Background: Because interleukin-22 is pathogenic in autoimmune inflammation its regulation and blockage by immunomodulation is crucial.
Results: Interleukin-22 promoter activation in a T cell model is supported by CREB, NF-AT, and IKKα. Largely by action on NF-AT, cyclosporin impairs interleukin-22 expression.
Conclusion: Interleukin-22 is a direct cyclosporin target in T cells.
Significance: Observations made likely contribute to cyclosporin efficacy in autoimmunity such as psoriasis.
Keywords: Cytokines Induction, Gene Regulation, Immunosuppression, Inflammation, Interleukin, T Cell Biology, Cyclosporin A, Interleukin-22
Abstract
IL-22 is an immunoregulatory cytokine displaying pathological functions in models of autoimmunity like experimental psoriasis. Understanding molecular mechanisms driving IL-22, together with knowledge on the capacity of current immunosuppressive drugs to target this process, may open an avenue to novel therapeutic options. Here, we sought to characterize regulation of human IL22 gene expression with focus on the established model of Jurkat T cells. Moreover, effects of the prototypic immunosuppressant cyclosporin A (CsA) were investigated. We report that IL-22 induction by TPA/A23187 (T/A) or αCD3 is inhibited by CsA or related FK506. Similar data were obtained with peripheral blood mononuclear cells or purified CD3+ T cells. IL22 promoter analysis (−1074 to +156 bp) revealed a role of an NF-AT (−95/−91 nt) and a CREB (−194/−190 nt) binding site for gene induction. Indeed, binding of CREB and NF-ATc2, but not c-Rel, under the influence of T/A to those elements could be proven by ChIP. Because CsA has the capability to impair IκB kinase (IKK) complex activation, the IKKα/β inhibitor IKKVII was evaluated. IKKVII likewise reduced IL-22 induction in Jurkat cells and peripheral blood mononuclear cells. Interestingly, transfection of Jurkat cells with siRNA directed against IKKα impaired IL22 gene expression. Data presented suggest that NF-AT, CREB, and IKKα contribute to rapid IL22 gene induction. In particular the crucial role of NF-AT detected herein may form the basis of direct action of CsA on IL-22 expression by T cells, which may contribute to therapeutic efficacy of the drug in autoimmunity.
Introduction
IL-22 (1) has been identified as one key component of cytokine profiles associated with differentiation of crucial T cell lineages, namely Th1, Th17, and Th22 (2–6). In addition, there is increasing evidence that IL-22 is likewise a vital product of activated CD8+ T cells (7–9). IL-22 is derived from either T cells or innate sources, specifically natural killer cells, CD11+ dendritic cells, and heterogeneous lineages of innate lymphoid cell populations (10, 11) and apparently fulfills a fundamental function as messenger between leukocytic and non-leukocytic cell compartments. This observation is based on restricted cell type-specific expression of IL-22R1, which is predominantly detected on non-leukocytic cells, in particular cells of epithelial origin. Once bound to its heterodimeric receptor, namely IL-22R1/IL-10R2, IL-22 will engage signal transduction dominated by activation of STAT3 (3) with subsequent context-specific amplification of anti-apoptotic, tissue protective, anti-bacterial, immunoregulatory, or proinflammatory genes. B cell lymphoma-2 and B cell lymphoma-xl (12), mucin-1 (13), β-defensins (14) and lipocalin (15), suppressor of cytokine signaling-3 (16), as well as CXCL8 (17), CXCL5, matrix metalloproteinase-3 (18), and iNOS (19) shall be quoted herein exemplarily. Immunoregulatory properties of IL-22 are likewise strictly context specific and range from protective functions seen in models of infection/microbe-driven inflammation (15, 20), hepatitis (12), and ventilator-induced lung injury (16) to a clear pathogenic function seen in models of autoimmunity, specifically collagen-induced arthritis (21), and experimental psoriasis (22).
Cytokine networks that determine human T cell differentiation and associated production of IL-22 have attracted considerable attention in recent years. Whereas IL-12 and IFNγ pave the way toward Th1, IL-1, IL-6, and IL-23 are supposed to be critical for generating the human Th17 subset (23–25). Currently, circumstances favoring differentiation of skin-homing Th22 cells, a distinct CD4+ cell type producing IL-22 but not IL-17 or IFNγ (5, 7), are only insufficiently defined, although IL-6 and TNFα have been implicated in induction of this T cell species (5).
Whereas external signals supporting T cell-derived IL-22 have been characterized in quite detail, gene expression studies that investigate regulation of the human IL22 promoter are currently lacking. Here, we studied IL22 gene induction on a molecular level using the well established cell culture model of human Jurkat T cells. Data are complemented by experiments on peripheral blood mononuclear cells (PBMC)2 and isolated CD3+ T cells. Because control of IL-22 by pharmacological means is supposed to be a most relevant task that relates to pathogenesis and treatment of inflammatory/autoimmune diseases like psoriasis, a further focus of the current study lies on modulatory mechanisms implemented by the immunosuppressive calcineurin/nuclear factor of activated T cells (NF-AT) (26, 27) inhibitor cyclosporin A (CsA). T cells are obviously the major target of CsA action, although effects on diverse cell types, including keratinocytes, have been observed in past years (28–30). In fact, CsA is one major pillar of psoriasis therapy and successfully employed to control exacerbations in severe disease (31).
EXPERIMENTAL PROCEDURES
Reagents
Phorbol 12-myristate 13-acetate (T), A23187 (A), 6-formylindolo[3,2-b]carbazole (FICZ), and FK506 were purchased from Alexis/Enzo Life Sciences (Lörrach, Germany) or AppliChem (Darmstadt, Germany), respectively. CsA, U0126, and IκB kinase (IKK) inhibitor VII (IKKVII) were from Calbiochem-Novabiochem (Bad Soden, Germany). αCD3 was from eBioscience (Frankfurt, Germany). Cycloheximide (CHX), forskolin, and β-naphthoflavone (β-NF) were from Sigma.
Cultivation of Jurkat T Cells
Jurkat T cells were obtained from the American Type Culture Collection (Manassas, VA). For maintenance, Jurkat T cells were cultured in RPMI 1640 (Invitrogen) supplemented with 100 units/ml of penicillin, 100 μg/ml of streptomycin, and 10% heat-inactivated FCS (Invitrogen). For experiments, Jurkat T cells were seeded on 6-well polystyrene plates (Greiner) at a density of 3 × 106 cells/ml. All cell cultures in the current study were performed at 37 °C and 5% CO2.
Isolation and Cultivation of Human PBMC Obtained from Healthy Volunteers
For isolation of PBMC, blood was taken from healthy donors. This procedure and the respective consent documents were approved by the “Ethik Kommission” of the University Hospital Goethe-University Frankfurt (Geschäfts number 170/1998). Informed consent was obtained from volunteers. Healthy donors had abstained from taking drugs for 2 weeks prior to the study. PBMC were freshly isolated from peripheral blood using Histopaque®-1077 (Sigma) according to the manufacturer's instructions. For cultivation, PBMC were routinely resuspended in RPMI 1640 supplemented with 10 mm HEPES, 100 units/ml of penicillin, 100 μg/ml of streptomycin, and 1% human serum (Invitrogen) and seeded at 3 × 106 cells/ml in round-bottom polypropylene tubes (Greiner).
Isolation of CD3+ Cells from PBMC
CD3+ T cells were isolated by using the Pan-T-cell isolation kit II according to the manufacturer's instructions (Miltenyi, Bergisch Gladbach, Germany). Briefly, up to 8 × 107 PBMC were used per column and were counted again after completion of the isolation procedure. Cells were resuspended in the aforementioned PBMC medium and seeded at 3 × 106 cells/ml in round-bottom polypropylene tubes. To assess successful isolation, FACS analysis (FACS Canto, BD Biosciences) was performed with mouse monoclonal anti-human CD3-PerCP/Cy5.5 (Biozol, Eching, Germany). FACS data were analyzed by gating on lymphocytes. CD3+ T cell isolation resulted in a mean purity of 98.2 ± 0.46% (n = 14).
Cloning of the Human IL22 Promoter, Transient Transfection of Jurkat T Cells, and Luciferase Reporter Assays
Using genomic DNA isolated from human KG1 cells, we amplified 5′-flanking regions of the IL22 gene (NM_020525) using Pfu polymerase (Invitrogen). The following forward primers (excluding an additional flanking BglII cloning/restriction site) were used: Prom1 (1230 bp), forward 5′-CAATAGGTATTTGCATTTTGATAC-3′; Prom2 (557 bp), forward 5′-GATCACCTCCAATGAGATAAG-3′; Prom3 (457 bp), forward 5′-CTAAATCTGAACTCTACTAAGAC-3′; and Prom4 (299 bp), forward 5′-GTTTTGTGGGCTCCTGTG-3′. The reverse primer for all fragments (excluding an additional flanking HindIII cloning/restriction site) was: 5′-TGCAGACAATTCTAACTCGAG-3′. Each promoter fragment ends 5′ adjacent to the adenine nucleotide of the IL22 translational start site. Fragments were cloned into pGL3-Basic (Promega, Mannheim, Germany) and sequenced thereafter (Seqlab, Göttingen, Germany). Site-directed mutagenesis was performed using the QuikChange site-directed mutagenesis kit (Stratagene) to generate promoter fragments that show dysfunctional putative proximal NF-AT, CREB, or STAT5 binding sites. The following primers were used for that purpose: pGL3-NF-ATS1 (−242/−235 nt relative to IL22 transcriptional start site), forward, 5′-GAAAAATATGTAGGGTTTTTAAAATTTCTGGGATTTGTCTGTAAAATACC-3′; pGL3-NF-ATS2 (−183/−179 nt), forward, 5′-GGCTCTAATAGTGACGTTTTAGTTAAACACTTGCATCTCAAGG-3′; pGL3-NF-ATS3 (−161/−157 nt), forward, 5′-ACACTTGCATCTCAAGGTTTAAAGGATAGAGGTGGTGT-3′; pGL3-NF-ATS4 (−95/−91 nt), forward, 5′-GGTCGTTCTCAGAAGACAGTACTTTAAATTAGATAATTGCTGATGTC-3′; pGL3-CRE (−194/−190 nt), forward, 5′-CCCTCCGGGCTCTAATAGTTACATTTTAGGGAAACACTTGC-3′; pGL3-STAT5S1 (−266/−258 nt), forward 1, 5′-CTCTACTAAGACAAAACAATTGTGTTTTTTTTAAAATATGTAGGGTTTAG-3′, forward 2, 5′-CTCTACTAAGACAAAACAATTGTGAACTTTGAAAAATATGTAGGGTTTAG-3′; pGL3-STAT5-S2 (−113/−105 nt), forward 1, 5′-CCTGTGGTGGTTAGGTCGTTTTCATTAGACAGTACTGGAAATTAG-3′, forward 2, 5′-CCTGTGGTGGTTAGGTCGAACTCAGAAGACAGTACTGGAAATTAG-3′. The identity of the mutants was confirmed by sequencing (Seqlab). pGL3-Plasmids or pNFAT-Luc Reporter Plasmid (Agilent Technologies, Böblingen, Germany) were transiently transfected into Jurkat T cells using DMRIE-C reagent (Invitrogen). For each reaction, 4 μg of the indicated plasmids were transfected into 2.5 × 106 Jurkat T cells according to the manufacturer's instructions. 0.1 μg of pRL-TK (Promega) coding for Renilla luciferase were cotransfected. The transfection was stopped after 5 h by adding 2 ml of Jurkat culture medium (as mentioned above) supplemented with 5% heat-inactivated FCS. After 15 h of resting, cells were stimulated as described in the figure legends. Thereafter, cells were harvested and luciferase activity was determined using the dual reporter gene system (Promega) and an automated chemiluminescence detector (Berthold, Bad Wildbad, Germany).
