Abstract
Cys is much different from other common amino acids in proteins. Being one of the least abundant residues, Cys is often observed in functional sites in proteins. This residue is reactive, polarizable, and redox-active; has high affinity for metals; and is particularly responsive to the local environment. A better understanding of the basic properties of Cys is essential for interpretation of high-throughput data sets and for prediction and classification of functional Cys residues. We provide an overview of approaches used to study Cys residues, from methods for investigation of their basic properties, such as exposure and pKa, to algorithms for functional prediction of different types of Cys in proteins.
Keywords: Computational Biology, Cysteine-mediated Cross-linking, Disulfide, Protein Structure, Redox Regulation, Selenocysteine, Thioredoxin
Introduction
Among the 20 common amino acids in proteins, Cys is often an outlier credited with several unique features. It is one of the least abundant residues (often the least abundant) in organisms, yet it is frequently observed in functionally important sites of proteins, where it serves catalytic, regulatory, structure-stabilizing, cofactor-binding, and other functions. Although Cys is thought to be a later addition to the genetic code (1), it has also been shown to accumulate more than any other amino acid in present day organisms (2), implying that the usage of Cys may further expand in their descendents. The functional importance of Cys in biology is also highlighted by the observation that, in humans, Cys mutations lead to genetic diseases more often than expected on the basis of its abundance (3). With respect to aging, several studies showed that Cys content in mitochondrially encoded proteins inversely correlates with life span in animals (4). Although the biological interpretation of this relationship is debated (5–7), it highlights the fact that Cys residues in proteins appear to be under strict evolutionary control. This feature has to be associated with several unique biological functions of Cys, as detected by genome-wide analyses of its tendency to form functional clusters, such as structural disulfides and metal-binding sites (8, 9).
Another unique property of Cys is its ability to functionally interchange with Sec. Sec, known as the 21st amino acid in the genetic code, is a selenium-containing amino acid that differs from Cys by a single atom (i.e. selenium versus sulfur). Sec is the only natural amino acid thought to be located exclusively in catalytic sites (although a possible exception to this rule has been reported (10)), and its function can be partially preserved when Cys, but not any other amino acid, replaces Sec. This functional interplay is unique in the protein world; for example, the relationship between pyrrolysine (the 22nd amino acid) and lysine is not functional (11). A recent study made the relation between Cys and Sec even more intriguing. It has been found that Cys can be inserted in proteins in place of Sec (12). The unexpected aspect is that incorporation of Cys occurs specifically through the Sec insertion machinery: Cys is synthesized by Sec synthase directly on Sec-tRNA (from serine and thiophosphate) and inserted at UGA codons.
What are the physicochemical features of Cys that make this residue such a unique case? The main feature appears to be the high reactivity and chemical plasticity of its sulfur-based functional group. First of all, Cys thiols are capable of unique reactivity in the protein world: covalent interactions with other thiols create intra- and intermolecular disulfide bonds. In addition, Cys can coordinate a variety of metals and metalloids: along with His, Cys is the most frequently employed residue in metal-binding sites of proteins. The side chain of Cys can also directly react with many oxidants or oxidized cellular products under physiological and pathophysiological conditions: reversible oxidation of Cys thiols is known to play a role in redox regulation of proteins via the formation of sulfenic acid intermediates (13–15), mixed disulfides with glutathione (16), and overoxidation to sulfinic acids (17). Last, but certainly not least, Cys is the main target of nitrosative stress, leading to the formation of reversible S-nitrosothiols (18). The susceptibility of Cys to these modifications is largely dependent on the reactivity of each specific thiol. Thus, understanding Cys properties is not only very important per se but is also critical to understanding the nature and function of thiol-mediated redox processes in the cell.
For this reason, we decided to structure this minireview in two main parts. First, we provide an overview of the current understanding of the general properties and physicochemical features of Cys residues as well as theoretical approaches used to study them. Second, we shift our attention to various functional types of reactive Cys in proteins. Following a discussion of the biological relevance of Cys reactivity and function, we introduce and review the bioinformatics tools currently available to study various types of reactive redox thiols.
Properties of Cys Residues in Proteins: General Comments on Thiol Reactivity
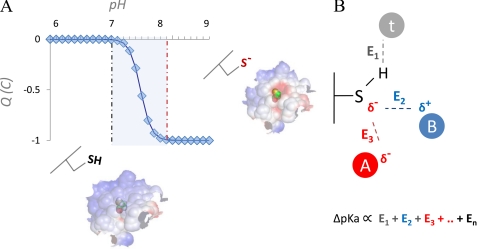
The general properties of Cys, from either physicochemical or biological points of view, are difficult to categorize. A deeply buried Cys may behave as a hydrophobic residue (due to the hydrophobic nature of amino acid packing inside the protein), whereas an exposed Cys (i.e. accessible to the solvent either on molecular surfaces or in the solvated polar microenvironment found in protein pockets and cavities) may interact with H-bond partners (e.g. water molecules) and other titratable groups of polar residues, which are abundant on protein surfaces. These interactions may considerably polarize exposed Cys, influencing its pKa (e.g. decrease it). Moreover, exposed Cys residues have been estimated to have, on average and in respect to all other titratable amino acids, the closest pKa to the physiological pH (9). The latter observation implies that, even for small variations within the physiological range of local pH values, exposed Cys residues may (i) easily switch their ability to function as nucleophiles and (ii) experience sudden charge shifts and significant electrostatic changes, which can extend to the proximal portions of the molecular surface (Fig. 1A). In some circumstances, similar electrostatic changes can affect the ability of a protein to interact with the environment, for instance, with other proteins and charged molecules. This observation highlights the intrinsically high responsiveness of exposed Cys to changes in physiological states and environmental conditions, an aptitude that may provide a biological (more so than chemical or physical) explanation for why Cys residues are found much less frequently (than expected) on molecular surfaces. Unless employed for a specific function, exposed Cys residues tend to be removed from protein surfaces (9). All of these considerations highlight two particularly important descriptors of Cys reactivity: exposure to solvent and the protonation status of its functional group.
