Background: Heme is an essential cofactor yet toxic in free form, necessitating strict intracellular control.
Results: A heme sensor regulates the conserved hrtBA genes in Lactococcus lactis, whose products mediate heme efflux.
Conclusion: L. lactis controls heme homeostasis by sensing intracellular heme and activating heme efflux.
Significance: The use of an intracellular heme sensor to control heme efflux constitutes a novel paradigm for bacterial heme homeostasis.
Keywords: Bacteria, Heme, Microbiology, Transcription Repressor, Transport Metals, Commensal Bacteria, Heme Efflux, Heme Sensor, Homeostasis, Lactococcus Lactis
Abstract
Most commensal and food bacteria lack heme biosynthesis genes. For several of these, the capture of environmental heme is a means of activating aerobic respiration metabolism. Our previous studies in the Gram-positive bacterium Lactococcus lactis showed that heme exposure strongly induced expression of a single operon, called here hrtRBA, encoding an ortholog of the conserved membrane hrt (heme-regulated transporter) and a unique transcriptional regulator that we named HrtR. We show that HrtR expressed as a fusion protein is a heme-binding protein. Heme iron interaction with HrtR is non-covalent, hexacoordinated, and involves two histidines, His-72 and His-149. HrtR specifically binds a 15-nt palindromic sequence in the hrtRBA promoter region, which is needed for hrtRBA repression. HrtR-DNA binding is abolished by heme addition, which activates expression of the HrtB-HrtA (HrtBA) transporter in vitro and in vivo. The use of HrtR as an intracellular heme sensor appears to be conserved among numerous commensal bacteria, in contrast with numerous Gram-positive pathogens that use an extracellular heme-sensing system, HssRS, to regulate hrt. Finally, we show for the first time that HrtBA permease controls heme toxicity by its direct and specific efflux. The use of an intracellular heme sensor to control heme efflux constitutes a novel paradigm for bacterial heme homeostasis.
Introduction
Heme2 uptake is accepted widely as a mechanism for iron acquisition by bacterial pathogens. Remarkably, however, numerous bacteria lacking heme biosynthesis genes use heme directly as a cofactor to activate aerobic respiration. A well studied example is Lactococcus lactis, a lactic acid bacterium used widely for industrial fermentation. When heme is added to an aerated culture, these bacteria activate a terminal cytochrome oxidase, causing a shift to an energetically favorable respiratory metabolism (1). The switch to respiration has a major positive impact on biomass and long term survival, and as such, respiration growth has been implemented in the large-scale production of lactococcal starter cultures (1–3). Numerous other lactic acid bacteria, including several opportunist pathogens such as Streptococcus agalactiae and Enterococcus faecalis, activate respiration growth in the presence of heme (4–8). Although the importance of heme as a cofactor for respiration and numerous bacterial functions is well established (9–11), the mechanisms involved in controlling intracellular homeostasis remain largely unknown. In numerous bacteria, intracellular heme availability is managed by regulating expression of heme-degrading heme oxygenases or deferrochelatases (12–16). An alternative mechanism intervenes at the heme synthesis level, e.g. via iron regulatory (irr) protein-mediated regulation of ferrochelatase activity as in Rhizobium (17–19). In S. agalactiae, which does not synthesize its own heme, we showed that heme efflux is used to manage intracellular heme and protoporphyrin IX (the iron-free precursor of heme; PPIX3), based on a novel regulon called Pef comprising two multi-drug resistance efflux pumps and a MarR-type heme-responsive regulator; homologs of at least part of the Pef regulon are present in other bacteria (20). Studies in Staphylococcus aureus established the existence of another locus involved in preventing heme toxicity, based on HrtB (permease) and HrtA (ATPase), the Hrt (heme regulated transporter) proteins. Inactivation of this locus resulted in heme hypersensitivity (21). Expression of hrtBA is controlled by adjacent hssRS genes, encoding a two-component heme sensor and response regulator. Analogs of the entire system are also present in several other Gram-positive pathogens such as Bacillus anthracis and Corynebacterium diphteriae (21–26). Permease-defective mutants were heme hypersensitive, but a direct role of HrtB-HrtA (HrtBA) in heme efflux was not demonstrated (24).
In L. lactis, previous proteomic and transcriptome studies revealed that components of the ygfCBA operon, encoding a putative transcriptional regulator (YgfC), a predicted permease (YgfB), and an ATPase (YgfA) were induced ∼40-fold by hemin (Fe3+PPIX) (27). YgfB and YgfA are HrtB and HrtA orthologs (21, 22). However, no genes encoding a two component HssRS system were found adjacent to the ygfCBA operon and the predicted transcriptional regulator, YgfC, was unique to L. lactis, leading us to hypothesize that YgfC is implicated in the regulation of ygfBA genes. Accordingly, ygfC was renamed hrtR (heme-regulated transporter regulator); ygfB and ygfA were renamed hrtB and hrtA, respectively (see Fig. 1A). Here, we report the identification of HrtR as a high affinity heme-binding protein and transcriptional regulator that senses intracellular heme to regulate its intracellular homeostasis by efflux in L. lactis.
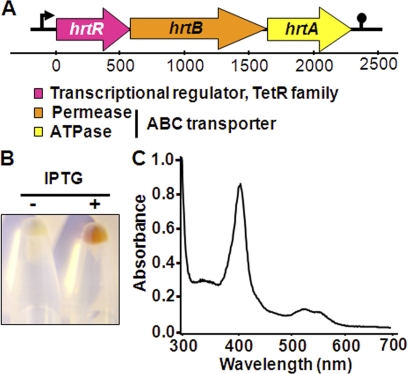
FIGURE 1.
