Background: The SH3 domain of IRSp53 interacts with several proteins that control actin dynamics.
Results: IRSp53 interacts with mDia1 and WAVE2 within filopodia, and knocking down either protein reduces IRSp53-driven filopodium formation.
Conclusion: IRSp53-mediated filopodium formation involves the actin regulators mDia1 and WAVE2.
Significance: Identifying proteins involved in filopodium formation enables an understanding of how these structures arise in mammalian cells.
Keywords: Actin, Cdc42, Cell Biology, Cytoskeleton, Fluorescence Resonance Energy Transfer (FRET), Formin, Filopodia, IRSp53, WAVE2, mDia
Abstract
Filopodia are dynamic actin-rich cell surface protrusions involved in cell migration, axon guidance, and wound healing. The RhoGTPase Cdc42 generates filopodia via IRSp53, a multidomain protein that links the processes of plasma membrane deformation and actin dynamics required for their formation in mammalian cells. The Src homology 3 domain of IRSp53 binds to the actin regulators Mena, Eps8, WAVE1, WAVE2, mDia1, and mDia2. We show that mDia1 and WAVE2 synergize with IRSp53 to form filopodia. IRSp53 also interacts directly with these two proteins within filopodia, as observed in acceptor photobleaching FRET studies. Measurement of filopodium formation by time-lapse imaging of live cells also revealed that depleting neuronal cells of either mDia1 or WAVE2 protein decreases the ability of IRSp53 to induce filopodia. In contrast, IRSp53 does not appear to partner WAVE1 or mDia2 to give rise to these structures. In addition, although all three isoforms of mDia are capable of inducing filopodia, IRSp53 requires only mDia1 to do so. These findings suggest that mDia1 and WAVE2 are important Src homology 3 domain partners of IRSp53 in forming filopodia.
Introduction
Mammalian cells extend thin cylindrical protrusions known as filopodia during processes such as migration, axon guidance, angiogenesis, phagocytosis, and pathogen invasion (1–4). These dynamic structures are tubular extensions of the plasma membrane filled with bundles of linear actin filaments, and the cytoskeletal rearrangements leading to their formation are controlled by RhoGTPases such as Cdc42 (5–7) and Rif (8, 9).
IRSp53 (insulin receptor substrate protein 53 kDa) is a key effector of Cdc42 that couples the two events essential for filopodium formation, plasma membrane protrusion and actin dynamics (7). Its N-terminal inverse bin-amphiphysin-Rvs (I-BAR)3 domain binds to and deforms the plasma membrane, whereas its C-terminal Src homology 3 (SH3) domain interacts with various actin regulators such as neural Wiskott-Aldrich syndrome protein (N-WASP) (7), mammalian enabled (Mena) (6, 10), EGF receptor kinase substrate 8 (Eps8) (11), the WASP family verprolin homology (WAVE) isoforms WAVE1 (7) and WAVE2 (7, 12), and the mammalian Diaphanous (mDia) isoforms mDia1 (7, 13) and mDia2 (7). In the inactive state, the SH3 domain of IRSp53 is hidden by an intramolecular interaction between the N and C termini of the protein. Binding of Cdc42 to a partial Cdc42/Rac interactive binding motif located between the I-BAR and SH3 domains is believed to activate IRSp53 by exposing the SH3 domain (7).
mDia1 and mDia2 are members of the formin family of proteins, which are known for their ability to nucleate and polymerize linear actin filaments (14). They contain a proline-rich formin homology 1 domain that binds SH3 and WW domain-containing proteins and profilin and a formin homology 2 domain that binds actin monomers (15). Interaction between an N-terminal Diaphanous inhibitory domain and a C-terminal Diaphanous autoregulatory domain locks these formin proteins in an inactive conformation. Transition to an active state occurs when binding of a RhoGTPase to the GTPase binding domain (GBD) disrupts the autoinhibitory interaction (16). In addition to these domains, mDia1 and mDia2 also have an N-terminal basic domain that binds phospholipids and allows them to localize to the plasma membrane (17). Both of these formins have been linked to filopodium formation downstream of Rif, with mDia1 binding to this RhoGTPase within the protrusions (9). Under the control of RhoA, mDia1 also plays a role in stress fiber formation (18), neurite outgrowth (19), cell polarity (20), and adherens junction integrity (21). mDia2 too has been shown to partner other RhoGTPases in regulating various types of cytoskeletal rearrangements as follows: with RhoA it forms the actin scaffold required for constriction of the contractile ring during cytokinesis (22), with RhoB it controls actin dynamics required for vesicle movement (23), and with Rac it forms the contractile actin ring needed for enucleation of erythroblasts during erythropoiesis (24).
WAVE proteins function in actin dynamics by activating the actin-nucleating Arp2/3 (actin-related proteins 2 and 3) complex downstream of the RhoGTPase Rac (25). They contain a WAVE homology domain, a basic region, a proline-rich region, and a verprolin homology domain, central domain and acidic domain (VCA) region (25). The WAVE homology domain allows WAVE proteins to function as a multiprotein WAVE complex by binding directly to Abelson-interacting protein and hematopoietic stem/progenitor cell protein C300 (HSPC300), indirectly to Nck-associated protein 1 (NAP1), and specifically to Rac1-associated protein 1 (SRA1), while the VCA region brings together the Arp2/3 complex and an actin monomer to facilitate actin polymerization (26). Interaction of IRSp53 with the proline-rich region of WAVE2 is believed to mediate WAVE2 activity downstream of Rac (27). WAVE1 has been linked to the formation of dorsal ruffles, while WAVE2 has been implicated in both peripheral ruffle (28) and filopodial protrusion (29, 30) and demonstrated to be essential for lamellipodia (31, 32). WAVE2 has also been postulated to limit mDia2-induced filopodial activity by sequestering it together with the Arp2/3 complex (33).
N-WASP (7), Mena (7), and Eps8 (11) have been demonstrated to be important for IRSp53 to form filopodia, based on the lack of IRSp53-induced filopodia in knock-out cells devoid of the respective proteins. These three proteins have also been shown to interact with IRSp53 within filopodia (7). In contrast, WAVE1 does not appear to be involved in IRSp53-driven filopodium formation (7). However, the importance of other IRSp53 SH3 domain interactors, WAVE2 (7, 12), mDia1 (7, 13), and mDia2 (7), in filopodial protrusion mediated by IRSp53 has not been fully investigated. In this study, we show that mDia1 and WAVE2 can synergize with IRSp53 and induce filopodium formation. Acceptor photobleaching FRET (AP-FRET) experiments revealed that IRSp53 interacts with these two proteins in the filopodia of neuronal cells. In addition, we measured filopodium formation quantitatively using multichannel rapid time-lapse imaging of live cells as described in our previous work (7, 9) and found that silencing the expression of either mDia1 or WAVE2 reduced the number of IRSp53 filopodia formed. In contrast, knockdown of mDia2 did not affect IRSp53-induced filopodium formation. Furthermore, when mDia2 was coexpressed, there was a loss of the IRSp53 filopodial phenotype in neuronal cells. IRSp53 and mDia2 can colocalize and interact with each other in cytoplasmic puncta, which may explain the null filopodial phenotype observed in coexpression experiments. Taken together, these results suggest that mDia1 and WAVE2 are important SH3 domain partners of IRSp53 involved in filopodial protrusion.
EXPERIMENTAL PROCEDURES
Expression Constructs, Antibodies, and Reagents
pXJ40-mRFP-IRSp53 is described in Lim et al. (7). pXJ40-mRFP-IRSp53-FP/AA was created by subcloning the insert of pEGFP.C1-IRSp53-FP/AA (from Akiko Yamagishi, National Cancer Center Research Institute, Japan) into pXJ40-mRFP. GFP-Mena was from Klemens Rottner (Helmholtz Centre for Infection Research, Germany); EGFP-Eps8, GFP-WAVE1, and GFP-WAVE2 were from Giorgio Scita (FIRC Institute of Molecular Oncology Foundation-European Institute of Oncology, Italy); pEYFP.C1-mDia1 and pEYFP.C1-mDia2 were from Art Alberts (Van Andel Institute); pIRESpuro3-mCherry-Abp140p was from Philippe Chavrier (CNRS, France), and pEGFP-mDia3 was from Shuh Narumiya (Kyoto University, Japan). The primary antibodies used were as follows: mouse monoclonal anti-mDia1 (610848, 1:500, BD Transduction Laboratories); rabbit polyclonal anti-mDia2 N terminus (1:1000, from Shuh Narumiya, Kyoto University, Japan); goat polyclonal anti-mDia3 (sc-10889, 1:1000, Santa Cruz Biotechnology, Inc.); rabbit polyclonal anti-WAVE2 (sc-33548, 1:250, Santa Cruz Biotechnology, Inc.); mouse monoclonal anti-IRSp53 (1:1000, from S. Ahmed); mouse monoclonal anti-GFP (sc-9996, 1:500, Santa Cruz Biotechnology, Inc.), and mouse monoclonal anti-tubulin (1:5000, Sigma). The secondary antibodies used were as follows: HRP-conjugated goat anti-mouse IgG (sc-2005, 1:10,000); donkey anti-goat IgG (sc-2033, 1:2500), and goat anti-rabbit IgG (sc-2004, 1:5000), all from Santa Cruz Biotechnology, Inc.
