Background: Ndc10 is a DNA-binding protein in yeast that is responsible for centromere formation.
Results: The structure of the protein unexpectedly shows that it contains a type IB topoisomerase/λ-integrase fold.
Conclusion: The structure demonstrates how the IB/Int fold has been adapted to a new cellular role.
Significance: The structure is the first example of the IB/Int fold being used in a non-catalytic protein.
Keywords: Centromeres, Integrase, Kinetochore, Structural Biology, X-ray Crystallography
Abstract
We have solved the x-ray structure of the N-terminal half of the yeast kinetochore protein Ndc10 at 1.9 Å resolution. This essential protein is a key constituent of the budding yeast centromere and is essential for the recruitment of the centromeric nucleosome and establishment of the kinetochore. The fold of the protein shows unexpected similarities to the tyrosine recombinase/λ-integrase family of proteins, most notably Cre, with some variation in the relative position of the subdomains. This finding offers new insights into kinetochore evolution and the adaptation of a well studied protein fold to a novel role. By comparison with tyrosine recombinases and mutagenesis studies, we have been able to define some of the key DNA-binding motifs.
Introduction
The kinetochore is the large, multiprotein assembly that serves to connect condensed sister chromatids to the mitotic spindle (1). Proteins in the kinetochore are responsible for several functions, including transmission of mechanical tension, mitotic checkpoint control, and modulation of microtubule dynamics. The kinetochore binds to a chromosomal locus known as the centromere, which in budding yeast consists of a short ∼125-bp DNA sequence that is both necessary and sufficient to support kinetochore formation (2). This sequence may be subdivided into three sections, CDEI, CDEII, and CDEIII (3). CDEI binds the non-essential Cbf1 protein (4). CDEII is thought to be involved in interactions with the Cse4-containing centromeric nucleosome (5), whereas CDEIII is bound by the CBF3 complex (6) (see Fig. 1a). This ∼445-kDa complex contains four essential proteins, Ndc10, Cep3, Ctf13, and Skp1. Ctf13 and Skp1 appear to fulfill regulatory roles in the assembly of the complex (7–10), whereas Cep3 and Ndc10 can directly bind DNA. Cep3 has a Gal4-type DNA-binding domain that contacts a conserved CCG triplet in CDEIII, whereas Ndc10 has been proposed to bind the centromere both in the context of the intact CBF3 complex and independently at the CDEII element (11, 12). Recent work suggests that the primary function of CBF3/Ndc10 is to recruit the centromere-specific Cse4 nucleosome to DNA after S-phase, possibly by interactions with the histone chaperone Scm3 (13). Ndc10 has also been observed to relocate to the spindle midzone during anaphase, via interactions with the yeast survivin homolog, Bir1, indicating that it also plays an important role in coordination of cell division (14–17). Currently, the only proteins in the CBF3 complex for which we have structural data are Cep3 (12, 18) and Skp1 (19). In this study, we describe the x-ray structure of a large fragment of Ndc10, responsible for DNA binding.
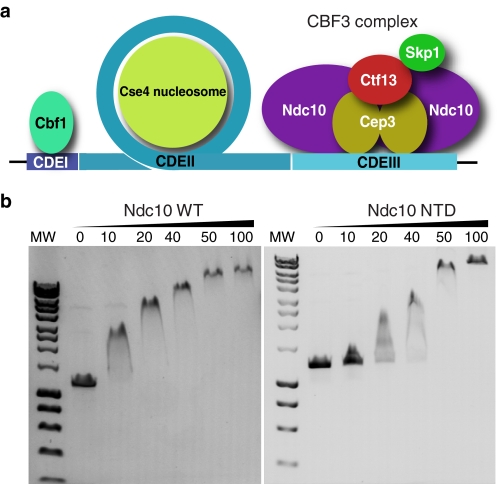
FIGURE 1.
Function of Ndc10 and CBF3 complex. a, schematic diagram of the budding yeast centromere, showing the three sequence elements and the proteins that bind them. b, DNA gel-shift assay showing the binding of the Ndc10 full-length and NTD to a 1.2-kb non-centromeric DNA sequence. Final protein concentrations in μm are shown above the gel. MW, molecular weight markers.