FIGURE 1.
Effect of pH perturbation on net charge of exposed Cys and electrostatic properties of molecular surface. Exposed Cys residues are significantly more polar than buried Cys residues, a consequence of the high degree of Cys polarizability. For many exposed and polar Cys residues, pKa is close to the physiological pH (A, blue, shaded). In such cases, for a typical monoprotic acid in a physiological solution (assuming Henderson-Hasselbalch behavior), sudden negative charge switches can occur in the response to even very limited local pH shifts (A, neutral Cys for lower pH values and anionic Cys for higher pH values; note the steep transition between the two Cys forms in the shaded area). For any Cys in the protein, its interactions with other titratable groups and solvent determine the degree of reactivity of that Cys and influence its pKa. From a computational perspective, a common way to quantify the pKa of a residue is to calculate its deviation (ΔpKa) from the reference pKa value (pKa(REF)) for that amino acid type; ΔpKa is derived by properly accounting for all interactions with the titratable functional group, e.g. E1, E2, … En in B, where interactions with the generic titratable residues A (red circle; indicating an interacting acidic residue), B (blue circle, indicating an interacting basic residue), and t (gray circle; indicating a generic non-charged titratable residue, e.g. Thr) are shown. The ΔpKa then allows the pKa of a residue to be expressed as pKa = pKa(REF) + ΔpKa.
Computation of amino acid exposure requires structural information. A common approach for estimation of protein exposure is to use a molecular probe (usually, a sphere whose radius is variable, e.g. 1.4 Å to mimic the dimensions of the water molecule) that is rolled over the protein body. Proteins are usually treated as rigid bodies: the probe cannot penetrate the surface and just touches its residues. Commonly used (and free for download) programs include Naccess (version 2.1.1) and Surface Racer. The standalone versions of these programs are available, making them useful tools for automated large-scale analysis.
With regard to pKa, no definitive protocols for its prediction for Cys residues have been established as of yet. The acid dissociation constant (pKa) of different Cys residues greatly correlates with the reactivity of these residues: thiolates are considerably better nucleophiles than their protonated counterparts. Additionally, thiolates generally react more rapidly with oxidants, such as H2O2, than thiols (19), although variations can occur depending on the protein environment (20). Consequently, reliable prediction of Cys pKa, especially for Cys residues that undergo reversible redox conversions under physiological conditions, would be extremely valuable in the area of Cys biology.
Different approaches have been used to study reactive Cys. One method makes use of density functional theory (DFT)2 calculations to calculate pKa through natural population analysis charge on Cys sulfur atoms (21). The method, when benchmarked against different thiol oxidoreductases (Table 1), proved to work well (linear correlation between theoretical and experimental values, R2 = 0.96). To date, a limitation of quantum mechanics (QM) approaches resides in their speed. Given the intrinsic complexity of the analysis, even a small protein cannot be analyzed in full, and the use of a reduced protein model is necessary.
TABLE 1.
Comparison of pKa prediction methods
PDB codes are reported for each protein analyzed. If a PDB structure contains more than one protein, the name of the protein analyzed is specified in the Cys residue column. Experimental pKa values are from the cited literature. QM pKa refers to the DFT-based approach developed by Roos et al. (21). PROPKA refers to the program maintained by Jensen and co-workers (23). Trx, thioredoxin.
| PDB code (Ref.) | Cys residue | Experimental pKa | QM pKa | PROPKA | Genea |
|---|---|---|---|---|---|
| 1xob (69) | Cys32 | 7.1 | 6.5 | 6.6 | trxAb |
| 2ppt (70) | Cys73 | 5.2 | 4.8 | 5.7 | trxCb |
| 1su9 (71) | Cys76 | 8.2 | 8.1 | 10 | resAb |
| 2o89 (21) | Cys29 | 6.4 | 6.5 | 4 | trxCb |
| 1ljl (21) | Cys89 | 9.5 | 10 | 9.2 | arsC |
| 1ljl (21) | Cys10 | 6.8 | 6.9 | 6.8 | arsC |
| 2ipa (30) | Cys82 (ArsC) | 6.3 | 6 | trxA,barsC | |
| 2ipa (30) | Cys32 (Trx) | 8.3 | 8.1 | trxA,barsC | |
| 2gzy (30) | Cys29 (Trx) | 5.5 | 5.9 | trxAb | |
| 2gzy (W28A) (30) | Cys29 (Trx) | 5.5 | 5.9 | trxAb | |
| 1jbb (37) | Cys87 | 11.1 | 10 | UBC13 | |
| 1jas (37) | Cys88 | 10.2 | 9 | UBE2B | |
| 1i7k (37) | Cys114 | 10.9 | 10.5 | UBE2C | |
| 1l1d (58) | Cys117 | 9.3 | 9.5 | msrAB |
a Associated gene symbols are reported (retrieved via UniProt).
b Thioredoxin from different species: 1xob, Escherichia coli; 2ppt, Rhodobacter capsulatus; 2o89, Staphylococcus aureus; and 2ipa and 2gzy, Bacillus subtilis.