Cell Culture, Transfection, and Microinjection
N1E115, N-WASP WT and KO, and CHO cells were cultured as described in Lim et al. (7). 293T, B16F1, HeLa, and NIH3T3 cells were grown in DMEM supplemented with 4500 mg/liter glucose, 10% FBS, and 1% penicillin/streptomycin. Transient transfections were done according to the manufacturer's protocol using Lipofectamine 2000 (Invitrogen) and, in the case of CHO cells, FuGENE 6 (Roche Applied Science). For imaging experiments involving N1E115 cells, glass coverslips or glass bottom dishes (MatTek Corp.) were coated with 10 μg/ml laminin (Invitrogen).
Protein Knockdown by RNAi
Transient knockdown of mDia1, mDia2, and WAVE2 proteins was performed, and a decrease in target protein levels assessed by Western blot of treated cells as described in Goh et al. (9).
Immunofluorescence
Cells grown on glass coverslips were washed once with PBS, fixed with 4% paraformaldehyde in PBS for 30 min, and then washed twice with PBS for 10 min. Cells were then permeabilized with 0.5% (v/v) Triton X-100 in PBS for 1 min, and washed twice with PBS for 10 min. After treatment with Alexa FluorTM 647-conjugated phalloidin (Molecular Probes) at 37 °C for 1 h, cells were washed twice with PBS containing 0.1% Tween 20 for 10 min, and coverslips mounted onto glass slides. All steps were carried out at room temperature unless otherwise specified. Cells were viewed using a multilaser scanning confocal microscope (Olympus FV1000) equipped with argon (488 nm), diode pumped solid state (561 nm), and helium neon (633 nm) lasers.
Live Cell Imaging and Analysis of Cell Morphology
Cells were cultured, transfected, and imaged, and images compiled into movies as described in Goh et al. (9). For images of fixed cells, as well as time-lapse imaging of live cells, morphological characteristics, including filopodial length and lifetime, were defined and scored as explained in Goh et al. (9). Protrusions that met the following characteristics of filopodia as defined by live imaging studies done on this cell line (length of up to 15 μm, width between 0.6 and 1.2 μm, nontapered i.e. of even width along their length, unbranched and emerging at random angles relative to the cell perimeter) (7) were scored as filopodium-like protrusions. This set of criteria was used to distinguish them from neurites, which typically span the length of at least one cell body in N1E115 cells (greater than 15 μm in length), as well as retraction fibers, which we have observed by live imaging to often be tapered (wider at the base than at the distal end closer to the tip) and branched, and sometimes appear in clusters or with several of them emerging from the same angle with respect to the cell perimeter (aligned parallel to one another). Fascin and myosin X were not used as markers for identifying filopodia as they have been shown to be present in retraction fibers (34). Only peripheral filopodia were scored, as the dorsal protrusions observed in both fixed and live N1E115 cells overexpressing activated Rif (9) were not seen in the same type of cells overexpressing IRSp53. We believe that at least in this cell line, the Rif pathway drives both apical and peripheral filopodium formation, while the IRSp53 pathway is responsible for only peripheral filopodia. For cotransfection experiments, mean GFP (or EYFP) and mRFP fluorescence intensities of cotransfected cells were semi-quantified, and only cells with a GFP/mRFP or EYFP/mRFP ratio of mean fluorescence intensities in the range of 0.5 to 2.0 were scored.
AP-FRET
AP-FRET was carried out using a Zeiss LSM 510 confocal microscope with a C-Apochromat ×63 water 1.2 NA objective. The filter settings used for mRFP/GFP and mRFP/EYFP FRET pairs are given in Lim et al. (7) and Goh et al. (9), respectively.
Statistical Analysis
Values in bar charts and tables are stated as mean values ± S.E. unless otherwise indicated. t test (two-tailed distribution, unpaired, equal variance) was calculated using Microsoft Excel, and the resulting p values are represented as follows: * denotes p < 0.05 and ** denotes p < 0.01.
RESULTS
Phenotypes of IRSp53 Partners in Neuronal Cells
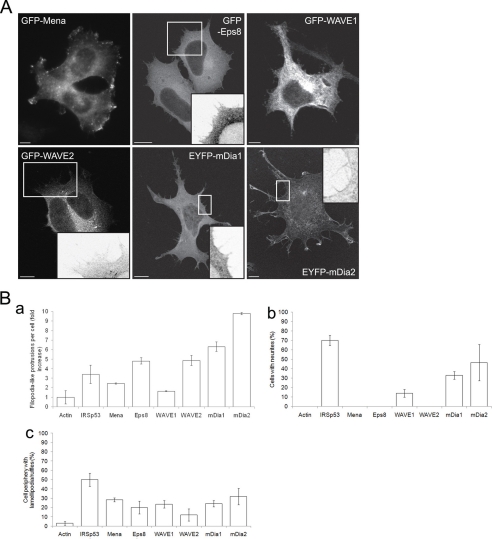
Mena (6), Eps8 (11), WAVE1 and WAVE2 (12), and mDia1 and mDia2 were identified as putative interacting partners of IRSp53 by mass spectrometry identification of brain and T-cell lysate proteins that bound to the IRSp53 SH3 domain (7). All of these proteins have been linked to actin cytoskeleton dynamics (25, 35). To determine which of these proteins are important for filopodium formation in neuronal cells, we overexpressed each of them in N1E115 cells. Cells transfected with fluorescent-tagged cDNA constructs were fixed at 20–38 h post-transfection and imaged. Stimulation of filopodium-like protrusions was seen in cells overexpressing GFP-Eps8, GFP-WAVE2, EYFP-mDia1, and EYFP-mDia2 (Fig. 1). In addition, GFP-WAVE1, EYFP-mDia1, and EYFP-mDia2 also induced neurite outgrowth (Fig. 1B, panel b). Most of the putative interacting partners tested also induced lamellipodia and ruffles (Fig. 1B, panel c). GFP-WAVE1 was found within the cell body but not seen in filopodium-like protrusions (Fig. 1A), a pattern of cellular distribution similar to that observed by Nozumi et al. (29) in both fixed and live NG108-15 neuroblastoma-glioma hybrid cells overexpressing EGFP-WAVE1.
FIGURE 1.
Phenotype of putative IRSp53-interacting partners in neuronal cells. N1E115 cells transfected with fluorescence-tagged cDNA constructs were fixed at 20–38 h post-transfection. A, examples of cells overexpressing GFP-Mena, GFP-Eps8, GFP-WAVE1, GFP-WAVE2, EYFP-mDia1, and EYFP-mDia2. Magnified sections of cells are shown in the insets. Bar, 10 μm. B, quantification of morphological characteristics of transfected N1E115 cells. Panel a, fold increase in number of filopodium-like protrusions formed per transfected cell; panel b, percentage of transfected cells with neurites, and panel c, average percentage of cell periphery with lamellipodia/ruffles. Data are presented as mean ± S.E. (n = 15–58).
Phenotypes Induced by Coexpression of IRSp53 and Its Interacting Partners
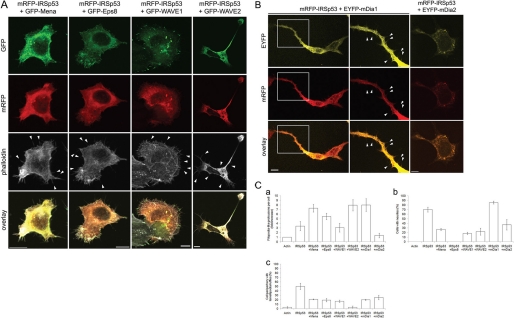
To investigate whether any of the putative interacting partners affect IRSp53 in giving rise to the morphological structures observed, we coexpressed them with mRFP-IRSp53 in N1E115 cells. Expression of full-length fluorescence-tagged IRSp53 and its putative interacting partners was verified by Western blot of lysates of cotransfected cells (supplemental Fig. S1).4 Transfected cells were fixed at 20–38 h post-transfection and stained with Alexa FluorTM 647-conjugated phalloidin. Cells expressing both mRFP-IRSp53 and GFP-Mena, GFP-Eps8, GFP-WAVE1, GFP-WAVE2, or EYFP-mDia1 had filopodium-like protrusions along their periphery (Fig. 2, A and B). In particular, Mena, WAVE2, and mDia1 synergized with IRSp53 in forming these protrusions (Fig. 2C, panel a). However, no filopodial projections were seen in cells coexpressing IRSp53 and mDia2. Instead, both proteins appeared in the cytoplasm and sometimes in cytoplasmic puncta (Fig. 2B). Neurite outgrowth was observed in cells positive for both mRFP-IRSp53 and GFP-Mena or GFP-WAVE2 (Fig. 2C, panel b), which was not seen when either GFP-Mena or GFP-WAVE2 was overexpressed alone. In the case of EYFP-mDia1, the presence of mRFP-IRSp53 resulted in enhanced neurite protrusion as compared with expressing either protein on its own (Figs. 1B, panel b, and 2C, panel b). However, no increase in neurite formation was seen when mRFP-IRSp53 was coexpressed with either GFP-WAVE1 or EYFP-mDia2 (Fig. 2C, panel b). mRFP-IRSp53 also appeared to hinder the ability of GFP-Mena, GFP-WAVE1, and GFP-WAVE2 to trigger lamellipodial protrusion and ruffling (Fig. 2C, panel c).