EXPERIMENTAL PROCEDURES
Molecular Biology
A DNA fragment encoding amino acid residues 1–551 (Ndc10 N-terminal domain (NTD))2 of the Saccharomyces cerevisiae Ndc10 (CBF3A) gene was generated by PCR and then subcloned into a pET28a plasmid that also encodes an N-terminal SUMO tag and SUMO protease site. Site-directed mutagenesis was carried out using standard PCR-based techniques.
Protein Expression and Purification
BL21-RIL (DE3)-competent cells (Agilent Technologies) were transformed with the Ndc10 NTD expression vector and grown in LB broth to A600 of 0.6 at 37 °C. Protein expression was induced overnight at 16 °C by the addition of isopropyl β-d-1-thiogalactopyranoside to a final concentration of 1 mm. Cells were harvested by centrifugation, resuspended in buffer A (60 mm Hepes, pH 7.5, 300 mm NaCl, 40 mm imidazole, 5% glycerol, 10 mm benzamidine, 1 mm PMSF, 10 mm β-mercaptoethanol), and immediately lysed by sonication in an ice bath. The cell lysate was centrifuged at 33,000 × g, and the clarified supernatant was loaded onto a pre-equilibrated His-Trap column (HisTrap FF, GE Healthcare) in buffer A. The SUMO-Ndc10 NTD protein was eluted in buffer A by a 40–500 mm imidazole gradient and immediately incubated overnight with a SUMO protease. Following removal of the tag, the protein was further purified by anion exchange chromatography (MonoQ 10/100GL, GE Healthcare) followed by size exclusion chromatography (HiLoad 16/60 Superdex 200, GE Healthcare). The purified protein was stored in buffer B (20 mm Hepes, 100 mm NaCl, 1 mm DTT). The protein purity and size was assessed by SDS-PAGE. Selenomethionine-labeled Ndc10 NTD was produced by expression in a methionine auxotroph in LeMaster minimal medium supplemented with specific amino acids as well as selenomethionine and purified as described above.
DNA Binding Assay
Ndc10 NTD was prepared at a range of concentrations in 20 mm Hepes, pH 7.5, 50 mm NaCl, 1 mm DTT and added to 300 ng of a 1.2-kb random sequence DNA fragment. The protein/DNA mixes were incubated for 10 min at 4 °C and then electrophoresed on a 0.8% agarose gel in Tris-borate-EDTA buffer. DNA was visualized using ethidium bromide under UV light.
Analytical Ultracentrifugation
Solution masses of the Ndc10 NTD and full-length Ndc10 were determined by sedimentation velocity studies. Proteins were concentrated to between 0.5 and 1 mg/ml in a buffer containing 10 mm Hepes, pH 7.5, 150 mm NaCl, 1 mm DTT. Samples were analyzed using a Beckman XL-I analytical centrifuge running at 25,000 rpm (NTD) or 20,000 rpm (full-length) in an An-60Ti rotor. Data were analyzed using the program DCDT+.
Protein Crystallization and Structure Determination
Purified Ndc10 NTD was exchanged against a buffer solution containing 20 mm Hepes (pH 7.5), 50 mm NaCl, 1 mm DTT and concentrated to 15 mg/ml. The protein was crystallized at 16 °C by a hanging-drop vapor diffusion method where the sample and the crystallization buffer (200 mm potassium bromide, 100 mm sodium acetate (pH 5.5), and 10% PEG 4000) were mixed in a 1:1 (v/v) ratio. Rectangular crystals appeared overnight and were cryo-protected with 25% PEG 400 before being flash-frozen in liquid nitrogen. Diffraction data on native and selenomethionine proteins were collected at the Diamond synchrotron beamline I24. Crystals were of space group P212121 with cell dimensions a = 55.7 Å, b = 87.8 Å, c = 104.6 Å. All programs were from the CCP4 suite unless otherwise stated (20). Diffraction data were indexed, integrated, and scaled using XDS (21) and SCALA (22). The structure was solved using single-wavelength anomalous dispersion (SAD) phasing as implemented in the AutoSHARP system (23), and an initial model was auto-traced using BUCCANEER (24). Subsequent rounds of rebuilding in Coot (25) were iterated with refinement using REFMAC (26). Later stages of refinement were carried out against the native data to an R-factor of 19.4% (Rfree = 23.2%). The final structural model was validated using Coot and Molprobity (27) tools. In the Ramachandran plot, 98.9 and 1.1% of residues are in allowed and outlier regions, respectively. Structure figures were prepared with CCP4mg (28).