Determination of Human IL22 Transcriptional Start Site by RNA Ligase-mediated Rapid Amplification of 5′-cDNA Ends (5′-RACE)
To identify the IL22 transcriptional start site in Jurkat T cells, the Gene Racer and TOPO TA Cloning kits (Invitrogen) were used according to the manufacturer's instructions. We identified a transcriptional start site that was almost identical to that previously published (see Ref. 32, NCBI accession number AJ277248.1) with only a 1-nt shift in the 3′ direction. Thus, the human IL22 transcriptional start site as detected herein is at nt 516 within the sequence given in AJ277248.1. All topographic information concerning the human IL22 gene locus refers to this transcriptional start site.
Detection of IL-22 mRNA by Standard and Real Time PCR
Total RNA was isolated and transcribed using TriReagent (Sigma), random hexameric primers, and Moloney virus reverse transcriptase (Applied Biosystems, Weiterstadt, Germany) according to the manufacturer's instructions. The following sequences were performed for standard PCR: 95 °C for 10 min (1 cycle); 95 °C for 1 min, 60 °C (GAPDH, IL-22) for 30 s, and 72 °C for 1 min (with the indicated numbers of cycles); final extension phase was at 72 °C for 7 min. Primer sequences, length of amplicons, and numbers of cycles were: GAPDH, forward 5′-ACCACAGTCCATGCCATCAC-3′ and reverse 5′-TCCACCACCCTGTTGCTGTA-3′, 452 bp, 25 cycles; IL-22, forward 5′-CACGGAGTCAGTATGAGTGAG-3′ and reverse 5′-CAAATGCAGGCATTTCTCAGAGA-3′, 299 bp, 34 cycles. Identity of amplicons was confirmed by sequencing (Seqlab). During real time PCR, changes in fluorescence were caused by the Taq polymerase degrading the probe that contains a fluorescent dye (FAM used for IL-22, VIC for GAPDH) and a quencher (TAMRA). For IL-22 (number Hs00220924_m1) and GAPDH (number 4310884E) pre-developed assay reagents were obtained (Applied Biosystems). The assay mixture used was from Thermo Scientific. Real time PCR was performed on AbiPrism 7500 Fast Sequence Detector (Applied Biosystems). One initial step at 95 °C for 5 min was followed by 40 cycles at 95 °C for 2 s and 60 °C for 25 s. Detection of the dequenched probe, calculation of threshold cycles (Ct values), and data analysis were performed by the Sequence Detector software. Relative changes in IL-22 mRNA expression compared with unstimulated control and normalized to GAPDH were quantified by the 2−ΔΔCt method.
Detection of c-Rel, IκBα, pCREB/pATF, IKKα, and IKKβ by Immunoblot Analysis
To detect c-Rel and activated cAMP responsive element-binding protein (pCREB)/activating transcription factor-1 (pATF-1) (in nuclear extracts) as well as IκBα, IKKα, and IKKβ (in total extracts), cell lysates were generated as previously described (33). To detect histone deacetylase-1 (HDAC1) or β-tubulin on the same blot, the blots were cut in two parts. Antibodies used were: c-Rel, rabbit polyclonal antibody; pCREB-Ser-133/pATF-1, rabbit polyclonal antibody (Cell Signaling, Frankfurt Germany); IKKα, mouse monoclonal antibody; IKKβ, mouse monoclonal antibody (BD Bioscience, Heidelberg, Germany); HDAC1, goat polyclonal antibody; IκBα, rabbit polyclonal antibody; β-tubulin, mouse monoclonal antibody (Santa Cruz Biotechnology).
Chromatin Immunoprecipitation (ChIP Analysis)
DNA/protein samples were generated using an “Enzymatic Shearing Kit” according to the manufacturer's instructions (Active Motif, Rixensart, Belgium). Briefly, cells were fixed (7 min, 1% formaldehyde), centrifuged (800 × g, RT, 5 min), and resuspended in glycine buffer (5 min). After a further centrifugation (800 × g, 5 min), cells were resuspended in PBS/PMSF (1 mm) followed by incubation in lysis buffer (supplemented with PMSF (1 mm) and proteinase inhibitor mixture). After 30 min on ice, douncing, and centrifugation (16,000 × g, 4 °C, 10 min), nuclei were resuspendend in digestion buffer. Genomic DNA was digested by adding enzymatic shearing mixture solution (200 units/ml). After addition of 20 μl of EDTA (0.5 m), 10 min on ice, and centrifugation (16,000 × g, 4 °C, 10 min), 1/10 (volume) of the digested genomic DNA was removed for control of shearing efficiency and PCR input. 9/10 (volume) were immunoprecipitated and analyzed by ChIP. For ChIP analysis of DNA binding of NF-ATc1, NF-ATc2 or c-Rel isolates were incubated for 12 h at 4 °C under slight rotation with either control antibody (mouse control IgG for NF-ATc1/NF-ATc2 (at 2 μg), rabbit control IgG for c-Rel (1:50 dilution)) or antibodies specific for NF-ATc1 (2 μg of mouse monoclonal antibody, Santa Cruz Biotechnology), NF-ATc2 (2 μg of mouse monoclonal antibody, Santa Cruz Biotechnology), or c-Rel (rabbit polyclonal antibody (1:50 dilution), Cell Signaling). For the pCREB-ChIP the same general protocol was used along with a pCREB/pATF-1 antibody (rabbit polyclonal antibody, Cell Signaling) or a corresponding IgG control (rabbit, Cell Signaling), both in a dilution of 1:50. Subsequently, the complexes were coupled to Protein G beads (Roche Diagnostics) at 4 °C for 1.5 h (under slight rotation). After washing in PBS/RIPA buffer (10 mm Tris-HCl, pH 8.0, 140 mm NaCl, 1% Triton X-100, 0.1% SDS), DNA-protein complexes were eluted from the beads twice using 200 μl of 1% SDS (in TE buffer) and 200 μl of 0.67% SDS (in TE buffer) at 65 °C (10 min). After RNase (4 h, 65 °C) and proteinase K (2 h, 42 °C) digestion, sheared genomic DNA was precipitated. To amplify the IL22 promoter fragment containing the NF-AT (located −95/−91 nt relative to the IL22 transcriptional start site) or the CREB binding site (located −194/−190 nt) under investigation, the following primers were used: NF-ATS4, forward, 5′-CGTTCTCAGAAGACAGTACTGG-3′; reverse, 5′-GGAAAGAGCTCACAGATTTCTGC-3′; CREB binding site: forward, 5′-GCAGAGGATATAGGACATGGG-3′; reverse, 5′-CAGGAGCCCACAAAACACC-3′; PCR conditions were 95 °C for 10 min (1 cycle); 95 °C for 30 s, 60 °C for 30 s, and 72 °C for 1 min (NF-ATS4: 38 cycles; CREB binding site: 40 cycles); and a final extension phase at 72 °C for 7 min. Identity of amplicons for the NF-AT (305 bp) and CREB sites (307 bp) were confirmed by sequencing (Seqlab).
Reduction of IKKα and IKKβ Expression by siRNA Technology
For experiments, 3.5 × 106 Jurkat cells were electroporated by Nucleofector Technology (Amaxa Cell Line Nucleofector Kit V, Lonza, Cologne, Germany) according to the manufacturer's instructions. The following three siRNA molecules were used: IKKα- and IKKβ-directed siRNA (both Santa Cruz Biotechnology) as well as control siRNA (Silencer Negative Control siRNA#1, Ambion, Cambridgeshire, UK). All conditions without siRNA or control siRNA were mock-transfected by electroporation under the same conditions. After electroporation cells were seeded in 6-well polystyrene plates (Greiner). After 48 h in culture medium, cells were stimulated with TPA/A23187 for 4 h and harvested thereafter. For each siRNA experiment performed, the reduction of IKKα/β expression was verified by immunoblot analysis. In all cases down-regulation of the siRNA target protein (IKKα/β) was observed.
Analysis of Cytokine Release by Enzyme-linked Immunosorbent Assay (ELISA)
Levels of IL-22 (R&D Systems) and IL-2 (eBioscience) in cell-free culture supernatants were determined by ELISA according to the manufacturer's instructions.
Cytotoxicity Assays
Viability of Jurkat T cells treated with TPA, A23187, forskolin, CsA, IKKVII, and CHX in comparison to unstimulated cells was monitored by using either the WST-1 assay (Roche Diagnostics GmbH) or trypan blue (Sigma) according to the manufacturer's instructions. In all assays, no significant cytotoxicity was observed within the incubation periods selected in the current study.