FIGURE 2.
Phenotypes induced by coexpression of IRSp53 and its putative interacting partners. N1E115 cells cotransfected with mRFP-IRSp53 and fluorescence-tagged cDNA constructs were fixed at 20–38 h post-transfection and stained with Alexa FluorTM 647-conjugated phalloidin. A, examples of cells coexpressing mRFP-IRSp53 and GFP-Mena, GFP-Eps8, GFP-WAVE1, or GFP-WAVE2. Arrowheads indicate individual filopodium-like protrusions. Bar, 10 μm. B, examples of cells coexpressing mRFP-IRSp53 and EYFP-mDia1 or EYFP-mDia2. A magnified section (middle column) of a cell coexpressing mRFP-IRSp53 and EYFP-mDia1 (left column) is shown with arrowheads indicating individual filopodium-like protrusions. Bar, 10 μm. C, quantification of morphological characteristics of transfected N1E115 cells. Panel a, fold increase in number of filopodium-like protrusions formed per transfected cell; panel b, percentage of transfected cells with neurites, and panel c, average percentage of cell periphery with lamellipodia/ruffles. Data are presented as mean ± S.E. (n = 13–35).
mDia2 and mDia3 Induce Filopodia in Neuronal Cells
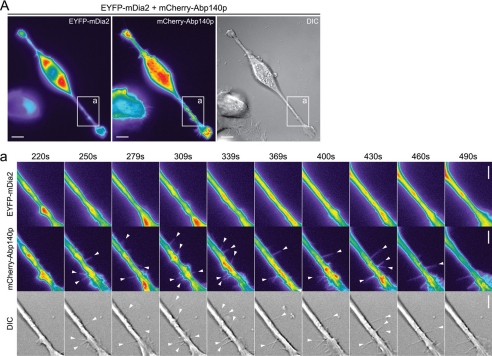
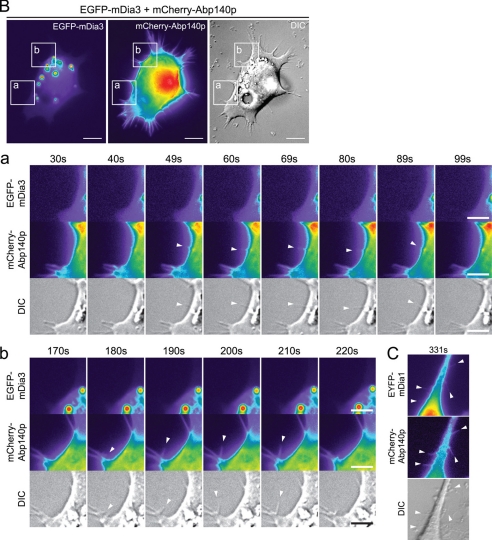
We have shown that mDia1 and mDia2 directly induce filopodium formation (9); however, mDia3 has not been investigated before. To compare the abilities of these three formins to induce filopodial protrusion, as well as the characteristics of the filopodia formed, N1E115 cells were transfected with EYFP-mDia2 or EGFP-mDia3 together with mCherry-Abp140p as a label for endogenous actin filaments. Cells positive for both mCherry and EYFP or EGFP were observed by time-lapse imaging at 18 h post-transfection. Filopodia were seen in cells expressing EYFP-mDia2, confirming that this construct is functional in terms of ability to trigger filopodial protrusion. Thus, the lack of filopodia seen earlier in cells coexpressing mRFP-IRSp53 and EYFP-mDia2 was not an artifact caused by the EYFP tag interfering with mDia2 protein function. The mDia2 filopodia were positive for mCherry but not EYFP along their lengths (Fig. 3A; supplemental Movies 1–3), showing that the protrusions contained actin but not mDia2. EGFP-mDia3 likewise induced mCherry-positive filopodia devoid of EGFP signal (Fig. 3B; supplemental Movies 4–9). The average length (4.59 μm) and lifetime (115 s) of the mDia2 filopodia were similar to that of mDia1 filopodia (p > 0.05). Although the filopodia induced by mDia3 had a lifetime similar to that of mDia1 and mDia2, they had a shorter average length (3.65 μm) than mDia2 filopodia (p < 0.01) (Table 1).
FIGURE 3.
mDia2 and mDia3 induce filopodia in neuronal cells. A, N1E115 cells were transfected with mCherry-Abp140p and EYFP-mDia2. Cells positive for both mCherry and EYFP were observed by time-lapse imaging at 18 h post-transfection. Bar, 10 μm. Panel a, series of time-lapse images shows a section of the transfected cell with arrowheads indicating individual filopodia (see supplemental Movies 1–3). Bar, 5 μm. B, N1E115 cells were transfected with mCherry-Abp140p and EGFP-mDia3. Cells positive for both mCherry and EGFP were observed by time-lapse imaging at 18 h post-transfection. Bar, 10 μm. Panels a and b, series of time-lapse images show sections of the transfected cell with arrowheads indicating individual filopodia (see supplemental Movies 4–9). Bar, 5 μm. C, section of an N1E115 cell with filopodia positive for both mCherry-Abp140p and EYFP-mDia1 from Goh et al. (9) is shown for comparison. For fluorescence images, fluorescence intensity is represented using a pseudocolor scale (from highest to lowest intensity: white, red, orange, yellow, green, blue, purple, and black) that allows structures with relatively low fluorescence intensity such as filopodia to be easily visualized. DIC, differential interference contrast.
TABLE 1.
Characteristics of filopodia induced by mDia isoforms
N1E115 cells were cotransfected with cDNA for mCherry-Abp140p and the protein of interest. Time-lapse imaging of fluorescent cells was done at 18 h post-transfection, and the number of filopodia formed per cell and the length and lifetime of such protrusions were measured (see “Experimental Procedures” for details). “Endogenous” refers to filopodia seen when cells were transfected with only mCherry-Abp140p; “EYFP-mDia1” refers to cells cotransfected with EYFP-mDia1 and mCherry-Abp140p; “myc-mDia2” refers to cells cotransfected with myc-mDia2 and GFP-actin; “EYFP-mDia2” refers to cells cotransfected with EYFP-mDia2 and mCherry-Abp140p, and “EGFP-mDia3” refers to cells cotransfected with EGFP-mDia3 and mCherry-Abp140p. Data are presented as mean ± S.E. * denotes p < 0.05 and ** denotes p < 0.01, all with respect to the values for endogenous in the same column; endogenous n = 22, EYFP-mDia2 n = 28, EGFP-mDia3 n = 30.
| Filopodia per cell | Length | Lifetime | |
|---|---|---|---|
| μm | s | ||
| Endogenous | 1.3 ± 1.03 | 4.68 ± 1.01 | 166 ± 17 |
| IRSp53a | 6.80 ± 1.88 | 187 ± 38 | |
| EYFP-mDia1b | 5.6 ± 1.27 | 3.99 ± 0.60 | 135 ± 19 |
| myc-mDia2b | 6.5 ± 0.73 | 4.61 ± 1.76 | 163 ± 78 |
| EYFP-mDia2 | 6.7 ± 0.67** | 4.59 ± 0.12 | 115 ± 31* |
| EGFP-mDia3 | 5.2 ± 1.37* | 3.65 ± 0.19 | 119 ± 31* |
a Data for IRSp53 filopodia in N1E115 cells from Lim et al. (7) are shown in italics for comparison but ar excluded from the statistical analysis.
b Data for mDia1 and myc-mDia2 filopodia in N1E115 cells from Goh et al. (9) are shown in italics for comparison but excluded from the statistical analysis.
