conducted experiments did not suggest a significant difference between the two isoforms. However, the fact that USP18-sf is fully functional seems to be desirable when taking into account that this truncated isoform is supposed to compensate for reduced USP18 levels when cap-dependent translation is impaired. Interestingly, the localization difference between USP18 and USP18-sf does suggest a more specific role of USP18-sf in the nucleus. Indeed, we found that levels of ISGylated protein in the nucleus are reduced when USP18-sf is present compared with USP18 (Fig. 5C and supplemental Fig. S4C). So far, no nuclear-specific function of USP18 is known but ISGylation of the transcription factor interferon regulatory factor 3 (IRF3) prevents its proteasomal degradation and increases its activity (61, 62). Therefore, nuclear USP18-sf might promote proteasomal degradation of activated IRF3 by deconjugating ISG15. This enzymatic activity toward ISGylated IRF3 may represent another, though indirect, way of USP18-mediated inhibition of type I IFN signaling.
Because none of the two isoforms contain a nuclear localization sequence it is possible that some unknown factor with high binding affinity to USP18 but not to USP18-sf retains the full-length protein in the cytosol. This unknown protein, however, is not IFN-induced since the localization difference was clearly observed in untreated HeLa and MCF-7 cells by immunofluorescence (Fig. 5A and supplemental Fig. S5A).
Herein, we report that translation of human USP18 is not initiated by AUG1 as previously thought but by the rare start codon CUG at codon position 16. Furthermore, we show that two independent mechanisms control expression of the newly identified isoform USP18-sf providing additional diversity and flexibility to its regulation. By quantitative studies we determined that inefficient translation initiation at CUG16 is the main regulating mechanism contributing to about 70% of USP18-sf expression, whereas the second mechanism, IRES mediated expression, contributes to about 30%. To our knowledge USP18-sf is the first reported protein isoform, whose expression is controlled by both leaky scanning and IRES. In particular, the involvement of IRES driven expression and the resulting resistance to down-regulation of cap-dependent translation shows the functional benefit of USP18-sf. However, the need for sustained USP18-sf protein levels upon inhibition of cap-dependent translation seems to be cell type-specific, which is probably due to varying amounts of ITAFs. Functional-wise, our data suggest a more dominant role of USP18-sf in the nucleus, which opens the door to a new field of USP18-related research. Future work will focus on identification and characterization of nuclear-specific functions of USP18-sf and the effect of ISGylation on nuclear proteins.
Supplementary Material
Acknowledgments
We thank Dr. Peter Sarnow for providing discistronic reporter constructs, and Dr. Keith Wilkinson and Dr. Yvonne Hoang as well as members of Zhang laboratory for valuable discussions.
This work was supported, in whole or in part, by National Institutes of Health Grant HL091549 (to D.-E. Z.).

This article contains supplemental Figs. S1–S6.
- IFN
- interferon
- IRES
- internal ribosome entry site
- ISG
- interferon-stimulated gene
- UBP
- ubiquitin-specific processing protease
- eIF
- eukaryotic initiation factor
- uORF
- upstream open reading frame.