HrtR is a hemin-binding protein. A, schematic representation of the hrtRBA operon in L. lactis (MG1363). The hrtR (llmg_0626) gene encodes a TetR family transcriptional regulator. The hrtB (llmg_0625) and hrtA (llmg_0624) genes encode a permease and an ATPase, respectively. The locus was named based on protein sequence alignments that identified HrtB and HrtA as orthologs of the heme regulated transporter components first described in S. aureus and present in numerous Gram-positive bacteria (21–26). B, E. coli cells overexpressing HrtR are colored red. E. coli were transformed with the HrtR expression vector pMBP-HrtR. Protein expression was induced with 0.5 mm isopropyl 1-thio-β-d-galactopyranoside (IPTG) (see “Experimental Procedures”). Bacteria were collected by centrifugation and photographed. C, UV-visible absorption spectrum of the MBP-HrtR-hemin complex. Holo-MBP-HrtR exhibits a Soret band at 414 nm and β and α bands at 530 and 558 nm, respectively.
EXPERIMENTAL PROCEDURES
Bacterial Strains, Growth Conditions, and Plasmids
The strains and plasmids used in this work are listed in supplemental Table 1. Growth of the bacterial strains and plasmid constructions are outlined in the supplemental data. Experiments in L. lactis were all performed in strain MG1363 and derivatives. Oligonucleotides used for plasmid constructions are listed in supplemental Table 2. Hemin (FePPIX) (Fluka) stock solution (10 mm) was prepared in 50 mm NaOH. Stocks of 10 mm protoporphyrin (PPIX), gallium-protoporphyrin (GaPPIX), cobalt-protoporphyrin (CoPPIX), manganese-protoporphyrin (MnPPIX), magnesium-protoporphyrin (MgPPIX), and zinc-protoporphyrin (ZnPPIX) (Frontier Scientific) were prepared in dimethyl sulfoxide.
Recombinant MBP-HrtR Purification
Escherichia coli Top10 transformed with pMBP-HrtR was used for the recombinant production of MBP-HrtR. The protein was prepared by affinity chromatography on amylose resin (see supplemental Fig. 1A). The MBP-HrtR-hemin complex was prepared by incubating 50 μg of purified MBP-HrtR with a 2-fold excess of hemin in 50 mm Tris-HCl, pH 7.5. Free hemin was removed by chromatography on a Sephadex G-15 gel filtration column (Sigma).
β-Gal Assay
Liquid cultures were generally grown to A600 = 0.5 and then incubated for 1 h with the test porphyrin at the indicated concentration. β-Galactosidase (β-gal) activity was assayed on bacteria permeabilized as described (28). β-Gal activity was quantified by luminescence in an Infinite M200 spectroluminometer (Tecan) using the β-Glo assay system as recommended by manufacturer (Promega). β-Gal activity was revealed on solid medium by streaking bacteria on LB agar containing 80 μg/ml X-Gal.
EMSA
A 352-bp DNA fragment containing the hrt promoter region was PCR-amplified from MG1363 genomic DNA with primers (O17-O18) (supplemental Table 2). A 352-bp DNA fragment located in the hrtR-coding region was generated similarly with primers (O19-O20) (supplemental Table 2) and used as a negative control. The palindrome was mutated by using overlapping PCR using primers (O17-O22) and (O21-O18) as described (29). O22 and its reverse counterpart O21 were designed to encompass the 15-nt palindromic sequence and in which the 15 nucleotides were randomly altered to produce the sequence: 5′-ATTATATAGAGAGAA-3′ (instead of 5′-ATGACACAGTGTCAT-3′) (supplemental Table 2). The two fragments were annealed and PCR-amplified with primers (O17-O18). Fragments were gel-purified (Invitrogen gel extraction and purification kit) and quantified. MBP-HrtR-DNA binding studies were performed in 20 mm Tris-HCl, pH 8, 50 mm KCl, 0.2 mm MgCl2, 1 mm EDTA, 0.2 mm DTT, and 5% glycerol. Reaction mixtures were incubated for 1 h at 30 °C. Binding was analyzed by gel electrophoresis on an 8% polyacrylamide gel in TBE buffer stained with ethidium bromide following electrophoresis.
Intracellular Iron 57Fe Determinations
Strains ΔhrtRBA(pCtrl) and ΔhrtRBA(phrtBA) were grown in the absence and presence of 57Fe-hemin. Both strains were grown to A600 = 0.5 prior to addition or not of 3 μm 57FePPIX (Frontier Scientific) for an additional hour. Cells were washed three times in H2O supplemented with 0.5 mm EDTA. Cell pellets were dessicated and mineralized by successive incubations in 65% nitric acid solution. 57Fe was quantified by inductively coupled plasma mass spectroscopy (ICP-MS) (Agilent 7700X).