IRSp53 Interacts with mDia1 and WAVE2 in Filopodia
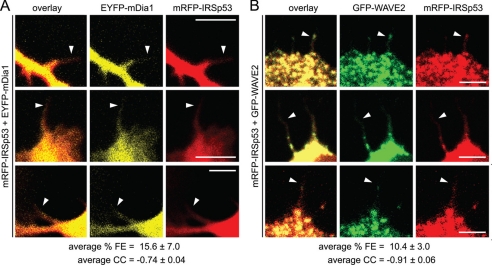
We have previously used AP-FRET to show protein-protein interaction in filopodia (7, 36, 37). Here, the same technique was employed to determine which of its SH3 domain binding partners IRSp53 interacts with within filopodia. N1E115 cells were cotransfected with mRFP-IRSp53 and GFP- or EYFP-tagged proteins of interest and fixed at 24 h post-transfection. mRFP-IRSp53 (acceptor) was bleached in selected regions of interest, and resultant changes in GFP or EYFP (donor) and mRFP (acceptor) fluorescence were measured. FRET occurs when donor and acceptor molecules are no further than 10 nm apart and is expressed in terms of percentage of FRET efficiency (% FE). FRET also gives rise to a negative correlation in the rates of change of fluorescence between the acceptor and donor, which is quantified and expressed as a correlation coefficient (CC) (7). Positive control (mRFP-GFP and mRFP-EYFP tandem constructs) and negative control (mRFP and GFP, mRFP-IRSp53 and GFP, mRFP and EYFP-mDia1, and mRFP and EYFP-mDia2) data were taken from previous studies done by Lim et al. (7) for mRFP/GFP pairs and by Goh et al. (9) for mRFP/EYFP pairs. % FE values greater than 3% with corresponding CC values of −1.0 to −0.7 were taken as an indication of positive FRET and protein-protein interaction. Positive FRET between IRSp53 and mDia1, as well as between IRSp53 and WAVE2, was observed within filopodium-like protrusions in N1E115 cells (Fig. 4; Table 2), showing that these two pairs of proteins interact within these structures in vivo. This is similar to the results obtained for the IRSp53/Eps8 and IRSp53/Mena FRET pairs (Table 2) (7). In contrast, IRSp53 interacted with mDia2 and WAVE1 only in the cytoplasm of CHO (supplemental Table S1) and N1E115 cells, respectively (Table 2). IRSp53 also interacted with WAVE2, Eps8, and Mena in cytoplasmic puncta and with mDia1 in neurites (data not shown). In control experiments where wild-type IRSp53 was replaced by IRSp53-FP/AA, a mutant with a nonfunctional SH3 domain (38), no positive FRET was detected for all of the IRSp53 SH3 domain-binding partners studied (Table 2).
FIGURE 4.
IRSp53 interacts with mDia1 and WAVE2 in filopodia. N1E115 cells transfected with mRFP-IRSp53 and EYFP-mDia1 (A) or GFP-WAVE2 (B) were fixed at 24 h post-transfection. Individual filopodia were selected as regions of interest, and the fluorescence intensities of the regions of interest were monitored in both mRFP and EYFP or GFP channels for the entire duration of the experiment. mRFP was bleached using a 561 nm laser once base line intensities of both mRFP (acceptor) and EYFP or EGFP (donor) fluorescence were established. Subsequent changes in these fluorescence intensities were measured and expressed as % FE, and the correlation between the rates of change of these values was expressed as CC (see “Experimental Procedures” for details). % FE and CC values obtained for the various control and experimental setups are given in Table 2. Positive FRET was defined as % FE > 3% with CC values of −1.0 to −0.7. Arrowheads indicate individual filopodia used for FRET measurements. Bar, 5 μm. Data are presented as mean ± S.D. (mRFP-IRSp53/EYFP-mDia1 n = 3; mRFP-IRSp53/GFP-WAVE2 n = 4.)
TABLE 2.
FRET analysis of interaction between IRSp53 and its SH3 domain binding partners in N1E115 cells
N1E115 cells transfected with the following combinations of cDNA constructs encoding the various controls, mRFP-IRSp53, mRFP-IRSp53-FP/AA, and IRSp53 SH3 domain binding proteins, were fixed at 24 h post-transfection. Fluorescence intensities of selected regions of interest were monitored in both mRFP and GFP or EYFP channels for the entire duration of the experiment. mRFP was bleached using a 561-nm laser once base-line intensities of both mRFP (acceptor) and GFP or EYFP (donor) fluorescence were established. Subsequent changes in these fluorescence intensities were measured and expressed as % FE, and the correlation between the rates of change of these values were expressed as CC (see “Experimental Procedures” for details). Positive FRET was defined as % FE > 3% with CC values of −1.0 to −0.7. Data are presented as mean ± S.D. (n = 6–14).
| FRET pairs | % FE | CC |
|---|---|---|
| mRFP/GFP controls | ||
| mRFP-GFP (tandem positive control)a | 22.3 ± 5.3 | −0.99 ± 0.001 |
| mRFP and GFP (negative control)a | 2.1 ± 1.9 | 0.09 ± 0.69 |
| mRFP-IRSp53 and GFP (negative control)a | 1.4 ± 1.2 | 0.29 ± 0.40 |
| IRSp53 and GFP-tagged partners | ||
| mRFP-IRSp53 and GFP-Mena | 8.6 ± 2.2 | −0.94 ± 0.06 |
| mRFP-IRSp53-FP/AA and GFP-Mena | 2.3 ± 1.4 | −0.24 ± 0.37 |
| mRFP-IRSp53 and GFP-Eps8 | 9.1 ± 4.3 | −0.89 ± 0.08 |
| mRFP-IRSp53-FP/AA and GFP-Eps8 | 1.8 ± 1.7 | −0.13 ± 0.48 |
| mRFP-IRSp53 and GFP-WAVE1 | 11.2 ± 2.9 | −0.94 ± 0.06 |
| mRFP-IRSp53-FP/AA and GFP-WAVE1 | 3.7 ± 2.5 | −0.16 ± 0.45 |
| mRFP-IRSp53 and GFP-WAVE2 | 8.5 ± 3.5 | −0.89 ± 0.06 |
| mRFP-IRSp53-FP/AA and GFP-WAVE2 | 2.7 ± 1.8 | −0.41 ± 0.34 |
| mRFP/EYFP controls | ||
| mRFP-EYFP (tandem positive control)b | 17.8 ± 3.6 | −0.97 ± 0.04 |
| mRFP and EYFP-mDia1 (negative control)b | 4.0 ± 2.3 | −0.41 ± 0.42 |
| mRFP and EYFP-mDia2 (negative control)b | 1.4 ± 1.7 | 0.05 ± 0.37 |
| IRSp53 and EYFP-tagged partners | ||
| mRFP-IRSp53 and EYFP-mDia1 | 13.3 ± 1.5 | −0.96 ± 0.02 |
| mRFP-IRSp53-FP/AA and EYFP-mDia1 | 1.8 ± 1.8 | −0.49 ± 0.39 |
| mRFP-IRSp53 and EYFP-mDia2 | 2.3 ± 1.4 | −0.16 ± 0.52 |
| mRFP-IRSp53-FP/AA and EYFP-mDia2 | 2.4 ± 2.0 | −0.19 ± 0.53 |
Expression of mDia Isoforms in Various Cell Lines
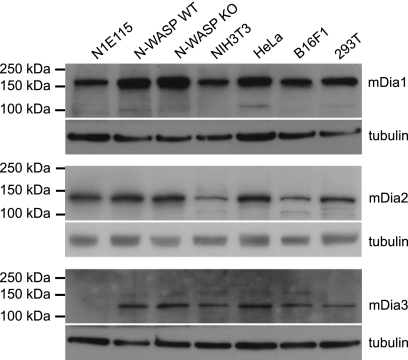
To determine whether mDia1, -2, or -3 are required for IRSp53-mediated filopodium formation, we first examined the expression of these three proteins in various cell lines that have been used in previous studies to investigate filopodium formation. The antibodies against mDia1 and mDia2 used in this study have been demonstrated by Western blot to be specific for the respective isoforms based on their ability to detect exogenous mDia1 or mDia2 protein. The anti-mDia1 antibody picked up overexpressed mutant mDia1 lacking the GBD domain (24), while the anti-mDia2 antibody detected overexpressed EYFP-mDia2 (9). These antibodies also picked up the reduction in endogenous mDia1 or mDia2 protein levels in cells treated with the respective specific RNAi but not in those treated with control RNAi (9). To verify the specificity of the anti-mDia3 antibody, N1E115 cells were transfected with EGFP-mDia3, and lysate obtained at 21 h post-transfection was probed with the antibody. A single band with an apparent molecular mass that corresponds to the predicted value of ∼152 kDa (EGFP 27 kDa and mDia3 125 kDa) for EGFP-mDia3 was seen (supplemental Fig. S2). Lysates from untransfected 293T and N1E115 cells were included for comparison, and while a single band of lower molecular mass (between 100 and 150 kDa) was seen for 293T cells, no band was detected for N1E115 (supplemental Fig. S2).
Next, the antibodies specific for mDia1, mDia2, or mDia3 were used to probe Western blots of whole cell lysates from the following cell lines: N1E115 mouse neuroblastoma; N-WASP WT and KO mouse fibroblasts; NIH3T3 mouse embryonic fibroblasts; HeLa human epithelial carcinoma; B16F1 mouse melanoma, and 293T human embryonic kidney. mDia1 was detected in all of the above cell lines (Fig. 5), although mDia3 was present in all except N1E115 (Fig. 5). For mDia2, a band corresponding to the 134-kDa molecular mass of the protein was clearly seen in all cell lines (Fig. 5).
FIGURE 5.
Expression of mDia isoforms in various cell lines. Antibodies specific for mDia1 (top panels), mDia2 N terminus (middle panels), or mDia3 (bottom panels) were used to probe Western blots of whole cell lysates from the following cell lines: N1E115 mouse neuroblastoma; N-WASP WT and KO mouse fibroblasts; NIH3T3 mouse embryonic fibroblasts; HeLa human epithelial carcinoma; B16F1 mouse melanoma, and 293T human embryonic kidney.