REFERENCES
- 1. Pestka S., Krause C. D., Walter M. R. (2004) Interferons, interferon-like cytokines, and their receptors. Immunol. Rev. 202, 8–32 [DOI] [PubMed] [Google Scholar]
- 2. Leaman D. W., Chawla-Sarkar M., Jacobs B., Vyas K., Sun Y., Ozdemir A., Yi T., Williams B. R., Borden E. C. (2003) Novel growth and death related interferon-stimulated genes (ISGs) in melanoma: greater potency of IFN-beta compared with IFN-α2. J. Interferon. Cytokine Res. 23, 745–756 [DOI] [PubMed] [Google Scholar]
- 3. Borden E. C., Williams B. R. (2011) Interferon-stimulated genes and their protein products: what and how? J. Interferon. Cytokine Res. 31, 1–4 [DOI] [PubMed] [Google Scholar]
- 4. Platanias L. C., Fish E. N. (1999) Signaling pathways activated by interferons. Exp. Hematol. 27, 1583–1592 [DOI] [PubMed] [Google Scholar]
- 5. Biron C. A. (2001) Interferons α and β as immune regulators–a new look. Immunity 14, 661–664 [DOI] [PubMed] [Google Scholar]
- 6. Chawla-Sarkar M., Lindner D. J., Liu Y. F., Williams B. R., Sen G. C., Silverman R. H., Borden E. C. (2003) Apoptosis and interferons: role of interferon-stimulated genes as mediators of apoptosis. Apoptosis 8, 237–249 [DOI] [PubMed] [Google Scholar]
- 7. Liu L. Q., Ilaria R., Jr., Kingsley P. D., Iwama A., van Etten R. A., Palis J., Zhang D. E. (1999) A novel ubiquitin-specific protease, UBP43, cloned from leukemia fusion protein AML1-ETO-expressing mice, functions in hematopoietic cell differentiation. Mol. Cell. Biol. 19, 3029–3038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Schwer H., Liu L. Q., Zhou L., Little M. T., Pan Z., Hetherington C. J., Zhang D. E. (2000) Cloning and characterization of a novel human ubiquitin-specific protease, a homologue of murine UBP43 (Usp18). Genomics 65, 44–52 [DOI] [PubMed] [Google Scholar]
- 9. Malakhov M. P., Malakhova O. A., Kim K. I., Ritchie K. J., Zhang D. E. (2002) UBP43 (USP18) specifically removes ISG15 from conjugated proteins. J. Biol. Chem. 277, 9976–9981 [DOI] [PubMed] [Google Scholar]
- 10. Kim K. I., Zhang D. E. (2005) UBP43, an ISG15-specific deconjugating enzyme: expression, purification, and enzymatic assays. Methods Enzymol. 398, 491–499 [DOI] [PubMed] [Google Scholar]
- 11. Ritchie K. J., Hahn C. S., Kim K. I., Yan M., Rosario D., Li L., de la Torre J. C., Zhang D. E. (2004) Role of ISG15 protease UBP43 (USP18) in innate immunity to viral infection. Nat. Med. 10, 1374–1378 [DOI] [PubMed] [Google Scholar]
- 12. Randall G., Chen L., Panis M., Fischer A. K., Lindenbach B. D., Sun J., Heathcote J., Rice C. M., Edwards A. M., McGilvray I. D. (2006) Silencing of USP18 potentiates the antiviral activity of interferon against hepatitis C virus infection. Gastroenterology 131, 1584–1591 [DOI] [PubMed] [Google Scholar]
- 13. Malakhova O. A., Kim K. I., Luo J. K., Zou W., Kumar K. G., Fuchs S. Y., Shuai K., Zhang D. E. (2006) UBP43 is a novel regulator of interferon signaling independent of its ISG15 isopeptidase activity. EMBO J. 25, 2358–2367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sarasin-Filipowicz M., Wang X., Yan M., Duong F. H., Poli V., Hilton D. J., Zhang D. E., Heim M. H. (2009) α interferon induces long-lasting refractoriness of JAK-STAT signaling in the mouse liver through induction of USP18/UBP43. Mol. Cell. Biol. 29, 4841–4851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kozak M. (1999) Initiation of translation in prokaryotes and eukaryotes. Gene 234, 187–208 [DOI] [PubMed] [Google Scholar]