RESULTS
HrtR Is Heme-binding Protein
HrtR was purified as a fusion to the maltose-binding-protein (MBP-HrtR) in E. coli (supplemental Fig. 1A). Cells carrying the MBP-HrtR expression plasmid were pink when isopropyl 1-thio-β-d-galactopyranoside inducer was added, suggesting that HrtR could scavenge and bind to endogenous heme (which is red) (Fig. 1B). Purified MBP-HrtR was also colored pink. The protein was saturated with excess hemin and then purified from free hemin by gel filtration chromatography. The purified complex was red, and its UV-visible spectrum at pH 7.4 revealed a Soret band at A414 and β/α bands at 530 and 558 nm, respectively, consistent with ferric hexacoordinate low spin binding through the iron center (Fig. 1C and see below). Reduction of the complex with dithionite resulted in a shift in the Soret band from 414 nm to 426 nm, whereas oxidation with ammonium persulfate did not change the profile, suggesting that HrtR binds ferric hemin in our experimental conditions (supplemental Fig. 1B). The saturation point, as determined by titration, was ∼10 μm hemin, indicating that HrtR binds one molecule of hemin per monomer (supplemental Fig. 1C and inset). This conclusion was substantiated further using the pyridine hemochrome assay to assess heme content: 8 μm MBP-HrtR bound 6.8 ± 0.3 μm hemin (mean ± S.E.; n = 3) (supplemental Fig. 1D). Titration curves were fitted to a one binding site model (see supplemental Materials and Methods). The dissociation constant Kd value of the MBP-HrtR-hemin complex was 0.4 ± 0.2 μm (mean ± S.E.; n = 4). Taken together, these results demonstrate that HrtR is a hemin-binding protein.
HrtR Controls hrtRBA Transcription by Binding to 15-nt Palindromic Sequence
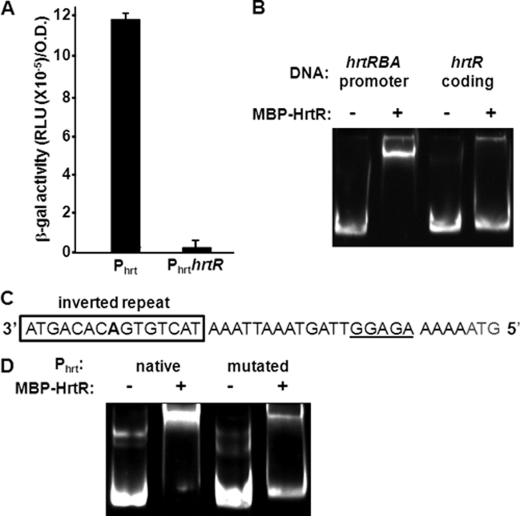
Members of the TetR family generally repress their own transcription together with the transporters they regulate (30). We first verified that HrtR was a repressor of the hrtRBA operon in vivo, using fusions between the hrtRBA promoter region (Phrt), or the promoter and hrtR coding region (PhrthrtR), and the lacZ gene reporter. These constructs were established in a ΔhrtRBA strain. In this context, the Phrt-lac construct was constitutively expressed (Fig. 2A). In contrast, β-gal activity expressed from pPhrthrtR-lac was greatly diminished (Fig. 2A), illustrating that HrtR is a transcriptional repressor of the hrtRBA operon. EMSA confirmed that HrtR specifically binds the hrtRBA promoter region: MBP-HrtR incubated with a 352-bp DNA comprising the hrtRBA promoter region resulted in a DNA shift (Fig. 2B, lanes 1 and 2). A similarly sized DNA fragment corresponding to an internal hrtR region did not shift under the same conditions (Fig. 2B, lanes 3 and 4), indicating specificity of the interaction. TetR family operators usually comprise a 15-nucleotide palindromic sequence with internal palindromic symmetry around a central base pair (30). One such palindrome, 5′-ATGACACAGTGTCAT-3′, was identified upstream of the hrtR start codon (Fig. 2C). Its replacement by a 15-nt non-palindromic sequence (5′-ATTATATAGAGAGAA-3′) significantly diminished HrtR binding (Fig. 2D), suggesting that the initial sequence is targeted by HrtR. This conclusion was further substantiated by the deletion of the central adenine of the 15-nt palindrome, which alleviated HrtR binding to the hrt promoter region deleted for the central adenine (supplemental Fig. 2A) and in vivo HrtR-mediated inhibition of PhrtΔAhrtR-lac (supplemental Fig. 2B). We conclude that the 15-nt palindrome is the HrtR DNA binding site.
FIGURE 2.
HrtR controls hrtRBA transcription via binding to a 15-nt palindromic sequence in the hrtRBA promoter. A, HrtR is a transcriptional repressor of the hrtRBA operon. Transcriptional fusions between the hrtRBA promoter region, including or not the coding region of HrtR (Phrt-lac and PhrthrtR-lac, respectively), were expressed in the ΔhrtRBA L. lactis strain. β-Gal activity was quantified by luminescence (see “Experimental Procedures”; results shown for at least three experiments) B, HrtR binds the promoter region of hrtRBA. EMSA shows specific binding of HrtR to the hrtRBA promoter. 30 pmol of the hrtRBA promoter fragment (lanes 1 and 2) and of the hrtR-coding region (lanes 3 and 4) were incubated in the presence of 200 pmol (lanes 2 and 4) of MBP-HrtR. C, a 15-nucleotide palindromic motif (in boldface type and boxed; the central adenine is in boldface type) is present upstream of hrtRBA. The start codon is in gray, and the RBS sequence is underlined. D, the palindrome is needed for HrtR-DNA binding. EMSA was done using a native DNA fragment encompassing the promoter region, or a fragment in which the 15-nt palindromic sequence was substituted with a random sequence. A gel representative of three experiments is shown. RLU, relative light units.