IRSp53-induced Filopodium Formation Involves mDia1 and WAVE2 but Not mDia2
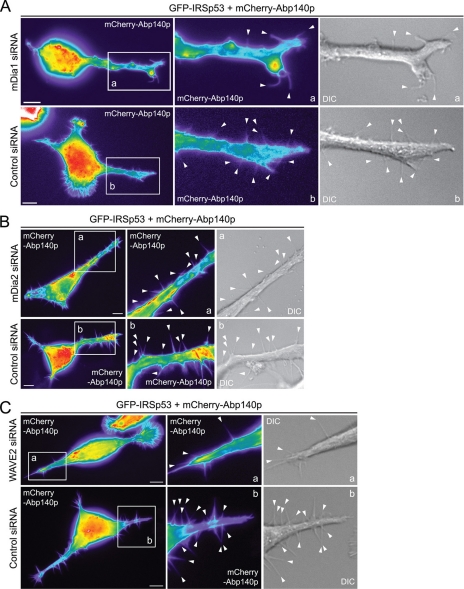
IRSp53 interacts with both mDia1 and mDia2 as shown by mass spectrometry identification of protein partners pulled down by IRSp53 (7). It also interacts with mDia1 and WAVE2 within filopodia based on results of the AP-FRET experiments (Table 2). To determine which of these three proteins are involved in IRSp53-mediated filopodium formation, we knocked down each of them separately using siRNA as described previously in Goh et al. (9). Endogenous mDia1, mDia2, and WAVE2 protein levels were reduced by 81, 87, and 66%, respectively (9). N1E115 cells were cotransfected with GFP-IRSp53, mDia1 or control siRNA, and mCherry-Abp140p, and cells positive for both GFP and mCherry were observed by time-lapse imaging at 24 h post-transfection. There was a statistically significant decrease (p < 0.01) in the number of filopodia formed per cell when mDia1 was knocked down (Fig. 6A; Table 3), although the average length and lifetime of the filopodia formed did not differ. A statistically significant reduction (p < 0.05) in number of IRSp53 filopodia was also seen when WAVE2 was silenced (Fig. 6C; Table 3). In contrast, no loss of IRSp53-induced filopodia occurred when the experiment was carried out using siRNA targeting mDia2 (Fig. 6B; Table 3).
FIGURE 6.
IRSp53 filopodium formation involves mDia1 and WAVE2. A, N1E115 cells were cotransfected with GFP-IRSp53, mCherry-Abp140p, and either mDia1 or nontargeting control siRNA, and time-lapse imaging of GFP-positive cells done at 24 h post-transfection. Bar, 10 μm. Magnified sections of transfected cells are shown in panels a and b with arrowheads indicating individual filopodia. B, N1E115 cells were cotransfected with GFP-IRSp53, mCherry-Abp140p, and either mDia2 or nontargeting control siRNA, and time-lapse imaging of GFP-positive cells done at 40 h post-transfection. Bar, 10 μm. Magnified sections of transfected cells are shown in panels a and b with arrowheads indicating individual filopodia. C, N1E115 cells were transfected with GFP-IRSp53, mCherry-Abp140p, and either WAVE2 or nontargeting control siRNA, and time-lapse imaging of GFP-positive cells done at 42 h post-transfection. Bar, 10 μm. Magnified sections of transfected cells are shown in panels a and b with arrowheads indicating individual filopodia. For fluorescence images, fluorescence intensity is represented using a pseudocolor scale (from highest to lowest intensity: white, red, orange, yellow, green, blue, purple, and black) that allows structures with relatively low fluorescence intensity such as filopodia to be easily visualized.
TABLE 3.
Effect of mDia1, mDia2, and WAVE2 knockdown on IRSp53-driven filopodium formation
N1E115 cells were transfected with GFP-IRSp53, mCherry-Abp140p, and RNAi targeting the protein of interest or nontargeting control RNAi, and time-lapse imaging was done on GFP- and mCherry-positive cells. The number of filopodia formed per cell and the length and lifetime of such protrusions were measured (see “Experimental Procedures” for details). Data are presented as mean ± S.E. * denotes p < 0.05, and ** denotes p < 0.01 when compared with the respective controls; mDia1 siRNA n = 24; control siRNA n = 19; mDia2 siRNA n = 21; control siRNA n = 28; WAVE2 siRNA n = 23; control siRNA n = 17.
| Filopodia per cell | Length | Lifetime | |
|---|---|---|---|
| μm | s | ||
| mDia1 siRNA | 4.5 ± 1.05** | 3.91 ± 0.72 | 157 ± 20 |
| Control siRNA | 9.9 ± 1.33 | 5.01 ± 0.67 | 186 ± 10 |
| mDia2 siRNA | 8.8 ± 1.02 | 5.38 ± 0.64 | 150 ± 16 |
| Control siRNA | 9.2 ± 1.50 | 5.36 ± 0.35 | 166 ± 26 |
| WAVE2 siRNA | 6.1 ± 1.19* | 4.53 ± 0.13 | 153 ± 8b |
| Control siRNA | 10.1 ± 1.55 | 4.41 ± 0.50 | 174 ± 8 |
DISCUSSION
Filopodia are dynamic actin-containing cell surface protrusions that can extend and retract over a time span of a few minutes (9). We have defined mammalian filopodia using a robust and quantitative time-lapse imaging assay in living cells (7, 9). Endogenous filopodia in mammalian cells are 8–15 μm long, with a uniform tubular morphology and a lifetime of 79–142 s (7). This approach to investigating filopodium formation has allowed us to confirm that the protrusions generated by Cdc42, IRSp53, and N-WASP (7), Rif and mDia1 (9), and Toca-1 (transducer of Cdc42-dependent actin assembly 1) (36) are similar to endogenous filopodia. Furthermore, the quantitative analysis allows us to probe the molecular mechanism of filopodium formation. Here, we have used the same technique to characterize the filopodia induced by mDia2 and mDia3, as well as to study the involvement of WAVE2, mDia1, and mDia2 in IRSp53-driven filopodium formation in neuronal cells.
In this study, we have used overexpression of IRSp53 to dissect the Cdc42/IRSp53/N-WASP signaling pathway to filopodium formation. Unlike stimuli such as growth factors or phorbol esters that have the potential to activate multiple signaling pathways, this overexpression approach allows us to investigate the specific pathway(s) downstream of IRSp53. This specificity is pertinent in light of our previous findings that more than one independent pathway to filopodium formation exists (9), indicating a need to discriminate between pathways. In future work, to confirm and extend the conclusions reached here, other approaches will need to be taken to complement these overexpression studies.
We have previously shown that EYFP-mDia1 and myc-mDia2 induce filopodia in N1E115 neuroblastoma cells (9). Here, we present three further observations on the mDia filopodial phenotype in N1E115 cells. First, EYFP-mDia2 has the same capability as myc-mDia2 to induce filopodia. In the earlier study, we were unable to verify the presence or absence of mDia2 protein within the filopodia as observed by time-lapse imaging because of the lack of a fluorescence label (9). In this study, EYFP-tagged mDia2 was seen to induce a similar number of filopodia per cell, and the structures had a similar length and lifetime. This verifies our earlier findings and shows that tagging the N terminus of mDia2 does not interfere with its filopodial phenotype. EGFP-mDia3 is also able to drive filopodium formation. This indicates that all three mDia isoforms can form filopodia, suggesting a prominent role for mDia proteins in this process. Given the function of mDia1 (18, 39) and possibly mDia2 (40) in stress fiber formation, the dual role of these mDia isoforms might allow them to determine the balance in the antagonistic relationship between filopodia and stress fibers found in most cell types. The involvement of mDia proteins in generating other types of structures such as invadopodia (41), lamellipodia (42), and the filopodial precursors of dendritic spines (43) also suggests that they control the formation of a wide range of actin-based cellular protrusions.
Second, mDia1 but not mDia2 or mDia3 can be detected within filopodia as observed by live cell imaging. This implies that mDia2 and mDia3 might be involved in the initiation stage of filopodium formation and not the elongation stage or that they might play an indirect role. Third, mDia1 and mDia2 are strong inducers of lamellipodia/membrane ruffles and neurite outgrowth. This agrees with the findings of earlier studies where mDia1 localized to lamellae and membrane ruffles (44, 45), and mDia2 was found to be essential for lamellipodia based on the ability of exogenous mDia2 to rescue the loss of the lamellipodial phenotype in mDia2 knockdown cells (42). Other groups have also linked mDia1 to axon elongation downstream of Rho (19) and have shown mDia2 to be involved in neuritogenesis (46).