- 16. Kozak M. (2002) Pushing the limits of the scanning mechanism for initiation of translation. Gene 299, 1–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Harding H. P., Novoa I., Zhang Y., Zeng H., Wek R., Schapira M., Ron D. (2000) Regulated translation initiation controls stress-induced gene expression in mammalian cells. Mol. Cell 6, 1099–1108 [DOI] [PubMed] [Google Scholar]
- 18. Dever T. E. (2002) Gene-specific regulation by general translation factors. Cell 108, 545–556 [DOI] [PubMed] [Google Scholar]
- 19. Sudhakar A., Ramachandran A., Ghosh S., Hasnain S. E., Kaufman R. J., Ramaiah K. V. (2000) Phosphorylation of serine 51 in initiation factor 2 α (eIF2α) promotes complex formation between eIF2 α(P) and eIF2B and causes inhibition in the guanine nucleotide exchange activity of eIF2B. Biochemistry 39, 12929–12938 [DOI] [PubMed] [Google Scholar]
- 20. Roth W., Wagenknecht B., Dichgans J., Weller M. (1998) Interferon-α enhances CD95L-induced apoptosis of human malignant glioma cells. J. Neuroimmunol. 87, 121–129 [DOI] [PubMed] [Google Scholar]
- 21. Marissen W. E., Lloyd R. E. (1998) Eukaryotic translation initiation factor 4G is targeted for proteolytic cleavage by caspase 3 during inhibition of translation in apoptotic cells. Mol. Cell. Biol. 18, 7565–7574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Dhar D., Roy S., Das S. (2007) Translational control of the interferon regulatory factor 2 mRNA by IRES element. Nucleic Acids Res. 35, 5409–5421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Oumard A., Hennecke M., Hauser H., Nourbakhsh M. (2000) Translation of NRF mRNA is mediated by highly efficient internal ribosome entry. Mol. Cell. Biol. 20, 2755–2759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kim K. I., Giannakopoulos N. V., Virgin H. W., Zhang D. E. (2004) Interferon-inducible ubiquitin E2, Ubc8, is a conjugating enzyme for protein ISGylation. Mol. Cell. Biol. 24, 9592–9600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Papa F. R., Hochstrasser M. (1993) The yeast DOA4 gene encodes a deubiquitinating enzyme related to a product of the human tre-2 oncogene. Nature 366, 313–319 [DOI] [PubMed] [Google Scholar]
- 26. Potu H., Sgorbissa A., Brancolini C. (2010) Identification of USP18 as an important regulator of the susceptibility to IFN-alpha and drug-induced apoptosis. Cancer Res. 70, 655–665 [DOI] [PubMed] [Google Scholar]
- 27. Wegrzyn J. L., Drudge T. M., Valafar F., Hook V. (2008) Bioinformatic analyses of mammalian 5′-UTR sequence properties of mRNAs predicts alternative translation initiation sites. BMC Bioinformatics 9, 232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Schwab S. R., Shugart J. A., Horng T., Malarkannan S., Shastri N. (2004) Unanticipated antigens: translation initiation at CUG with leucine. PLoS Biol. 2, e366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Gerashchenko M. V., Su D., Gladyshev V. N. (2010) CUG start codon generates thioredoxin/glutathione reductase isoforms in mouse testes. J. Biol. Chem. 285, 4595–4602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Baird S. D., Turcotte M., Korneluk R. G., Holcik M. (2006) Searching for IRES. RNA 12, 1755–1785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Pelletier J., Sonenberg N. (1985) Insertion mutagenesis to increase secondary structure within the 5′-noncoding region of a eukaryotic mRNA reduces translational efficiency. Cell 40, 515–526 [DOI] [PubMed] [Google Scholar]