Hemin Alleviates HrtR Repression of hrtRBA Operon
Members of the TetR-like family of transcriptional regulators act as chemical sensors (30, 31). Binding to their ligands alleviates their interaction with their respective operators, leading to promoter induction (30, 31). These characteristics, together with our findings that HrtR 1) controls hrtRBA operon expression and 2) binds hemin, strongly suggested that hemin is the signal that relieves hrtRBA transcriptional repression. We investigated the in vivo effects of hemin on hrtRBA expression using PhrthrtR-lac as reporter and following β-gal activity (Fig. 3A). PhrthrtR-lac was virtually off in the absence of hemin, establishing the hemin requirement for hrtRBA operon induction. Expression of β-gal was induced at 1 μm and saturated at ∼5 μm hemin. We next studied the role of hemin in HrtR DNA binding activity in vitro with EMSA (Fig. 3B). Addition of hemin to MBP-HrtR abolished formation of the DNA-HrtR complex in a concentration-dependent manner, as seen by the disappearance of the band shift. Complete inhibition was obtained when the MBP-HrtR to hemin ratio was 1:1, consistent with the finding that one HrtR protein binds one molecule of hemin.
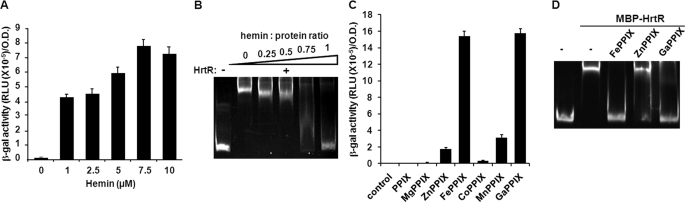
FIGURE 3.
Hemin alleviates HrtR repression of the hrtRBA operon. A, in vivo induction of hrtRBA operon by hemin. The strain carrying PhrthrtR-lac was grown in the presence of the indicated concentrations of hemin to A600 = 1. β-Gal activity was quantified by luminescence (see “Experimental Procedures”). Results represent the mean ± S.D. from the triplicate sample. B, effect of hemin on binding of HrtR to the hrtRBA promoter by EMSA. Thirty pmol of hrtRBA promoter fragment were incubated in the presence of 200 pmol of MBP-HrtR together with increasing amounts of hemin. C, in vivo induction of Phrt with PPIX and metal porphyrins. β-Gal activity of the pPhrthrtR-lac plasmid, established in WT L. lactis was evaluated on bacteria incubated without or with 5 μm of the indicated PPIX or metal-conjugated porphyrins. D, EMSA was used to test ligand specificity of HrtR. Thirty pmol of hrtRBA promoter fragment were incubated in the presence of 200 pmol of MBP-HrtR and 200 pmol of the indicated metalloporphyrin. β-Gal activity was quantified by luminescence (see “Experimental Procedures”). Results represent the mean ± S.D. from triplicate experiments. RLU, relative light units.
Several TetR family regulators including E. coli AcrR, the closest HrtR homolog recognize a variety of structurally unrelated compounds (30, 31). We investigated the possibility that HrtR might interact with other porphyrins. The capacity of HrtR to bind PPIX and other metal-substituted porphyrins was tested. PPIX, GaPPIX, CoPPIX, MnPPIX, MgPPIX, and ZnPPIX, containing metals that have all coordination chemistry similar to iron (32). These porphyrins were assayed in vivo for induction of hrtRBA in bacteria harboring pPhrthrtR-lac (Fig. 3C). Surprisingly, in these conditions, hrt induction was strongly dependent on the metal replacing the iron ion of the porphyrin. Only GaPPIX and to a slight extent MnPPIX induced hrtRBA. To validate that these porphyrins were internalized, a plasmid expressing the heme catalase KatA from E. faecalis was established in L. lactis and used as an in vivo indicator; KatA protein was stabilized in the presence of all the PPIX molecules (supplemental Fig. 3) (29, 33, 34). ZnPPIX and GaPPIX, non-inducing and inducing pPhrthrtR-lac in vivo (Fig. 3C), respectively, were assayed by EMSA (Fig. 3D). Interestingly ZnPPIX had virtually no effect on the DNA shift (Fig. 3D, lane 4), whereas GaPPIX, similar to hemin, abolished the shift (Fig. 3D, lanes 3 and 5). These results confirm that HrtR does not bind all metal porphyrins reinforce our conclusion that FePPIX and GaPPIX specifically bind HrtR and deregulate the hrt promoter .
HrtR Interaction with Hemin and DNA Implicates Two Histidines
The spectroscopic profile of the MBP-HrtR-heme complex (Figs. 1C and 4A) was characteristic of a low spin hexacoordination of the heme iron as seen by the position of the β and α bands (530 and 558 nm, respectively; Fig. 1C). Low spin coordination implies that two strong amino acids are involved in binding.
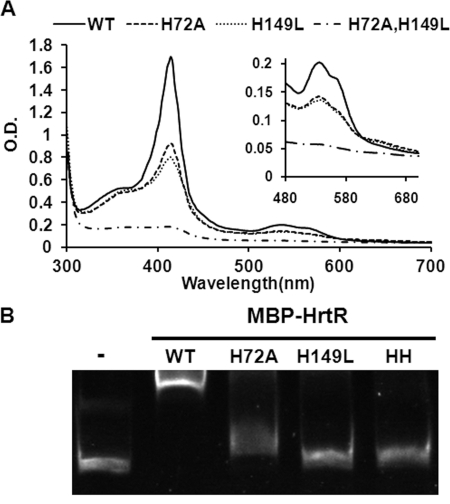
FIGURE 4.
Role of histidines His-72 and His-149 in HrtR binding to heme and to DNA. A, UV-visible spectra of 30 μm ferric heme (hemin) and MBP-HrtR WT, H72A/H149L, and H72A/H149L (HH). WT and mutant HrtR were bound with 3-fold excess hemin. Unbound hemin was then removed by size exclusion chromatography. UV-visible spectra of the complexes (30 μm in 200 μl) were obtained in a microplate spectrophotometer (Infinite M200, Tecan, Austria). Inset, magnification of the 500–700 nm area. Results are representative of three experiments. B, EMSA of HrtR and histidine mutants binding to the hrtRBA promoter region. Thirty pmol of the hrtRBA promoter fragment were incubated in the presence of 200 pmol of WT and mutated variants as above. Lane 1 was a control without protein. Results are representative of three experiments.