RESULTS
Structure Determination
Like many kinetochore proteins, the sequence of Ndc10 contains no recognizable structural motifs and does not appear to be related to any other proteins in the sequence database (29, 30). The 111.9-kDa protein has 956 residues and is predicted to contain a large number of intrinsically disordered sequences, particularly in the C-terminal. During purifications of the full-length protein, we identified a 64.6-kDa proteolytically derived fragment of Ndc10, which was determined to contain residues 1–551 of the N-terminal. Both the full-length protein and this fragment were found to be competent for DNA binding against a random (non-centromeric) DNA sequence (Fig. 1b). We crystallized this fragment and determined the x-ray structure at 1.9 Å resolution using the SAD phasing technique. The final model was refined to an R-factor of 19.4% (Rfree = 23.2%) and has excellent geometry (Table 1). No clear electron density was seen for residues 1–43 or 538–551, and loops between residues 65–72, 97–108, 168–182, 257–264, and 414–423 were also disordered and not included in the final model.
TABLE 1.
Crystallographic statistics
Native and selenomethionine (SAD) data statistics are presented. Figures given in parentheses refer to data in highest resolution shell. Rsym is as defined by Evans (22).
| Native | SAD | |
|---|---|---|
| Wavelength (Å) | 0.970 | 0.9787 |
| Resolution range (Å) | 47.05–1.89 | 52.4–1.79 |
| Total reflections | 135,352 | 210,835 |
| Unique reflections | 41,242 | 48,942 |
| Multiplicity | 3.3 (3.3) | 4.3 (4.3) |
| Completeness (%) | 98.70 (98.80) | 99.10 (99.50) |
| I/σ(I) | 13.40 (2.20) | 13.70 (2.30) |
| Wilson B-factor | 25.11 | 23.05 |
| Rsym | 0.051 (0.496) | 0.063 (0.659) |
| R-factor | 0.194 | |
| Rfree | 0.232 | |
| Number of atoms | 3879 | |
| Protein residues | 452 | |
| Water molecules | 86 | |
| r.m.s. bonds (Å) | 0.021 | |
| r.m.s. angles (degrees) | 2.02 | |
| Ramachandran allowed (%) | 98.9 | |
| Ramachandran outliers (%) | 1.1 | |
| Model B-factor | 27.9 |
Overall Features
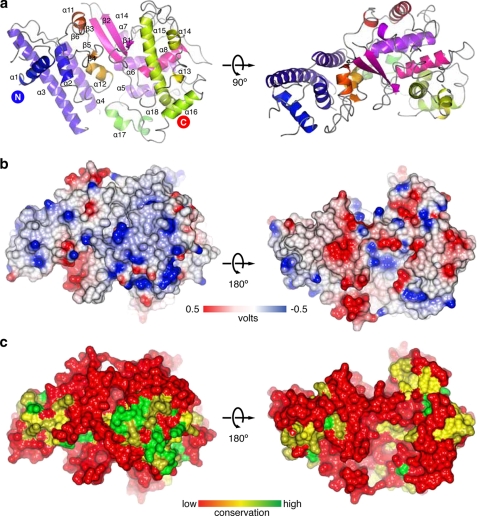
The structure of the protein is shown as a ribbon diagram in Fig. 2a. The overall fold may be divided into two α-helical lobes, which sandwich a central β-sheet formed by residues 239–370. The N-lobe of the protein consists of a distinct antiparallel four-helix bundle, whereas the C-lobe consists of several long loops interspersed with short helices and wraps back around the core of the protein. Analysis of the electrostatic molecular surface (Fig. 2b) shows that the convex “top” of the molecule is predominantly positively charged and is also one of the most highly conserved sections of the protein (Fig. 2c).
FIGURE 2.
Structural features of Ndc10 N-terminal domain. a, ribbon diagram of the Ndc10 NTD with secondary structural elements labeled. The positions of the N and C termini are indicated. b, electrostatic surface of the Ndc10 NTD. The first figure is in the same orientation as the second panel of a. Positive electrostatic potential is shown in blue, and negative electrostatic potential is in red. Note the large positively charged patch on the bottom right half of the protein. c, surface conservation plot between five Ndc10 orthologs. The most conserved area on the surface corresponds to the positively charged patch in b.