Statistics
Data are shown as mean ± S.E. (PBMC) or mean ± S.D. (Jurkat T cells) and are presented as pg/ml, ng/ml, fold-induction, or fold of control (% of αCD3 or % of T/A). Statistical analysis was performed on raw data either by unpaired Student's t test, paired Student's t test, or one-way ANOVA with post hoc Bonferroni correction (GraphPad 4.0) as indicated in the figure legends.
RESULTS
IL22 Gene Expression in Jurkat T Cells
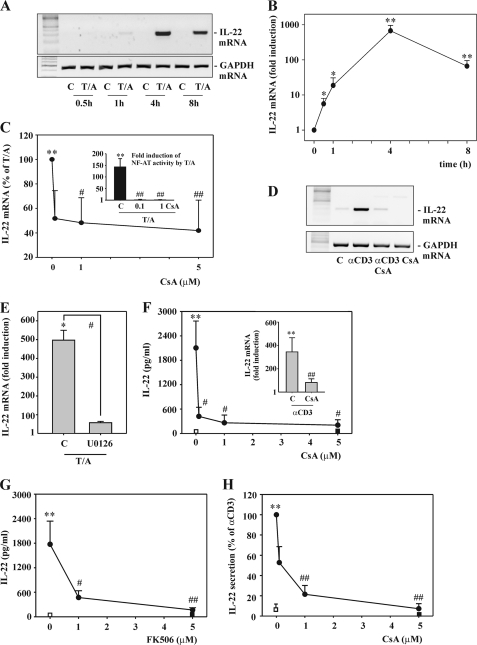
For molecular analysis of human IL-22 expression in T cells, we chose to use the well established model of CD3+ CD4+ CD8− Jurkat T cells. Those were activated by TPA in combination with the Ca2+ ionophore A23187 (T/A). This regime mediates strong PKC activation along with a robust Ca2+ signal and is regarded as a standard biochemical model of T cell receptor (TCR) activation (34, 35). As shown by standard (Fig. 1A) and real time PCR (Fig. 1B), fast induction of IL-22 mRNA was detectable in response to T/A that reached a maximum at a 4-h incubation period and declined thereafter. mRNA induction of IL-22 by T/A was inhibited under the influence of CsA (Fig. 1C). We confirm previous observations concerning modulation of NF-AT and IKK/NF-κB signaling by CsA in the context of the present experimental setting (34). In fact, CsA potently blocked degradation of IκBα, nuclear translocation of c-Rel (data not shown), as well as NF-AT-dependent gene expression as detected by luciferase reporter assays (Fig. 1C, inset). Significant inhibition of IL-22 expression was also obtained by coincubation with the related calcineurin inhibitor FK506 (data not shown). To use an alternative stimulatory condition, Jurkat T cells were exposed to a soluble antibody targeting the CD3 TCR complex (αCD3) (36). In agreement with T/A, exposure of Jurkat T cells to αCD3 mediated CsA-sensitive IL-22 induction (Fig. 1D). Signal transduction pathways activated in Jurkat T cells exposed to phorbol esters and Ca2+-mobilizing agents such as A23187 or ionomycin and being modulated by calcineurin inhibitors include in particular NF-AT and the IKK/canonical NF-κB axis (26, 27, 34, 35, 37). Both of these pivotal signaling pathways are amplified by activation of the MEK/ERK1/2 kinase pathway (38, 39). In fact, expression of IL-22 in response to T/A was suppressed by coincubation of cells with the MEK1/2 inhibitor U0126 (Fig. 1E).
FIGURE 1.
Rapid IL22 gene expression as detected in Jurkat and primary T cells under the influence of T/A or αCD3. A and B, Jurkat T cells were either kept as unstimulated control or stimulated with T (100 ng/ml)/A (10 μm). After the indicated time periods, IL-22 mRNA was assessed by standard (A) or real time PCR (B). A, one representative of three independently performed experiments is shown. B, IL-22 mRNA was normalized to that of GAPDH and is shown as mean fold-induction compared with unstimulated control (at the respective time point) ± S.D. (n = 3); *, p < 0.05, **, p < 0.01 compared with unstimulated control of the respective time point; raw data were analyzed by Student's t test. C, Jurkat T cells were either kept as unstimulated control or stimulated with T (100 ng/ml)/A (10 μm) for 4 h. Where specified, cells had been preincubated for 1 h with the indicated concentrations of CsA before T/A addition. After 5 h, IL-22 mRNA was determined by real time PCR analysis. IL-22 mRNA was normalized to that of GAPDH and is shown as percentage of induction by T/A alone ± S.D. (n = 11); **, p < 0.01 compared with unstimulated control; #, p < 0.05; ##, p < 0.01 compared with induction by T/A alone; raw data were analyzed by one-way ANOVA with post hoc Bonferroni correction. Inset, Jurkat T cells were transfected for 5 h with pNFAT-Luc together with Renilla luciferase as described under “Experimental Procedures.” After 15 h of rest, cells were either kept as unstimulated control or stimulated with T (100 ng/ml)/A (10 μm). Where specified, cells had been preincubated for 1 h with 0.1 or 1 μm CsA before T/A addition. After another 8 h, cells were harvested and luciferase reporter assays were performed. Means of luciferase activity are shown as fold-induction compared with unstimulated control ± S.D. (n = 3); **, p < 0.01 compared with unstimulated control; ##, p < 0.01 compared with T/A alone; raw data were analyzed by one-way ANOVA with post hoc Bonferroni correction. A–C, all cultures were adjusted to a final concentration of 0.1% DMSO (vehicle for T/A, CsA). D, Jurkat T cells were either kept as unstimulated control or stimulated with αCD3 (50 μg/ml) for 12 h. Where indicated, cells had been preincubated for 1 h with CsA (5 μm) before αCD3 addition. After 13 h, IL-22 mRNA was assessed by standard PCR analysis. One representative of three independently performed experiments is shown. All cultures were adjusted to a final concentration of 0.01% DMSO (vehicle for CsA). E, Jurkat T cells were either kept as unstimulated control or stimulated with T (100 ng/ml)/A (10 μm) for 4 h. Where indicated, cells had been preincubated for 1 h with U0126 (10 μm) before T/A addition. All cultures were adjusted to a final concentration of 0.3% DMSO (vehicle for T/A, U0126). After 5 h, IL-22 mRNA was assessed by real time PCR analysis. IL-22 mRNA was normalized to that of GAPDH and is shown as mean fold-induction compared with unstimulated control ± S.D. (n = 3); *, p < 0.05 compared with unstimulated control; #, p < 0.05 compared with T/A alone; raw data were analyzed by one-way ANOVA with post hoc Bonferroni correction. F, PBMC were either kept as unstimulated control or stimulated with αCD3 (7 μg/ml) for 24 h. Where specified, cells had been preincubated for 1 h with the indicated concentrations of CsA before αCD3 addition. All cultures were adjusted to a final concentration of 0.01% DMSO (vehicle for CsA). After 25 h, IL-22 release was determined by ELISA. Data are expressed as mean ± S.E. (n = 3); open square denotes unstimulated control, closed square denotes CsA (5 μm) alone; **, p < 0.01 compared with unstimulated control; #, p < 0.05 compared with αCD3 alone; raw data were analyzed by one-way ANOVA with post hoc Bonferroni correction. Inset, PBMC were either kept as unstimulated control or stimulated with αCD3 (7 μg/ml) for 24 h. Where indicated, cells had been preincubated for 1 h with CsA (5 μm) before αCD3 addition. All cultures were adjusted to a final concentration of 0.01% DMSO (vehicle for CsA). After 25 h, IL-22 mRNA was assessed by real time PCR. IL-22 mRNA was normalized to that of GAPDH and is shown as mean fold-induction compared with unstimulated control ± S.E. (n = 4); **, p < 0.01 compared with unstimulated control; ##, p < 0.01 compared with αCD3 alone; raw data were analyzed by one-way ANOVA with post hoc Bonferroni correction. G, PBMC were either kept as unstimulated control or stimulated with αCD3 (7 μg/ml) for 24 h. Where specified, cells had been preincubated for 1 h with the indicated concentrations of FK506 before αCD3 addition. All cultures were adjusted to a final concentration of 0.05% DMSO (vehicle for FK506). After 25 h, IL-22 release was determined by ELISA. Data are expressed as mean ± S.E. (n = 6); open square denotes unstimulated control, closed square denotes FK506 (5 μm) alone; **, p < 0.01 compared with unstimulated control; #, p < 0.05; ##, p < 0.01 compared with αCD3 alone; raw data were analyzed by one-way ANOVA with post hoc Bonferroni correction. H, primary CD3+ T cells were either kept as unstimulated control or stimulated with αCD3 (7 μg/ml) for 24 h. Where specified, cells were preincubated for 1 h with the indicated concentrations of CsA before αCD3 addition. All cultures were adjusted to a final concentration of 0.01% DMSO (vehicle for CsA). After 25 h, IL-22 release was determined by ELISA. Data are expressed as percent of αCD3 stimulation ± S.E. (n = 5–13); open square denotes unstimulated control, closed square denotes CsA (5 μm) alone; **, p < 0.01 compared with unstimulated control; ##, p < 0.01 compared with αCD3 alone. Raw data were analyzed by one-way ANOVA with post hoc Bonferroni correction.
Ligands of the AhR enhance production of IL-22 during human Th17 development (40). Subsequent studies also demonstrated induction of IL-22 by this pathway in the context of naive and memory CD4+ T cells as well as PBMC (6, 41). Accordingly, prototypic AhR ligands were tested in the present model of Jurkat T cells. For that purpose, cells were cultured/activated in the presence or absence of β-NF or FICZ. As shown in Table 1, experiments revealed that those AhR ligands did not up-regulate T/A-induced IL-22 mRNA induction in Jurkat T cells. AhR ligands used as the sole stimulus were likewise not capable of mediating induction of IL-22 compared with unstimulated control. Those present results shall not call into question that AhR ligands are crucial mediators of human T cell-derived IL-22. However, these data indicate that AhR ligands may not act as rapid and direct activators of IL-22 mRNA transcription. Notably, in aforementioned previous reports up-regulation of IL-22 production has been demonstrated after exposure of T cells to AhR ligands for days (6, 40, 41).