Likely candidates for axial ligands are histidine, methionine, and cysteine residues. Based on the general structure of TetR transcriptional regulators (30), only a single histidine (His-149) was present in the predicted substrate binding C-terminal region of HrtR (supplemental Fig. 4A). Another potential candidate His residue was identified at position 72 of the amino acid chain (His-72), near the border of the predicted DNA-binding region (supplemental Fig. 4A). To test the role of these two histidines in heme binding, we substituted both with aliphatic residues, thus generating the mutant proteins H149L and H72A, and a double mutant H149L/H72A. In both single mutant proteins, the ferric form bound considerably less heme than did the wild type, and the double mutant showed virtually no heme binding (Fig. 4A). For both single mutants, the ferric spectra exhibit reduced and shifted Soret peaks (413, 408, and 410 nm for WT, H72A, and H149L, respectively) and show a reduced α band ∼558 nm, indicating that oxidized heme lost its strong ligand hexacoordinate binding and is most likely pentacoordinate (Fig. 4A, inset). The high spin state of the complex is also highlighted by a characteristic small shoulder above 630 nm (Fig. 4A, inset) (36). The HrtR H72A/H149L mutant was virtually incapable of binding hemin (Fig. 4A). In the ferrous state, spectral profiles of the WT and mutant HrtR proteins were also typical of pentacoordinate heme coordination (data not shown). Consistently, addition of imidazole to the mutant proteins restored hemin binding, suggesting that imidazole could replace the His ligand substitution (supplemental Fig. 4B) (37, 38). Circular dichroism spectra of the native and mutant proteins were indistinguishable, indicating that the double histidine mutation did not modify HrtR folding; both WT and mutant proteins contained negative bands at 209 and 220 nm (supplemental Fig. 4C), indicative of α helix-rich proteins, as reported for TetR-like family regulators. Altogether, these data identify His-72 and His-149 as the axial heme ligands.
EMSA was performed with HrtR histidine variants (Fig. 4B). Strikingly, both HrtR His variants, and the double-mutated HrtR lost their ability to bind DNA, indicating the importance of the two His residues for binding to both the hrt promoter and hemin.
HrtBA Effluxes Heme
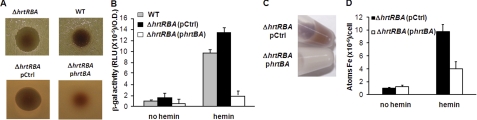
Orthologs of the heme-regulated HrtBA transporter were shown to protect numerous Gram-positive bacteria against heme toxicity (22). An obvious role for HrtBA is to export intracellular heme; however, demonstration has proven difficult, and the substrate for the putative HrtBA transporter remains unidentified (26). To address this question, we separated HrtBA expression from its regulator, and examined its role when constitutively expressed from the p23 promoter (on plasmid phrtBA; (39)) in an L. lactis ΔhrtRBA strain. The latter strain exhibited hemin hypersensitivity on solid medium (Fig. 5A). In comparison, the ΔhrtRBA strain carrying phrtBA was insensitive to the same hemin concentration (Fig. 5A) (22).
FIGURE 5.
The HrtBA transporter controls environmental heme toxicity through efflux in L. lactis. A, HrtBA controls heme toxicity. Stationary phase cultures of WT, ΔhrtRBA, and ΔhrtRBA strains carrying either a control plasmid (pG+host8, designated pCtrl) or phrtBA were plated on solid medium. Hemin (10 μl of a 10 mm stock solution) was pipetted directly onto plates, which were incubated at 30 °C for 24 h. Inhibition zones appear as dark clearing in the center of each panel. Results are representative of at least three experiments. B, HrtBA regulates intracellular heme content. β-Gal activity of the pPhrthrtR-lac plasmid, established in WT, ΔhrtRBA(pCtrl), or ΔhrtRBA(phrtBA) strains, was evaluated on bacteria cultivated without or with 5 μm hemin. β-Gal activity was quantified by luminescence. Results represent the mean ± S.D. from triplicate experiments. C, color phenotype of ΔhrtRBA transformed with pCtrl and ΔhrtRBA carrying phrtBA. Bacteria were grown to A600 = 0.5 prior to addition or not of 10 μm hemin in the culture medium for an additional hour. Bacteria were pelleted by centrifugation. Control bacteria exhibited a strong red color, whereas bacteria overexpressing HrtBA did not. D, iron ([57Fe] content determination of ΔhrtRBA(pCtrl) and ΔhrtRBA(phrtBA) strains grown as in C in the absence and presence of 57Fe-hemin. Results represent the mean ± S.D. from triplicate experiments.