- 32. Kozak M. (1986) Influences of mRNA secondary structure on initiation by eukaryotic ribosomes. Proc. Natl. Acad. Sci. U.S.A. 83, 2850–2854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Van Eden M. E., Byrd M. P., Sherrill K. W., Lloyd R. E. (2004) Demonstrating internal ribosome entry sites in eukaryotic mRNAs using stringent RNA test procedures. RNA 10, 720–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Holcik M., Graber T., Lewis S. M., Lefebvre C. A., Lacasse E., Baird S. (2005) Spurious splicing within the XIAP 5' UTR occurs in the Rluc/Fluc but not the betagal/CAT bicistronic reporter system. RNA 11, 1605–1609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Spriggs K. A., Bushell M., Mitchell S. A., Willis A. E. (2005) Internal ribosome entry segment-mediated translation during apoptosis: the role of IRES-trans-acting factors. Cell Death Differ 12, 585–591 [DOI] [PubMed] [Google Scholar]
- 36. Lewis S. M., Holcik M. (2008) For IRES trans-acting factors, it is all about location. Oncogene 27, 1033–1035 [DOI] [PubMed] [Google Scholar]
- 37. Semler B. L., Waterman M. L. (2008) IRES-mediated pathways to polysomes: nuclear versus cytoplasmic routes. Trends Microbiol. 16, 1–5 [DOI] [PubMed] [Google Scholar]
- 38. Rutkowski D. T., Kaufman R. J. (2004) A trip to the ER: coping with stress. Trends Cell Biol. 14, 20–28 [DOI] [PubMed] [Google Scholar]
- 39. Warnakulasuriyarachchi D., Cerquozzi S., Cheung H. H., Holcík M. (2004) Translational induction of the inhibitor of apoptosis protein HIAP2 during endoplasmic reticulum stress attenuates cell death and is mediated via an inducible internal ribosome entry site element. J. Biol. Chem. 279, 17148–17157 [DOI] [PubMed] [Google Scholar]
- 40. Gradi A., Svitkin Y. V., Imataka H., Sonenberg N. (1998) Proteolysis of human eukaryotic translation initiation factor eIF4GII, but not eIF4GI, coincides with the shutoff of host protein synthesis after poliovirus infection. Proc. Natl. Acad. Sci. U.S.A. 95, 11089–11094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Fernandez J., Yaman I., Sarnow P., Snider M. D., Hatzoglou M. (2002) Regulation of internal ribosomal entry site-mediated translation by phosphorylation of the translation initiation factor eIF2α. J. Biol. Chem. 277, 19198–19205 [DOI] [PubMed] [Google Scholar]
- 42. Robert F., Kapp L. D., Khan S. N., Acker M. G., Kolitz S., Kazemi S., Kaufman R. J., Merrick W. C., Koromilas A. E., Lorsch J. R., Pelletier J. (2006) Initiation of protein synthesis by hepatitis C virus is refractory to reduced eIF2.GTP.Met-tRNA(i)(Met) ternary complex availability. Mol. Biol. Cell 17, 4632–4644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kim J. H., Park S. M., Park J. H., Keum S. J., Jang S. K. (2011) eIF2A mediates translation of hepatitis C viral mRNA under stress conditions. EMBO J. 30, 2454–2464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Zhu Y., Lambert K., Corless C., Copeland N. G., Gilbert D. J., Jenkins N. A., D'Andrea A. D. (1997) DUB-2 is a member of a novel family of cytokine-inducible deubiquitinating enzymes. J. Biol. Chem. 272, 51–57 [DOI] [PubMed] [Google Scholar]
- 45. Duex J. E., Sorkin A. (2009) RNA interference screen identifies Usp18 as a regulator of epidermal growth factor receptor synthesis. Mol. Biol. Cell 20, 1833–1844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Bonnet M. E., Erbacher P., Bolcato-Bellemin A. L. (2008) Systemic delivery of DNA or siRNA mediated by linear polyethylenimine (L-PEI) does not induce an inflammatory response. Pharm. Res. 25, 2972–2982 [DOI] [PubMed] [Google Scholar]
- 47. Kim J. H., Luo J. K., Zhang D. E. (2008) The level of hepatitis B virus replication is not affected by protein ISG15 modification but is reduced by inhibition of UBP43 (USP18) expression. J. Immunol. 181, 6467–6472 [DOI] [PubMed] [Google Scholar]