TABLE 1.
T (100 mg/ml)/A (10 μm)-induced IL-22 mRNA expression under the influence of AhR ligands
mRNA expression after a 4-h incubation period was determined by real time PCR. Data are expressed as mean ± S.D. (n = 4).
| Stimulation | IL-22 mRNA |
|---|---|
| Fold-induction compared to unstimulated control | |
| T/A | 219.7 ± 36.0a |
| T/A plus β-NF (1 μm) | 212.5 ± 53.2a |
| T/A plus β-NF (3 μm) | 220.5 ± 50.7a |
| T/A plus FICZ (0.3 μm) | 222.4 ± 28.5a |
| T/A plus FICZ (0.5 μm) | 184.4 ± 19.7a |
| β-NF (3 μm) | 1.4 ± 0.8 |
| FICZ (0.5 μm) | 1.3 ± 0.4 |
a p < 0.01 versus unstimulated control.
CsA Inhibits IL-22 Expression in PBMC and Primary T Cells
To extend observations related to CsA to primary cells, experiments with freshly isolated PBMC or primary CD3+ T cells were performed. We and others have recently reported on IL-22 production by PBMC under the influence of polyclonal T cell stimulation as achieved by phytohemagglutinin (42) or αCD3 (43). Heterogenous T cell subsets of the memory cell phenotype have been linked to release of IL-22 under those experimental conditions (43). Here, we report that the calcineurin inhibitors CsA (Fig. 1F) and FK506 (Fig. 1G) dramatically reduce IL-22 production by αCD3-activated PBMC. This was associated with significant reduction of IL-22 mRNA expression (Fig. 1F, inset). Similar results were likewise obtained using primary T cells (Fig. 1H).
An NF-AT Binding Site Close to the Transcriptional Start Site of the IL22 Promoter Contributes to Gene Induction in T/A-activated Jurkat T Cells
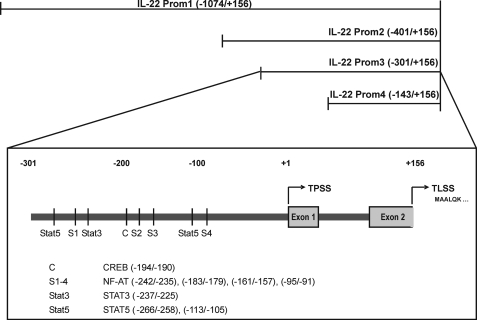
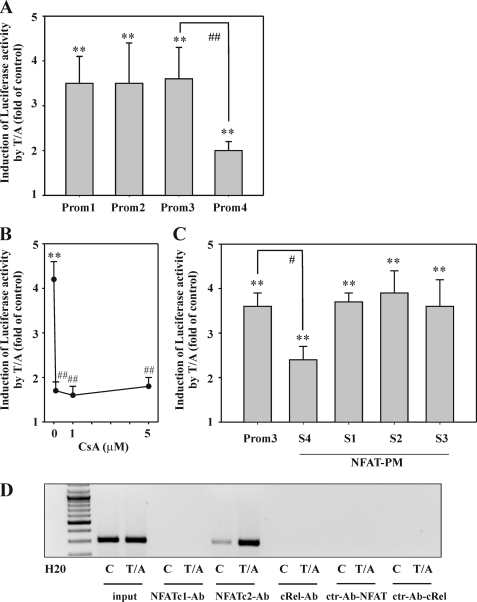
To further delineate molecular mechanisms driving IL-22 in Jurkat T cells, a human IL22 promoter fragment was cloned that spans from nt −1074 to +156 (immediately adjacent to the translational start site at +157) relative to the transcriptional start site. Based on this primary fragment (Prom1), deletion mutants (Prom2, Prom3, and Prom4) were generated (Fig. 2), and fragments were analyzed in luciferase reporter assays performed in the context of T/A-activated Jurkat T cells. No differences were detectable between fragments Prom1 to -3, whereas a significant reduction of luciferase activity was observed with the shortest fragment, Prom4 (Fig. 3A). Consequently, further studies focused on the Prom3 fragment. In accord with its capability to inhibit IL-22 mRNA and protein expression by T cells, Prom3-directed induction of luciferase activity was likewise suppressed by coincubation of cells with CsA (Fig. 3B). Sequence analysis actually disclosed the presence of four potential NF-AT binding sites in the Prom3 sequence. Those were denoted NF-ATS1–4 (Fig. 2). Mutational analysis of each site revealed that loss of NF-ATS1, NF-ATS2, or NF-ATS3 did not affect IL22 promoter activity in T/A-activated Jurkat T cells. In contrast, mutation of NF-ATS4 (−95/−91 nt) resulted in a significant 47.8% decrease of luciferase inducibility (Fig. 3C). In fact, enhanced binding of NF-ATc2, but not of NF-ATc1, to this specific promoter site was detected by ChIP (Fig. 3D). A conserved Rel homology domain is used by c-Rel and NF-AT proteins. Accordingly, both transcription factors have the potential to couple to identical DNA motifs at promoter sites (44, 45). Because c-Rel in Jurkat T cells is, along with NF-AT, being activated in response to T/A, we also tested for c-Rel binding to the NF-ATS4 binding site. However, we were unable to demonstrate binding of c-Rel to NF-ATS4 by ChIP (Fig. 3D).
FIGURE 2.
Schematic illustration of human IL22 promoter fragments used in the current study. The transcriptional start site (TPSS) was identified by 5′-RACE and is indicated as +1. Cloned promoter fragments and transcription factor binding sites investigated in the current study are indicated. TLSS denotes the IL22 translational start site.
FIGURE 3.
The role of NF-AT for IL-22 expression as detected in T/A-stimulated Jurkat T cells. A, Jurkat T cells were transfected for 5 h with the indicated pGL3-IL22 promoter constructs (Prom1–4) together with Renilla luciferase as described under “Experimental Procedures.” After 15 h of rest, cells were either kept as unstimulated control or stimulated with T (100 ng/ml)/A (10 μm). After another 8 h, cells were harvested and luciferase assays were performed. Means of luciferase activity are shown as fold-induction compared with unstimulated control (transfected with the same plasmid) ± S.D. (n = 5); **, p < 0.01 compared with unstimulated control (transfected with the same plasmid); ##, p < 0.01 compared with pGL3-Prom3 upon T/A stimulation; raw data were analyzed by one-way ANOVA with post hoc Bonferroni correction. B, Jurkat T cells were transfected for 5 h with pGL3-Prom3 together with Renilla luciferase as described under “Experimental Procedures.” After 15 h of rest, cells were either kept as unstimulated control or stimulated with T (100 ng/ml)/A (10 μm) for another 8 h. Where specified, cells were preincubated for 1 h with the indicated concentrations of CsA before T/A addition. Thereafter, cells were harvested and luciferase assays were performed. Means of luciferase activity are shown as fold-induction compared with unstimulated control ± S.D. (n = 4); **, p < 0.01 compared with unstimulated control; ##, p < 0.01 compared with T/A alone; raw data were analyzed by one-way ANOVA with post hoc Bonferroni correction. C, Jurkat T cells were transfected for 5 h with pGL3-Prom3 or pGL3-Prom3 displaying mutated NF-ATS1–4 (PM) together with Renilla luciferase as described under “Experimental Procedures.” After 15 h of rest, cells were either kept as unstimulated control or stimulated with T (100 ng/ml)/A (10 μm) for 8 h. Thereafter, cells were harvested and luciferase assays were performed. Means of luciferase activity are shown as fold-induction compared with unstimulated control (transfected with the same plasmid) ± S.D. (n = 3); **, p < 0.01 compared with unstimulated control (transfected with the same plasmid); #, p < 0.05 compared with unmutated pGL3-Prom3 stimulated with T/A; raw data were analyzed by one-way ANOVA with post hoc Bonferroni correction. D, Jurkat T cells were either kept as unstimulated control or stimulated with T (100 ng/ml)/A (10 μm). After 4 h, cells were harvested followed by ChIP analysis for detection of NF-ATc1, NF-ATc2, and c-Rel binding to NF-ATS4 as described under “Experimental Procedures.” One representative of three independently performed experiments is shown. All cultures were adjusted to a final concentration of 0.1% DMSO (vehicle for T/A, CsA).
A CREB Binding Site Contributes to Gene Induction in T/A-activated Jurkat T Cells
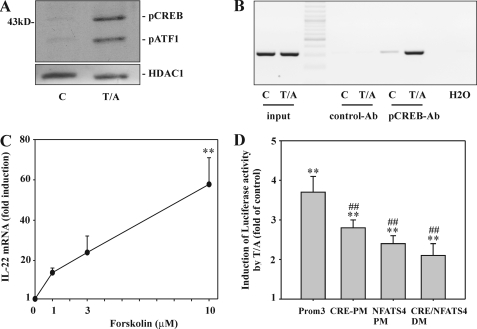
Thorough inspection of the Prom3 fragment brought to light a specific CRE site (−194/−190 nt) (Fig. 2) with an unusual imperfect nucleotide sequence (46). In fact, phosphorylated nuclear CREB became detectable in T/A-stimulated Jurkat T cells (Fig. 4A), an observation agreeing with activation of this transcription factor by PKC (46). T/A-mediated physical binding of CREB to this particular CRE in the IL22 promoter was confirmed by ChIP (Fig. 4B). Besides PKC, CREB activation is achieved by the cAMP/PKA axis. To further strengthen a proposed role of CREB for IL-22 induction, Jurkat T cells were incubated with forskolin, a cAMP elevating agent directly acting on the adenylate cyclase. Notably, forskolin as the sole stimulus was sufficient to mediate up-regulation of IL-22 mRNA, although induction was modest compared with that achieved by T/A (Fig. 4C). Mutational analysis of this specific CRE element (−194/−190 nt) displayed a significant 31.5% reduction of IL22 promoter activity in response to T/A. A Prom3 fragment containing a CRE/NF-ATS4 double mutation reduced IL22 promotor activity by 60.3%. However, this total reduction achieved by the double mutation actually came out relatively modest compared with both single mutations that particularly applied to NF-ATS4-PM (Fig. 4D). These results may be read as an indication that CREB and NF-ATc2 act in a coordinated or interrelated manner on the IL22 promoter. Promoter analysis presented herein altogether implies that those specific CRE and NF-ATS4 sites to a significant part mediate rapid induction of IL-22 in the experimental context of T/A-activated Jurkat T cells.