Oligomerization
Although Ndc10 has been proposed to exist as a dimer in the intact CBF3 complex (9), the crystal packing of the isolated NTD does not suggest any biologically relevant oligomerization. To analyze this further, we determined the molecular masses of the NTD and full-length protein by analytical ultracentrifugation. The NTD was determined to have a mass of ∼61 kDa, whereas the full-length protein was 222 kDa. These results are consistent with the NTD being monomeric and the full-length protein being dimeric. This strongly suggests that the C terminus of the protein (residues 551–957) is solely responsible for dimerization.
Structural Homologies
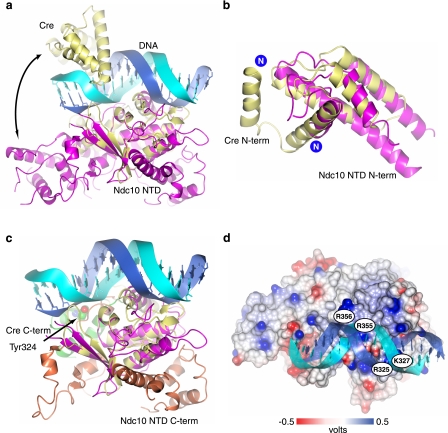
Analysis of the fold of the Ndc10 NTD using the DALI server (31) unexpectedly revealed that it belongs to the type IB topoisomerase/λ-integrase (IB/Int) family. This large group of proteins includes viral and eukaryotic type IB topoisomerases, phage integrases, and recombinases (32–34). All these enzymes utilize a conserved nucleophilic tyrosine residue that forms a covalent enzyme-nucleic acid intermediate during cleavage of the DNA phosphodiester backbone (35). The most significant structural similarity was against Cre recombinase (DALI z-score = 8.3). A superimposition of the Ndc10 NTD and the Cre recombinase bound to duplex DNA (36) is shown in Fig. 3a. The proteins may be superimposed with a Cα r.m.s. deviation of 2.6 Å over 127 residues. As can be seen, the domain spanning residues 139–274 in Cre, which is structurally equivalent to residues 182–354 in Ndc10, forms the key protein-DNA backbone contacts. This domain includes the aforementioned β-sheet and positively charged helix.
FIGURE 3.
Comparison of Ndc10 NTD to Cre recombinase. a, structure-based superposition of the Ndc10 NTD to a Cre-DNA structure (Protein Data Bank (PDB) I.D. 1xo0). Ndc10 is shown in purple, and Cre is in yellow. The similarity between the folds is particularly evident in the overlap of the β-strands. The differing locations of the N-terminal helical bundle are indicated with a curved arrow. b, alignment of the Cre N-terminal (N-term) four-helix bundle (yellow) to the N terminus of Ndc10 (purple). The extreme N-terminal helix in Cre is not resolved in the Ndc10 crystal structure. c, a structural alignment of the Ndc10 NTD and Cre, showing the divergent C-terminal (C-term) domains of the two proteins. The helical fold containing the catalytic tyrosine in Cre is shown in green with Tyr-324 depicted in space-filling mode. The C terminus of the Ndc10 NTD (orange) can be seen to fold back along the structure. The N terminus of both proteins has been omitted for clarity. d, electrostatic surface of the Ndc10 NTD, colored as in Fig. 1b. The DNA from the aligned Cre structure (protein not shown) lies directly over the conserved positively charged patch on Ndc10.
Closer analysis of the N-terminal four-helix bundle of Ndc10 revealed that it is structurally homologous to the N-terminal helical bundle of Cre but occupies a different position in the intact structure (Fig. 3, a and b). This difference is accommodated by a flexible loop that connects the domain to the rest of the protein. In Cre, this domain is required for both DNA interactions and tetramerization of the protein, suggesting that both are substantially different in Ndc10, although we cannot exclude the possibility that the domain might adopt a substantially different conformation upon DNA binding. Sequence analysis of the Ndc10 four-helix bundle shows little obvious similarities to the equivalent Cre domain, although there is weak conservation of some of the DNA-contacting basic residues. However, the surface of the Ndc10 domain is considerably less electropositive than that of Cre (data not shown), making it unclear whether it is capable of binding DNA in the same manner. Interestingly, an atomic force microscopy study on the intact CBF3 complex (37) showed that a fraction of the complexes examined bound two DNA duplexes in a non-covalent manner, giving rise to four-armed structures. These show a marked resemblance to the recombination synapses formed by Cre, which also links two duplexes to form a four-way junction. However, it is not clear whether Ndc10, when in the intact CBF3 complex, could mediate formation of such a structure or what the functional significance of this might be.