TetR regulators characteristically bind the substrates of the transporter they regulate (30). Our findings that HrtR binds hemin to regulate hrtRBA operon transcription gave a first indication that HrtBA indeed transports heme. To substantiate this role, we first used HrtR as a cytoplasmic heme sensor in ΔhrtRBA cells that constitutively expressed hrtBA genes on plasmid phrtBA. In this system, hrtR expression was a valid heme sensor, as it was dissociated from hrtBA expression. HrtR activation was followed using the PhrthrtR-lac fusion in cells grown with hemin (Fig. 5B). Addition of 5 μm hemin to the ΔhrtRBA strain resulted in a slight increase in β-gal expression levels compared with the WT strain. Remarkably, constitutive HrtBA expression in the complemented strain resulted in nearly complete extinction of reporter protein expression, indicating that cytoplasmic heme availability was too low to allow HrtR binding. In keeping with these results, More direct evidence for the role of HrtBA as a heme transporter was provided by the cell pellets of the ΔhrtRBA strain grown in hemin showing a red color, whereas pellets of the HrtBA overexpression strain were white (Fig. 5C). The presence of hemin in the two strains was confirmed and quantified by ICP-MS. We used 57FePPIX, which represents only 2.12% of the four stable isotopes of natural terrestrial isotopes. Thus, the 57Fe cellular content is representative of the hemin added to cells independently of environmental iron. Results (Fig. 5D) show ∼2.5-fold lower intracellular 57Fe accumulation in the ΔhrtRBA mutant expressing the HrtBA overproducer plasmid compared with ΔhrtRBA carrying a control plasmid. Taken together, these results indicate that the HrtB permease acts as a heme-specific efflux pump.
DISCUSSION
In this work, we elucidated how intracellular heme homeostasis is managed in the heme auxotroph L. lactis. Our main findings are that HrtR is an intracellular heme sensor that regulates transcription of hrtBA and that HrtBA constitutes a heme efflux pump.
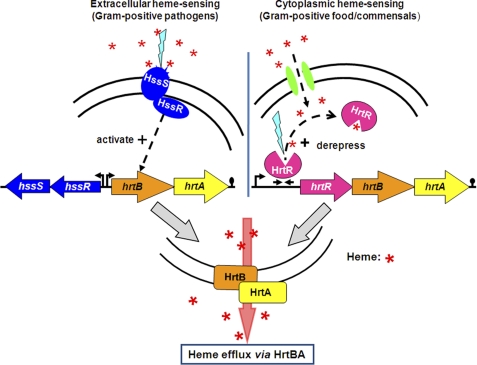
Interestingly, as first characterized in S. aureus and in all bacteria studied except for L. lactis, HrtBA is regulated by adjacent genes encoding the two-component heme sensor histidine kinase (HssS) and the response regulator HssR (21, 22, 25, 26). HssS sensing of extracellular hemin leads to induction of HrtBA, now elucidated to be the heme efflux pump (schematically depicted in Fig. 6, see left panel) (21, 26). In contrast, in light of our results, lactococci have evolved a specific induction system that relies on intracellular hemin (Fig. 6, right panel). A notable difference between these bacteria is that lactococci are non-pathogenic, whereas the bacteria using the extracellular HssS sensor are Gram-positive pathogens that may be in contact with heme-rich blood. Interestingly, other food and commensal bacteria carry HrtR, HrtB, and HrtA homologs, including the 15-nt inverted repeat 5′-ATGACACAGTGTCAT-3′, supporting the notion that a preference for external versus internal heme sensing might be related to the bacterial biotope (supplemental Table 3).
FIGURE 6.
Schematic representation showing two distinct bacterial heme-sensing systems, one extracellular and one intracellular, that regulate heme homeostasis by HrtBA-mediated efflux. Left panel, extracellular heme-sensing induces the two-component HssSR system. Once stimulated, HssR activates hrtBA expression, as reported (25). Right panel, in L. lactis, heme is taken up by fhuDBA gene products (green ovals) (35).4 Internalized heme binds to available HrtR protein to relieve repression of the hrtRBA operon. Activation of hrtBA results in heme efflux. Red asterisk, heme. Blue lightning represents sensor activation.
Deciphering the role of HrtR in L. lactis as a heme-modulated transcriptional repressor of hrtBA helped elucidate HrtBA function as a heme efflux transporter. Indeed, TetR family regulators of efflux permeases generally bind the substrate that is transported (30); in the case of HrtR and the adjacent HrtBA permease, that substrate would be heme. Several strong lines of experimental evidence then led us to conclude that heme is the specific substrate for HrtBA-mediated export. This conclusion may be generalized to other Gram-positive bacteria.
Interestingly, in contrast to other heme-binding proteins (14), HrtR shows high binding specificity for heme. Among other metal porphyrins tested, only GaPPIX and to a lesser extent MnPPIX could substitute for hemin. In a recent in vitro study, only porphyrin metals with oxidation state III bound IsdH-NEAT3 with high affinity, suggesting that electrostatic forces play a role in the interaction (40). This is not the case for HrtR, as reduced hemin (Fe(II)PPIX) bound the protein. Other parameters such as the ionic radius of iron and gallium, which are very similar (0.62 versus 0.64A), might participate in the strict ligand recognition between HrtR and porphyrin metals (40).
Our findings that HrtBA is functional in L. lactis expands the suggested role and evolution of the hrt system beyond bacterial pathogens where it would protect cells from heme toxicity in conditions of high heme availability (as might be encountered in bacteremia) (22, 25). The existence of heme-responsive genes raises questions concerning the natural environments that are home to lactococci. The much improved growth and survival of lactococci in the presence of heme (6), and the expression of several heme-binding proteins may indicate that these bacteria are in contact with heme sources in their ecosystems. We suggest that heme homeostasis may be an important consideration for non-pathogens present, for example, in the gut, where heme stores vary according to contributions from flora, foods, and the host. In this setting, the hrtRBA operon would respond to keep intracellular heme at non-toxic level.
Supplementary Material
Acknowledgments
We thank Céline Henry (Plateforme d'Analyse Protéomique de Paris Sud-Ouest, Institut National de la Recherche Agronomique) for mass spectrometry, Human Rezaei (Virologie et Immunologie Moléculaire, Institut National de la Recherche Agronomique) for circular dichroism, Chantal Doucet (Université de Montpellier II) for ICP-MS, and Lars Hederstedt (Lund University, Sweden) for the generous gift of pLUMB5 plasmid and anti-KatA antibody.