FIGURE 4.
The role of CREB for IL-22 expression as detected in T/A-stimulated Jurkat T cells. A, Jurkat T cells were either kept as unstimulated control or stimulated with T (100 ng/ml)/A (10 μm). After 4 h, nuclear pCREB/pATF1 and HDAC1 content was evaluated by Western blot analysis. One representative of five independently performed experiments is shown. B, Jurkat T cells were either kept as unstimulated control or stimulated with T (100 ng/ml)/A (10 μm). After 4 h, cells were harvested followed by ChIP analysis as described under “Experimental Procedures.” One representative of three independently performed experiments is shown. C, Jurkat T cells were either kept as unstimulated control or stimulated with the indicated concentrations of forskolin. After 4 h, IL-22 mRNA was assessed by real time PCR. IL-22 mRNA was normalized to that of GAPDH and is shown as mean fold-induction compared with unstimulated control ± S.D. (n = 5); **, p < 0.01 compared with unstimulated control; raw data were analyzed by one-way ANOVA with post hoc Bonferroni correction. D, Jurkat T cells were transfected for 5 h with either pGL3-Prom3 or pGL3-Prom3 displaying a mutated CRE (−194/−190 nt) (PM) or a mutated NF-ATS4 (−95/−91 nt) (PM) or with pGL3-Prom3 displaying a CRE/NF-ATS4 double mutation (DM) together with Renilla luciferase as described under “Experimental Procedures.” After 15 h of rest, cells were either kept as unstimulated control or stimulated with T (100 ng/ml)/A (10 μm) for another 8 h. Thereafter, cells were harvested and luciferase assays were performed. Means of luciferase activity are shown as fold-induction compared with unstimulated control (transfected with the same plasmid) ± S.D. (n = 3); **, p < 0.01 compared with unstimulated control (transfected with the same plasmid); ##, p < 0.01 compared with unmutated pGL3-Prom3 stimulated with T/A; raw data were analyzed by one-way ANOVA with post hoc Bonferroni correction. All cultures were adjusted to a final concentration of 0.1% DMSO (vehicle for T/A, forskolin).
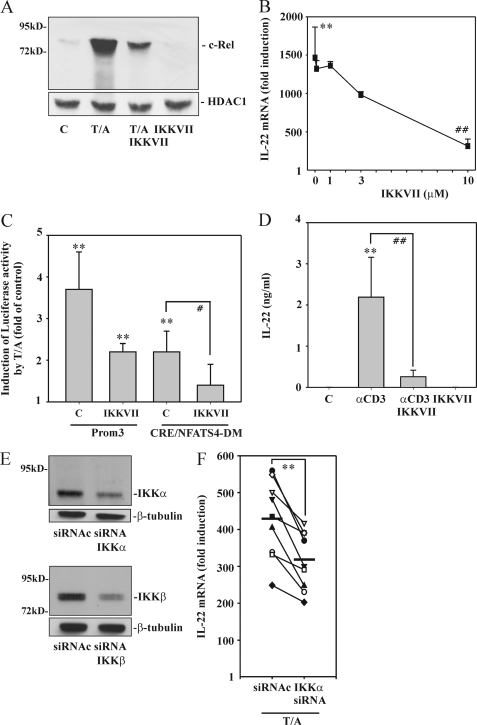
IKK Activation Contributes to IL-22 Induction
As already alluded to, IKK/NF-κB activation in T cells is impaired upon calcineurin blockage (34, 37). Therefore, besides using CsA, consequences of IKK suppression were investigated in Jurkat T cells by exposure to the IKKα/β inhibitor IKKVII. As expected, IKKVII efficiently reduced nuclear translocation of the prototypic NF-κB family member c-Rel (Fig. 5A). This was associated with reduction of T/A-induced IL-22 expression in a dose-dependent manner (Fig. 5B). Furthermore, coincubation with IKKVII likewise resulted in significant reduction of the IL22-Prom3 fragment inducibility by T/A (Fig. 5C). Interestingly, IKKVII also significantly inhibited promoter activation in the context of a Prom3 fragment bearing the CRE/NF-ATS4 double mutation by a further 69.6%. Notably, this double-mutated Prom3-CRE/NF-ATS4 fragment lacks obvious NF-κB binding sites. In total, a significant 79.2% reduction of T/A-induced IL22 promoter activity was observed in the context of the CRE/NF-ATS4 double mutation and blockage of IKK activity (Fig. 5C). IKKVII data obtained from T/A-activated Jurkat T cells did translate into the experimental setting of primary cells. Accordingly, Fig. 5D demonstrates that IL-22 secretion mediated by αCD3 is suppressed by incubation of PBMC with IKKVII. Similar data were obtained by using isolated primary CD3+ T cells (data not shown).
FIGURE 5.
The role of IKK for IL-22 expression as detected in T/A-stimulated Jurkat T cells. A, Jurkat T cells were either kept as unstimulated control or stimulated with T (100 ng/ml)/A (10 μm) for 4 h. Where indicated, cells were preincubated for 1 h with IKKVII (10 μm) before T/A addition. After 5 h, nuclear c-Rel and HDAC1 content was evaluated by Western blot analysis. One representative of three independently performed experiments is shown. B, Jurkat T cells were either kept as unstimulated control or stimulated with T (100 ng/ml)/A (10 μm) for 4 h. Where specified, cells were preincubated for 1 h with the indicated concentrations of IKKVII before T/A addition. After 5 h, IL-22 mRNA was assessed by real time PCR. IL-22 mRNA was normalized to that of GAPDH and is shown as mean fold-induction compared with unstimulated control ± S.D. (n = 5); **, p < 0.01 compared with unstimulated control; ##, p < 0.01 compared with T/A alone; raw data were analyzed by one-way ANOVA with post hoc Bonferroni correction. C, Jurkat T cells were transfected for 5 h with either pGL3-Prom3 or pGL3-Prom3 displaying the aforementioned CRE/NF-ATS4 double mutation (DM) together with Renilla luciferase as described under “Experimental Procedures.” After 15 h of rest, cells were either kept as unstimulated control or stimulated with T (100 ng/ml)/A (10 μm) for another 8 h. Where indicated, cells were preincubated for 1 h with IKKVII (5 μm) before T/A addition. Thereafter, cells were harvested and luciferase assays were performed. Means of luciferase activity are shown as fold-induction compared with unstimulated control ± S.D. (n = 5); **, p < 0.01 compared with unstimulated control (transfected with the same plasmid); #, p < 0.05 compared with T/A-activated cells (without IKKVII and transfected with the same plasmid); raw data were analyzed by one-way ANOVA with post hoc Bonferroni correction. A–C, all cultures were adjusted to a final concentration of 0.2% DMSO (vehicle for T/A, IKKVII). D, PBMC were either kept as unstimulated control or stimulated with αCD3 (7 μg/ml) for 24 h. Where indicated, cells were preincubated for 1 h with IKKVII (5 μm) before αCD3 addition. All cultures were adjusted to a final concentration of 0.05% DMSO (vehicle for IKKVII). After 25 h, IL-22 release was determined by ELISA. Data are expressed as mean ± S.E. (n = 6); **, p < 0.01 compared with unstimulated control; ##, p < 0.01 compared with stimulation with αCD3 alone; raw data were analyzed by one-way ANOVA with post hoc Bonferroni correction. E and F, silencing of IKKα/β in T/A-activated (4 h) Jurkat cells was performed as described under “Experimental Procedures.” E, after the transfection/stimulation period, expression of IKKα/β under the influence of control siRNA (siRNAc) or siRNA-IKKα (upper panel) or siRNA-IKKβ (lower panel) was verified by immunoblot analysis. One representative experiment for either IKKα or IKKβ silencing is shown (n = 9 for each IKK). F, expression of IL-22 mRNA under the influence of siRNA-IKKα is shown. mRNA expression was assessed by real time PCR in the same set of experiments. IL-22 mRNA was normalized to that of GAPDH and is displayed as absolute values of fold-induction (n = 9); **, p < 0.01 for siRNAc compared with siRNA-IKKα; raw data were analyzed by paired Student's t test. E and F, all cultures were adjusted to a final concentration of 0.1% DMSO (vehicle for T/A).
To further verify the connection between IKKVII and activation of IKK kinases in the context of IL-22 expression, IKKα and IKKβ were down-regulated in Jurkat T cells using siRNA technology. Notably, both kinases are activated in T cells upon stimulation (35). Fig. 5E demonstrates that silencing of both kinases was achieved although significant residual IKKα/β expression endured in all experiments performed. Notably, despite residual expression, reduction of IKKα by siRNA was associated with reduction of T/A-induced IL-22 mRNA expression in each experiment performed (Fig. 5F). In contrast, silencing of IKKβ failed to mediate a significant effect on IL-22 inducibility (418.2 ± 101.4-fold induction versus 394.6 ± 113.6-fold induction for transfection with control siRNA (siRNAc) versus transfection with IKKβ-siRNA). However, interpretation of IKKβ-siRNA data must take into account that residual IKKβ expression/activity may enhance IL-22 in an indirect manner via the canonical NF-κB pathway. In fact, IκB degradation was still detectable in cells transfected with IKKβ-siRNA (data not shown).