Catalytic Domain
The IB/Int family of proteins has catalytic activity, whereas none has been reported for Ndc10 or CBF3. IB/Int proteins catalyze DNA rearrangements by a phosphoryl transfer reaction. The reaction involves a nucleophilic attack on the DNA backbone by a conserved tyrosine residue to form covalent 3′-phosphotyrosine linkage and release the free 5′-hydroxyl group. The active site is composed of several conserved residues including one tyrosine, one lysine, and two arginine residues (35, 38). In known IB/Int structures, the catalytic tyrosine and the basic residues responsible for transition state stabilization are located on a short helix located in the C terminus of the protein, after the core DNA-binding fold. This helix is entirely absent in Ndc10, and no spatially equivalent residues are seen. Instead, the C terminus of the NTD loops back around the main body of the protein, on the reverse face to the putative DNA-binding residues, then continues in the opposite direction to the N-terminal four-helix bundle (Fig. 3c).
DNA Binding
Superimposition of the Cre structure on Ndc10 (Fig. 3, a and d) shows that the basic patch, which includes Arg-325, Lys-327, Lys-354, Arg-355, and Arg-356 in the two conserved Ndc10 sequence motifs RGKS and YKRR, is perfectly positioned to make contact with the DNA phosphodiester backbone (Fig. 4, a and b). We prepared versions of the Ndc10 NTD with both Arg-325 and Lys-327 or Arg-355 and Arg-356 mutated to alanine and tested the DNA binding properties (Fig. 4c). The Arg-325/Lys-327 mutations totally abolished binding, whereas it was substantially reduced in the Arg-355/Arg-356 mutant. Mutating all four residues to alanine totally eliminated binding.
FIGURE 4.
DNA-binding residues. a, partial sequence alignment of seven Ndc10 orthologs showing the conserved basic motifs (red bars) that are proposed to interact with the DNA backbone. The secondary structure elements comprising this sequence are shown above the alignment. The sequence of the corresponding helix in Cre is indicated; however, the adjacent residues cannot be aligned by either sequence or structure. Amino acids are colored according to the Clustal default. b, expanded view of the basic residues Arg-325, Lys-327, Arg-355, and Arg-356 proposed to bind DNA based on the modeled Cre duplex. c, DNA gel-shift assay showing the binding of the basic patch mutants to a non-centromeric DNA sequence. Final protein concentrations in μm are shown above the gel. MW, molecular weight markers.
Phosphorylation Sites
Kinetochore assembly is tightly regulated by several kinases, notably Aurora B (Ipl1 in budding yeast) (39). Studies have suggested that Ndc10 is a target of both Ipl1 and casein kinase 2 (CK2) (40, 41). Phosphorylations of multiple serine and threonine residues influence Ndc10 localization on the anaphase spindle and targeting to kinetochore. Two of these residues lie within the crystallized construct. Threonine 106 is disordered in the crystal but is situated on a solvent-exposed flexible loop that would be accessible to a kinase. However, serine 189 is buried deep inside the structure, and so it is hard to see how such a phosphorylation event might occur.
DISCUSSION
The data presented here provide new insights into the formation and evolution of an essential complex in the budding yeast kinetochore. Recent work (13) has lead to the development of a model in which the CBF3 complex is the key positional marker of the centromere. Binding of the complex to CDEIII allows recruitment of the centromeric nucleosome upon which the assembly of the rest of the kinetochore depends. We have shown that Ndc10 will bind non-centromeric sequences, and we predict that the protein-nucleic acid interactions are likely to occur through the DNA backbone. This would suggest that binding occurs in a non-sequence-specific manner. It is possible that the centromere specificity of the CBF3 complex is entirely due to the Cep3 protein, whereas Ndc10 may increase the overall affinity for DNA. It is possible that the binding of Ndc10 is important for local alteration of DNA structure, for example overwinding and/or bending that might be important for the nucleosome loading process.