Cdc42 induces filopodia through IRSp53. The IRSp53 I-BAR domain can deform membranes but does not induce bona fide filopodia. Full-length IRSp53, with a functional I-BAR domain, Cdc42-binding site, and SH3 domain, is required to generate filopodia (7, 47). Binding partners of the IRSp53 SH3 domain that have been implicated in IRSp53-driven filopodium formation include N-WASP (7), Mena (6), and Eps8 (11). Two-hybrid studies first showed the ability of WAVE1 and mDia1 to interact with the IRSp53 SH3 domain (12, 38). Subsequently, mass spectrometry analysis revealed that GST-tagged IRSp53 SH3 domain can pull down WAVE1, WAVE2, mDia1, and mDia2 (12). In this study we looked at the role of IRSp53-WAVE1, IRSp53-WAVE2, IRSp53-mDia1, and IRSp53-mDia2 interactions in the formation of filopodia and other morphological structures (lamellipodia, membrane ruffles, and neurites) in N1E115 cells. IRSp53 synergized with mDia1 and WAVE2 in filopodium formation but not with mDia2, and the IRSp53-mDia2 combination resulted in a null filopodial phenotype instead. WAVE1 caused a decrease in IRSp53-induced neurite outgrowth and lamellipodia/ruffle protrusion. For both the IRSp53-WAVE1 and IRSp53-mDia2 cotransfection experiments, there was strong expression of full-length mRFP-IRSp53 protein in the cotransfected cells as shown by Western blot. This rules out the possibility that the reduction in phenotypes was due to low mRFP-IRSp53 expression and indicates that the changes in phenotype were due to GFP-WAVE1 and EYFP-mDia2. In a study by Peng et al. (48), knocking out mDia1 suggested that mDia2 functions downstream of Cdc42 in actin microspike formation. In contrast, Pellegrin and Mellor (8) found that mDia2 on its own or in combination with Cdc42 did not induce actin microspikes. Because time-lapse experiments were not performed in either study, it is not possible to draw from them conclusions on the role of mDia2 in filopodium formation. Peng et al. (48) also showed by sensitized emission FRET a direct interaction between Cdc42 and mDia2. This interaction was predominantly in the cytoplasm at sites of γ-tubulin staining (at the microtubule organizing center) or at the leading edge but not significantly within actin microspikes (48). We found that in CHO cells, IRSp53 localizes and interacts with mDia2 in the cell cytoplasm and that mDia2 appears to recruit IRSp53 to cytoplasmic puncta (data not shown).
To examine whether IRSp53 interacts with mDia1 and WAVE2 in filopodia, we carried out FRET experiments. We have previously used AP-FRET to show that IRSp53 interacts with Mena, N-WASP, and Eps8. With this approach, we found that IRSp53 interacted with both mDia1 and WAVE2 in filopodia. Both IRSp53 and mDia1 were seen along the length of filopodia; this is consistent with the idea that IRSp53 establishes and maintains the curved membrane protrusion of the structure (49), but in contrast to the tip nucleation model of filopodium formation where formins are localized to the filopodial tip (2). In vivo evidence for formins localizing at the tips of mammalian filopodia comes from studies that overexpressed constitutively active truncated mutants of mDia2 missing either the GBD or Diaphanous autoregulatory domain (42, 48, 50). Abnormal club-shaped filopodia were observed with the use of these mutants (42, 50), and these structures might not be fully representative of filopodia that form under normal physiological conditions. Full-length endogenous mDia2 has been shown to be at the tips of filopodia only in B16F1 melanoma cells (42). As for mDia1, full-length exogenous mDia1 has been found to localize to filopodial tips only in the presence of overexpressed constitutively active Rif in HeLa cells (39). The exact role(s) played by mDia1 and mDia2 in filopodium formation in mammalian cells is unclear. In our live imaging studies, we found that only mDia1 and not mDia2 was seen within the filopodia that they induced (this study) and with mDia1 distributed along the entire length of the structures (9). Although mDia1 was shown to be poor at bundling actin filaments relative to mDia2 under specific conditions in vitro (51), it is possible that mDia1 might play a role in bundling filopodial actin filaments in vivo, in which case its distribution within filopodia would be uniform and not restricted to the tips. Further work is necessary to define the mechanistic roles of mDia1 in filopodium formation in mammalian cells. IRSp53 did not interact with mDia2 in N1E115 cells. However, these two proteins did interact in the cytoplasm and cytoplasmic puncta of CHO cells but not at the cell periphery or in filopodia. Interestingly, IRSp53 also interacted with mDia1, WAVE1, Eps8, and Mena in cytoplasmic puncta (data not shown). We believe the cytoplasmic puncta sites of IRSp53 interaction with these partners could be physiologically relevant, reflecting a possible membrane trafficking role for these complexes. Further work will be required to determine their exact function.
A critical part of the analysis was to investigate whether knockdown of mDia1, mDia2, or WAVE2 affected IRSp53-mediated filopodium formation. To establish the knockdown of mDia proteins, it was crucial to verify the specificity of the antibodies used. The specificity of the anti-mDia1 and -mDia2 antibodies has been demonstrated by their ability to detect the following: (i) exogenous overexpressed Flag-mDia1ΔGBD (an N-terminal truncated mutant) (24) or EYFP-mDia2 (9), as well as the endogenous proteins (9, 24), and (ii) reduction in endogenous protein levels in cells treated with specific RNAi but not those treated with control RNAi (9). Here, we carried out a similar verification for the anti-mDia3 antibody using EGFP-mDia3 and found it to be specific. Using these antibodies specific for mDia1, mDia2, and mDia3, we screened cell lines for the expression of these isoforms. Interestingly, mDia3 protein was not detected in N1E115 cells and is thus not believed to be essential for IRSp53-mediated filopodium formation. For this reason, mDia3 was not investigated further. mDia1 and mDia2 were detected in N1E115 as well as other cell lines, with protein bands corresponding to the expected molecular mass.
Knockdown of mDia1 and WAVE2 but not mDia2 inhibited IRSp53-mediated filopodium formation. This indicates that mDia2 is not required for IRSp53 to form filopodia. This is in contrast to Rif-induced filopodia, where both mDia isoforms but not WAVE2 are involved (9). Rif has been shown to induce filopodia independently of N-WASP, Mena, and WAVE2 (9). As such, mDia1 appears to be the only effector demonstrated to be common between the Rif- and IRSp53-driven pathways for filopodium formation. In NG108 neuroblastoma cells, WAVE2 was localized to the initiation sites of microspikes at the leading edge and also at the tips of filopodia (29). It has also been seen at the tips of filopodia in B16 melanoma cells (30) and at the edges of microspikes in Jurkat T cells treated with Arp2 shRNA (52). A study that attempted to recapitulate filopodium formation in vitro has proposed that the initiation and elongation stages of filopodial protrusion involve Arp2/3- and formin-mediated actin polymerization, respectively (53). In view of this, WAVE2 could potentially play a role in Arp2/3 complex recruitment and activation during the initiation of IRSp53 filopodia, while mDia1 drives the elongation of the nascent structures into fully formed filopodia.
We have shown that the three isoforms of mDia are capable of inducing filopodia. However, only mDia1 was seen within the structures. It was also the only isoform to synergize with IRSp53 to form filopodia and to interact with IRSp53 within them. In neuroblastoma cells, silencing mDia1 expression significantly reduced IRSp53-induced filopodium formation, although knockdown of mDia2 did not. We conclude that all three mDia isoforms are able to play a role in filopodia, but only mDia1 appears to be important for the IRSp53-mediated pathway for forming these protrusions. Like mDia1, WAVE2 was established as a partner of IRSp53 in filopodium formation, based on its interaction with IRSp53 within filopodia and the decrease in IRSp53 filopodia in neuroblastoma cells depleted of WAVE2 protein. Taken together with previously published work, the data suggest that at least five proteins (Mena, Eps8, N-WASP, WAVE2, and mDia1) play a role controlling actin dynamics associated with IRSp53-mediated filopodium formation (see Table 4 for summary of data from different studies). Examining the specific localization of IRSp53 and its targets within filopodia (to the base, shaft, and/or tip) will aid in identifying their functions in driving the initiation, extension, and retraction of these structures. For example, we have found Toca-1 at the base of filopodia, indicating that it could play a role (via N-WASP) in forming short actin filaments that might develop further into filopodia (36). This is in contrast to Mena and Eps8, which are located at filopodial tips (6).5 Although IRSp53 is sometimes seen in the tips of filopodia, it is also distributed along the entire length of the structures (5). However, mDia1 is not found in filopodial tips. We are currently using super-resolution microscopy to reveal the detailed architecture of filopodia.
TABLE 4.