Another method that has been applied to the investigation of reactive Cys is the empirical pKa predictor PROPKA (22). For a titratable residue, a pKa shift is evaluated as a function of the sum of energy contributions provided by surrounding residues (Fig. 1B). Although the theory of the approach is relatively simple, it has been praised for its balance of speed and performance. We tested its performance against the data set of proteins previously evaluated by a QM approach (Table 1). Overall, the two methods yielded consistent results (R2 = 0.602, linear correlation of their respective results). Moreover, the PROPKA prediction correlated sufficiently well (R2 = 0.74, average deviation from the experimental value of 0.88 ± 0.8) with experimental pKa values, showing performances not too far from those of more sophisticated approaches.
A third category of methods, which has been applied to the analysis of redox Cys (24, 25), is based on the numerical solution of the Poisson-Boltzmann equation: electrostatic calculations provide (free) energies of each of the protonation microstates in the system. This allows the probability of protonation to be calculated over a range of pH values for each titratable residue (i.e. the titration curve and its associated pK½ can be computed) (Fig. 1), a feature that can be very informative (26, 27). As a general note, each approach has its unique features, e.g. PROPKA is ultrafast while still providing acceptable results; if properly set up, the QM-based methods can be very accurate; and Poisson-Boltzmann methods can compute, besides the pKa, the whole titration curve in proximity to the pK½. Thus, these methods can be considered complementary rather than in competition, as they can provide insights from different perspectives and ultimately help establish a more complete picture of Cys reactivity. Besides pKa prediction, QM investigations can be useful in unraveling other aspects of Cys properties and reactivity. For example, they have been applied to investigate the structural determinants of Cys oxidation to sulfenic acid (28). Other QM-based studies have provided interesting insights into nitrosylation of Cys residues (29) and the role of substituents in disulfide exchange reactions (30). Currently, DFT-based methods for the calculation of sulfenic acid/thiol reduction potential are in preliminary stages of development (31). Once available to the community, these methods would offer a significant addition to the current arsenal of computational tools in the redox biology area.
Bioinformatics Approaches Used for Prediction of Reactive Cys in Proteins
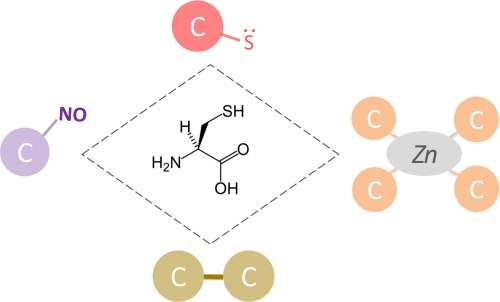
Cys may serve different functions, ranging from structural stabilization to catalytic activity and including a variety of post-translational modifications (PTMs) and associated regulatory roles. Therefore, it is useful to classify Cys residues on the basis of their function. Different functional categories of Cys include (i) structural cystine residues (i.e. stable disulfide-bonded Cys), (ii) metal-coordinating Cys residues, (iii) catalytic Cys residues, and (iv) Cys residues that serve as sites of PTMs (regulatory Cys), as shown schematically in Fig. 2. It has to be noted that not all functional Cys residues can be unambiguously classified. One example is the bacterial chaperone Hsp33, in which some Cys residues serve both structural and regulatory functions depending on the redox state (32, 33). Notably, this functional switch is reversible (Fig. 2). It is also possible that additional, currently undiscovered, functional categories of Cys exist. In the following paragraphs, we briefly introduce the relevant biological aspects of each Cys functional category; then, for each category, a brief discussion of how bioinformatics approaches can be used to investigate the subject and with which tools is provided.
FIGURE 2.
Different functional categories of Cys residues. A schematic representation of Cys functionality is shown. Starting from the top of the rhombus, which gives the molecular structure of Cys, and going clockwise: catalytic residues (red circle; representing a nucleophilic thiolate), metal-binding Cys (orange circles; representing a zinc-Cys4-binding site), structural disulfides (yellow circles; representing a covalent bond between two Cys residues), and regulatory Cys (violet circle; representing an S-nitrosylation site). As discussed in text, not all functional Cys can be reliably categorized. For example, some catalytic Cys residues can also be S-nitrosylated or oxidized to sulfenic acid, and some metal-binding Cys residues can, in certain situations, turn to cystine residues. To represent this complexity and an occasional interplay of functions, the rhombus connecting different functional Cys categories is shown with a dashed line.
Structural Disulfides
Disulfide bond formation is a major mechanism employed by proteins to stabilize their structure. As a norm, the formation of structural disulfides during the folding process (often discussed as oxidative folding) involves a specialized cellular machinery (34, 35). Structural disulfides are common in proteins residing in oxidizing cellular environments, such as the bacterial periplasm, eukaryotic endoplasmic reticulum, and mitochondrial intermembrane space, as well as in secreted proteins. However, transient disulfides are also common in the reducing environments of cellular compartments. Computational approaches used to predict structural disulfides can be divided into sequence-based and structure-based. Among the latter, the simplest method is to examine protein structures for sulfur-to-sulfur (S-S) distances between the two Cys residues. A distance of 2.5 Å is commonly considered a safe cutoff discerning disulfide bonds from other types of functional Cys clusters (36). Often, disulfide bonds, detected by analyzing the S-S distance, can be found already annotated in the Protein Data Bank (PDB) repository (e.g. by directly accessing the PDB file header). A modification of this method envisages the use of a distance between α-carbons of Cys residues. Although less specific (it increases the false positive rate), the computational advantage is remarkable, as only backbone trace coordinates are required. This makes the approach well suited for large-scale comparative structure-based analyses (8).
In some cases, however, it would be desirable to work without structural information, as the structural coverage of natural proteins is still largely incomplete. Here, several methods have been developed. A common underlying concept of these methods is that the majority of disulfides show recurring motifs in the primary structure and thus can be predicted once these patterns are discovered. The simplest approach is the use of curated sequence patterns and profiles, such as those found in the PROSITE database (38). A PROSITE pattern is an annotated regular expression that describes a relatively short portion of protein sequence that may have a biological meaning or function. The PROSITE web server provides a simple interface (ScanProsite) (39) to browse for S-S patterns in any input sequence. Although many S-S patterns with perfect specificity have been compiled, PROSITE profiles and regular expressions can detect only a minority of disulfide-bonded Cys residues (40).