The structure of the DNA-binding domain of Ndc10 provides the first example of the co-option of a tyrosine recombinase DNA-binding domain to the chromosome segregation apparatus, and as far as we are aware, is the first example of the fold being used in a non-catalytic role. Given the extremely weak sequence homology between Ndc10 and other members of the IB/Int family, it seems probable that the fold might also be utilized by other, as yet uncharacterized, proteins in unrelated pathways. It also demonstrates the remarkable reuse of protein domains from other pathways by the CBF3 complex. Ctf13, an F-box protein, and Skp1 are more commonly found as components of E3 ubiquitin ligases, again in catalytically active complexes, whereas in CBF3, they appear to fulfill structural functions. The Cep3 protein has made use of a sequence-specific DNA-binding domain from the Gal4 family, albeit in a very different way from the canonical transcription factor. This structure of Ndc10 shows the incorporation of yet another motif, more closely associated with DNA topology and transposition, and demonstrates the versatility of the fold. It has been proposed that the point centromere of Saccharomyces and related yeasts is a relatively recent evolutionary adaptation (42), and this scavenging of pre-existing protein motifs to build the key DNA-binding component of the centromere demonstrates how such adaptation might be accelerated.
The structure also allows us to make predictions about the likely way that DNA is bound. The conservation of basic residues involved in DNA backbone interactions suggests that Ndc10 binds duplex DNA in a manner similar to other members of the IB/Int family. However, the possible oligomerization of Ndc10 within the CBF3 complex and DNA interactions with other CBF3 proteins, notably Cep3, may well further alter the DNA structure. Our data suggest that the dimerization of Ndc10 is mediated through the C terminus of the protein, although it is unclear whether this domain is also involved in the formation of the CBF3 complex.
While this manuscript was in revision, the structure of the equivalent domain of Ndc10 from Kluyveromyces lactis was solved, bound to a 30-mer DNA duplex (43). The protein-DNA interactions seen in the complex were broadly similar to those inferred from our model based on the Cre-DNA structure. Interestingly, the authors described a putative dimerization interface in the N-terminal domain, based on crystal packing analysis. We see no similar dimerization in our structure, consistent with our hydrodynamic studies. It may be that the C terminus-induced dimerization is necessary to stabilize this interface, or alternatively, it might represent a species-specific adaptation. Further biochemical and structural studies should hopefully resolve some of these issues.
Acknowledgments
We thank Andrew Purkiss-Trew for assistance with data collection and Neil McDonald and Ian Grainge for helpful discussions.
This work was supported by a grant from Cancer Research UK.
The atomic coordinates and structure factors (code 4aco) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- NTD
- N-terminal domain
- SUMO
- small ubiquitin-like modifier
- SAD
- single-wavelength anomalous dispersion
- Ib/Int
- type IB topoisomerase/λ-integrase
- r.m.s.
- root mean square deviation.
REFERENCES
- 1. Santaguida S., Musacchio A. (2009) The life and miracles of kinetochores. EMBO J. 28, 2511–2531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Clarke L., Carbon J. (1980) Isolation of a yeast centromere and construction of functional small circular chromosomes. Nature 287, 504–509 [DOI] [PubMed] [Google Scholar]
- 3. Carbon J., Clarke L. (1984) Structural and functional analysis of a yeast centromere (CEN3). J. Cell Sci. Suppl. 1, 43–58 [DOI] [PubMed] [Google Scholar]
- 4. Baker R. E., Fitzgerald-Hayes M., O'Brien T. C. (1989) Purification of the yeast centromere-binding protein CP1 and a mutational analysis of its binding site. J. Biol. Chem. 264, 10843–10850 [PubMed] [Google Scholar]