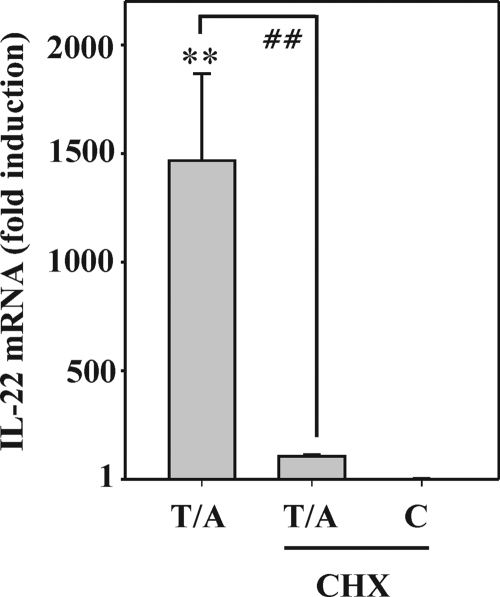
To investigate whether induction of a presently unknown gene product indirectly promotes IL-22 expression, Jurkat T cells were activated by T/A in the presence or absence of cycloheximide. This hypothesis is actually supported by the observation of almost complete suppression of T/A-induced IL-22 expression upon blockage of translation by coincubation with cycloheximide (Fig. 6).
FIGURE 6.
Suppression of IL-22 mRNA expression upon blockage of translation. Jurkat T cells were either kept as unstimulated control or stimulated with T (100 ng/ml)/A (10 μm) for 3 h. Where indicated, cells were preincubated for 1 h with CHX (10 μg/ml) before T/A addition. All cultures were adjusted to a final concentration of 0.1% DMSO (vehicle for T/A, CHX). After 4 h, IL-22 mRNA was determined by real time PCR analysis. IL-22 mRNA was normalized to that of GAPDH and is shown as mean fold-induction compared with unstimulated control ± S.D. (n = 6); **, p < 0.01 compared with unstimulated control; ##, p < 0.01 compared with T/A alone; raw data were analyzed by one-way ANOVA with post hoc Bonferroni correction.
Accordingly, studies were initiated to identify candidate factors that may support IL22 promoter activation and whose expression is induced by T/A as well as sensitive to IKK inhibition. One obvious candidate was IL-2. In fact, two potential STAT5 binding sites are present in the IL22-Prom3 fragment (at −266/−258 and −113/−105 nt). Production of IL-2 was detectable by T (100 ng/ml)/A (10 μm)-activated Jurkat T cells within 4 h. Coincubation with IKKVII efficiently suppressed this response (1555 ± 945.3 versus 249.8 ± 121.1 pg/ml for T/A activation without and with IKKVII at 5 μm, n = 3). However, we also confirm previous observations (47) that Jurkat T cells lack responsiveness to IL-2 (data not shown). In addition, mutational analysis revealed that both STAT5 binding sites do not contribute to T/A-mediated IL22 gene activation as detected in Jurkat T cells in the context of transfected Prom3 promoter fragments (data not shown). Another cytokine efficiently expressed by T/A-activated Jurkat T cells is LIF (48). Notably, a STAT3 binding site is present in the IL22-Prom3 fragment at −237/−225 nt. Despite robust STAT3 activation in Jurkat T cells in response to the cytokine, LIF did not affect expression of IL-22 by Jurkat T cells, either alone or in combination with T/A (data not shown).
DISCUSSION
By focusing on the well established model of Jurkat T cells, we set out to characterize molecular mechanisms directing instant induction of the human IL22 gene upon polyclonal stimulation. Promoter and mRNA expression studies identified a specific NF-AT binding site (−95/−91 nt) and a CRE site (−194/−190 nt), coupling to NF-ATc2 and pCREB, which along with IKK activity turned out to be essential for rapid and the most efficient onset of IL-22 induction. In fact, mutation of those two specific transcription factor binding sites in combination with pharmacological suppression of IKK activity resulted in 79.2% reduction of T/A-induced IL22 promoter activity. Coordinated activation of p65 (data not shown), c-Rel, NF-ATc2, and CREB as detected herein has regained considerable interest after recent characterization of a c-Rel-enhanceosome that apparently determines Foxp3 expression and thus development of regulatory T cells (45). Whether formation of a similar regulatory complex also applies to the IL22 locus is currently uncertain. As already pointed out, physical binding of c-Rel to NF-ATS4 was not observed in the current study.
The present observation of CREB being a significant determinant of IL-22 expression agrees with the view that this transcription factor promotes diverse aspects of T cell activation and proliferation, among others, production of IL-2 and IFNγ as well as Th1-directed immunity in general (46). In this context, it is noteworthy that IL-22 has been identified as part of the Th1 cytokine response early on (3). Subsequent to TCR activation, CREB is supposed to be activated foremost by PKC (46). In the current study, forskolin was used as a pharmacological tool to alternatively activate CREB via cAMP/PKA. Although cAMP is generally regarded as an efficient T cell deactivating messenger (49), it should be mentioned at this point that vasoactive intestinal peptide, apparently via cAMP, enhances release of IL-22 by murine Th17 cells (50).
Diminished IL-22 induction by T/A was observed herein under the influence of the IKKα/β inhibitor IKKVII. In fact, both IKK kinases are being activated in stimulated T cells (35). Using the siRNA approach we sought to discriminate potential functions of IKKα and IKKβ, respectively. Notably, reduction of IKKβ failed to influence IL-22 expression in T/A-activated Jurkat T cells. However, residual activation of the canonical NF-κB pathway in IKKβ-siRNA-treated cells unfortunately persisted and may have indirectly promoted IL-22 expression by immediate induction of regulatory factors. In the present study, we investigated two such candidates with the potential of mediating CsA-sensitive indirect NF-κB effects, namely IL-2 and LIF. Particularly the LIF/STAT3 axis appears interesting because hyper-IgE patients displaying a missense STAT3 mutation lack IL-22 production by activated CD4+ T cells (51). However, IL-2 and LIF failed to affect IL-22 expression in Jurkat T cells.
In contrast, we specifically demonstrate herein that modulating IKKα by siRNA significantly decreases IL-22 expression in activated Jurkat T cells. Notably, T/A is not capable to efficiently activate the IKKα/non-canonical NF-κB axis in Jurkat T cells (52). Because blockage of IKKα moreover fails to impair biological activity of the canonical NF-κB pathway in this cell type (35), data imply that IKKα promotes IL-22 expression by a mechanism independent on NF-κB. Such a mechanism is unlikely related to NF-AT and CREB because IKKVII inhibited the IL22 promoter in the context of mutated NF-AT and CREB binding sites. Recently, IKKα was shown to promote IL-17 production by histone H3 phosphorylation (53). However, epigenetic principles are not supposed to determine promoter activation as detected by luciferase reporter plasmids. Experiments are currently being designed to further clarify in detail the basis for up-regulation of IL-22 by IKKα. Notably, whereas CsA certainly interferes with initiation of the active heterotrimeric IKK complex (37), IKKα phosphorylation and activation per se should not be affected by the drug. Although molecular details of IKKα activation are still cloudy it is noteworthy that MEKK3 is supposed to be located upstream of IKKs (54). A recent report in fact documents the pivotal role of MEKK3 for TCR-driven T cell activation and subsequent engagement of ERK1/2 as well as cytokine production (55). The remarkable potency of the MEK1/2 inhibitor U0126 detected herein to inhibit IL-22 expression is certainly of note in this context. Besides the proposed link to IKKα, MAK kinase signaling is crucial for NF-AT function. Notably, inhibition of MEK1 suppresses T/ionomycin-mediated activation of Jurkat T cells as detected by NF-AT function and subsequent IL-2 expression (38, 56). Likewise, overexpression of the MEK1 downstream kinase ERK1 associates with up-regulated NF-AT activity in Jurkat T cells (39).
Altogether data presented herein indicate that NF-AT, MAP kinase signaling, CREB, and IKKα activity determine rapid gene induction of IL-22 as detected in Jurkat cells. The crucial role of NF-AT and MAP kinase signaling shows striking similarity to the regulation of IL-17 in Jurkat T cells. Also for IL-17 those two signaling principles, NF-AT and MAP kinases, display a pivotal role (57). This correspondence of regulatory mechanisms may actually reflect partly overlapping functions of IL-17 and IL-22 in host defense and inflammation (11, 58, 59).
Psoriasis is an autoimmune, T cell-driven, inflammatory disease of the skin that is characterized by disturbances of keratinocyte terminal differentiation (60, 61). As already alluded to, IL-22 has been identified as a key pathogenic molecule in experimental psoriasis (22). In fact, IL-22 is up-regulated in psoriasis patients' sera (62) and highly expressed in lesional skin tissue (63). Biological functions possibly underlying disease promotion by IL-22 include modulation of keratinocyte differentiation and induction of proinflammatory cytokines, chemokines, and matrix metalloproteinases, among others IL-20, CXCL5, and matrix metalloproteinases-1/3 (18, 61, 62, 64, 65). Altogether, basic research and so far available clinical data suggest IL-22 as a prime target for neutralizing therapy in psoriasis patients. As IL-22 is not involved in leukocyte activation, such immunomodulatory intervention may not be associated with overt immunosuppression.
Exacerbations in hard-to-treat psoriasis patients are currently an indication for CsA therapy (31). Analysis of cutaneous cytokine expression profiles in such patients identified IL-22, besides IL-1β and IL-23 (p19/p40), as being down-regulated most efficiently after 14 days of oral CsA therapy. IL-17 likewise displayed down-regulation, albeit with a somewhat delayed kinetic (66). Whether suppression of IL-22 in vivo is secondary to a general CsA-mediated T cell deactivation and immunosuppression or possibly a direct effect of the drug on the IL22 promoter level remained open. In the current study, we provide evidence for direct action of CsA on the IL22 locus. CsA, like the related drug FK506, impaired expression of IL-22 as detected in activated Jurkat T cells and in the setting of primary cells, specifically PBMC or isolated CD3+ T cells. We demonstrate that by impairing NF-AT, CsA has the capability to directly suppress IL22 promoter activation in T cells. Thus, data presented herein may form the basis for effective modulation of IL-22 that is observed in psoriasis patients undergoing CsA therapy (66).
This work was supported by Deutsche Forschungsgemeinschaft (DFG) Grant GRK1172, Biologicals.