Improved sequence-based approaches have been developed in recent years that could manage an additional level of sequence information (e.g. nature of adjacent amino acids, conservation of flanking residues, etc.) besides the primary sequence (41–43). One such approach is DISULFIND, which uses support vector machines and neural networks to classify and rank different Cys residues in a protein sequence. The algorithm is fast and performs, overall, better than PROSITE (43). Another recently developed method is a structure-based machine learning approach called DBCP. Starting from a FASTA sequence, it automatically calculates a homology model for the protein using Modeler (44). A support vector machine-based algorithm then runs over the structural predictions and assigns a score to each Cys. Only those Cys residues with scores comparable to known structural disulfides are predicted as cystine residues.
It should be noted that prediction of transient disulfides, such as those present in reducing compartments of the cell, is currently challenging, in part due to conformational changes often associated with disulfide bond formation and reduction. These issues are typically addressed by examining the conformational mobility of various regions of proteins or, more directly, by solving protein structures in both reduced and oxidized states.
Metal-binding Cys Residues
Together with His, Cys is the most frequently employed amino acid for metal coordination (45). Metals in proteins have many functions. One example is the stabilization of protein structures. This is common in zinc-Cys4 complexes, where four thiolate-Zn2+ bridges act as stabilizing elements (46), particularly under the reducing conditions of the cytosol, where stable disulfides are disfavored. Other functions of metal ions in proteins include a direct involvement in catalysis and occurrence in regulatory sites. In this regard, Cys properties make this amino acid a preferred residue for redox-dependent regulation of metal binding. For example, the Zn2+-sulfur moiety permits zinc to be tightly bound yet available for release upon oxidation (46). This is a prominent feature of Cys-based metal-binding sites, often referred to as the redox switching of Cys residues (32, 33, 46).
Considering that metal-binding sites are often highly conserved, the presence of conserved proximal Cys residues can be a good indication that these residues may bind metals. Therefore, one approach is the use of manually curated sequence patterns and profiles, such as those found in PROSITE (30, 41). As in the case of disulfide prediction, PROSITE patterns allow fast and easy implementation (e.g. ScanProsite (39)) but lack the ability to properly recognize many metal-binding sites (i.e. have low true positive and high false negative rates) (40, 47). More sophisticated approaches have been developed based on machine learning (40, 48) and nonlinear statistical methods (49). An example is provided by the method called MetalDetector, freely available as a web-accessible service. Methods like this have been tested against and outperformed PROSITE pattern-based analysis while maintaining many of its advantages.
Structure-based approaches can be a valuable alternative for the prediction of metal-binding Cys residues. One interesting method involves the use of the empirical force field FoldX (50). The searching algorithm uses geometric information typically found in metal-binding sites as a starting point to predict new sites. After analyzing the typical arrangement of Cys ligands around zinc coordination sites, the method can recognize similar structural patterns in terms of both the nature of ligands clustered in space and their relative geometries and therefore predict new metal-binding sites (50). A standalone program implementing the algorithm for prediction of metal-binding sites (as well as several other algorithms for energy-based evaluation and protein design) is available at the FoldX web site.
Regulatory Cys Residues
Common reversible PTMs of Cys include sulfenic acid (Cys-SOH), disulfide bonds (both intramolecular and intermolecular), S-nitrosylation (NO-Cys), and glutathionylation. Additionally, Cys can react with endogenous hydrogen sulfide (H2S), a modification that can lead to various physiological (51–53) and structural (54) effects. All of the above can be classified as redox-based PTMs and are reversible. However, other important Cys modifications that are stable and do not involve a change in the redox state occur, for example, the formation of a thioether bond with farnesyl or geranylgeranyl groups, leading to protein lipidation and membrane anchoring (55) or covalent binding of protein cofactors, such as heme. These Cys modifications may be classified into a separate category of functional Cys residues. Below, we focus on the reversible Cys modifications. These PTMs can affect protein properties (local structure, electrostatic interactions, etc.) and, ultimately, protein function or its network of interactions; for these reasons, they are often referred to as regulatory Cys residues (56).
From a computational perspective, regulatory Cys residues are challenging to investigate. Recent progress in proteomics approaches provided a substantial improvement in both quantity and quality of the data produced. In turn, this allows bioinformaticians to start tackling the fundamental issue of the defining features of regulatory Cys. To date, no reliable sequence-based predictive patterns have been developed for different types of regulatory Cys. Instead, high heterogeneity of sequence features was detected. However, structure-based analyses have provided important insights, particularly in the case of NO-Cys (18, 57, 59) and Cys-SOH (60). As for the latter, an important role of uncharged H-bond donors, particularly Thr, was revealed (60). Spatial (but not sequence) proximity to these residues can lead to activation of Cys, mainly by lowering its pKa. In the case of NO-Cys, sequence-based bioinformatics analyses also revealed high heterogeneity around modification sites (57, 59). However, structural analyses provided new insights. First, a QM-based study demonstrated that NO modification can induce a significant charge redistribution in the side chain of Cys, with only marginal effects on the backbone atoms (29). In this study, specific force field parameters and charge schemes for NO-Cys were developed, paving the way for setting up docking experiments with NO-Cys-containing substrates, such as S-nitrosoglutathione (59). Indeed, docking calculations could be a valid computational alternative for detection of specific Cys residues amenable to modification with different nitrosylated substrates (NO-Cys, S-nitrosylated small peptides, etc.). Particularly challenging but certainly feasible is the investigation of the role of protein-protein interactions in the transfer of NO groups from one protein to another (the so-called protein interaction-based transnitrosylation). This process has so far escaped detailed computational studies, partly due to the complexity of the system. However, the steady development of suitable docking software (e.g. RosettaDock) and the information gained from previous studies (29, 59) may make it soon possible to investigate protein interaction-based transnitrosylation.