Protein involvement in the Cdc42-IRSp53 pathway to filopodium formation
The following four criteria were used to determine whether IRSp53 SH3 domain-binding partners play a role in filopodium formation: (i) ability to induce filopodia and evidence of synergy with IRSp53 (increase in number of filopodia generated when coexpressed with IRSp53 as compared with overexpression of IRSp53 alone); (ii) localization to and within filopodia (at the base, shaft, and/or tip); (iii) direct interaction with IRSp53 in filopodia (as seen by AP-FRET); and (iv) effect of KD/KO on IRSp53-mediated filopodium formation. The role of the various proteins in actin dynamics is given in the last column. ND means not determined.
| Protein | Filopodial induction (synergy with IRSp53) | Filopodial localization | IRSp53 interaction in filopodia (as seen by AP-FRET) | Effect of KD/KO on IRSp53-driven filopodium formation | Function in actin dynamics during IRSp53-mediated filopodium formation |
|---|---|---|---|---|---|
| IRSp53 | Yes (6) | Yes, tip (7) | Scaffold protein localizing actin dynamics to sites of membrane deformation | ||
| N-WASP | Yes (54) (ND) | Yes, shaft and base (36) | Yes (7) | Yes (KO) (7) | Stimulation of actin nucleation by Arp2/3 complex |
| Mena | Yes (Yes) | Yes, tip (7, 10, 55) and shaft (7) | Yes (7) | Yes (KO) (7) | Anti-capping |
| Eps8 | Yes (56) (Yes) | Yes, tipa and shaft (7, 11, 56) | Yes (7) | Yes (KO) (11) | Capping, bundling |
| WAVE1 | No (No) | No | No | No (KD)(7) | |
| WAVE2 | Yes (Yes) | Yes, tip (29, 30) and shaft | Yes | Yes (KD) | Stimulation of actin nucleation by Arp2/3 complex? |
| mDia1 | Yes (9) (Yes) | Yes, shaft (9) | Yes | Yes (KD) | Anti-capping, filament elongation, bundling? |
| mDia2 | Yes (No) | No | No | No (KD) | |
| mDia3b | Yes (ND) | No | ND | ND |
a See Footnote 4.
b N1E115 cells do not express mDia3, and it is therefore not required for IRSp53-mediated filopodium formation.
Supplementary Material
Acknowledgments
We thank Shuh Narumiya and Sadanori Watanabe for their generous gift of anti-mDia2 antibodies and EGFP-mDia3 cDNA.

This article contains supplemental Figs. S1 and S2, Table S1, Movies 1–9, and additional references.
In cotransfections of N1E115 cells using Lipofectamine 2000, an average of 45% double transfected cells (expressing both cDNA constructs), 5% single transfected cells (expressing only one of either two cDNA constructs), and 50% untransfected cells was obtained. When taking only transfected cells into account, an average of 89% of the transfected cells expressed both cDNA constructs.
W. Bu and S. Ahmed, unpublished data.
- I-BAR
- inverse bin-amphiphysin-Rvs
- SH3
- Src homology 3
- N-WASP
- neural Wiskott-Aldrich syndrome protein
- Mena
- mammalian enabled
- WAVE
- WASP family verprolin homology
- GBD
- GTPase binding domain
- AP-FRET
- acceptor photobleaching FRET
- mRFP
- monomeric red fluorescent protein
- EGFP
- enhanced green fluorescent protein
- EYFP
- enhanced yellow fluorescent protein
- % FE
- percentage FRET efficiency
- CC
- correlation coefficient
- KD
- knocked down
- KO
- knocked out.
REFERENCES
- 1. Mattila P. K., Lappalainen P. (2008) Filopodia: molecular architecture and cellular functions. Nat. Rev. Mol. Cell Biol. 9, 446–454 [DOI] [PubMed] [Google Scholar]
- 2. Mellor H. (2010) The role of formins in filopodia formation. Biochim. Biophys. Acta 1803, 191–200 [DOI] [PubMed] [Google Scholar]
- 3. Dixit R., Tiwari V., Shukla D. (2008) Herpes simplex virus type 1 induces filopodia in differentiated p19 neural cells to facilitate viral spread. Neurosci. Lett. 440, 113–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Berger C. N., Crepin V. F., Jepson M. A., Arbeloa A., Frankel G. (2009) The mechanisms used by enteropathogenic Escherichia coli to control filopodia dynamics. Cell. Microbiol. 11, 309–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Govind S., Kozma R., Monfries C., Lim L., Ahmed S. (2001) Cdc42Hs facilitates cytoskeletal reorganization and neurite outgrowth by localizing the 58-kDa insulin receptor substrate to filamentous actin. J. Cell Biol. 152, 579–594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Krugmann S., Jordens I., Gevaert K., Driessens M., Vandekerckhove J., Hall A. (2001) Cdc42 induces filopodia by promoting the formation of an IRSp53-ena complex. Curr. Biol. 11, 1645–1655 [DOI] [PubMed] [Google Scholar]
- 7. Lim K. B., Bu W., Goh W. I., Koh E., Ong S. H., Pawson T., Sudhaharan T., Ahmed S. (2008) The Cdc42 effector IRSp53 generates filopodia by coupling membrane protrusion with actin dynamics. J. Biol. Chem. 283, 20454–20472 [DOI] [PubMed] [Google Scholar]
- 8. Pellegrin S., Mellor H. (2005) The Rho family GTPase Rif induces filopodia through mDia2. Curr. Biol. 15, 129–133 [DOI] [PubMed] [Google Scholar]
- 9. Goh W. I., Sudhaharan T., Lim K. B., Sem K. P., Lau C. L., Ahmed S. (2011) Rif-mDia1 interaction is involved in filopodium formation independent of Cdc42 and Rac effectors. J. Biol. Chem. 286, 13681–13694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Tokuo H., Ikebe M. (2004) Myosin X transports Mena/VASP to the tip of filopodia. Biochem. Biophys. Res. Commun. 319, 214–220 [DOI] [PubMed] [Google Scholar]
- 11. Disanza A., Mantoani S., Hertzog M., Gerboth S., Frittoli E., Steffen A., Berhoerster K., Kreienkamp H. J., Milanesi F., Di Fiore P. P., Ciliberto A., Stradal T. E., Scita G. (2006) Regulation of cell shape by Cdc42 is mediated by the synergic actin-bundling activity of the Eps8-IRSp53 complex. Nat. Cell Biol. 8, 1337–1347 [DOI] [PubMed] [Google Scholar]
- 12. Miki H., Yamaguchi H., Suetsugu S., Takenawa T. (2000) IRSp53 is an essential intermediate between Rac and WAVE in the regulation of membrane ruffling. Nature 408, 732–735 [DOI] [PubMed] [Google Scholar]
- 13. Fujiwara T., Mammoto A., Kim Y., Takai Y. (2000) Rho small G-protein-dependent binding of mDia to an Src homology 3 domain-containing IRSp53/BAIAP2. Biochem. Biophys. Res. Commun. 271, 626–629 [DOI] [PubMed] [Google Scholar]
- 14. Faix J., Grosse R. (2006) Staying in shape with formins. Dev. Cell 10, 693–706 [DOI] [PubMed] [Google Scholar]
- 15. Wallar B. J., Alberts A. S. (2003) The formins. Active scaffolds that remodel the cytoskeleton. Trends Cell Biol. 13, 435–446 [DOI] [PubMed] [Google Scholar]
- 16. Young K. G., Copeland J. W. (2010) Formins in cell signaling. Biochim. Biophys. Acta 1803, 183–190 [DOI] [PubMed] [Google Scholar]
- 17. Ramalingam N., Zhao H., Breitsprecher D., Lappalainen P., Faix J., Schleicher M. (2010) Phospholipids regulate localization and activity of mDia1 formin. Eur J. Cell Biol. 89, 723–732 [DOI] [PubMed] [Google Scholar]
- 18. Watanabe N., Kato T., Fujita A., Ishizaki T., Narumiya S. (1999) Cooperation between mDia1 and ROCK in Rho-induced actin reorganization. Nat. Cell Biol. 1, 136–143 [DOI] [PubMed] [Google Scholar]
- 19. Arakawa Y., Bito H., Furuyashiki T., Tsuji T., Takemoto-Kimura S., Kimura K., Nozaki K., Hashimoto N., Narumiya S. (2003) Control of axon elongation via an SDF-1α/Rho/mDia pathway in cultured cerebellar granule neurons. J. Cell Biol. 161, 381–391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Yamana N., Arakawa Y., Nishino T., Kurokawa K., Tanji M., Itoh R. E., Monypenny J., Ishizaki T., Bito H., Nozaki K., Hashimoto N., Matsuda M., Narumiya S. (2006) The Rho-mDia1 pathway regulates cell polarity and focal adhesion turnover in migrating cells through mobilizing Apc and c-Src. Mol. Cell. Biol. 26, 6844–6858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Carramusa L., Ballestrem C., Zilberman Y., Bershadsky A. D. (2007) Mammalian diaphanous-related formin Dia1 controls the organization of E-cadherin-mediated cell-cell junctions. J. Cell Sci. 120, 3870–3882 [DOI] [PubMed] [Google Scholar]
- 22. Watanabe S., Okawa K., Miki T., Sakamoto S., Morinaga T., Segawa K., Arakawa T., Kinoshita M., Ishizaki T., Narumiya S. (2010) Rho and anillin-dependent control of mDia2 localization and function in cytokinesis. Mol. Biol. Cell 21, 3193–3204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wallar B. J., Deward A. D., Resau J. H., Alberts A. S. (2007) RhoB and the mammalian Diaphanous-related formin mDia2 in endosome trafficking. Exp. Cell Res. 313, 560–571 [DOI] [PubMed] [Google Scholar]