- PBMC
- peripheral blood mononuclear cells
- A
- A23187
- AhR
- aryl hydrocarbon receptor
- CRE
- cAMP response element
- CREB
- cAMP response element-binding protein
- CsA
- cyclosporin A
- HDAC1
- histone deacetylase-1
- IKK
- IκB kinase
- LIF
- leukemia inhibitory factor
- NF-AT
- nuclear factor of activated T cells
- T
- phorbol 12-myristate 13-acetate
- TCR
- T cell receptor
- ATF1
- activating transcription factor-1
- FICZ
- 6-formylindolo[3,2-b]carbazole
- β-NF
- β-naphthoflavone
- CHX
- cycloheximide
- nt
- nucleotide(s)
- ANOVA
- analysis of variance
- DMSO
- dimethyl sulfoxide.
REFERENCES
- 1. Dumoutier L., Van Roost E., Colau D., Renauld J. C. (2000) Human interleukin-10-related T cell-derived inducible factor. Molecular cloning and functional characterization as an hepatocyte-stimulating factor. Proc. Natl. Acad. Sci. U.S.A. 97, 10144–10149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Steinman L. (2007) A brief history of T(H)17, the first major revision in the T(H)1/T(H)2 hypothesis of T cell-mediated tissue damage. Nat. Med. 13, 139–145 [DOI] [PubMed] [Google Scholar]
- 3. Wolk K., Witte E., Witte K., Warszawska K., Sabat R. (2010) Biology of interleukin-22. Semin. Immunopathol. 32, 17–31 [DOI] [PubMed] [Google Scholar]
- 4. Blaschitz C., Raffatellu M. (2010) Th17 cytokines and the gut mucosal barrier. J. Clin. Immunol. 30, 196–203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Duhen T., Geiger R., Jarrossay D., Lanzavecchia A., Sallusto F. (2009) Production of interleukin 22 but not interleukin 17 by a subset of human skin-homing memory T cells. Nat. Immunol. 10, 857–863 [DOI] [PubMed] [Google Scholar]
- 6. Trifari S., Kaplan C. D., Tran E. H., Crellin N. K., Spits H. (2009) Identification of a human helper T cell population that has abundant production of interleukin 22 and is distinct from T(H)-17, T(H)1, and T(H)2 cells. Nat. Immunol. 10, 864–871 [DOI] [PubMed] [Google Scholar]
- 7. Nograles K. E., Zaba L. C., Shemer A., Fuentes-Duculan J., Cardinale I., Kikuchi T., Ramon M., Bergman R., Krueger J. G., Guttman-Yassky E. (2009) IL-22-producing “T22” T cells account for up-regulated IL-22 in atopic dermatitis despite reduced IL-17-producing TH17 T cells. J. Allergy Clin. Immunol. 123, 1244–1252.e2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bachmann M., Horn K., Rudloff I., Goren I., Holdener M., Christen U., Darsow N., Hunfeld K. P., Koehl U., Kind P., Pfeilschifter J., Kraiczy P., Mühl H. (2010) Early production of IL-22 but not IL-17 by peripheral blood mononuclear cells exposed to live Borrelia burgdorferi. The role of monocytes and interleukin-1. PLoS Pathog. 6, e1001144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Res P. C., Piskin G., de Boer O. J., van der Loos C. M., Teeling P., Bos J. D., Teunissen M. B. (2010) Overrepresentation of IL-17A and IL-22 producing CD8 T cells in lesional skin suggests their involvement in the pathogenesis of psoriasis. PLoS One 5, e14108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Pickert G., Neufert C., Leppkes M., Zheng Y., Wittkopf N., Warntjen M., Lehr H. A., Hirth S., Weigmann B., Wirtz S., Ouyang W., Neurath M. F., Becker C. (2009) STAT3 links IL-22 signaling in intestinal epithelial cells to mucosal wound healing. J. Exp. Med. 206, 1465–1472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Sonnenberg G. F., Fouser L. A., Artis D. (2011) Border patrol. Regulation of immunity, inflammation, and tissue homeostasis at barrier surfaces by IL-22. Nat. Immunol. 12, 383–390 [DOI] [PubMed] [Google Scholar]
- 12. Radaeva S., Sun R., Pan H. N., Hong F., Gao B. (2004) Interleukin 22 (IL-22) plays a protective role in T cell-mediated murine hepatitis. IL-22 is a survival factor for hepatocytes via STAT3 activation. Hepatology 39, 1332–1342 [DOI] [PubMed] [Google Scholar]
- 13. Sugimoto K., Ogawa A., Mizoguchi E., Shimomura Y., Andoh A., Bhan A. K., Blumberg R. S., Xavier R. J., Mizoguchi A. (2008) IL-22 ameliorates intestinal inflammation in a mouse model of ulcerative colitis. J. Clin. Invest. 118, 534–544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wolk K., Kunz S., Witte E., Friedrich M., Asadullah K., Sabat R. (2004) IL-22 increases the innate immunity of tissues. Immunity 21, 241–254 [DOI] [PubMed] [Google Scholar]
- 15. Aujla S. J., Chan Y. R., Zheng M., Fei M., Askew D. J., Pociask D. A., Reinhart T. A., McAllister F., Edeal J., Gaus K., Husain S., Kreindler J. L., Dubin P. J., Pilewski J. M., Myerburg M. M., Mason C. A., Iwakura Y., Kolls J. K. (2008) IL-22 mediates mucosal host defense against Gram-negative bacterial pneumonia. Nat. Med. 14, 275–281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hoegl S., Bachmann M., Scheiermann P., Goren I., Hofstetter C., Pfeilschifter J., Zwissler B., Muhl H. (2011) Protective properties of inhaled IL-22 in a model of ventilator-induced lung injury. Am. J. Respir. Cell Mol. Biol. 44, 369–376 [DOI] [PubMed] [Google Scholar]
- 17. Brand S., Beigel F., Olszak T., Zitzmann K., Eichhorst S. T., Otte J. M., Diepolder H., Marquardt A., Jagla W., Popp A., Leclair S., Herrmann K., Seiderer J., Ochsenkühn T., Göke B., Auernhammer C. J., Dambacher J. (2006) IL-22 is increased in active Crohn disease and promotes proinflammatory gene expression and intestinal epithelial cell migration. Am. J. Physiol. Gastrointest. Liver Physiol. 290, G827–838 [DOI] [PubMed] [Google Scholar]
- 18. Boniface K., Bernard F. X., Garcia M., Gurney A. L., Lecron J. C., Morel F. (2005) IL-22 inhibits epidermal differentiation and induces proinflammatory gene expression and migration of human keratinocytes. J. Immunol. 174, 3695–3702 [DOI] [PubMed] [Google Scholar]
- 19. Ziesché E., Bachmann M., Kleinert H., Pfeilschifter J., Mühl H. (2007) The interleukin-22/STAT3 pathway potentiates expression of inducible nitric-oxide synthase in human colon carcinoma cells. J. Biol. Chem. 282, 16006–16015 [DOI] [PubMed] [Google Scholar]
- 20. Zheng Y., Valdez P. A., Danilenko D. M., Hu Y., Sa S. M., Gong Q., Abbas A. R., Modrusan Z., Ghilardi N., de Sauvage F. J., Ouyang W. (2008) Interleukin-22 mediates early host defense against attaching and effacing bacterial pathogens. Nat. Med. 14, 282–289 [DOI] [PubMed] [Google Scholar]
- 21. Geboes L., Dumoutier L., Kelchtermans H., Schurgers E., Mitera T., Renauld J. C., Matthys P. (2009) Proinflammatory role of the Th17 cytokine interleukin-22 in collagen-induced arthritis in C57BL/6 mice. Arthritis Rheum. 60, 390–395 [DOI] [PubMed] [Google Scholar]
- 22. Ma H. L., Liang S., Li J., Napierata L., Brown T., Benoit S., Senices M., Gill D., Dunussi-Joannopoulos K., Collins M., Nickerson-Nutter C., Fouser L. A., Young D. A. (2008) IL-22 is required for Th17 cell-mediated pathology in a mouse model of psoriasis-like skin inflammation. J. Clin. Invest. 118, 597–607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Miossec P., Korn T., Kuchroo V. K. (2009) Interleukin-17 and type 17 helper T cells. N. Engl. J. Med. 361, 888–898 [DOI] [PubMed] [Google Scholar]
- 24. Louten J., Boniface K., de Waal Malefyt R. (2009) Development and function of TH17 cells in health and disease. J. Allergy Clin. Immunol. 123, 1004–1011 [DOI] [PubMed] [Google Scholar]
- 25. O'Connor W., Jr., Zenewicz L. A., Flavell R. A. (2010) The dual nature of T(H)17 cells. Shifting the focus to function. Nat. Immunol. 11, 471–476 [DOI] [PubMed] [Google Scholar]
- 26. Macian F. (2005) NFAT proteins. Key regulators of T-cell development and function. Nat. Rev. Immunol. 5, 472–484 [DOI] [PubMed] [Google Scholar]
- 27. Müller M. R., Rao A. (2010) NFAT, immunity and cancer. A transcription factor comes of age. Nat. Rev. Immunol. 10, 645–656 [DOI] [PubMed] [Google Scholar]
- 28. Al-Daraji W. I., Grant K. R., Ryan K., Saxton A., Reynolds N. J. (2002) Localization of calcineurin/NFAT in human skin and psoriasis and inhibition of calcineurin/NFAT activation in human keratinocytes by cyclosporin A. J. Invest. Dermatol. 118, 779–788 [DOI] [PubMed] [Google Scholar]
- 29. Mühl H., Kunz D., Rob P., Pfeilschifter J. (1993) Cyclosporin derivatives inhibit interleukin 1β induction of nitric-oxide synthase in renal mesangial cells. Eur. J. Pharmacol. 249, 95–100 [DOI] [PubMed] [Google Scholar]
- 30. Hämäläinen M., Korhonen R., Moilanen E. (2009) Calcineurin inhibitors down-regulate iNOS expression by destabilizing mRNA. Int. Immunopharmacol. 9, 159–167 [DOI] [PubMed] [Google Scholar]
- 31. Maza A., Montaudié H., Sbidian E., Gallini A., Aractingi S., Aubin F., Bachelez H., Cribier B., Joly P., Jullien D., Le Maître M., Misery L., Richard M. A., Ortonne J. P., Paul C. (2011) J. Eur. Acad. Dermatol. Venereol. 25, Suppl. 2, 19–27 [DOI] [PubMed] [Google Scholar]