Catalytic Cys Residues
In many enzymes, Cys plays a critical role as a nucleophile in enzyme-catalyzed reactions. Such Cys residues represent a functional category of catalytic Cys residues. This category can be further subdivided into redox and non-redox Cys functions based on whether the redox state of Cys changes during catalysis. Examples of enzymes with non-redox catalytic Cys residues are protein-tyrosine phosphatases, Cys peptidases, various members of the deubiquitination system, and dCMP hydroxymethylases. Other enzymes called thiol oxidoreductases present catalytic redox Cys in the active sites; here, the catalytic Cys function involves substrate oxidation or reduction, disulfide bond isomerization, and detoxification of various compounds. To our knowledge, no computational approaches for the detection of catalytic non-redox Cys residues have been developed. Thus, we focus further on thiol oxidoreductases, for which better progress has been made.
Thiol oxidoreductases are the only known enzymes that also use Sec as the catalytic residue. This very unique feature was used to develop a bioinformatics strategy for the prediction of thiol oxidoreductases. The method allows high-throughput identification of catalytic redox Cys in protein sequences by searching for sporadic Cys/Sec pairs in homologous sequences (61). It initially identifies unique Cys/Sec pairs flanked by homologous sequences within a pool of translated nucleotide sequences. These pairs then serve as seeds for sequence analysis at the level of protein families and subfamilies. Application of this method identified the majority of known thiol oxidoreductases and indicated the identity of the catalytic Cys. A key advantage of this approach, together with sensitivity, is its speed. High-throughput analyses are possible in a reasonable amount of time, allowing genome-wide analyses of thiol oxidoreductases (62).
Bioinformatics approaches applied to the study of thiol oxidoreductases are not limited to their prediction. Structural and functional adaptations have been examined for different classes of thiol oxidoreductases (63, 64), e.g. evolution of the thioredoxin fold, which led to the emergence of different functions, such as thiol redox regulation, glutathione transfer to electrophilic compounds, etc. In addition, a tool based on structural profiles of reactive Cys sites was developed and employed for functional classification of different subfamilies of peroxiredoxins (65, 66). By using active site signatures, this method allowed researchers to define six peroxiredoxin subfamilies, each of them with specific functionalities (66, 67). Although employed only with peroxiredoxins, it can be extended to analyses of other families of thiol oxidoreductases.
Concluding Remarks
In this minireview, we focused on the properties and functions of reactive Cys residues and methods used to analyze them. A better understanding of basic chemical and physical features of Cys seems to be crucial to improve currently available tools for recognition and functional annotation of reactive thiols in proteins. With this aim, we first focused on two important descriptors of Cys reactivity: exposure and pKa. We then shifted the discussion to the various functional roles played by reactive Cys residues in proteins and reviewed the current status of computational methods used to investigate and predict Cys functions. In some cases, bioinformatics has provided important insights and tools, especially for catalytic redox Cys, metal-binding Cys, and disulfide bonds. In other cases, progress has been limited, e.g. regulatory Cys, sites of stable PTMs and catalytic non-redox Cys.
Difficulties in computational redox biology are linked to the complexity of the subject. Despite the recent dramatic increase in the studies that addressed this question experimentally, many aspects of thiol-based redox regulation and signaling are still not well understood. However, experimental advances, especially in proteomics and structural and post-translational data sets, have provided researchers with the opportunity to both analyze certain Cys categories on a genome-wide scale and address the general principles of various Cys functions from a computational perspective. For example, a new proteomics approach called isoTOP-ABPP (68) allows high-throughput identification of reactive Cys residues in proteins and quantification of their reactivity. This method permits unambiguous identification of reactive Cys residues and also scores them by assigning a reactivity value (R) to each Cys. This feature offers a range of future applications in computational analyses of reactive Cys. For example, as R values provide a measure of Cys nucleophilicity, it would be interesting to analyze how R scores correlate with the results obtained with different theoretical methods for Cys pKa prediction. We may expect further improvements in the understanding of Cys properties in proteins, and it is likely that a better theoretical description of reactive Cys will be vital to improve the predictive power of computational methods that target Cys functions in proteins.
This work was supported, in whole or in part, by National Institutes of Health Grant GM065204 (to V. N. G.). This is the fourth article in the Thematic Minireview Series on Redox Sensing and Regulation.
- DFT
- density functional theory
- QM
- quantum mechanics
- PTM
- post-translational modification
- PDB
- Protein Data Bank.