- 24. Ji P., Jayapal S. R., Lodish H. F. (2008) Enucleation of cultured mouse fetal erythroblasts requires Rac GTPases and mDia2. Nat. Cell Biol. 10, 314–321 [DOI] [PubMed] [Google Scholar]
- 25. Takenawa T., Suetsugu S. (2007) The WASP-WAVE protein network. Connecting the membrane to the cytoskeleton. Nat. Rev. Mol. Cell Biol. 8, 37–48 [DOI] [PubMed] [Google Scholar]
- 26. Kurisu S., Takenawa T. (2010) WASP and WAVE family proteins. Friends or foes in cancer invasion? Cancer Sci. 101, 2093–2104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Abou-Kheir W., Isaac B., Yamaguchi H., Cox D. (2008) Membrane targeting of WAVE2 is not sufficient for WAVE2-dependent actin polymerization. A role for IRSp53 in mediating the interaction between Rac and WAVE2. J. Cell Sci. 121, 379–390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Suetsugu S., Yamazaki D., Kurisu S., Takenawa T. (2003) Differential roles of WAVE1 and WAVE2 in dorsal and peripheral ruffle formation for fibroblast cell migration. Dev. Cell 5, 595–609 [DOI] [PubMed] [Google Scholar]
- 29. Nozumi M., Nakagawa H., Miki H., Takenawa T., Miyamoto S. (2003) Differential localization of WAVE isoforms in filopodia and lamellipodia of the neuronal growth cone. J. Cell Sci. 116, 239–246 [DOI] [PubMed] [Google Scholar]
- 30. Nakagawa H., Miki H., Nozumi M., Takenawa T., Miyamoto S., Wehland J., Small J. V. (2003) IRSp53 is colocalized with WAVE2 at the tips of protruding lamellipodia and filopodia independently of Mena. J. Cell Sci. 116, 2577–2583 [DOI] [PubMed] [Google Scholar]
- 31. Yamazaki D., Suetsugu S., Miki H., Kataoka Y., Nishikawa S., Fujiwara T., Yoshida N., Takenawa T. (2003) WAVE2 is required for directed cell migration and cardiovascular development. Nature 424, 452–456 [DOI] [PubMed] [Google Scholar]
- 32. Yan C., Martinez-Quiles N., Eden S., Shibata T., Takeshima F., Shinkura R., Fujiwara Y., Bronson R., Snapper S. B., Kirschner M. W., Geha R., Rosen F. S., Alt F. W. (2003) WAVE2 deficiency reveals distinct roles in embryogenesis and Rac-mediated actin-based motility. EMBO J. 22, 3602–3612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Beli P., Mascheroni D., Xu D., Innocenti M. (2008) WAVE and Arp2/3 jointly inhibit filopodium formation by entering into a complex with mDia2. Nat. Cell Biol. 10, 849–857 [DOI] [PubMed] [Google Scholar]
- 34. Lin L., McCroskery S., Ross J. M., Chak Y., Neuhuber B., Daniels M. P. (2010) Induction of filopodium-like protrusions by transmembrane agrin. Role of agrin glycosaminoglycan chains and Rho family GTPases. Exp. Cell Res. 316, 2260–2277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Chesarone M. A., DuPage A. G., Goode B. L. (2010) Unleashing formins to remodel the actin and microtubule cytoskeletons. Nat. Rev. Mol. Cell Biol. 11, 62–74 [DOI] [PubMed] [Google Scholar]
- 36. Bu W., Chou A. M., Lim K. B., Sudhaharan T., Ahmed S. (2009) The Toca-1-N-WASP complex links filopodial formation to endocytosis. J. Biol. Chem. 284, 11622–11636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Sudhaharan T., Liu P., Foo Y. H., Bu W., Lim K. B., Wohland T., Ahmed S. (2009) Determination of in vivo dissociation constant, KD, of Cdc42-effector complexes in live mammalian cells using single wavelength fluorescence cross-correlation spectroscopy. J. Biol. Chem. 284, 13602–13609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Yamagishi A., Masuda M., Ohki T., Onishi H., Mochizuki N. (2004) A novel actin bundling/filopodium-forming domain conserved in insulin receptor tyrosine kinase substrate p53 and missing in metastasis protein. J. Biol. Chem. 279, 14929–14936 [DOI] [PubMed] [Google Scholar]
- 39. Fan L., Pellegrin S., Scott A., Mellor H. (2010) The small GTPase Rif is an alternative trigger for the formation of actin stress fibers in epithelial cells. J. Cell Sci. 123, 1247–1252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Tominaga T., Sahai E., Chardin P., McCormick F., Courtneidge S. A., Alberts A. S. (2000) Diaphanous-related formins bridge Rho GTPase and Src tyrosine kinase signaling. Mol. Cell 5, 13–25 [DOI] [PubMed] [Google Scholar]
- 41. Lizárraga F., Poincloux R., Romao M., Montagnac G., Le Dez G., Bonne I., Rigaill G., Raposo G., Chavrier P. (2009) Diaphanous-related formins are required for invadopodia formation and invasion of breast tumor cells. Cancer Res. 69, 2792–2800 [DOI] [PubMed] [Google Scholar]
- 42. Yang C., Czech L., Gerboth S., Kojima S., Scita G., Svitkina T. (2007) Novel roles of formin mDia2 in lamellipodia and filopodia formation in motile cells. PLoS Biol. 5, e317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Hotulainen P., Llano O., Smirnov S., Tanhuanpää K., Faix J., Rivera C., Lappalainen P. (2009) Defining mechanisms of actin polymerization and depolymerization during dendritic spine morphogenesis. J. Cell Biol. 185, 323–339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Zaoui K., Honoré S., Isnardon D., Braguer D., Badache A. (2008) Memo-RhoA-mDia1 signaling controls microtubules, the actin network, and adhesion site formation in migrating cells. J. Cell Biol. 183, 401–408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Chhabra E. S., Higgs H. N. (2007) The many faces of actin. Matching assembly factors with cellular structures. Nat. Cell Biol. 9, 1110–1121 [DOI] [PubMed] [Google Scholar]
- 46. Dent E. W., Kwiatkowski A. V., Mebane L. M., Philippar U., Barzik M., Rubinson D. A., Gupton S., Van Veen J. E., Furman C., Zhang J., Alberts A. S., Mori S., Gertler F. B. (2007) Filopodia are required for cortical neurite initiation. Nat Cell Biol. 9, 1347–1359 [DOI] [PubMed] [Google Scholar]
- 47. Robens J. M., Yeow-Fong L., Ng E., Hall C., Manser E. (2010) Regulation of IRSp53-dependent filopodial dynamics by antagonism between 14-3-3 binding and SH3-mediated localization. Mol. Cell. Biol. 30, 829–844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Peng J., Wallar B. J., Flanders A., Swiatek P. J., Alberts A. S. (2003) Disruption of the Diaphanous-related formin Drf1 gene encoding mDia1 reveals a role for Drf3 as an effector for Cdc42. Curr. Biol. 13, 534–545 [DOI] [PubMed] [Google Scholar]
- 49. Ahmed S., Goh W. I., Bu W. (2010) I-BAR domains, IRSp53 and filopodium formation. Semin. Cell Dev. Biol. 21, 350–356 [DOI] [PubMed] [Google Scholar]
- 50. Block J., Stradal T. E., Hänisch J., Geffers R., Köstler S. A., Urban E., Small J. V., Rottner K., Faix J. (2008) Filopodia formation induced by active mDia2/Drf3. J. Microsc. 231, 506–517 [DOI] [PubMed] [Google Scholar]
- 51. Harris E. S., Rouiller I., Hanein D., Higgs H. N. (2006) Mechanistic differences in actin bundling activity of two mammalian formins, FRL1 and mDia2. J. Biol. Chem. 281, 14383–14392 [DOI] [PubMed] [Google Scholar]
- 52. Gomez T. S., Kumar K., Medeiros R. B., Shimizu Y., Leibson P. J., Billadeau D. D. (2007) Formins regulate the actin-related protein 2/3 complex-independent polarization of the centrosome to the immunological synapse. Immunity 26, 177–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Lee K., Gallop J. L., Rambani K., Kirschner M. W. (2010) Self-assembly of filopodium-like structures on supported lipid bilayers. Science 329, 1341–1345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Miki H., Sasaki T., Takai Y., Takenawa T. (1998) Induction of filopodium formation by a WASP-related actin-depolymerizing protein N-WASP. Nature 391, 93–96 [DOI] [PubMed] [Google Scholar]
- 55. Lanier L. M., Gates M. A., Witke W., Menzies A. S., Wehman A. M., Macklis J. D., Kwiatkowski D., Soriano P., Gertler F. B. (1999) Mena is required for neurulation and commissure formation. Neuron 22, 313–325 [DOI] [PubMed] [Google Scholar]
- 56. Roffers-Agarwal J., Xanthos J. B., Miller J. R. (2005) Regulation of actin cytoskeleton architecture by Eps8 and Abi1. BMC Cell Biol. 6, 36. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.