- 5. Furuyama S., Biggins S. (2007) Centromere identity is specified by a single centromeric nucleosome in budding yeast. Proc. Natl. Acad. Sci. U.S.A. 104, 14706–14711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lechner J., Carbon J. (1991) A 240-kDa multisubunit protein complex, CBF3, is a major component of the budding yeast centromere. Cell 64, 717–725 [DOI] [PubMed] [Google Scholar]
- 7. Lingelbach L. B., Kaplan K. B. (2004) The interaction between Sgt1p and Skp1p is regulated by HSP90 chaperones and is required for proper CBF3 assembly. Mol. Cell. Biol. 24, 8938–8950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Rodrigo-Brenni M. C., Thomas S., Bouck D. C., Kaplan K. B. (2004) Sgt1p and Skp1p modulate the assembly and turnover of CBF3 complexes required for proper kinetochore function. Mol. Biol. Cell 15, 3366–3378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Russell I. D., Grancell A. S., Sorger P. K. (1999) The unstable F-box protein p58-Ctf13 forms the structural core of the CBF3 kinetochore complex. J. Cell Biol. 145, 933–950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Stemmann O., Neidig A., Köcher T., Wilm M., Lechner J. (2002) Hsp90 enables Ctf13p/Skp1p to nucleate the budding yeast kinetochore. Proc. Natl. Acad. Sci. U.S.A. 99, 8585–8590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Espelin C. W., Kaplan K. B., Sorger P. K. (1997) Probing the architecture of a simple kinetochore using DNA-protein cross-linking. J. Cell Biol. 139, 1383–1396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Purvis A., Singleton M. R. (2008) Insights into kinetochore-DNA interactions from the structure of Cep3Δ. EMBO Rep. 9, 56–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Camahort R., Li B., Florens L., Swanson S. K., Washburn M. P., Gerton J. L. (2007) Scm3 is essential to recruit the histone h3 variant cse4 to centromeres and to maintain a functional kinetochore. Mol Cell 26, 853–865 [DOI] [PubMed] [Google Scholar]
- 14. Bouck D. C., Bloom K. S. (2005) The kinetochore protein Ndc10p is required for spindle stability and cytokinesis in yeast. Proc. Natl. Acad. Sci. U.S.A. 102, 5408–5413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Gillis A. N., Thomas S., Hansen S. D., Kaplan K. B. (2005) A novel role for the CBF3 kinetochore-scaffold complex in regulating septin dynamics and cytokinesis. J. Cell Biol. 171, 773–784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Romao M., Tanaka K., Sibarita J. B., Ly-Hartig N. T., Tanaka T. U., Antony C. (2008) Three-dimensional electron microscopy analysis of ndc10-1 mutant reveals an aberrant organization of the mitotic spindle and spindle pole body defects in Saccharomyces cerevisiae. J. Struct. Biol. 163, 18–28 [DOI] [PubMed] [Google Scholar]
- 17. Widlund P. O., Lyssand J. S., Anderson S., Niessen S., Yates J. R., 3rd, Davis T. N. (2006) Phosphorylation of the chromosomal passenger protein Bir1 is required for localization of Ndc10 to the spindle during anaphase and full spindle elongation. Mol. Biol. Cell 17, 1065–1074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bellizzi J. J., 3rd, Sorger P. K., Harrison S. C. (2007) Crystal structure of the yeast inner kinetochore subunit Cep3p. Structure 15, 1422–1430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Schulman B. A., Carrano A. C., Jeffrey P. D., Bowen Z., Kinnucan E. R., Finnin M. S., Elledge S. J., Harper J. W., Pagano M., Pavletich N. P. (2000) Insights into SCF ubiquitin ligases from the structure of the Skp1-Skp2 complex. Nature 408, 381–386 [DOI] [PubMed] [Google Scholar]
- 20. Collaborative Computational Project, Number 4 (1994) The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 50, 760–763 [DOI] [PubMed] [Google Scholar]
- 21. Kabsch W. (2010) XDS. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Evans P. (2006) Scaling and assessment of data quality. Acta Crystallogr. D Biol. Crystallogr. 62, 72–82 [DOI] [PubMed] [Google Scholar]
- 23. Bricogne G., Vonrhein C., Flensburg C., Schiltz M., Paciorek W. (2003) Generation, representation, and flow of phase information in structure determination: recent developments in and around SHARP 2.0. Acta Crystallogr. D Biol. Crystallogr. 59, 2023–2030 [DOI] [PubMed] [Google Scholar]