REFERENCES
- 1. Trifonov E. N. (2004) The triplet code from first principles. J. Biomol. Struct. Dyn. 22, 1–11 [DOI] [PubMed] [Google Scholar]
- 2. Jordan I. K., Kondrashov F. A., Adzhubei I. A., Wolf Y. I., Koonin E. V., Kondrashov A. S., Sunyaev S. (2005) A universal trend of amino acid gain and loss in protein evolution. Nature 433, 633–638 [DOI] [PubMed] [Google Scholar]
- 3. Wu H., Ma B. G., Zhao J. T., Zhang H. Y. (2007) How similar are amino acid mutations in human genetic diseases and evolution. Biochem. Biophys. Res. Commun. 362, 233–237 [DOI] [PubMed] [Google Scholar]
- 4. Moosmann B., Behl C. (2008) Mitochondrially encoded cysteine predicts animal life span. Aging Cell 7, 32–46 [DOI] [PubMed] [Google Scholar]
- 5. Jobson R. W., Dehne-Garcia A., Galtier N. (2010) Apparent longevity-related adaptation of mitochondrial amino acid content is due to nucleotide compositional shifts. Mitochondrion 10, 540–547 [DOI] [PubMed] [Google Scholar]
- 6. Moosmann B. (2011) Respiratory chain cysteine and methionine usage indicate a causal role for thiyl radicals in aging. Exp. Gerontol. 46, 164–169 [DOI] [PubMed] [Google Scholar]
- 7. Schindeldecker M., Stark M., Behl C., Moosmann B. (2011) Differential cysteine depletion in respiratory chain complexes enables the distinction of longevity from aerobicity. Mech. Ageing Dev. 132, 171–179 [DOI] [PubMed] [Google Scholar]
- 8. Beeby M., O'Connor B. D., Ryttersgaard C., Boutz D. R., Perry L. J., Yeates T. O. (2005) The genomics of disulfide bonding and protein stabilization in thermophiles. PloS Biol. 3, e309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Marino S. M., Gladyshev V. N. (2010) Cysteine function governs its conservation and degeneration and restricts its utilization on protein surfaces. J. Mol. Biol. 404, 902–916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lee B. C., Lobanov A. V., Marino S. M., Kaya A., Seravalli J., Hatfield D. L., Gladyshev V. N. (2011) A 4-selenocysteine, 2-selenocysteine insertion sequence (SECIS) element methionine sulfoxide reductase from Metridium senile reveals a non-catalytic function of selenocysteines. J. Biol. Chem. 286, 18747–18755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zhang Y., Baranov P. V., Atkins J. F., Gladyshev V. N. (2005) Pyrrolysine and selenocysteine use dissimilar decoding strategies. J. Biol. Chem. 280, 20740–20751 [DOI] [PubMed] [Google Scholar]
- 12. Xu X. M., Turanov A. A., Carlson B. A., Yoo M. H., Everley R. A., Nandakumar R., Sorokina I., Gygi S. P., Gladyshev V. N., Hatfield D. L. (2010) Targeted insertion of cysteine by decoding UGA codons with mammalian selenocysteine machinery. Proc. Natl. Acad. Sci. U.S.A. 107, 21430–21434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Leonard S. E., Reddie K. G., Carroll K. S. (2009) Mining the thiol proteome for sulfenic acid modifications reveals new targets for oxidation in cells. ACS Chem. Biol. 4, 783–799 [DOI] [PubMed] [Google Scholar]
- 14. Poole L. B., Karplus P. A., Claiborne A. (2004) Protein sulfenic acids in redox signaling. Annu. Rev. Pharmacol. Toxicol. 44, 325–347 [DOI] [PubMed] [Google Scholar]
- 15. Shenton D., Grant C. M. (2003) Protein S-thiolation targets glycolysis and protein synthesis in response to oxidative stress in the yeast Saccharomyces cerevisiae. Biochem. J. 374, 513–519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Cabiscol E., Levine R. L. (1996) The phosphatase activity of carbonic anhydrase III is reversibly regulated by glutathiolation. Proc. Natl. Acad. Sci. U.S.A. 93, 4170–4174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wood Z. A., Schröder E., Robin Harris J., Poole L. B. (2003) Structure, mechanism, and regulation of peroxiredoxins. Trends Biochem. Sci. 28, 32–40 [DOI] [PubMed] [Google Scholar]
- 18. Hess D. T., Matsumoto A., Kim S. O., Marshall H. E., Stamler J. S. (2005) Protein S-nitrosylation: purview and parameters. Nat. Rev. Mol. Cell Biol. 6, 150–166 [DOI] [PubMed] [Google Scholar]
- 19. Winterbourn C. C., Metodiewa D. (1999) Reactivity of biologically important thiol compounds with superoxide and hydrogen peroxide. Free Radic. Biol. Med. 27, 322–328 [DOI] [PubMed] [Google Scholar]
- 20. Winterbourn C. C., Hampton M. B. (2008) Thiol chemistry and specificity in redox signaling. Free Radic. Biol. Med. 45, 549–561 [DOI] [PubMed] [Google Scholar]
- 21. Roos G., Foloppe N., Van Laer K., Wyns L., Nilsson L., Geerlings P., Messens J. (2009) How thioredoxin dissociates its mixed disulfide. PLoS Comput. Biol. 5, e1000461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sanchez R., Riddle M., Woo J., Momand J. (2008) Prediction of reversibly oxidized protein cysteine thiols using protein structure properties. Protein Sci. 17, 473–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Li H., Robertson A. D., Jensen J. H. (2005) Very fast empirical prediction and rationalization of protein pKa values. Proteins 61, 704–721 [DOI] [PubMed] [Google Scholar]
- 24. Tosatto S. C., Bosello V., Fogolari F., Mauri P., Roveri A., Toppo S., Flohé L., Ursini F., Maiorino M. (2008) The catalytic site of glutathione peroxidases. Antioxid. Redox Signal. 10, 1515–1526 [DOI] [PubMed] [Google Scholar]
- 25. Foloppe N., Sagemark J., Nordstrand K., Berndt K. D., Nilsson L. (2001) Structure, dynamics, and electrostatics of the active site of glutaredoxin 3 from Escherichia coli: comparison with functionally related proteins. J. Mol. Biol. 310, 449–470 [DOI] [PubMed] [Google Scholar]