This work was supported in part by the “StrepRespire” project by the French “Agence Nationale de la Recherche” and Chr. Hansen.

This article contains supplemental “Materials and Methods,” Tables 1–3, Figs. 1–4, and additional references.
In this report, heme refers to iron protoporphyrin IX regardless of the iron redox state, whereas hemin refers to ferric iron protoporphyrin IX.
P. Gaudu, E. Van West, and A. Gruss, unpublished observations.
- PPIX
- protoporphyrin IX
- MBP
- maltose-binding protein
- ICP-MS
- inductively coupled plasma mass spectroscopy.
REFERENCES
- 1. Duwat P., Sourice S., Cesselin B., Lamberet G., Vido K., Gaudu P., Le Loir Y., Violet F., Loubière P., Gruss A. (2001) Respiration capacity of the fermenting bacterium Lactococcus lactis and its positive effects on growth and survival. J. Bacteriol. 183, 4509–4516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Gaudu P., Vido K., Cesselin B., Kulakauskas S., Tremblay J., Rezaïki L., Lamberret G., Sourice S., Duwat P., Gruss A. (2002) Respiration capacity and consequences in Lactococcus lactis. Antonie Van Leeuwenhoek 82, 263–269 [PubMed] [Google Scholar]
- 3. Kaneko T., Takahashi M., Suzuki H. (1990) Acetoin fermentation by citrate-positive Lactococcus lactis subsp. lactis 3022 grown aerobically in the presence of hemin or Cu. Appl. Environ. Microbiol. 56, 2644–2649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Christel Garrigues Johansen E., Pedersen M. B., Møllgaard H., Sørensen K. I., Gaudu P., Gruss A., Lamberet G. (2006) Nat. Rev. Microbiol. 4, c2; author reply c3 [DOI] [PubMed] [Google Scholar]
- 5. Høier E., Janzen T., Rattray F., Sørensen K., Børsting M. W., Brockmann E., Johansen E. T. (2010) in Technology of Cheesemaking (Law B. A.., Tamime A. Y., eds) 2nd Ed., pp. 193–230, Wiley-Blackwell, Oxford, UK [Google Scholar]
- 6. Lechardeur D., Cesselin B., Fernandez A., Lamberet G., Garrigues C., Pedersen M., Gaudu P., Gruss A. (2011) Using heme as an energy boost for lactic acid bacteria. Curr. Opin. Biotechnol. 22, 143–149 [DOI] [PubMed] [Google Scholar]
- 7. Winstedt L., Frankenberg L., Hederstedt L., von Wachenfeldt C. (2000) Enterococcus faecalis V583 contains a cytochrome bd-type respiratory oxidase. J. Bacteriol. 182, 3863–3866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Winstedt L., von Wachenfeldt C. (2000) Terminal oxidases of Bacillus subtilis strain 168: one quinol oxidase, cytochrome aa3 or cytochrome bd, is required for aerobic growth. J. Bacteriol. 182, 6557–6564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Anzaldi L. L., Skaar E. P. (2010) Overcoming the heme paradox: heme toxicity and tolerance in bacterial pathogens. Infect. Immun. 78, 4977–4989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Reniere M. L., Torres V. J., Skaar E. P. (2007) Intracellular metalloporphyrin metabolism in Staphylococcus aureus. Biometals 20, 333–345 [DOI] [PubMed] [Google Scholar]
- 11. Schultz I. J., Chen C., Paw B. H., Hamza I. (2010) Iron and porphyrin trafficking in heme biogenesis. J. Biol. Chem. 285, 26753–26759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Skaar E. P., Gaspar A. H., Schneewind O. (2004) IsdG and IsdI, heme-degrading enzymes in the cytoplasm of Staphylococcus aureus. J. Biol. Chem. 279, 436–443 [DOI] [PubMed] [Google Scholar]
- 13. Wilks A., Schmitt M. P. (1998) Expression and characterization of a heme oxygenase (Hmu O) from Corynebacterium diphtheriae. Iron acquisition requires oxidative cleavage of the heme macrocycle. J. Biol. Chem. 273, 837–841 [DOI] [PubMed] [Google Scholar]
- 14. Létoffé S., Heuck G., Delepelaire P., Lange N., Wandersman C. (2009) Bacteria capture iron from heme by keeping tetrapyrrol skeleton intact. Proc. Natl. Acad. Sci. U.S.A. 106, 11719–11724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kaur A. P., Lansky I. B., Wilks A. (2009) The role of the cytoplasmic heme-binding protein (PhuS) of Pseudomonas aeruginosa in intracellular heme trafficking and iron homeostasis. J. Biol. Chem. 284, 56–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Bibb L. A., Schmitt M. P. (2010) The ABC transporter HrtAB confers resistance to hemin toxicity and is regulated in a hemin-dependent manner by the ChrAS two-component system in Corynebacterium diphtheriae. J. Bacteriol. 192, 4606–4617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Small S. K., Puri S., O'Brian M. R. (2009) Heme-dependent metalloregulation by the iron response regulator (Irr) protein in Rhizobium and other Alpha proteobacteria. Biometals 22, 89–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Yang J., Sangwan I., Lindemann A., Hauser F., Hennecke H., Fischer H. M., O'Brian M. R. (2006) Bradyrhizobium japonicum senses iron through the status of haem to regulate iron homeostasis and metabolism. Mol. Microbiol. 60, 427–437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Qi Z., O'Brian M. R. (2002) Interaction between the bacterial iron response regulator and ferrochelatase mediates genetic control of heme biosynthesis. Mol. Cell 9, 155–162 [DOI] [PubMed] [Google Scholar]