- 48. Fitzgerald K. D., Semler B. L. (2009) Bridging IRES elements in mRNAs to the eukaryotic translation apparatus. Biochim. Biophys. Acta 1789, 518–528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Morris D. R., Geballe A. P. (2000) Upstream open reading frames as regulators of mRNA translation. Mol. Cell. Biol. 20, 8635–8642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Calkhoven C. F., Müller C., Leutz A. (2000) Translational control of C/EBPα and C/EBPβ isoform expression. Genes Dev. 14, 1920–1932 [PMC free article] [PubMed] [Google Scholar]
- 51. Vattem K. M., Wek R. C. (2004) Reinitiation involving upstream ORFs regulates ATF4 mRNA translation in mammalian cells. Proc. Natl. Acad. Sci. U.S.A. 101, 11269–11274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Peabody D. S. (1989) Translation initiation at non-AUG triplets in mammalian cells. J. Biol. Chem. 264, 5031–5035 [PubMed] [Google Scholar]
- 53. Hann S. R., Dixit M., Sears R. C., Sealy L. (1994) The alternatively initiated c-Myc proteins differentially regulate transcription through a noncanonical DNA-binding site. Genes Dev. 8, 2441–2452 [DOI] [PubMed] [Google Scholar]
- 54. Spotts G. D., Patel S. V., Xiao Q., Hann S. R. (1997) Identification of downstream-initiated c-Myc proteins which are dominant-negative inhibitors of transactivation by full-length c-Myc proteins. Mol. Cell. Biol. 17, 1459–1468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Jang S. K., Kräusslich H. G., Nicklin M. J., Duke G. M., Palmenberg A. C., Wimmer E. (1988) A segment of the 5' nontranslated region of encephalomyocarditis virus RNA directs internal entry of ribosomes during in vitro translation. J. Virol. 62, 2636–2643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Mokrejs M., Vopálensky V., Kolenaty O., Masek T., Feketová Z., Sekyrová P., Skaloudová B., Kríz V., Pospísek M. (2006) IRESite: the database of experimentally verified IRES structures (www.iresite.org). Nucleic Acids Res. 34, D125–D130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Stoneley M., Subkhankulova T., Le Quesne J. P., Coldwell M. J., Jopling C. L., Belsham G. J., Willis A. E. (2000) Analysis of the c-myc IRES; a potential role for cell-type specific trans-acting factors and the nuclear compartment. Nucleic Acids Res. 28, 687–694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Komar A. A., Hatzoglou M. (2011) Cellular IRES-mediated translation: the war of ITAFs in pathophysiological states. Cell Cycle 10, 229–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Palam L. R., Baird T. D., Wek R. C. (2011) Phosphorylation of eIF2 facilitates ribosomal bypass of an inhibitory upstream ORF to enhance CHOP translation. J. Biol. Chem. 286, 10939–10949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Sasaki A., Inagaki-Ohara K., Yoshida T., Yamanaka A., Sasaki M., Yasukawa H., Koromilas A. E., Yoshimura A. (2003) The N-terminal truncated isoform of SOCS3 translated from an alternative initiation AUG codon under stress conditions is stable due to the lack of a major ubiquitination site, Lys-6. J. Biol. Chem. 278, 2432–2436 [DOI] [PubMed] [Google Scholar]
- 61. Shi H. X., Yang K., Liu X., Liu X. Y., Wei B., Shan Y. F., Zhu L. H., Wang C. (2010) Positive regulation of interferon regulatory factor 3 activation by Herc5 via ISG15 modification. Mol. Cell. Biol. 30, 2424–2436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Lu G., Reinert J. T., Pitha-Rowe I., Okumura A., Kellum M., Knobeloch K. P., Hassel B., Pitha P. M. (2006) ISG15 enhances the innate antiviral response by inhibition of IRF-3 degradation. Cell Mol. Biol. 52, 29–41 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.