- 26. Ondrechen M. J., Clifton J. G., Ringe D. (2001) THEMATICS: a simple computational predictor of enzyme function from structure. Proc. Natl. Acad. Sci. U.S.A. 98, 12473–12478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Marino S. M., Gladyshev V. N. (2009) A structure-based approach for detection of thiol oxidoreductases and their catalytic redox-active cysteine residues. PLoS Comput. Biol. 5, e1000383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Cardey B., Enescu M. (2007) Selenocysteine versus cysteine reactivity: a theoretical study of their oxidation by hydrogen peroxide. J. Phys. Chem. A 111, 673–678 [DOI] [PubMed] [Google Scholar]
- 29. Han S. (2008) Force field parameters for S-nitrosocysteine and molecular dynamics simulations of S-nitrosated thioredoxin. Biochem. Biophys. Res. Commun. 377, 612–616 [DOI] [PubMed] [Google Scholar]
- 30. Roos G., Geerlings P., Messens J. (2010) The conserved active site tryptophan of thioredoxin has no effect on its redox properties. Protein Sci. 19, 190–194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Roos G., Messens J. (2011) Protein sulfenic acid formation: from cellular damage to redox regulation. Free Radic. Biol. Med. 51, 314–326 [DOI] [PubMed] [Google Scholar]
- 32. Jakob U., Muse W., Eser M., Bardwell J. C. (1999) Chaperone activity with a redox switch. Cell 96, 341–352 [DOI] [PubMed] [Google Scholar]
- 33. Ilbert M., Horst J., Ahrens S., Winter J., Graf P. C., Lilie H., Jakob U. (2007) The redox-switch domain of Hsp33 functions as dual stress sensor. Nat. Struct. Mol. Biol. 14, 556–563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Collet J. F., Bardwell J. C. (2002) Oxidative protein folding in bacteria. Mol. Microbiol. 44, 1–8 [DOI] [PubMed] [Google Scholar]
- 35. Tu B. P., Weissman J. S. (2004) Oxidative protein folding in eukaryotes: mechanisms and consequences. J. Cell Biol. 164, 341–346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Overington J., Donnelly D., Johnson M. S., Sali A., Blundell T. L. (1992) Environment-specific amino acid substitution tables: tertiary templates and prediction of protein folds. Protein Sci. 1, 216–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Tolbert B. S., Tajc S. G., Webb H., Snyder J., Nielsen J. E., Miller B. L., Basavappa R. (2005) The active site cysteine of ubiquitin-conjugating enzymes has a significantly elevated pKa: functional implications. Biochemistry 44, 16385–16391 [DOI] [PubMed] [Google Scholar]
- 38. Sigrist C. J., Cerutti L., Hulo N., Gattiker A., Falquet L., Pagni M., Bairoch A., Bucher P. (2002) PROSITE: a documented database using patterns and profiles as motif descriptors. Brief. Bioinform. 3, 265–274 [DOI] [PubMed] [Google Scholar]
- 39. de Castro E., Sigrist C. J., Gattiker A., Bulliard V., Langendijk-Genevaux P. S., Gasteiger E., Bairoch A., Hulo N. (2006) ScanProsite: detection of PROSITE signature matches and ProRule-associated functional and structural residues in proteins. Nucleic Acids Res. 34, W362–W365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Passerini A., Frasconi P. (2004) Learning to discriminate between ligand-bound and disulfide-bound cysteines. Protein Eng. Des. Sel. 17, 367–373 [DOI] [PubMed] [Google Scholar]
- 41. Chen Y. C., Lin Y. S., Lin C. J., Hwang J. K. (2004) Prediction of the bonding states of cysteines using the support vector machines based on multiple feature vectors and cysteine state sequences. Proteins 55, 1036–1042 [DOI] [PubMed] [Google Scholar]
- 42. Cheng J., Saigo H., Baldi P. (2006) Large-scale prediction of disulfide bridges using kernel methods, two-dimensional recursive neural networks, and weighted graph matching. Proteins 62, 617–629 [DOI] [PubMed] [Google Scholar]
- 43. Ceroni A., Passerini A., Vullo A., Frasconi P. (2006) DISULFIND: a disulfide bonding state and cysteine connectivity prediction server. Nucleic Acids Res. 34, W177–W181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Lin H. H., Tseng L. Y. (2010) DBCP: a web server for disulfide bonding connectivity pattern prediction without the prior knowledge of the bonding state of cysteines. Nucleic Acids Res. 38, W503–W507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Dokmani I., Siki M., Tomi S. (2008) Metals in proteins: correlation between the metal-ion type, coordination number, and the amino acid residues involved in the coordination. Acta Crystallogr. D Biol. Crystallogr. 64, 257–263 [DOI] [PubMed] [Google Scholar]
- 46. Kröncke K. D., Klotz L. O. (2009) Zinc fingers as biologic redox switches? Antioxid. Redox Signal. 11, 1015–1027 [DOI] [PubMed] [Google Scholar]
- 47. Andreini C., Bertini I., Rosato A. (2004) A hint to search for metalloproteins in gene banks. Bioinformatics 20, 1373–1380 [DOI] [PubMed] [Google Scholar]
- 48. Passerini A., Punta M., Ceroni A., Rost B., Frasconi P. (2006) Identifying cysteines and histidines in transition metal-binding sites using support vector machines and neural networks. Proteins 65, 305–316 [DOI] [PubMed] [Google Scholar]
- 49. Lin C. T., Lin K. L., Yang C. H., Chung I. F., Huang C. D., Yang Y. S. (2005) Protein metal-binding residue prediction based on neural networks. Int. J. Neural Syst. 15, 71–84 [DOI] [PubMed] [Google Scholar]