- 24. Cowtan K. (2006) The Buccaneer software for automated model building. 1. Tracing protein chains. Acta Crystallogr. D Biol. Crystallogr. 62, 1002–1011 [DOI] [PubMed] [Google Scholar]
- 25. Emsley P., Lohkamp B., Scott W. G., Cowtan K. (2010) Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 27. Lovell S. C., Davis I. W., Arendall W. B., 3rd, de Bakker P. I., Word J. M., Prisant M. G., Richardson J. S., Richardson D. C. (2003) Structure validation by Cα geometry: φ, ψ, and Cβ deviation. Proteins 50, 437–450 [DOI] [PubMed] [Google Scholar]
- 28. McNicholas S., Potterton E., Wilson K. S., Noble M. E. (2011) Presenting your structures: the CCP4mg molecular graphics software. Acta Crystallogr. D Biol. Crystallogr. 67, 386–394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Goh P. Y., Kilmartin J. V. (1993) NDC10: a gene involved in chromosome segregation in Saccharomyces cerevisiae. J. Cell Biol. 121, 503–512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Jiang W., Lechner J., Carbon J. (1993) Isolation and characterization of a gene (CBF2) specifying a protein component of the budding yeast kinetochore. J. Cell Biol. 121, 513–519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Holm L., Sander C. (1993) Protein structure comparison by alignment of distance matrices. J. Mol. Biol. 233, 123–138 [DOI] [PubMed] [Google Scholar]
- 32. Cheng C., Kussie P., Pavletich N., Shuman S. (1998) Conservation of structure and mechanism between eukaryotic topoisomerase I and site-specific recombinases. Cell 92, 841–850 [DOI] [PubMed] [Google Scholar]
- 33. Guo F., Gopaul D. N., van Duyne G. D. (1997) Structure of Cre recombinase complexed with DNA in a site-specific recombination synapse. Nature 389, 40–46 [DOI] [PubMed] [Google Scholar]
- 34. Redinbo M. R., Stewart L., Kuhn P., Champoux J. J., Hol W. G. (1998) Crystal structures of human topoisomerase I in covalent and noncovalent complexes with DNA. Science 279, 1504–1513 [DOI] [PubMed] [Google Scholar]
- 35. Gibb B., Gupta K., Ghosh K., Sharp R., Chen J., Van Duyne G. D. (2010) Requirements for catalysis in the Cre recombinase active site. Nucleic Acids Research 38, 5817–5832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ghosh K., Lau C. K., Guo F., Segall A. M., Van Duyne G. D. (2005) Peptide trapping of the Holliday junction intermediate in Cre-loxP site-specific recombination. J. Biol. Chem. 280, 8290–8299 [DOI] [PubMed] [Google Scholar]
- 37. Pietrasanta L. I., Thrower D., Hsieh W., Rao S., Stemmann O., Lechner J., Carbon J., Hansma H. (1999) Probing the Saccharomyces cerevisiae centromeric DNA (CEN DNA)-binding factor 3 (CBF3) kinetochore complex by using atomic force microscopy. Proc. Natl. Acad. Sci. U.S.A. 96, 3757–3762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Nunes-Düby S. E., Radman-Livaja M., Kuimelis R. G., Pearline R. V., McLaughlin L. W., Landy A. (2002) γ-Integrase complementation at the level of DNA binding and complex formation. J. Bacteriol. 184, 1385–1394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Cheeseman I. M., Anderson S., Jwa M., Green E. M., Kang J., Yates J. R., 3rd, Chan C. S., Drubin D. G., Barnes G. (2002) Phospho-regulation of kinetochore-microtubule attachments by the Aurora kinase Ipl1p. Cell 111, 163–172 [DOI] [PubMed] [Google Scholar]
- 40. Glover C. V., 3rd (1998) On the physiological role of casein kinase II in Saccharomyces cerevisiae. Prog. Nucleic Acid Res. Mol. Biol. 59, 95–133 [DOI] [PubMed] [Google Scholar]
- 41. Peng Y., Wong C. C., Nakajima Y., Tyers R. G., Sarkeshik A. S., Yates J., 3rd, Drubin D. G., Barnes G. (2011) Overlapping kinetochore targets of CK2 and Aurora B kinases in mitotic regulation. Mol. Biol. Cell 22, 2680–2689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Meraldi P., McAinsh A. D., Rheinbay E., Sorger P. K. (2006) Phylogenetic and structural analysis of centromeric DNA and kinetochore proteins. Genome Biol. 7, R23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Cho U. S., Harrison S. C. (2011) Ndc10 is a platform for inner kinetochore assembly in budding yeast. Nat. Struct. Mol. Biol. 19, 48–55 [DOI] [PMC free article] [PubMed] [Google Scholar]