- 32. Dumoutier L., Van Roost E., Ameye G., Michaux L., Renauld J. C. (2000) IL-TIF/IL-22. Genomic organization and mapping of the human and mouse genes. Genes Immun. 1, 488–494 [DOI] [PubMed] [Google Scholar]
- 33. Sadik C. D., Bachmann M., Pfeilschifter J., Mühl H. (2009) Nucleic Acids Res. 37, 5041–5056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Frantz B., Nordby E. C., Bren G., Steffan N., Paya C. V., Kincaid R. L., Tocci M. J., O'Keefe S. J., O'Neill E. A. (1994) Calcineurin acts in synergy with PMA to inactivate IκB/MAD3, an inhibitor of NF-κB. EMBO J. 13, 861–870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Trushin S. A., Pennington K. N., Algeciras-Schimnich A., Paya C. V. (1999) Protein kinase C and calcineurin synergize to activate IκB kinase and NF-κB in T lymphocytes. J. Biol. Chem. 274, 22923–22931 [DOI] [PubMed] [Google Scholar]
- 36. Ledbetter J. A., Gentry L. E., June C. H., Rabinovitch P. S., Purchio A. F. (1987) Mol. Cell. Biol. 7, 650–656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Palkowitsch L., Marienfeld U., Brunner C., Eitelhuber A., Krappmann D., Marienfeld R. B. (2011) The Ca2+-dependent phosphatase calcineurin controls the formation of the Carma1-Bcl10-Malt1 complex during T cell receptor-induced NF-κB activation. J. Biol. Chem. 286, 7522–7534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Whitehurst C. E., Geppert T. D. (1996) MEK1 and the extracellular signal-regulated kinases are required for the stimulation of IL-2 gene transcription in T cells. J. Immunol. 156, 1020–1029 [PubMed] [Google Scholar]
- 39. Park J. H., Levitt L. (1993) Overexpression of mitogen-activated protein kinase (ERK1) enhances T-cell cytokine gene expression. Role of AP1, NF-AT, and NF-KB. Blood 82, 2470–2477 [PubMed] [Google Scholar]
- 40. Veldhoen M., Hirota K., Westendorf A. M., Buer J., Dumoutier L., Renauld J. C., Stockinger B. (2008) The aryl hydrocarbon receptor links TH17 cell-mediated autoimmunity to environmental toxins. Nature 453, 106–109 [DOI] [PubMed] [Google Scholar]
- 41. Ramirez J. M., Brembilla N. C., Sorg O., Chicheportiche R., Matthes T., Dayer J. M., Saurat J. H., Roosnek E., Chizzolini C. (2010) Activation of the aryl hydrocarbon receptor reveals distinct requirements for IL-22 and IL-17 production by human T helper cells. Eur. J. Immunol. 40, 2450–2459 [DOI] [PubMed] [Google Scholar]
- 42. Ziesché E. (2009) Untersuchungen zur Bildung und Funktion von IL-21 bei entzündungsbedingter Immunaktivierung. Ph.D. Dissertation, Goethe-University Frankfurt, Frankfurt, Germany [Google Scholar]
- 43. Liu Y., Yang B., Zhou M., Li L., Zhou H., Zhang J., Chen H., Wu C. (2009) Memory IL-22-producing CD4+ T cells specific for Candida albicans are present in humans. Eur. J. Immunol. 39, 1472–1479 [DOI] [PubMed] [Google Scholar]
- 44. Rao A., Luo C., Hogan P. G. (1997) Transcription factors of the NFAT family. Regulation and function. Annu. Rev. Immunol. 15, 707–747 [DOI] [PubMed] [Google Scholar]
- 45. Ruan Q., Kameswaran V., Tone Y., Li L., Liou H. C., Greene M. I., Tone M., Chen Y. H. (2009) Development of Foxp3(+) regulatory T cells is driven by the c-Rel enhanceosome. Immunity 31, 932–940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Wen A. Y., Sakamoto K. M., Miller L. S. (2010) The role of the transcription factor CREB in immune function. J. Immunol. 185, 6413–6419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Granelli-Piperno A., Nolan P. (1991) Nuclear transcription factors that bind to elements of the IL-2 promoter. Induction requirements in primary human T cells. J. Immunol. 147, 2734–2739 [PubMed] [Google Scholar]
- 48. Bamberger A. M., Erdmann I., Bamberger C. M., Jenatschke S. S., Schulte H. M. (1997) Transcriptional regulation of the human “leukemia inhibitory factor” gene. Modulation by glucocorticoids and estradiol. Mol. Cell. Endocrinol. 127, 71–79 [DOI] [PubMed] [Google Scholar]
- 49. Mosenden R., Taskén K. (2011) Cyclic AMP-mediated immune regulation. Overview of mechanisms of action in T cells. Cell Signal. 23, 1009–1016 [DOI] [PubMed] [Google Scholar]
- 50. Yadav M., Rosenbaum J., Goetzl E. J. (2008) Cutting edge. Vasoactive intestinal peptide (VIP) induces differentiation of Th17 cells with a distinctive cytokine profile. J. Immunol. 180, 2772–2776 [DOI] [PubMed] [Google Scholar]
- 51. Ma C. S., Chew G. Y., Simpson N., Priyadarshi A., Wong M., Grimbacher B., Fulcher D. A., Tangye S. G., Cook M. C. (2008) Deficiency of Th17 cells in hyper IgE syndrome due to mutations in STAT3. J. Exp. Med. 205, 1551–1557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Xiao G., Cvijic M. E., Fong A., Harhaj E. W., Uhlik M. T., Waterfield M., Sun S. C. (2001) Retroviral oncoprotein Tax induces processing of NF-κB2/p100 in T cells. Evidence for the involvement of IKKα. EMBO J. 20, 6805–6815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Li L., Ruan Q., Hilliard B., Devirgiliis J., Karin M., Chen Y. H. (2011) Transcriptional regulation of the Th17 immune response by IKK(α). J. Exp. Med. 208, 787–796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Israël A. (2010) The IKK complex. A central regulator of NF-κB activation. Cold Spring Harb. Perspect. Biol. 2, a000158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Wang X., Zhang F., Chen F., Liu D., Zheng Y., Zhang Y., Dong C., Su B. (2011) MEKK3 regulates IFN-γ production in T cells through the Rac1/2-dependent MAPK cascades. J. Immunol. 186, 5791–5800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Kempiak S. J., Hiura T. S., Nel A. E. (1999) The Jun kinase cascade is responsible for activating the CD28 response element of the IL-2 promoter. Proof of cross-talk with the IκB kinase cascade. J. Immunol. 162, 3176–3187 [PubMed] [Google Scholar]
- 57. Liu X. K., Lin X., Gaffen S. L. (2004) Crucial role for nuclear factor of activated T cells in T cell receptor-mediated regulation of human interleukin-17. J. Biol. Chem. 279, 52762–52771 [DOI] [PubMed] [Google Scholar]
- 58. Mühl H., Bachmann M., Pfeilschifter J. (2011) Inducible NO synthase and antibacterial host defence in times of Th17/Th22/T22 immunity. Cell Microbiol. 13, 340–348 [DOI] [PubMed] [Google Scholar]
- 59. Gaffen S. L. (2008) An overview of IL-17 function and signaling. Cytokine 43, 402–407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Schön M. P., Boehncke W. H. (2005) Psoriasis. N. Engl. J. Med. 352, 1899–1912 [DOI] [PubMed] [Google Scholar]
- 61. Sabat R., Wolk K. (2011) Research in practice: IL-22 and IL-20: significance for epithelial homeostasis and psoriasis pathogenesis. J. Dtsch. Dermatol. Ges. 9, 518–523 [DOI] [PubMed] [Google Scholar]
- 62. Wolk K., Witte E., Wallace E., Döcke W. D., Kunz S., Asadullah K., Volk H. D., Sterry W., Sabat R. (2006) IL-22 regulates the expression of genes responsible for antimicrobial defense, cellular differentiation, and mobility in keratinocytes. A potential role in psoriasis. Eur. J. Immunol. 36, 1309–1323 [DOI] [PubMed] [Google Scholar]
- 63. Boniface K., Guignouard E., Pedretti N., Garcia M., Delwail A., Bernard F. X., Nau F., Guillet G., Dagregorio G., Yssel H., Lecron J. C., Morel F. (2007) A role for T cell-derived interleukin 22 in psoriatic skin inflammation. Clin. Exp. Immunol. 150, 407–415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Nograles K. E., Zaba L. C., Guttman-Yassky E., Fuentes-Duculan J., Suárez-Fariñas M., Cardinale I., Khatcherian A., Gonzalez J., Pierson K. C., White T. R., Pensabene C., Coats I., Novitskaya I., Lowes M. A., Krueger J. G. (2008) Th17 cytokines interleukin (IL)-17 and IL-22 modulate distinct inflammatory and keratinocyte-response pathways. Br. J. Dermatol. 159, 1092–1102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Wolk K., Witte E., Warszawska K., Schulze-Tanzil G., Witte K., Philipp S., Kunz S., Döcke W. D., Asadullah K., Volk H. D., Sterry W., Sabat R. (2009) The Th17 cytokine IL-22 induces IL-20 production in keratinocytes. A novel immunological cascade with potential relevance in psoriasis. Eur. J. Immunol. 39, 3570–3581 [DOI] [PubMed] [Google Scholar]
- 66. Haider A. S., Lowes M. A., Suárez-Fariñas M., Zaba L. C., Cardinale I., Khatcherian A., Novitskaya I., Wittkowski K. M., Krueger J. G. (2008) Identification of cellular pathways of “type 1,” Th17 T cells, and TNF and inducible nitric-oxide synthase-producing dendritic cells in autoimmune inflammation through pharmacogenomic study of cyclosporine A in psoriasis. J. Immunol. 180, 1913–1920 [DOI] [PubMed] [Google Scholar]