- 50. Schymkowitz J., Borg J., Stricher F., Nys R., Rousseau F., Serrano L. (2005) The FoldX web server: an online force field. Nucleic Acids Res. 33, W382–W388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Zanardo R. C., Brancaleone V., Distrutti E., Fiorucci S., Cirino G., Wallace J. L. (2006) Hydrogen sulfide is an endogenous modulator of leukocyte-mediated inflammation. FASEB J. 20, 2118–2120 [DOI] [PubMed] [Google Scholar]
- 52. Yang G., Sun X., Wang R. (2004) Hydrogen sulfide-induced apoptosis of human aorta smooth muscle cells via the activation of mitogen-activated protein kinases and caspase-3. FASEB J. 18, 1782–1784 [DOI] [PubMed] [Google Scholar]
- 53. Johansen D., Ytrehus K., Baxter G. F. (2006) Exogenous hydrogen sulfide (H2S) protects against regional myocardial ischemia-reperfusion injury: evidence for a role of KATP channels. Basic Res. Cardiol. 101, 53–60 [DOI] [PubMed] [Google Scholar]
- 54. Jiang B., Tang G., Cao K., Wu L., Wang R. (2010) Molecular mechanism for H2S-induced activation of KATP channels. Antioxid. Redox Signal. 12, 1167–1178 [DOI] [PubMed] [Google Scholar]
- 55. Zhang F. L., Casey P. J. (1996) Protein prenylation: molecular mechanisms and functional consequences. Annu. Rev. Biochem. 65, 241–269 [DOI] [PubMed] [Google Scholar]
- 56. Marino S. M., Gladyshev V. N. (2011) Redox biology: computational approaches to the investigation of functional cysteine residues. Antioxid. Redox Signal. 15, 135–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Greco T. M., Hodara R., Parastatidis I., Heijnen H. F., Dennehy M. K., Liebler D. C., Ischiropoulos H. (2006) Identification of S-nitrosylation motifs by site-specific mapping of the S-nitrosocysteine proteome in human vascular smooth muscle cells. Proc. Natl. Acad. Sci. U.S.A. 103, 7420–7425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Neiers F., Sonkaria S., Olry A., Boschi-Muller S., Branlant G. (2007) Characterization of the amino acids from Neisseria meningitidis methionine sulfoxide reductase B involved in the chemical catalysis and substrate specificity of the reductase step. J. Biol. Chem. 282, 32397–32405 [DOI] [PubMed] [Google Scholar]
- 59. Marino S. M., Gladyshev V. N. (2010) Structural analysis of cysteine S-nitrosylation: a modified acid-based motif and the emerging role of transnitrosylation. J. Mol. Biol. 395, 844–859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Salsbury F. R., Jr., Knutson S. T., Poole L. B., Fetrow J. S. (2008) Functional site profiling and electrostatic analysis of cysteines modifiable to cysteine sulfenic acid. Protein Sci. 17, 299–312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Fomenko D. E., Xing W., Adair B. M., Thomas D. J., Gladyshev V. N. (2007) High-throughput identification of catalytic redox-active cysteine residues. Science 315, 387–389 [DOI] [PubMed] [Google Scholar]
- 62. Fomenko D. E., Gladyshev V. N. (2012) Comparative genomics of thiol oxidoreductases reveals widespread and essential functions of thiol-based redox control of cellular processes. Antioxid. Redox Signal. 16, 193–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Atkinson H. J., Babbitt P. C. (2009) An atlas of the thioredoxin fold class reveals the complexity of function-enabling adaptations. PLoS Comput. Biol. 5, e1000541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Atkinson H. J., Babbitt P. C. (2009) Glutathione transferases are structural and functional outliers in the thioredoxin fold. Biochemistry 48, 11108–11116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Cammer S. A., Hoffman B. T., Speir J. A., Canady M. A., Nelson M. R., Knutson S., Gallina M., Baxter S. M., Fetrow J. S. (2003) Structure-based active site profiles for genome analysis and functional family subclassification. J. Mol. Biol. 334, 387–401 [DOI] [PubMed] [Google Scholar]
- 66. Nelson K. J., Knutson S. T., Soito L., Klomsiri C., Poole L. B., Fetrow J. S. (2011) Analysis of the peroxiredoxin family: using active-site structure and sequence information for global classification and residue analysis. Proteins 79, 947–964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Soito L., Williamson C., Knutson S. T., Fetrow J. S., Poole L. B., Nelson K. J. (2011) PREX: PeroxiRedoxin classification indEX, a database of subfamily assignments across the diverse peroxiredoxin family. Nucleic Acids Res. 39, D332–D337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Weerapana E., Wang C., Simon G. M., Richter F., Khare S., Dillon M. B., Bachovchin D. A., Mowen K., Baker D., Cravatt B. F. (2010) Quantitative reactivity profiling predicts functional cysteines in proteomes. Nature 468, 790–795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Dyson H. J., Jeng M. F., Tennant L. L., Slaby I., Lindell M., Cui D. S., Kuprin S., Holmgren A. (1997) Effects of buried charged groups on cysteine thiol ionization and reactivity in Escherichia coli thioredoxin: structural and functional characterization of mutants of Asp-26 and Lys-57. Biochemistry 36, 2622–2636 [DOI] [PubMed] [Google Scholar]
- 70. El Hajjaji H., Dumoulin M., Matagne A., Colau D., Roos G., Messens J., Collet J. F. (2009) The zinc center influences the redox and thermodynamic properties of Escherichia coli thioredoxin 2. J. Mol. Biol. 386, 60–71 [DOI] [PubMed] [Google Scholar]
- 71. Crow A., Acheson R. M., Le Brun N. E., Oubrie A. (2004) Structural basis of redox-coupled protein substrate selection by the cytochrome c biosynthesis protein ResA. J. Biol. Chem. 279, 23654–23660 [DOI] [PubMed] [Google Scholar]