- 20. Fernandez A., Lechardeur D., Derré-Bobillot A., Couvé E., Gaudu P., Gruss A. (2010) Two coregulated efflux transporters modulate intracellular heme and protoporphyrin IX availability in Streptococcus agalactiae. PLoS Pathog. 6, e1000860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Friedman D. B., Stauff D. L., Pishchany G., Whitwell C. W., Torres V. J., Skaar E. P. (2006) Staphylococcus aureus redirects central metabolism to increase iron availability. PLoS Pathog. 2, e87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Torres V. J., Stauff D. L., Pishchany G., Bezbradica J. S., Gordy L. E., Iturregui J., Anderson K. L., Dunman P. M., Joyce S., Skaar E. P. (2007) A Staphylococcus aureus regulatory system that responds to host heme and modulates virulence. Cell Host Microbe 1, 109–119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Stauff D. L., Torres V. J., Skaar E. P. (2007) Signaling and DNA-binding activities of the Staphylococcus aureus HssR-HssS two-component system required for heme sensing. J. Biol. Chem. 282, 26111–26121 [DOI] [PubMed] [Google Scholar]
- 24. Stauff D. L., Bagaley D., Torres V. J., Joyce R., Anderson K. L., Kuechenmeister L., Dunman P. M., Skaar E. P. (2008) Staphylococcus aureus HrtA is an ATPase required for protection against heme toxicity and prevention of a transcriptional heme stress response. J. Bacteriol. 190, 3588–3596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Stauff D. L., Skaar E. P. (2009) The heme sensor system of Staphylococcus aureus. Contrib. Microbiol. 16, 120–135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Stauff D. L., Skaar E. P. (2009) Bacillus anthracis HssRS signalling to HrtAB regulates haem resistance during infection. Mol. Microbiol. 72, 763–778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Vido K., Le Bars D., Mistou M. Y., Anglade P., Gruss A., Gaudu P. (2004) Proteome analyses of heme-dependent respiration in Lactococcus lactis: involvement of the proteolytic system. J. Bacteriol. 186, 1648–1657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Miller J. H. (1972) Experiments in Molecular Genetics, Cold Spring Harbor, NY [Google Scholar]
- 29. Lechardeur D., Fernandez A., Robert B., Gaudu P., Trieu-Cuot P., Lamberet G., Gruss A. (2010) The 2-Cys peroxiredoxin alkyl hydroperoxide reductase c binds heme and participates in its intracellular availability in Streptococcus agalactiae. J. Biol. Chem. 285, 16032–16041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ramos J. L., Martínez-Bueno M., Molina-Henares A. J., Terán W., Watanabe K., Zhang X., Gallegos M. T., Brennan R., Tobes R. (2005) The TetR family of transcriptional repressors. Microbiol. Mol. Biol. Rev. 69, 326–356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Routh M. D., Su C. C., Zhang Q., Yu E. W. (2009) Structures of AcrR and CmeR: insight into the mechanisms of transcriptional repression and multi-drug recognition in the TetR family of regulators. Biochim. Biophys. Acta 1794, 844–851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Stojiljkovic I., Kumar V., Srinivasan N. (1999) Non-iron metalloporphyrins: potent antibacterial compounds that exploit haem/Hb uptake systems of pathogenic bacteria. Mol. Microbiol. 31, 429–442 [DOI] [PubMed] [Google Scholar]
- 33. Brugna M., Tasse L., Hederstedt L. (2010) In vivo production of catalase containing haem analogues. FEBS J. 277, 2663–2672 [DOI] [PubMed] [Google Scholar]
- 34. Fernandez A., Lechardeur D., Derré-Bobillot A., Couvé E., Gaudu P., Gruss A. (2010) Two coregulated efflux transporters modulate intracellular heme and protoporphyrin IX availability in Streptococcus agalactiae. Plos Pathog. 6, e1000860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Gaudu P., Lamberet G., Poncet S., Gruss A. (2003) CcpA regulation of aerobic and respiration growth in Lactococcus lactis. Mol. Microbiol. 50, 183–192 [DOI] [PubMed] [Google Scholar]
- 36. Ran Y., Liu M., Zhu H., Nygaard T. K., Brown D. E., Fabian M., Dooley D. M., Lei B. (2010) Spectroscopic identification of heme axial ligands in HtsA that are involved in heme acquisition by Streptococcus pyogenes. Biochemistry 49, 2834–2842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Barrick D. (1994) Replacement of the proximal ligand of sperm whale myoglobin with free imidazole in the mutant His-93→Gly. Biochemistry 33, 6546–6554 [DOI] [PubMed] [Google Scholar]
- 38. Marletta M. A. (2006) Raising enzymes from the dead and the secrets they can tell. ACS Chem. Biol. 1, 73–74 [DOI] [PubMed] [Google Scholar]
- 39. van der Vossen J. M., van der Lelie D., Venema G. (1987) Isolation and characterization of Streptococcus cremoris Wg2-specific promoters. Appl. Environ. Microbiol. 53, 2452–2457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Moriwaki Y., Caaveiro J. M., Tanaka Y., Tsutsumi H., Hamachi I., Tsumoto K. (2011) Molecular basis of recognition of antibacterial porphyrins by heme-transporter IsdH-NEAT3 of Staphylococcus aureus. Biochemistry 50, 7311–7320 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.